Abstract
Influenza is a respiratory virus that alone or in combination with secondary bacterial pathogens can contribute to the development of acute pneumonia in persons >65 years of age. Host innate immune antiviral signaling early in response to influenza is essential to inhibit early viral replication and guide the initiation of adaptive immune responses. Using young adult (3 months) and aged adult mice infected with mouse adapted H1N1 or H3N2, the results of our study illustrate dysregulated and/or diminished activation of key signaling pathways in aged lung contribute to increased lung inflammation and morbidity. Specifically, within the first seven days of infection, there were significant changes in genes associated with TLR and RIG-I signaling detected in aged murine lung in response to H1N1 or H3N2. Taken together, the results of our study expand our current understanding of age-associated changes in antiviral signaling in the lung.
Keywords: antiviral, H3N2, H1N1, influenza, aging, lung
1. Introduction
Influenza A virus (IAV) is a respiratory virus that alone or in combination with secondary bacterial pathogens can contribute to the development of acute pneumonia in persons >65 years of age [1,2]. Historically, influenza has been recognized as one of the leading causes of respiratory tract infection and contributes to seasonal epidemics. Due to antigenic variation and interspecies transmission, influenza remains a threat to global health.
Production of type I interferons (IFN) represents one of the key lines of defense against influenza. Host innate immune antiviral signaling early in response to influenza is essential to inhibit early viral replication and guide the initiation of the adaptive immune response. Infection with influenza results in the production of pathogen-associated molecular patterns (PAMPs), which are recognized by pattern recognition receptors (PRR). Several PRRs, such as Toll like receptors (TLR), NOD-like receptors (NLR), and retinoic acid inducible gene-I (RIG-I) like receptors (RLR) play a key role in the activation of the antiviral immune response to influenza. TLR 3 and 7 recognize double stranded (dsRNA) and single stranded (ssRNA) RNA, respectively and can initiate TIR domain-containing adaptor inducing IFN-β (TRIF) or myeloid differentiation factor-88 (MyD88) dependent antiviral signaling [3,4,5,6,7,8]. In addition, RLR, which recognize invasive viral RNA produced during infection and, through induction of mitochondrial antiviral signaling (MAVS) molecule, can also induce type I IFN production [9,10,11]. Expression of additional signaling molecules, such as TRIM25 which induces MAVS complex formation with DExD/H-box helicase (DHX33), or MAVS mediated recruitment of TNF-receptor-associated factor 3 (TRAF3) and subsequent activation through TANK-binding kinase-1 (TBK1), can further amplify these signaling cascades in response to influenza [10,11,12]. Upregulation of type I IFN by multiple lung cell types, such as macrophages, dendritic cells (DC), plasmacytoid DC, and epithelial cells, and subsequent binding to IFN-α/β receptors (IFNAR) present on the surface of infected neighboring cells can further stimulate antiviral responses [13]. Interaction of type I IFNs with their cognate receptors contributes to the recruitment and activation of additional signaling molecules, such as signal transducer and activator of transcription (STAT) 1 and 2 [14,15].
Previous work has demonstrated a decline in TLR function and cellular immunity in aging that contributes to increased susceptibility of older persons to influenza [16,17,18,19,20,21,22,23,24,25,26]. Similarly, defects in RIG-I signaling, which contribute to cell-intrinsic defects in IFN induction, have been observed in older adults [27]. While various age-associated defects have been associated with immunosenescence and impaired CD4 and CD8 T cell mediated responses, increased basal levels of various inflammatory cytokines and chemokines also contribute to diminished and/or dysregulated immune capacity in the aged lung [16,26,28,29,30,31,32,33,34]. In addition to reduced T cell activation, experimental studies have illustrated age-associated defects in several key components of the immune response, such as reduced antigen-capture capacity, impaired migration of DCs, decreased IFN-γ production by natural killer (NK) cells, and reduced cytokine production [16,17,28,35,36,37,38,39,40]. The goal of our current work is to provide a greater insight of antiviral signaling at days 3, 5, and 7 of H1N1 or H3N2 infection in young and aged adult murine lung. Using a murine model of mouse adapted H1N1 and H3N2 infection, the results of our study illustrate dysregulated and/or diminished activation of key signaling pathways in aged lung contribute to increased lung inflammation and morbidity. Taken together, the findings presented expand our current understanding of age-associated changes in antiviral signaling in the lung.
2. Results
As innate antiviral signaling in the lung plays a pivotal role in mediating host innate immune responses to influenza, the purpose of our current study was to examine the impact of chronological aging on antiviral signaling in young and aged adult lung in response to mouse-adapted strains of H1N1 and H3N2.
2.1. Impact of Chronological Aging on Host Responses to Mouse-Adapted H1N1 and H3N2 Strains of Influenza
2.1.1. Morbidity and Histological Changes in the Young and Aged Murine Adult Lung
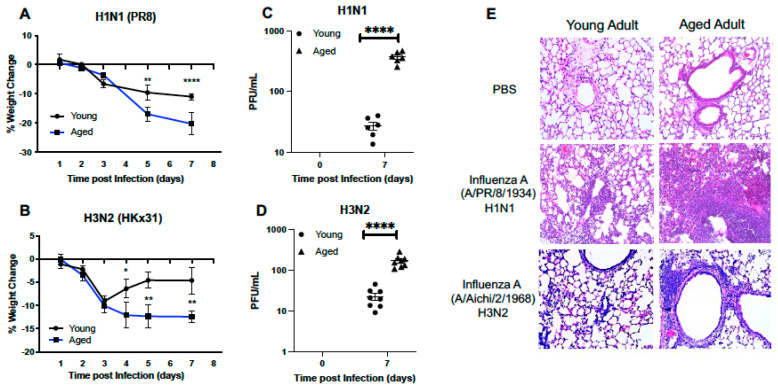
To understand if there were age-associated differences in the host response to influenza, we infected young (3 months) or aged-adult (18–20 months) mice with mouse-adapted strains of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). In response to both strains of influenza, significantly sustained loss of weight was observed in aged adult mice when compared to young (Figure 1A,B). When compared to young, there was also a significant increase in viral titer present in aged lung in response to either H1N1 (Figure 1C) or H3N2 (Figure 1D). Histological examination of lung tissue illustrated increased cellular infiltration, with a notable increase in inflammation and intra-alveolar edema detected in aged adult lung at day 7 of infection (Figure 1E). In response to H1N1, there was increased cellular recruitment to aged adult murine lung, with detectable damage to the alveolar capillary barrier (Figure 1E). Interestingly, when compared to lung tissue collected from aged H1N1 infected mice, there was less inflammation and inflammatory damage in aged H3N2 infected lung detected at day 7 post infection (Figure 1E).
Figure 1.
Morbidity and Histological Changes in the Young and Aged Murine Adult Lung. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Weight measurements were taken for young and aged mice at select time points post (A) H1N1 or (B) H3N2 infection. Viral titer in BAL was quantified and PFU/mL for (C) H1N1 and (D) H3N2. (E) Lung tissue was collected at day 7 post infection and H&E staining was performed to assess inflammation and cellular recruitment to lung. For H1N1 and H3N2 samples, each tissue section (20X magnification) shown represents a different mouse. Student’s t-test: * p < 0.05, ** p < 0.01, and **** p < 0.0001. Similar results were obtained from at least three independent experiments, with N = 10 per group. Data are expressed as the mean ± SD.
2.1.2. Cellular Infiltration and Lung Injury Is Increased in Aged Murine Lung in Response to Influenza
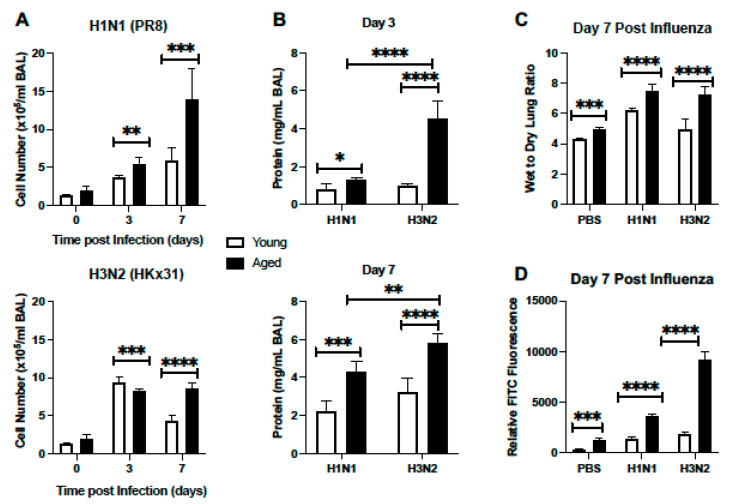
Based on these histological findings, we next examined cell numbers present in bronchoalveolar lavage (BAL) fluid isolated from young and aged adult lung at select time points post H1N1 or H3N2 infection. In response to H1N1, there was a significant increase in cells present in BAL detected at day 3 post infection (Figure 2A). Cell number continued to increase in aged lung, with significantly heightened quantities detected at day 7 post infection (Figure 2A). We next evaluated cellular numbers present in young and aged lung in response to H3N2 infection. At day 3 post infection, there was a marked increase in cellular recruitment, with significantly higher numbers of cells quantified in young lung when compared to aged (Figure 2A). By day 7 post infection, cell numbers remained significantly elevated in aged H3N2 infected lung (Figure 2A). We next examined protein levels in young and aged adult BAL samples collected from mice at day 3 and 7 post H1N1 or H3N2 infection. At day 3 post infection there was a significant increase in protein detected in aged adult lung in response to influenza, with heightened levels detected in response to H3N2 (Figure 2B). Similarly, at day 7 post influenza, there was significantly increased protein concentrations present in aged lung, with notably higher levels quantified in H3N2 lung (Figure 2B). We next investigated the amount of lung water accumulation in young and aged adult lung in response to H1N1 or H3N2 infection. At day 7 post H1N1 or H3N2 infection there was a significant increase in water accumulation in aged adult lung, as illustrated by increased wet to dry lung ratio (Figure 2C). To examine potential changes in alveolar epithelial and endothelial permeability in young and aged murine lung in response to H1N1 or H3N2, we quantified relative fluorescence levels in plasma post lung instillation with FITC. In response to influenza, there was a marked increase in permeability, as illustrated by increased FITC fluorescence in aged influenza infected lung, with significantly higher levels detected in response to H3N2 (Figure 2D).
Figure 2.
Cellular Infiltration and Lung Injury is Increased in Aged Murine Lung in Response to Influenza. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). (A,B) At select time points post infection BAL was collected from mice and (A) cell number or (B) protein concentration was quantified. (C) Lung tissue was collected at day 7 post infection. Wet weight (initial weight upon lung tissue removal) was quantified prior to incubation at 60 °C for 48 h to yield dry weight measurements. (D) Mice were instilled on day 7 with 3mg/mL of FITC-dextran and relative FITC fluorescence in plasma was assessed. Student’s t-test: * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
2.1.3. Dysregulated Type I IFN Signaling in Aged Lung
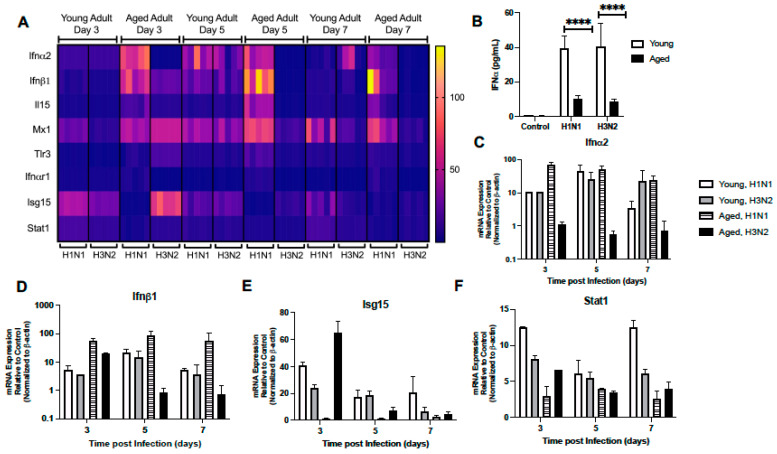
Given the importance of antiviral signaling on host immune responses to influenza, we next examined the expression of several genes associated with the type I IFN signaling response in young and aged adult murine lung treated with PBS or influenza (H1N1 or H3N2) at days 3, 5, and 7 post infection. As shown in Figure 3A, there was a distinct pattern of genes elevated in both young and aged adult lung in response to each strain of influenza. On day 3, despite high Ifnα2 gene expression in aged H1N1 infected lung, we noted significantly diminished levels of IFNα present in BAL collected from H1N1 and H3N2 infected aged murine lung (Figure 3A–C). At day 3 post infection there was an increase in Ifnα2, Isg15, and Stat1 gene expression detected in young adult lung in response to either H1N1 or H3N2 (Figure 3A,C,E, and F; Supplemental Table S1). At day 5 of H1N1 or H3N2 infection, there was elevated expression of Ifnα2, Ifnβ1, Il-15, Mx1, and Tlr3 detected in young lung (Figure 3A,C,D, Supplemental Table S1). In response to H1N1 or H3N2, there was increased expression of Il-15, Tlr3, and Stat1 detected in young lung at day 7 of infection (Figure 3A,F, Supplemental Table S1). Examination of aged lung demonstrated differential expression patterns of genes associated with type I IFN signaling. Specifically, at day 3 post H1N1, there was increased Ifnα2 and Ifnβ1 and decreased Isg15 expression detected in aged lung (Figure 3A,C–E, Supplemental Table S2). In contrast, there was elevated expression of Isg15, Stat1, Mx1, and Tlr3 genes detected in aged lung at day 3 post H3N2 (Figure 3A,E,F, Supplemental Table S2). By days 5 and 7 of infection, there was a marked difference in gene expression with heightened expression of Mx1, Tlr3, Ifnα2, Ifnαr1, Ifnβ1, and Il-15 detected in aged lung in response to H1N1 (Figure 3A,C,D, Supplemental Table S2). In contrast, a similar elevation of Ifnα2, Ifnαr1, Ifnβ1, and Il-15 was not detected in aged lung in response to H3N2 (Figure 3A,C,D, Supplemental Table S2).
Figure 3.
Dysregulated Type I IFN Signaling in Aged Lung. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). (A) Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Please see Table 1 for gene abbreviations and Supplemental Tables S1 and S2 for complete details. (B) IFN-α expression in BAL was assessed by ELISA on day 3 post infection (student’s t-test: **** p < 0.0001). Representative gene expression of (C) Ifnα2 (two-way ANOVA, p < 0.0001), (D) Ifnβ1 (two-way ANOVA, p = 0.0361), (E) Isg15 (two-way ANOVA, p < 0.0001), and (F) Stat1 (two-way ANOVA, p < 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
2.1.4. Altered Expression of TLR Signaling Responsive Genes
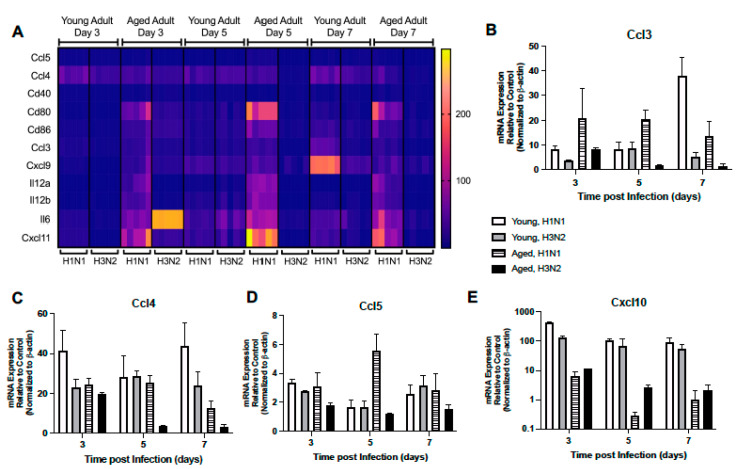
We next investigated the expression pattern of TLR receptor signaling responsive genes in young and aged lung during H1N1 or H3N2 infection. In response to H1N1 or H3N2, there was a marked elevation of Ccl3, Ccl4, Ccl5, and Cxcl10 in young lung at day 3 post infection (Figure 4A–E, Supplemental Table S1). While some expression patterns remained elevated during infection, by day 5 there was increased Il-12a, Il-12b, Il-6, Cd80, and Cxcl11 expression that corresponded with elevated Il-15 levels in young adult H1N1 and H3N2 infected lung (Figure 4A, Supplemental Table S1). By day 7, expression of Ccl3, Cxcl9, Cd86, Il-6, Cd80, and Cxcl11 remained elevated in young adult lung in response to H1N1, while expression of Ccl5, Ccl4, and Cd40 remained elevated in response to either strain of influenza (Figure 4A–D, Supplemental Table S1). In aged adult lung, there was a significant upregulation of multiple TLR signaling responsive genes at day 3 post H1N1, with a marked increase in Cd80, Cxcl11, and Il-6 expression being detected (Figure 4A, Supplemental Table S1). Interestingly, by days 5 and 7, there was heightened expression of multiple genes, such as Cxcl9, Ccl3, Ccl4, Il-12a, Il-12b, Cd86, Cxcl11, and Cd80, detected in aged lung in response to H1N1 (Figure 4A–C, Supplemental Table S2).
Figure 4.
Altered Expression of TLR Signaling Responsive Genes. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Please see Table 1 for gene abbreviations and Supplemental Tables S1 and S2 for complete list of results. Representative gene expression of (B) Ccl3 (two-way ANOVA, p < 0.0001), (C) Ccl4 (two-way ANOVA, p < 0.0001), (D) Ccl5 (two-way ANOVA, p < 0.0001), and (E) Cxcl10 (p < 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
2.1.5. Altered Expression of RIG-I-like Receptor Signaling
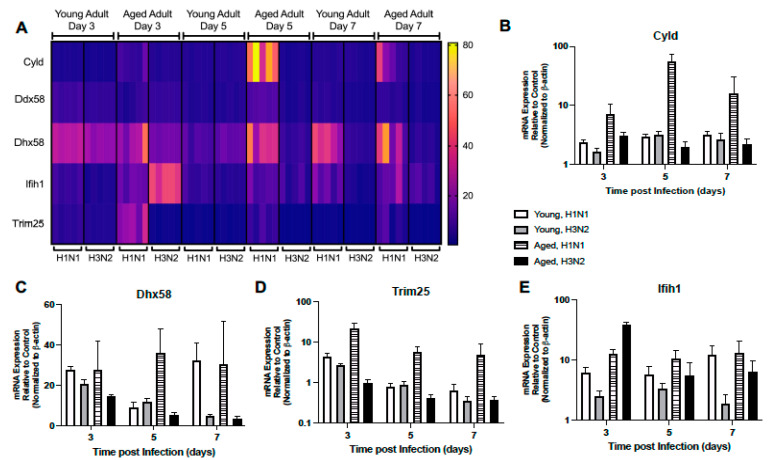
Given the importance of RIG-I signaling in host mediated immune response to influenza, we next examined the impact of age on lung responses to H1N1 or H3N2 infection. In young lung, there was similar expression patterns in H1N1 and H3N2 lung, with increased expression of Ddx58, Trim25, and Dhx58 detected on day 3 and Cyld expression on day 5 post infection (Figure 5A–D, Supplemental Table S1). When compared to H3N2, by day 7, Ddx58, Dhx58, and Ifih1 expression remained elevated in young H1N1 infected lung (Figure 5A,C,E, Supplemental Table S1). In contrast, there was altered expression in aged lung on day 3 post infection, with increased Dhx58, Cyld, Trim25, and Ddx58 detectable in aged H1N1 infected lung tissue (Figure 5A–D, Supplemental Table S2). Despite increased Ifih1 gene expression on day 3 post infection, diminished RIG-I like receptor signaling was observed in aged H3N2 infected lung on day 5 and 7 post infection (Figure 5A–E, Supplemental Table S2).
Figure 5.
Altered Expression of RIG-I-Like Receptor Signaling. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Please see Table 1 for gene abbreviations and Supplemental Tables S1 and S2 for complete list of results. Representative expression of (B) Cyld (two-way ANOVA, p < 0.0001), (C) Dhx58 (two-way ANOVA, p = 0.0058), (D) Trim25 (two-way ANOVA, p < 0.0001), and (E) Ifih1 (two-way ANOVA, p < 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
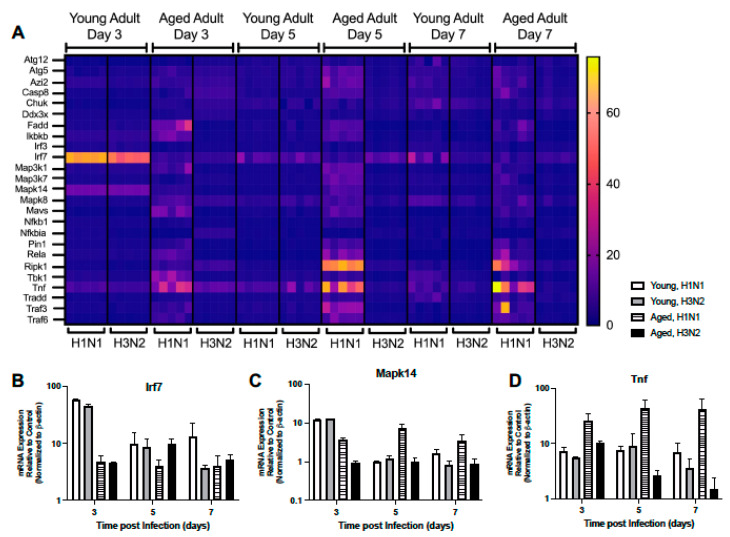
We investigated the impact of aging on the expression of downstream RIG-I-like receptor signaling molecules. In young lung, there were comparable Pin1, Iκbκb, Mapk14, Map3k1, Rela, Irf7, Map3k7, and Nfκb1 expression patterns at days 3 and 5 post H1N1 or H3N2 infection (Figure 6A–C, Supplemental Table S1). In response to H1N1 or H3N2 infection, on day 5 post infection there were also similar expression patterns of Mavs, Traf3, and Traf6 detected in young lung (Figure 6A, Supplemental Table S1). Interestingly, by day 7 of infection, multiple genes were elevated in young H1N1 infected lung, such as Atg5, Mapk8, Tbk1, Casp8, and Ripk1, that were not similarly expressed in response to H3N2 (Figure 6A, Supplemental Table S1). In aged adult lung, there was a differential pattern of gene expression observed in response to H1N1 or H3N2 infection (Figure 6A). Specifically, at day 3, while the expression of Tbk1, Tnf, Mavs, and Traf6 were upregulated in response to H1N1, only the expression of Ddx3x, Chuk, Irf7, and Nfκbia were highly expressed in aged lung in response to H3N2 (Figure 6A,B,D, Supplemental Table S2). By days 5 and 7, heightened expression of Pin1, Traf6, Ripk1, Map3k7, Map3k1, Mapk14, Atg5, and TNF were observed in aged lung in response to H1N1 (Figure 6A,D, Supplemental Table S2).
Figure 6.
Altered Expression of RIG-I-Like Receptor Signaling. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Please see Table 1 for gene abbreviations and Supplemental Tables S1 and S2 for complete list of results. Representative expression of (B) Irf7 (two-way ANOVA, p < 0.0001), (C) Mapk14 (two-way ANOVA, p < 0.0001), and (D) Tnf (two-way ANOVA, p = 0.00459). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
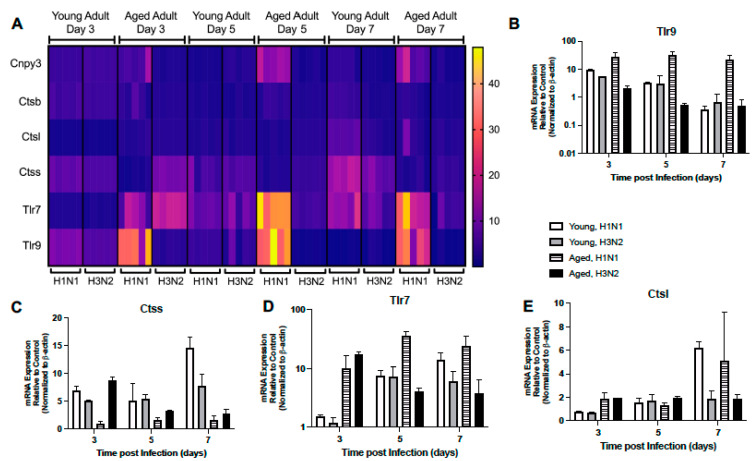
2.1.6. Dysregulated Expression of TLR Receptors and Chaperones
We investigated if there was an age-associated alteration in the expression of TLR receptors and chaperones in lung during H1N1 or H3N2 infection. In young adult lung, there was increased expression of Tlr9, Cnpy3, and Ctsb at day 3 post H1N1 or H3N2 infection (Figure 7A,B, Supplemental Table S1). Similar expression patterns of Tlr7, Ctsl, and Ctss were observed in young lung at day 5 post H1N1 or H3N2 infection (Figure 7A, C–E, Supplemental Table S1). However, by day 7, elevated Ctsl, Ctsb, and Cnpy3 only remained elevated in young, H1N1 infected lung tissue (Figure 7A,C,E, Supplemental Table S1). In aged lung, Tlr9 expression was remained highly elevated during H1N1 infection (Figure 7A,B, Supplemental Table S2). Despite similar levels of Tlr7 on day 3 post H1N1 or H3N2 infection, gene expression levels remained heightened in aged lung on day 5 and 7 post H1N1 (Figure 7A,D, Supplemental Table S2). By days 5 and 7, heightened expression of Ctsl and Cnpy3 were detected in aged lung in response to H1N1 infection (Figure 7A,E, Supplemental Table S2). Taken together, these data demonstrate that the expression of TLR receptors, chaperones, and signaling molecules was dysregulated in aged lung in response to influenza.
Figure 7.
Dysregulated Expression of TLR Receptors and Chaperones. Young (3 months) and aged (18–20 months) adult mice were intranasally instilled with 12.5 PFU of influenza (strain: A/Puerto Rico/8/1934, PR8, H1N1) or (strain: A/Aichi/2/1968, HKx31, H3N2). Lung tissue was collected from PBS treated or influenza infected young and aged mice at select time points post infection. (A) Gene expression was assessed using the RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z and results were quantified using analytical software provided by Qiagen Gene Globe. Please see Table 1 for gene abbreviations and Supplemental Tables S1 and S2 for complete list of results. Representative expression of (B) Tlr9 (two-way ANOVA, p = 0.0141), (C) Ctss (two-way ANOVA, p < 0.0001), (D) Tlr7 (two-way ANOVA, p = 0.00025), (E) Ctsl (two-way ANOVA, p = 0.0001). Similar results were obtained from at least three independent experiments, with N = 5 per group. Data are expressed as the mean ± SD.
3. Discussion
The purpose of our current study was to examine the impact of chronological aging on antiviral signaling in murine lung in response to mouse-adapted strains of influenza, such as H1N1 or H3N2. The results of our research demonstrate distinct influenza strain specific histopathological changes occur in young and aged adult murine lung. There was increased cellular infiltration and inflammation present in H1N1 or H3N2 infected aged lung when compared to young. Similarly, there was heightened permeability and increased fluid accumulation in aged H1N1 or H3N2 infected lung tissue. An examination of the antiviral signaling pathways that might contribute to this phenotype illustrated that there was dysregulated type I IFN signaling as well as altered expression of TLR signaling receptors and responsive genes. Taken together, the data in our current study demonstrate that age and strain dependent antiviral signaling contribute to differential outcomes in the lung during influenza infection.
Our results further demonstrate that decreased antiviral signaling in aged lung may contribute to increased permeability of aged lung and sustained weight loss during H3N2. Interferon production and expression of IFN stimulated genes play a critical role in the suppression of viral translocation and replication. Diminished antiviral signaling observed in aged lung in response to H3N2 infection can contribute to decreased transcriptional activation and impaired adaptive immune responses. By day 7 post H3N2 infection, there was a marked increase in lung injury and epithelial permeability in aged lung when compared to H1N1. As the expression of IFNs and their stimulated genes are critical for limiting viral replication, it is plausible that diminished expression of these genes in response to H3N2 may underlie this phenotype.
In agreement with previous work, we observed dysregulated TLR expression in aged lung in response to H1N1 or H3N2 infection, which may further contribute to increased susceptibility of older persons to influenza [16,17,18,19,20,21,22,23,24,25,26]. Interestingly, when compared to young, while the expression of TLR receptors and signaling complexes were dysregulated in aged lung in response to H1N1, these expression patterns were further reduced in aged lung during H3N2 infection. Given the essential role for TLR signaling in mediating host antiviral responses, future work to examine the mechanisms that contribute to dysregulated TLR expression and downstream signaling will need to be performed.
It has been well accepted that mitochondria play a critical role in not only maintaining cellular energetics and homeostasis, but also are essential for modulating innate antiviral signaling [41,42,43]. Expression of MAVS on the mitochondrial surface can be significantly altered in response to changes in mitochondrial metabolism and mitochondrial damage [44,45]. Our findings expand upon previous work and demonstrate diminished expression of RIG-I receptor signaling as well as decreased downstream RIG-I signaling molecules, such as MAVS, was present in aged murine lung in response to H3N2 [27]. Interestingly, in response to H1N1, there was detectable upregulation of RIG-I receptor signaling and MAVS expression in aged lung by day 3, with expression remaining elevated at day 5. It is therefore plausible that different influenza subtypes, in addition to the impact of chronological aging, may contribute to the upregulation of specific genes within the RIG-I signaling cascades and differential gene expression patterns observed in aged lung in response to H1N1 or H3N2 infection. Of note, our study demonstrates that expression of RIG-I/MAVS regulatory molecules, such as TRIM25, were disproportionality increased in aged lung in response to H1N1 and not H3N2 infection. While our findings examine gene expression in response to influenza, the role of mitophagy induction, reactive oxygen species, and MAVS degradation will need to be examined in future studies to fully differentiate the impact of aging on this pathway and diminished MAVS expression in aged lung in response to H3N2 infection. Taken together, our results illustrate a potential role for dysregulated RIG-I signaling in decreased IFN production and antiviral signaling in aged lung in response to influenza.
Mitogen-activated protein kinases (MAPK) cascades play an important role in mediating extracellular responses to regulatory proteins and mediate inflammatory signaling in response to infectious stimuli. When compared to young, there was diminished upregulation of several MAPK in aged lung in response to influenza. MAPK, such as MAPK14/MAPK p38 play a critical role IFN induction and expression of IFN-stimulated genes, such as STAT1 [46]. Diminished expression in aged lung in response to H1N1 or H3N2 may further contribute to decreased IFN induction and production of additional antiviral cytokines. Additional work has demonstrated that inhibition of MAPK p38 can contribute to viral particle retention [47]. Specifically, in the absence of MAPK p38, there was increased translation of cellular and viral protein [48]. Given our findings, delayed MAPK p38 expression might also contribute to increased influenza titers and heightened injury to the aged lung in response to infection.
There are several strengths and weaknesses of our current study. One strength of our study was that we used two distinct murine adapted strains of influenza to examine the impact of chronological aging on antiviral signaling. Another strength of our study was that chronologically aged animal models were utilized to investigate gene expression changes over a time course of infection. Similarly, our study, in addition to day 3, examined additional time points, such as days 5–7 of influenza infection that share similar clinical manifestations between mice and humans. One major limitation of our study was that we only present gene expression of antiviral immune signaling without protein levels or functional studies.
In summary, our current work provides a greater insight of antiviral signaling at days 3, 5, and 7 of H1N1 or H3N2 infection in young and aged adult murine lung. Using a murine model of mouse-adapted H1N1 and H3N2 infection, the results of our study demonstrate dysregulated and/or diminished activation of key signaling pathways in aged lung contribute to increased lung inflammation and morbidity and expand our current understanding of age-associated changes in antiviral signaling in the lung.
4. Materials and Methods
Mice: Young adult (3 months) and aged adult (18–20 months) male and female BALB/c mice were purchased from the NIA rodent facility (Charles River Laboratories). Upon receipt, mice were handled under identical husbandry conditions and fed certified commercial feed. Body weights were measured daily, and mice were humanely euthanized if they lost more than 15% of their starting body weight. The IACUC at Weill Cornell Medicine approved the use of animals in this study and methods were carried out in accordance with the relevant guidelines and regulations. No animals were used in the study if there was evidence of skin lesions, weight loss, or lymphadenopathy.
-
i.
Influenza: Viral stocks: H1N1 (strain: A/Puerto Rico/8/1934, PR8, material #: 10100374, batch #: 4XP170531, EID50 per ml: 1010.3) and H3N2 (strain: A/Aichi/2/1968, HKx31, material #:10100375, batch #: 4XX171019, EID50 per ml: 1010.5) were purchased from Charles River (Norwich, CT).
-
ii.
In Vivo Procedures and Tissue Collection: Influenza infection: All mice were anesthetized with isoflurane (5% for induction and 2% for maintenance) prior to intranasal instillation with 12.5 PFU of influenza (50 μL volume in PBS). Bronchoalveolar lavage (BAL): BAL was collected using previously published methods [49]. Briefly, 0.8-mL of PBS was slowly injected and aspirated 4 times prior to saving the recovered lavage fluid on ice. Lavage was clarified at 7000 rpm for 10 min at 4 °C. Viral titer assay of BAL: TCID50 in was calculated using the Viral ToxGlo Assay (Promega, Madison WI). Briefly, 3.16-fold serial dilutions of virus were plated for 24–48 h on >80% confluent MDCK cells. Upon visualization of cytopathic effect, ATP detection reagent was added, and luminescence was measured. Values were calculated by plotting net relative luminescence units (RLU) values after subtracting average blank wells against viral dilution. The TCID50 value is the reciprocal of the dilution that produced a 50% decline in ATP levels compared to untreated controls. Validated regression analysis was performed using GraphPad Prism. Protein quantification in BAL: Protein levels in clarified lavage were calculated using the BioRad protein assay (BioRad) per manufacturer’s instructions. IFNα ELISA: IFNα2 and 4 levels in clarified BAL were assessed by ELISA (ThermoFisher Scientific, Catalog # BMS6027) per manufacturer’s instructions. Lung tissue collection: At select time points of infection lung tissue was collected from control and influenza infected young and aged adult mice. Tissue was snap frozen or placed into Allprotect (Qiagen) for future analysis. FITC-Dextran Lung Permeability Assay: Young and aged adult mice were intranasally instilled with 50-μL of FITC-Dextran (3 mg/kg). After 1 h, blood was collected from euthanized mice, and plasma was isolated after centrifugation (7000 rpm, 10 min). Fluorescence was assessed (excitation 485, emission 528). Lung Wet to Dry Ratio: Lung tissue was collected from control and influenza infected young and aged adult mice. Lung tissue weight was assessed at harvest (wet weight) and after being placed in a 60 °C drying over for 48 h (dry weight). Histology: Mice were euthanized, and right lung tissue was collected for downstream analysis. To maintain architecture, left lung was distended with 1% low melting agarose and placed into cold formalin [50]. Tissue samples were processed, and H&E stained by the Translational Research Program at WCM Pathology and Laboratory of Medicine. Images were scanned using the EVOS FL Auto Imaging System (ThermoFisher Scientific). For all animal experiments, we used 5–10 mice per group and experiments were repeated at least three times.
-
iii.
RNA Purification and Real Time PCR: RNA samples were extracted using the automated Maxwell RNA extraction protocol (Madison, WI). Samples were quantified and A260/280 ratios were recorded. Samples were reverse transcribed using the First Stand Synthesis Kit and quantified RT2 ProfilerTM Assays (RT2 Profiler PCR Array, Mouse Antiviral Response, PAMM-122Z). Results were quantified using analytical software provided by Qiagen Gene Globe.
-
iv.
Statistical Analysis: Survival analysis between groups was calculated using the Mantel Cox test. Comparison of groups was performed using a two-tailed t-test and comparisons between groups were verified by one-way ANOVA. For two component comparisons (time post infection and age), two-way ANOVA was used to calculate statistical significance. All samples were independent and contained the same sample size for analysis. All data were analyzed using GraphPad Prism software (San Diego, CA). Statistical significance was considered by a * p < 0.05, **p < 0.01, *** p < 0.001, and **** p < 0.0001.
Table 1.
Symbol and gene names.
| Symbol | Gene Name |
|---|---|
| Atg12 | Autophagy-related 12 |
| Atg5 | Autophagy-related 5 |
| Azi2 | 5-azacytidine induced gene 2 |
| Casp8 | Caspase 8 |
| Ccl3 | Chemokine (C-C motif) ligand 3 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cd40 | CD40 antigen |
| Cd80 | CD80 antigen |
| Cd86 | CD86 antigen |
| Chuk | Conserved helix-loop-helix ubiquitous kinase |
| Cnpy3 | Canopy 3 homolog |
| Ctsb | Cathepsin B |
| Ctsl | Cathepsin L |
| Ctss | Cathepsin S |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Cyld | Cylindromatosis |
| Ddx3x | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 |
| Ddx58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| Dhx58 | DEXH (Asp-Glu-X-His) box polypeptide 58 |
| Fadd | Fas (TNFRSF6)-associated via death domain |
| Ifih1 | Interferon induced with helicase C domain 1 |
| Ifnα2 | Interferon alpha 2 |
| Ifnαr1 | Interferon (alpha and beta) receptor 1 |
| Ifnβ1 | Interferon beta 1 |
| Ikbkb | Inhibitor of kappaB kinase beta |
| Il12a | Interleukin 12A |
| Il12b | Interleukin 12b |
| Il15 | Interleukin 15 |
| Il6 | Interleukin 6 |
| Irf3 | Interferon regulatory factor 3 |
| Irf7 | Interferon regulatory factor 7 |
| Isg15 | ISG15 ubiquitin-like modifier |
| Map3k1 | Mitogen-activated protein 3 kinase 1 |
| Map3k7 | Mitogen-activated protein 3 kinase 7 |
| Mapk14 | Mitogen-activated protein kinase 14 |
| Mapk8 | Mitogen-activated protein kinase 8 |
| Mavs | Mitochondrial antiviral signaling protein |
| Mx1 | Myxovirus (Influenza virus) resistance 1 |
| Nfκb1 | Nuclear factor of kappa light polypeptide gene enhancer in B- cells 1, p105 |
| Nfκbia | Nuclear factor of kappa light polypeptide gene enhancer in B- cells inhibitor, alpha |
| Pin1 | Protein (peptidyl-proyl cis/trans isomerase) NIMA-interacting 1 |
| Rela | V-rel reticuloendotheliosis viral oncogene homolog A |
| Ripk1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| Stat1 | Signal transducer and activator of transcription 1 |
| Tbk1 | TANK-binding kinase 1 |
| Tlr3 | Toll-like receptor 3 |
| Tlr9 | Toll-like receptor 9 |
| Tnf | Tumor necrosis factor |
| Tradd | TNFRSF1A-associated via death domain |
| Traf3 | Tnf receptor-associated factor 3 |
| Traf6 | Tnf receptor-associated factor 6 |
| Trim25 | Tripartite motif-containing 25 |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222212097/s1.
Author Contributions
Conceptualization, R.H., S.J.C. and H.S.-D.; methodology, R.H., J.Y., S.J.C. and H.S.-D.; formal analysis, R.H. and H.S.-D.; data curation, R.H., J.Y., K.P. and H.S.-D.; writing—original draft preparation, R.H., S.J.C. and H.S.-D.; writing—review and editing, R.H., S.J.C., J.Y., K.P. and H.S.-D.; supervision, S.J.C. and H.S.-D.; project administration, S.J.C. and H.S.-D.; funding acquisition, H.S.-D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health, grant numbers: K08HL138285 (S.J.C.), R01AG052530 (H.S.-D.), and R01AG056699 (H.S.-D.).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Institutional Animal Care and Use Committee at Weill Cornell Medicine (protocol number 2016-0059, approved on 10 December 2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data presented in this study is provided within the article, with complete gene expression values listed in Supplemental Tables S1 and S2.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Global Seasonal Influenza-associated Mortality Collaborator, N. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F., Li H., Gu L., Liu M., Xue C.X., Cao B., Wang C. National Influenza Apdm09 Clinical Investigation Group of, C., Risk factors for nosocomial infection among hospitalised severe influenza A(H1N1)pdm09 patients. Respir. Med. 2018;134:86–91. doi: 10.1016/j.rmed.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Matsumoto M., Seya T. TLR3: Interferon induction by double-stranded RNA including poly(I:C) Adv. Drug Deliv. Rev. 2008;60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 5.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 6.Sato A., Iwasaki A. Induction of antiviral immunity requires Toll-like receptor signaling in both stromal and dendritic cell compartments. Proc. Natl. Acad Sci. USA. 2004;101:16274–16279. doi: 10.1073/pnas.0406268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki A., Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 8.Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., Flavell R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graef K.M., Vreede F.T., Lau Y.F., McCall A.W., Carr S.M., Subbarao K., Fodor E. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 2010;84:8433–8445. doi: 10.1128/JVI.00879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E., Farzan M., Inoue S., Jung J.U., Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koliopoulos M.G., Lethier M., van der Veen A.G., Haubrich K., Hennig J., Kowalinski E., Stevens R.V., Martin S.R., Reis e Sousa C., Cusack S., et al. Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat. Commun. 2018;9:1820. doi: 10.1038/s41467-018-04214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mibayashi M., Martinez-Sobrido L., Loo Y.M., Cardenas W.B., Gale M., Jr., Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannidis I., Ye F., McNally B., Willette M., Flano E. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J. Virol. 2013;87:3261–3270. doi: 10.1128/JVI.01956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz K.S., Mossman K.L. Viral Evasion Strategies in Type I IFN Signaling—A Summary of Recent Developments. Front. Immunol. 2016;7:498. doi: 10.3389/fimmu.2016.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schindler C., Levy D.E., Decker T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Vargas E.A., Wilk E., Canini L., Toapanta F.R., Binder S.C., Uvarovskii A., Ross T.M., Guzman C.A., Perelson A.S., Meyer-Hermann M. Effects of aging on influenza virus infection dynamics. J. Virol. 2014;88:4123–4131. doi: 10.1128/JVI.03644-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panda A., Qian F., Mohanty S., van Duin D., Newman F.K., Zhang L., Chen S., Towle V., Belshe R.B., Fikrig E., et al. Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J. Immunol. 2010;184:2518–2527. doi: 10.4049/jimmunol.0901022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haq K., McElhaney J.E. Immunosenescence: Influenza vaccination and the elderly. Curr. Opin. Immunol. 2014;29:38–42. doi: 10.1016/j.coi.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Mbawuike I.N., Acuna C.L., Walz K.C., Atmar R.L., Greenberg S.B., Couch R.B. Cytokines and impaired CD8+ CTL activity among elderly persons and the enhancing effect of IL-12. Mech. Ageing Dev. 1997;94:25–39. doi: 10.1016/S0047-6374(96)01855-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee N., Chan P.K., Hui D.S., Rainer T.H., Wong E., Choi K.W., Lui G.C., Wong B.C., Wong R.Y., Lam W.Y., et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J. Infect. Dis. 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang K.S., Lee N., Shin M.S., Kim S.D., Yu Y., Mohanty S., Belshe R.B., Montgomery R.R., Shaw A.C., Kang I. An altered relationship of influenza vaccine-specific IgG responses with T cell immunity occurs with aging in humans. Clin. Immunol. 2013;147:79–88. doi: 10.1016/j.clim.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Duin D., Allore H.G., Mohanty S., Ginter S., Newman F.K., Belshe R.B., Medzhitov R., Shaw A.C. Prevaccine determination of the expression of costimulatory B7 molecules in activated monocytes predicts influenza vaccine responses in young and older adults. J. Infect. Dis. 2007;195:1590–1597. doi: 10.1086/516788. [DOI] [PubMed] [Google Scholar]
- 23.Frasca D., Blomberg B.B., Garcia D., Keilich S.R., Haynes L. Age-related factors that affect B cell responses to vaccination in mice and humans. Immunol. Rev. 2020;296:142–154. doi: 10.1111/imr.12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bufan B., Arsenovic-Ranin N., Petrovic R., Zivkovic I., Stoiljkovic V., Leposavic G. Strain specificities in influence of ageing on germinal centre reaction to inactivated influenza virus antigens in mice: Sex-based differences. Exp. Gerontol. 2020;133:110857. doi: 10.1016/j.exger.2020.110857. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.A., Seong R.K., Shin O.S. Enhanced Viral Replication by Cellular Replicative Senescence. Immune Netw. 2016;16:286–295. doi: 10.4110/in.2016.16.5.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Geest K.S., Abdulahad W.H., Tete S.M., Lorencetti P.G., Horst G., Bos N.A., Kroesen B.J., Brouwer E., Boots A.M. Aging disturbs the balance between effector and regulatory CD4+ T cells. Exp. Gerontol. 2014;60:190–196. doi: 10.1016/j.exger.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Molony R.D., Nguyen J.T., Kong Y., Montgomery R.R., Shaw A.C., Iwasaki A. Aging impairs both primary and secondary RIG-I signaling for interferon induction in human monocytes. Sci. Signal. 2017;10:509. doi: 10.1126/scisignal.aan2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toapanta F.R., Ross T.M. Impaired immune responses in the lungs of aged mice following influenza infection. Respir. Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keef E., Zhang L.A., Swigon D., Urbano A., Ermentrout G.B., Matuszewski M., Toapanta F.R., Ross T.M., Parker R.S., Clermont G. Discrete Dynamical Modeling of Influenza Virus Infection Suggests Age-Dependent Differences in Immunity. J. Virol. 2017;91:e00395-17. doi: 10.1128/JVI.00395-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boianelli A., Nguyen V.K., Ebensen T., Schulze K., Wilk E., Sharma N., Stegemann-Koniszewski S., Bruder D., Toapanta F.R., Guzman C.A., et al. Modeling Influenza Virus Infection: A Roadmap for Influenza Research. Viruses. 2015;7:5274–5304. doi: 10.3390/v7102875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McElhaney J.E., Upshaw C.M., Hooton J.W., Lechelt K.E., Meneilly G.S. Responses to influenza vaccination in different T-cell subsets: A comparison of healthy young and older adults. Vaccine. 1998;16:1742–1747. doi: 10.1016/S0264-410X(98)00133-9. [DOI] [PubMed] [Google Scholar]
- 32.Sridharan A., Esposo M., Kaushal K., Tay J., Osann K., Agrawal S., Gupta S., Agrawal A. Age-associated impaired plasmacytoid dendritic cell functions lead to decreased CD4 and CD8 T cell immunity. Age. 2011;33:363–376. doi: 10.1007/s11357-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang I., Hong M.S., Nolasco H., Park S.H., Dan J.M., Choi J.Y., Craft J. Age-associated change in the frequency of memory CD4+ T cells impairs long term CD4+ T cell responses to influenza vaccine. J. Immunol. 2004;173:673–681. doi: 10.4049/jimmunol.173.1.673. [DOI] [PubMed] [Google Scholar]
- 34.Hong M.S., Dan J.M., Choi J.Y., Kang I. Age-associated changes in the frequency of naive, memory and effector CD8+ T cells. Mech. Ageing Dev. 2004;125:615–618. doi: 10.1016/j.mad.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Beli E., Clinthorne J.F., Duriancik D.M., Hwang I., Kim S., Gardner E.M. Natural killer cell function is altered during the primary response of aged mice to influenza infection. Mech. Ageing Dev. 2011;132:503–510. doi: 10.1016/j.mad.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogusa S., Ritz B.W., Kassim S.H., Jennings S.R., Gardner E.M. Characterization of age-related changes in natural killer cells during primary influenza infection in mice. Mech. Ageing Dev. 2008;129:223–230. doi: 10.1016/j.mad.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Agrawal A. Mechanisms and implications of age-associated impaired innate interferon secretion by dendritic cells: A mini-review. Gerontology. 2013;59:421–426. doi: 10.1159/000350536. [DOI] [PubMed] [Google Scholar]
- 38.Prakash S., Agrawal S., Cao J.N., Gupta S., Agrawal A. Impaired secretion of interferons by dendritic cells from aged subjects to influenza: Role of histone modifications. Age. 2013;35:1785–1797. doi: 10.1007/s11357-012-9477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plowden J., Renshaw-Hoelscher M., Gangappa S., Engleman C., Katz J.M., Sambhara S. Impaired antigen-induced CD8+ T cell clonal expansion in aging is due to defects in antigen presenting cell function. Cell. Immunol. 2004;229:86–92. doi: 10.1016/j.cellimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Lu J., Duan X., Zhao W., Wang J., Wang H., Zhou K., Fang M. Aged Mice are More Resistant to Influenza Virus Infection due to Reduced Inflammation and Lung Pathology. Aging Dis. 2018;9:358–373. doi: 10.14336/AD.2017.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker M.A., Volpi S., Sims K.B., Walter J.E., Traggiai E. Powering the immune system: Mitochondria in immune function and deficiency. J. Immunol. Res. 2014;2014:164309. doi: 10.1155/2014/164309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberg S.E., Sena L.A., Chandel N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills E.L., Kelly B., O’Neill L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017;18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 44.Lei Y., Moore C.B., Liesman R.M., O’Connor B.P., Bergstralh D.T., Chen Z.J., Pickles R.J., Ting J.P. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS ONE. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y., Sun X., Nie X., Sun L., Tang T.S., Chen D., Sun Q. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012;8:e1003086. doi: 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borgeling Y., Schmolke M., Viemann D., Nordhoff C., Roth J., Ludwig S. Inhibition of p38 mitogen-activated protein kinase impairs influenza virus-induced primary and secondary host gene responses and protects mice from lethal H5N1 infection. J. Biol. Chem. 2014;289:13–27. doi: 10.1074/jbc.M113.469239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchant D., Singhera G.K., Utokaparch S., Hackett T.L., Boyd J.H., Luo Z., Si X., Dorscheid D.R., McManus B.M., Hegele R.G. Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. J. Virol. 2010;84:11359–11373. doi: 10.1128/JVI.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luig C., Kother K., Dudek S.E., Gaestel M., Hiscott J., Wixler V., Ludwig S. MAP kinase-activated protein kinases 2 and 3 are required for influenza A virus propagation and act via inhibition of PKR. FASEB J. 2010;24:4068–4077. doi: 10.1096/fj.10-158766. [DOI] [PubMed] [Google Scholar]
- 49.Sun F., Xiao G., Qu Z. Murine Bronchoalveolar Lavage. Bio-Protoc. 2017;7:e2287. doi: 10.21769/BioProtoc.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halbower A.C., Mason R.J., Abman S.H., Tuder R.M. Agarose infiltration improves morphology of cryostat sections of lung. Lab. Investig. 1994;71:149–153. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this study is provided within the article, with complete gene expression values listed in Supplemental Tables S1 and S2.