Abstract
Laccase (LAC) plays important roles in different plant development and defense processes. In this study, we identified laccase genes (CsLACs) in Camellia sinensis cv ‘Longjing43′ cultivars, which were classified into six subclades. The expression patterns of CsLACs displayed significant spatiotemporal variations across different tissues and developmental stages. Most members in subclades II, IV and subclade I exhibited contrasting expression patterns during leaf development, consistent with a trade-off model for preferential expression in the early and late developmental stages. The extensive transcriptional changes of CsLACs under different phytohormone and herbivore treatment were observed and compared, with the expression of most genes in subclades I, II and III being downregulated but genes in subclades IV, V and VI being upregulated, suggesting a growth and defense trade-off model between these subclades. Taken together, our research reveal that CsLACs mediate multi-perspective trade-offs during tea plant development and defense processes and are involved in herbivore resistance in tea plants. More in-depth research of CsLACs upstream regulation and downstream targets mediating herbivore defense should be conducted in the future.
Keywords: Camellia sinensis, laccase gene family, trade-off, development, induced defense, insect herbivore
1. Introduction
As sessile organisms, plants cannot escape adverse biotic and abiotic stresses that have negative impacts on their growth and development. Under these pressures, plants have evolved a sophisticated network of defenses over millions of years [1]. In general, plant defenses can be classified as constitutive, which are always present, and induced, which are activated only upon attack [2]. In terms of induced defense, plants perceive and decode damage-associated and/or herbivore-associated molecular patterns (DAMP/HAMP) via receptors and then activate early signaling components such as Ca2+ influx, reactive oxygen species, and MAP kinases. Subsequently, specific defense responses are initiated and regulated by multiple phytohormone signaling pathways, including jasmonic acid (JA), salicylic acid (SA), ethylene (ET), hydrogen peroxide (H2O2) and other growth-related phytohormones, such as gibberellins, cytokinins, brassinosteroids and auxins [3]. Cross-talk between these signaling pathways activates a series of changes at the molecular, biochemical, and physiological levels, which may lead to the enhancement of plant resistance but the reduction of plant growth, development, and productivity [4]. Activation of defense responses generally comes at the expense of plant growth penalties, which could be in line with a delicate balance between growth and defense, known as the “growth-defense trade-off” phenomenon. Competition for limited available resources has long been considered as the force driving this trade-off, but it has recently been hypothesized to be the result of opposite molecular pathways regulating growth and defense [5].
Laccase (LAC) is a kind of polyphenol oxidase (PPO, EC 1.10.3.2) that occurs widely in bacteria, fungi, insects, and higher plants. Common LAC acting as multicopper oxidases, has four copper atoms, all forming their catalytic site, and contains three cupredoxin domains (domain 1, 2 and 3). One mononuclear copper site exists in domain 3 and one trinuclear copper cluster appears at the interface between domain 1 and 3. The domain 2 joins and positions domain 1 and 3 [6,7]. The important biological functions of LAC include the broad spectrum of substrate and using molecular oxygen as the final electron acceptor. Many studies have demonstrated that plant LAC play important roles, involving in polymerization of phenolic compounds, lignification of the cell wall structure, defense against various stresses, wound healing and iron metabolism, amongst others [8]. LAC catalyzed the final step of monolignol polymerization and involved in lignin biosynthesis, which has been demonstrated in LAC mutants. In Arabidopsis thaliana, lac4 and lac17 double mutants showed hypolignified fibres and collapsed xylem vessel phenotypes, and lac4, lac17, lac11 triple mutants displayed severe growth defects and failed to exhibit lignification either in stems or roots [9,10]. Overexpression of rice OsLAC10, pear PbLAC1 and cotton (Gossypium hirsutum) GhLAC15 in Arabidopsis and overexpression of GhLAC1 in cotton enhanced lignin accumulation [11,12,13,14]. Overall, these studies have demonstrated that LAC proteins were essential for lignin formation in plants. The LAC genes exhibited diverse temporal and spatial expression patterns in xylem, lignifying and non-lignifying tissues, which suggested that LAC genes might play certain roles in the development of roots, flowers and seeds. For example, AtLAC15 functions in root elongation and proanthocyanidin polymerization from its monomer epicatechin, while AtLAC8 regulates flowering and one OsLAC is involved in yield gains [15,16,17].
Plant LACs are induced upon exposure to both biotic and abiotic stresses and are implicated in plant defensive processes. Overexpression of GhLac1 in cotton enhanced broad-spectrum biotic defense against the fungal pathogen Verticillium dahlia and the insect pests cotton bollworm (Helicoverpa armigera) and cotton aphid (Aphis gossypii) via increased lignin deposition. Furthermore, suppression of GhLAC1 leads to a redirection of defense metabolic flux in the phenylpropanoid pathway and JA accumulation to confer resistance to V. dahliae and cotton bollworm but potentiates susceptibility to cotton aphids [12]. The VIGS-based transient silencing of wheat TaLAC4 increased susceptibility to Fusarium graminearum, mainly due to decrease pathogen-induced lignification of secondary cell walls [18]. Additionally, ZmLAC1 can be induced by salt stress, suggesting its possible role in Zea mays tolerance to salt stress [19]. However, a high degree of redundancy of LAC genes has been reported in many plant species [20]; thus, investigating the gene family and screening function-specific genes would be helpful for genetic engineering, which is especially important for plants with a larger number of LAC genes.
The tea plant (Camellia sinensis (L.) O. Kuntze) originated in Southwest China and is one of the most important woody cash crops. The tender buds and leaves of tea plants are the raw material for commercial tea, the most widely drunk nonalcoholic beverage in the world [21]. In nature, tea plants suffer from abiotic stresses such as drought, extreme temperature, and biotic stresses such as herbivore infestation and pathogen infection, all of which incur severe losses of yield and low-quality tea production. Thus, there has been considerable attention focused on investigating how tea plants resist multiple challenges. Due to the multiple functions of plant LACs as stated above and the few studies on tea laccase genes (CsLACs), investigation of the CsLAC gene family is extremely necessary. In this work, based on a transcriptome and genome database of tea plants, 43 candidates of CsLAC family were identified and validated in C. sinensis cv ‘Longjing43′ cultivars using reverse transcription polymerase chain reaction (RT-PCR) and RACE-PCR methods. The identified CsLACs were then further analyzed for gene architecture, conserved domain profile, and physical properties (protein size, isoelectric point and subcellular localization). Next, the expression levels of the total 43 CsLACs in mature leaves were analyzed from RNA-seq datasets. Furthermore, using quantitative real-time PCR (qRT-PCR), the expression profiling of important CsLACs in leaves at different developmental stages and different tissues were detected, and the transcriptional changes of CsLACs under abiotic and biotic stresses were also examined. The present study helps us to better understand the multiple layers of biological functions of CsLACs and provides theoretical references for studying the molecular mechanisms of LAC-mediated multiperspective trade-offs in tea plant during development and defense processes.
2. Results
2.1. Molecular Cloning, Phylogenetic Analysis and Chromosomal Distribution of CsLACs
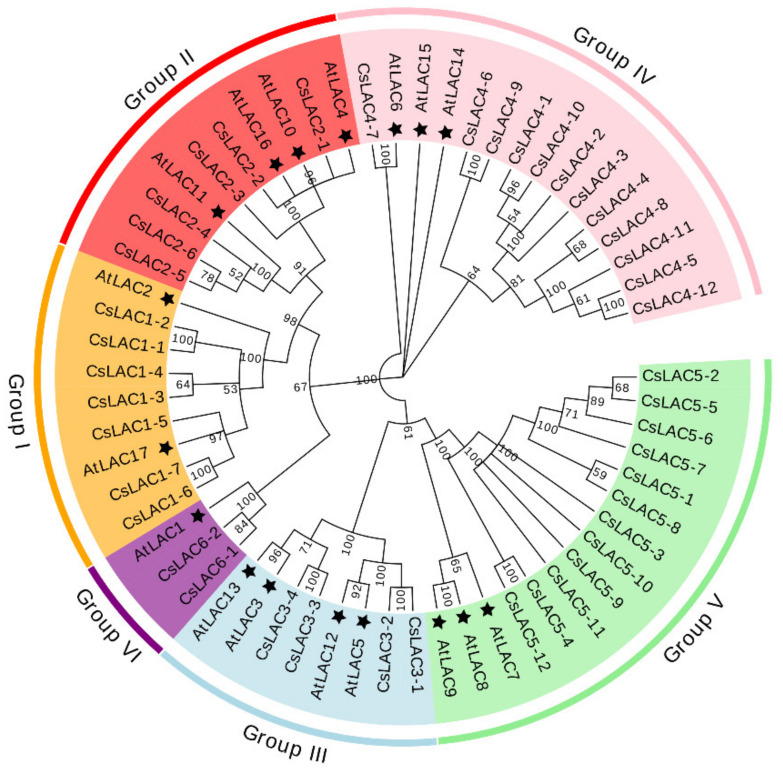
Through a genome-wide computational search (http://tpia.teaplant.org/Blast.html accessed on 30 November 2018) and subsequent gene model prediction, a total of 30 full-length CsLACs were obtained using molecular cloning approaches, including 5′ and 3′ RACE and PCR amplification of coding regions, and the other 13 CsLACs were identified from the second version of the tea genome (released in May 2020) [22]. A phylogenetic tree was constructed using the full-length amino acid residue sequences of 43 CsLACs and 17 AtLACs (Table S1). The 43 CsLACs were divided into six subclades based on the classification standard of AtLACs from Arabidopsis (Figure 1 and Figure S1). In detail, seven CsLACs in subclade I were clustered in Group I with two AtLACs, while six CsLACs in subclade II were gathered in Group II with four AtLACs. Interestingly, Group IV comprised 12 CsLACs along with three AtLACs. Subclade V contained 12 CsLACs and was distributed together with stress-induced AtLAC7, AtLAC8 and AtLAC9. In addition, Group III contained four CsLACs and four AtLACs with unknown functions. Group VI included two CsLACs with only AtLAC1. These results showed that the LAC gene family in tea plants underwent specific evolutionary events after the divergence of C. sinensis and Arabidopsis.
Figure 1.
Phylogenetic analysis of Arabidopsis and Camellia sinensis LAC genes. Phylogenetic tree was constructed using 60 protein sequences from Arabidopsis (17) and C. sinensis (43). Neighbor-joining method was used with boot strap replication of 1000 times to create the phylogenetic tree. Six subclades of the family are highlighted in different colors. ★, AtLAC1 to 17.
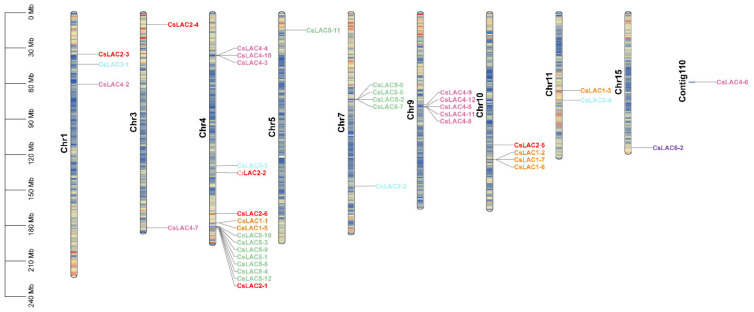
The Tea Plant Information Archive provides the exact coordinates and orientations of CsLACs in tea chromosomes. The identified 42 CsLACs were randomly distributed on 10 out of 15 chromosomes, while the chromosomal locations of the remaining 1 CsLACs remained unknown (Figure 2). In most cases, CsLACs are present in distinct clusters on each chromosome. Chromosome 4 contained the highest number of CsLACs (16), accounting for 36.3%, where seven CsLACs from subclade V formed a cluster. A similar phenomenon was observed on chromosomes 7 and 9. Additionally, we found four CsLACs on chromosome 10, 3 CsLACs on chromosome 1, and two CsLACs on chromosomes 11 and 3. However, chromosomes 5, 15 and contig110 contained the lowest number of CsLACs, with one on each.
Figure 2.
Chromosomal localization and clustering of CsLACs in tea genome. Some chromosomes e.g., 4, 7 and 9 contain higher number of CsLACs, but 5 and15 chromosomes each possesses only one CsLAC, presenting uneven distribution throughout the chromosomes.
2.2. Analysis of Conserved Domains, Gene Architecture and Promoter Elements of CsLACs
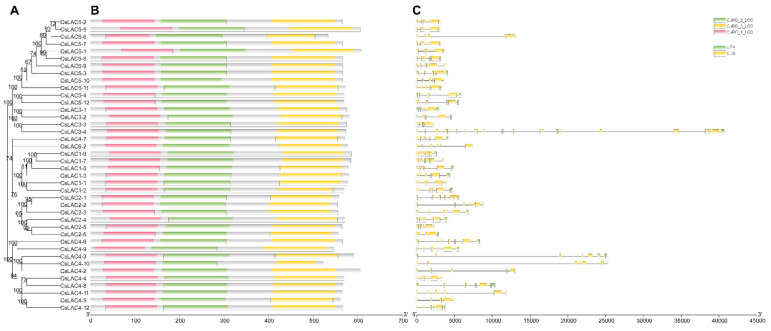
We analysed the conserved domains of the CsLAC proteins and their distribution, as shown in Figure 3. All of the identified proteins (except for CsLAC4-1) shared three characteristic domains [NCBI CDD (conserved domain database): LAC, CuRO_1_LCC, CuRO_2_LCC and CuRO_3_LCC)]. The genomic and physical characteristics of 43 CsLACs were further analysed in silico (Table 1). In total, the coding sequences ranged from 1425 to 1824 bp in length, as the exons of CsLACs varied from 4 to 8, with 6.12 on average. The predicted molecular weights were 53.109 to 67.871 kDa, and the isoelectric points were 5.05–9.54. The theoretical pI values differed extensively among six subclades. All CsLACs in subclades I and II were basic proteins (pI, 8.2–9.5), while 10 of 12 CsLACs in subclade V and 7 of 12 in subclade IV were acidic proteins (pI, 6.0–6.8). In summary, these findings suggest the relatively conserved gene structure of CsLACs and functional diversity either between different subclades (I, II and V, IV) or within the same subclade (IV).
Figure 3.
Phylogenetic tree of CsLACs as well as protein conserved domain and gene structure of corresponding CsLACs. (A) Phylogenetic tree of CsLACs, as shown in Figure 1. (B) Conserved protein motifs and their distribution. The boxes with different colors represent the conserved motifs. (C) Exon-intron structure. The CDS, UTR (untranslated region) and introns are represented by yellow boxes, green boxes and gray lines, respectively.
Table 1.
Identification and analyses of CsLACs.
| Name | Accession Number | Intron | Extron | ORF Size (bp) | Protein | Sub-Cellular Localization | ||
|---|---|---|---|---|---|---|---|---|
| Amino Acid Residues | MW (Da) | pI | ||||||
| CsLAC1-1 | CSS0050170 | 6 | 7 | 1734 | 578 | 64.230 | 9.54 | Secretory |
| CsLAC1-2 | CSS0030617 | 5 | 6 | 1710 | 569 | 62.934 | 9.47 | Secretory |
| CsLAC1-3 | CSS0040822 | 5 | 6 | 1740 | 580 | 63.814 | 9.30 | Secretory |
| CsLAC1-4 | TEA004738.1 | 5 | 6 | 1734 | 578 | 63.801 | 8.81 | Secretory |
| CsLAC1-5 | CSS0032027 | 5 | 6 | 1737 | 579 | 64.134 | 8.90 | Secretory |
| CsLAC1-6 | CSS0029337 | 6 | 7 | 1755 | 585 | 64.656 | 9.14 | Secretory |
| CsLAC1-7 | CSS0045289 | 7 | 8 | 1755 | 584 | 64.398 | 9.02 | Secretory |
| CsLAC2-1 | CSS0030904 | 6 | 7 | 1674 | 558 | 60.821 | 9.34 | Secretory |
| CsLAC2-2 | CSS0014129 | 6 | 7 | 1665 | 554 | 61.137 | 8.93 | Secretory |
| CsLAC2-3 | CSS0015036 | 5 | 6 | 1668 | 555 | 61.106 | 8.34 | Secretory |
| CsLAC2-4 | CSS0047304 | 6 | 7 | 1713 | 571 | 63.545 | 8.99 | Secretory |
| CsLAC2-5 | CSS0001101 | 5 | 6 | 1692 | 564 | 63.073 | 8.25 | Secretory |
| CsLAC2-6 | CSS0005481 | 5 | 6 | 1671 | 557 | 62.160 | 9.12 | Secretory |
| CsLAC3-1 | CSS0035921 | 5 | 6 | 1728 | 575 | 62.560 | 6.13 | Secretory |
| CsLAC3-2 | CSS0017559 | 4 | 5 | 1746 | 581 | 63.949 | 9.18 | Secretory |
| CsLAC3-3 | CSS0041657 | 5 | 6 | 1722 | 574 | 63.758 | 6.30 | Secretory |
| CsLAC3-4 | CSS0009670 | 5 | 6 | 1719 | 573 | 64.121 | 9.05 | Secretory |
| CsLAC4-1 | NOT FOUND | - | - | 1335 | 444 | 49.38 | 5.51 | Non-secretory |
| CsLAC4-2 | CSS0038356 | 4 | 5 | 1821 | 606 | 67.629 | 5.26 | Secretory |
| CsLAC4-3 | CSS0043663 | 6 | 7 | 1773 | 591 | 66.744 | 5.27 | Secretory |
| CsLAC4-4 | CSS0043918 | 5 | 6 | 1707 | 569 | 63.872 | 8.95 | Secretory |
| CsLAC4-5 | CSS0007135 | 5 | 6 | 1683 | 561 | 62.828 | 5.16 | Secretory |
| CsLAC4-6 | CSS0022921 | 5 | 6 | 1767 | 589 | 66.331 | 7.62 | Non-secretory |
| CsLAC4-7 | CSS0004662 | 5 | 6 | 1713 | 571 | 63.469 | 7.22 | Secretory |
| CsLAC4-8 | CSS0047533 | 3 | 4 | 1701 | 567 | 62.980 | 8.52 | Secretory |
| CsLAC4-9 | CSS0036236 | 5 | 6 | 1638 | 545 | 61.148 | 6.09 | Non-secretory |
| CsLAC4-10 | CSS0025249 | 6 | 7 | 1544 | 522 | 59.050 | 5.33 | Secretory |
| CsLAC4-11 | CSS0039645 | 5 | 6 | 1701 | 566 | 63.180 | 8.30 | Secretory |
| CsLAC4-12 | CSS0046037 | 5 | 6 | 1425 | 474 | 53.109 | 5.05 | Non-secretory |
| CsLAC5-1 | CSS0013370 | 5 | 6 | 1824 | 608 | 67.871 | 6.53 | Non-secretory |
| CsLAC5-2 | CSS0010920 | 5 | 6 | 1695 | 565 | 62.439 | 6.33 | Secretory |
| CsLAC5-3 | CSS0010479 | 5 | 6 | 1698 | 565 | 62.148 | 6.18 | Secretory |
| CsLAC5-4 | CSS0020412 | 5 | 6 | 1707 | 569 | 62.319 | 7.34 | Secretory |
| CsLAC5-5 | CSS0045107 | 5 | 6 | 1698 | 565 | 62.463 | 6.33 | Secretory |
| CsLAC5-6 | CSS0048878 | 6 | 7 | 1602 | 533 | 58.490 | 6.14 | Secretory |
| CsLAC5-7 | CSS0008882 | 5 | 6 | 1695 | 564 | 62.714 | 6.08 | Secretory |
| CsLAC5-8 | CSS0010391 | 5 | 6 | 1701 | 566 | 62.778 | 6.59 | Secretory |
| CsLAC5-9 | CSS0030703 | 5 | 6 | 1698 | 565 | 62.411 | 6.88 | Secretory |
| CsLAC5-10 | CSS0044116 | 5 | 6 | 1698 | 565 | 62.360 | 8.31 | Secretory |
| CsLAC5-11 | CSS0013475 | 5 | 6 | 1719 | 572 | 63.498 | 7.07 | Secretory |
| CsLAC5-12 | CSS0023848 | 5 | 6 | 1710 | 569 | 62.247 | 6.71 | Secretory |
| CsLAC6-1 | TEA021330 | 4 | 5 | 1737 | 579 | 64.576 | 6.82 | Secretory |
| CsLAC6-2 | CSS0019151 | 5 | 6 | 1725 | 574 | 63.731 | 6.36 | Secretory |
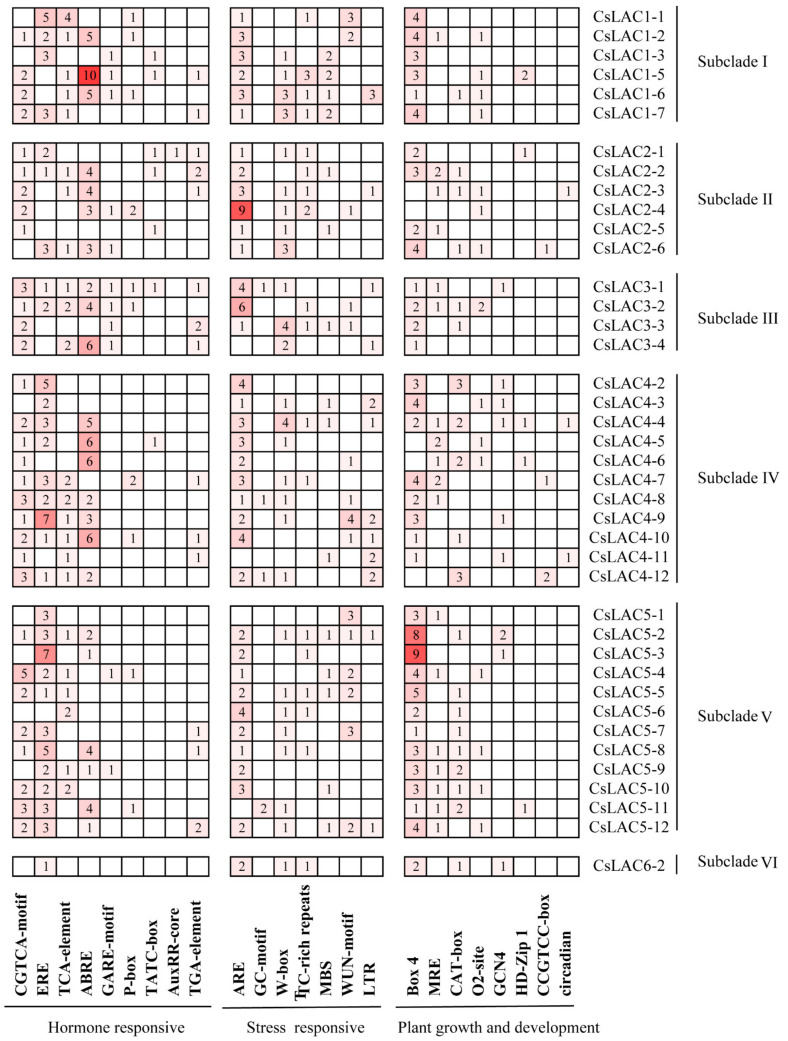
Cis-regulatory elements are DNA sequences located in the promoter region of target genes and interact with transcription factors to trigger target gene expression. To gain insight into the regulatory functions of the CsLAC promoters, a number of cis-acting elements were predicted in the promoters of 40 CsLACs. Many cis-elements including the plant hormone responsive elements, stresse responsive elements and plant growth and development responsive elements were observed in the CsLAC promoters, and which were divided into three categories and 24 types (Figure 4). Among them, the hormone responsive types were especially abundant. For instance, the most abundant cis-elements were JA-responsive CGTCA-motif in the 31 CsLAC promoters, followed by ethylene-responsive EREs (30 CsLACs), SA-responsive TCA-element (24 CsLACs), abscisic acid-responsive ABREs (23 CsLACs) and gibberellin-responsive elements, including GARE-motif, P-box and TATC-box. These results imply that CsLACs may play significant roles in plant response to different phytohormones. The stress-related cis-elements were also found in the promoter region of CsLACs. The most abundant cis-elements were anaerobic-inductive elements (ARE, cis-acting regulatory element essential for the anaerobic induction) in the 36 CsLAC promoters. In addition, W-box, TC-rich repeats (cis-acting element involved in defense and stress responsiveness), MBS (drought-inducible elements), WUN-motif (wound-inducible elements), LTR (low temperature-inducible elements) were identified in some CsLACs. These results suggest that CsLACs may be more generally involved in the responsiveness of different stresses. Moreover, the promoter regions of many CsLACs contained different binding sites involved in plant development and stress responses. Out of 40 CsLACs, 35 possessed Box 4 element (part of a conserved DNA module involved in light responsiveness), 18 possessed MRE element (MYB binding site involved in light responsiveness), and 20 had a CAT-box (cis-acting regulatory element related to meristem expression), which might be involved in plant development and stress responses.
Figure 4.
Analysis of cis-elements in CsLAC promoters. The numbers of cis-elements existed in the promoter sequences of CsLACs were identified involving in three types biological processes. The depth of red colour indicates the quantity of cis-elements of CsLACs.
2.3. Expression Patterns of CsLACs during Leaf Development and in Different Tissues
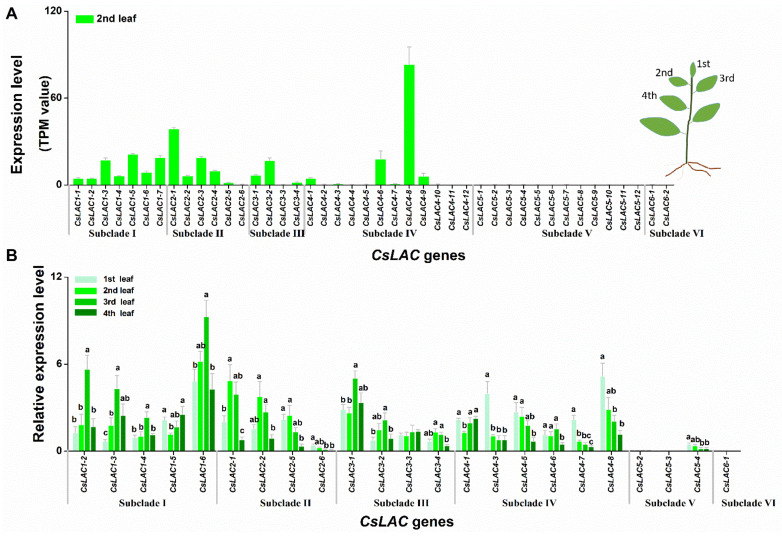
Tea leaves are important raw material of tea products, containing numerous bioactive components that are responsible for the distinctive taste and fragrance [23]. Firstly, we analysed the expression levels of 43 CsLACs in the 2nd leaf from the transcriptome database (Figure 5A). The results showed that the genes in subclades IV (except for CsLAC4-1, CsLAC4-6, CsLAC4-8, CsLAC4-9), V and VI were barely expressed, while genes in subclades I, II and III were highly or moderately expressed, exhibiting apparent differential expression patterns between these subclades. Then, by RT-qPCR, we investigated their development-specific expression patterns of CsLACs during leaf development process (1st, 2nd, 3rd and 4th leaves, shown as a schematic in Figure 5A). Due to primer design restrictions and lower expression levels, only 23 CsLACs were assessed in this study. Among these selected genes, the transcripts of nearly all members of subclade I (except for CsLAC1-5) gradually increased from the 1st to 3rd leaves and subsequently declined in the 4th leaves. In contrast, the transcripts of all genes in subclade II gradually decreased from 2nd to 4th leaves. Subclade IV members, except for CsLAC4-1 and CsLAC4-6, were highly expressed in the 1st leaves but reduced in pace with maturity. The mRNA levels of subclade III members were comparatively irregular. The genes in subclade V and VI were at extremely low levels in leaves at all different stages of maturity, and these genes were therefore considered to be silencing genes, pseudogenes or inducible expression genes under stresses in the leaf tissue (Figure 5B).
Figure 5.
Expression patterns of CsLACs in leaves of C. sinensis cv ‘Longjing43′ cultivars. (A) Expression levels of 43 CsLACs in the 2nd leaves and their abundance was evaluated by converting read count to TPM. Inset: Schematic illustration of sample collection. (B) Expression patterns of CsLACs in leaves at different stages of maturity (1st, 2nd, 3rd and 4th leaf). Different letters indicate significant differences among different maturity stages (p < 0.05, Turkey’s honest significant difference (HSD) post-hoc test).
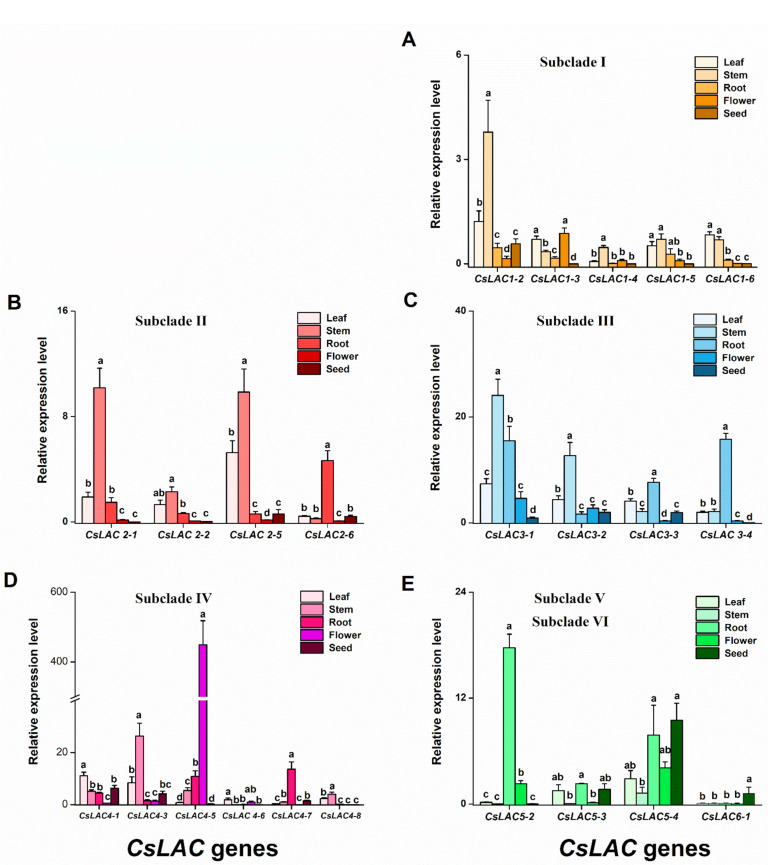
Lastly, the tissue-specific expression patterns of CsLACs were detected by RT-qPCR. Generally, the expression of CsLACs from subclades I, II, III and IV (except for CsLAC4-5, CsLAC4-7) showed the highest levels in leaves and stems, less in roots, and the lowest in flowers and seeds, whereas in subclades V and VI, CsLACs (CsLAC5-2, CsLAC5-3) showed high or moderate expression levels in roots relative to other tissues. Some CsLACs among the selected genes showed specific expression pattern. For example, only four CsLACs (CsLAC3-3, CsLAC4-1, CsLAC4-3, CsLAC4-7) showed moderate expression in seeds. Two CsLACs (CsLAC5-4, CsLAC6-1) were predominantly expressed in seeds, and CsLAC4-5 was specifically expressed in flowers (Figure 6).
Figure 6.
Transcript levels of CsLACs in leave, stem, root, flower and seed of C. sinensis cv ‘Longjing43′ cultivars. The relative expression level of CsLACs from subclades I (A), II (B), III (C), IV (D) and subclades V and VI (E). Results are expressed as mean +SE (n = 6). Different letters indicate significant differences among tissues (p < 0.05, Turkey’s honest significant difference (HSD) post-hoc test).
In general, CsLACs within the same subclade showed similar expression patterns. Conversely, different subclades, e.g., subclades I, II versus subclades V, VI, showed differential expression patterns. These findings suggest that CsLAC familiy may play important and different roles in tea plant development, but the role of individual CsLAC may be limited.
2.4. Expression Profiles of CsLACs in Response to Exogenous Application of JA and SA
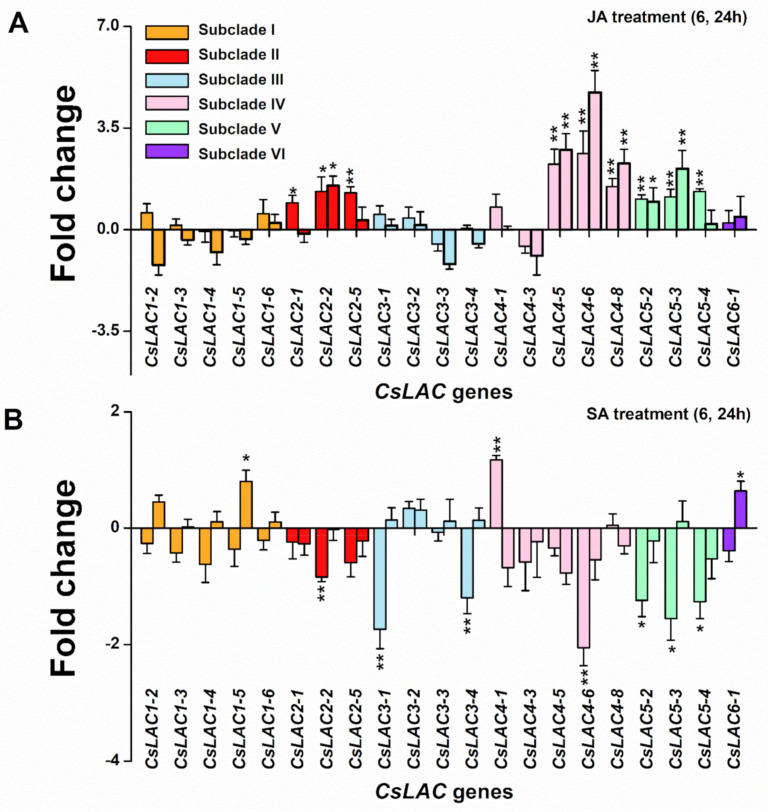
Plants respond to environmental stresses via the production of specific defense responses, which are regulated by multiple signaling pathways, including JA, SA, ethylene (ET), etc. Since JA and SA play important roles in the induced defense response of plants, including tea plants [24]. We investigated the induced effects of JA and SA treatment on CsLAC expression levels. Our results showed that under JA treatment, the 9 CsLACs in subclade II (CsLAC2-1, CsLAC2-2, CsLAC2-5), subclade IV (CsLAC4-5, CsLAC4-6, CsLAC4-8) and subclade V (CsLAC5-2, CsLAC5-3, CsLAC5-4) were significantly upregulated, particularly CsLAC4-5 and CsLAC4-6, which were 9.4 and 42.6 times higher, respectively, than the controls (Figure 7A). When tea plants were treated with SA, seven CsLACs (CsLAC2-2, CsLAC3-1, CsLAC3-4, CsLAC4-6, CsLAC5-2, CsLAC5-3, CsLAC5-4) and three CsLACs (CsLAC1-5, CsLAC4-1, CsLAC6-1) were significantly downregulated and upregulated, respectively (Figure 7B). In general, the expression analysis showed that nine CsLACs were up-regulated by JA, but seven CsLACs were down-regulated and three CsLACs were up-regulated by SA. Interestingly, among JA-/SA-responsive genes, the CsLACs (CsLAC2-2, CsLAC4-6) and subclade V (CsLAC5-2, CsLAC5-3, CsLAC5-4) were concomitantly regulated by JA and SA signaling, but exhibiting opposite expression pattern.
Figure 7.
Expression patterns of differentially expressed CsLACs from C. sinensis cv ‘Longjing43′ cultivars treated with jasmonic acid (JA) and salicylic acid (SA). The fold change in expression levels (treated group vs. control group) is shown on a log2 scale and the left and right bar graphs for each gene represent the fold change after 6h and 24h of treatment, respectively. (A) The change in expression levels of CsLACs in response to JA treatment by RT-qPCR. (B) The change in expression levels of CsLACs in response to SA treatment by RT-qPCR. The asterisks above lines indicate significant differences between the herbivore treatment and the control (*, p < 0.05, **, p < 0.01, Student’s t-test).
2.5. Expression Profiles of CsLACs in Response to Infestation by Herbivores
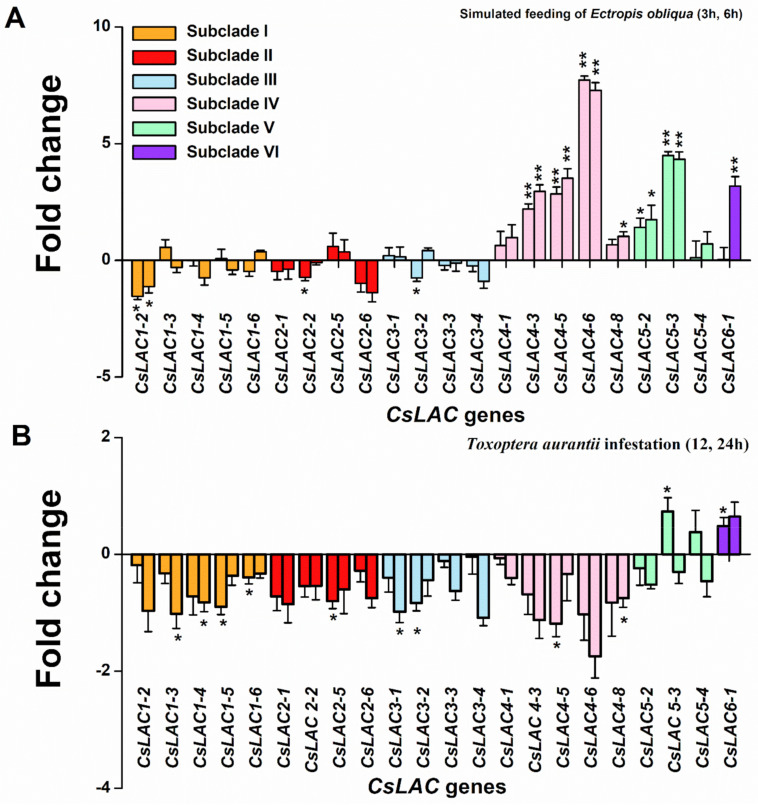
Changes in gene expression upon stress are considered as a basic component of plant defense [25]. Many studies have reported that expression levels have been used to analyse the differences in transcriptional profiles of candidate genes under various stress conditions. We also analyzed the expression patterns of CsLACs in leaves following attack by a chewing insect herbivore Ectropis obliqua and a pierce-sucking insect herbivore Toxoptera aurantii, respectively. Upon simulated feeding of E. obliqua at 3 h and 6 h after the start of treatment, the expression levels of 10 genes were changed, while the other 12 were not affected (Figure 8A). The genes in subclade IV (CsLAC4-3, CsLAC4-5, CsLAC4-6, CsLAC4-8), subclade V (CsLAC5-2, CsLAC5-3) and subclade VI (CsLAC6-1) were significantly more highly expressed, while the expressions of genes (CsLAC1-2, CsLAC2-2, CsLAC3-2) were downregulated compared to the untreated control. Upon the infestation of T. aurantii, the expression levels of genes in subclade I (CsLAC1-3, CsLAC1-4, CsLAC1-5, CsLAC1-6) and subclades II, III, IV (CsLAC2-5, CsLAC3-1, CsLAC3-2, CsLAC4-5, CsLAC4-8) were significantly downregulated, but expressions of CsLAC5-3 and CsLAC6-1 were upregulated (Figure 8B). From the above results, we identified the following two points. First, the results showed that seven CsLACs were up-regulated and three CsLACs were down-regulated under simulated E. obliqua feeding, but two CsLACs were up-regulated and nine CsLACs were down-regulated under the infestation of T. aurantii, suggesting that the infestation of chewing and pierce-sucking insect herbivores triggered different LAC-responsive defense pathways. Next, among herbivore-responsive genes, the CsLACs (CsLAC4-3, CsLAC4-6, CsLAC5-2, CsLAC5-3, CsLAC6-1) were upregulated in tea leaves attacked by chewing or/and pierce-sucking insect pests, suggesting their potential broad-spectrum roles in tea resistance against herbivorous pests. However, the expression levels of CsLACs in subclade I (CsLAC1-3, CsLAC1-4, CsLAC1-5, CsLAC1-6) and subclades II, III (CsLAC2-2, CsLAC3-1, CsLAC3-2) were downregulated, further suggesting their potential negative roles in tea resistance.
Figure 8.
Expression patterns of differentially expressed CsLACs from C. sinensis cv ‘Longjing43′ cultivars upon herbivory infestation. The fold change in expression levels (treated group vs. control group) is shown on a log2 scale. (A) The change in expression levels of CsLACs in response to simulated Ectropis obliqua feeding by RT-qPCR. The left and right bar graphs for each gene represent the fold change after treatment at 3h and 6h, respectively. (B) The change in expression levels of CsLACs in response to Toxoptera aurantii infestation by RT-qPCR. The left and right bar graphs for each gene represent the fold change after treatment at 12h and 24h, respectively. The asterisks above lines indicate significant differences between the herbivore treatment and the control (*, p < 0.05, **, p < 0.01, Student’s t-test).
3. Discussion
In recent decades, increasing amounts of evidence have demonstrated that plant LAC plays multifaceted roles in development and response to biotic and abiotic stresses. With the completion of whole-genome sequencing in numerous plant species, members of LAC gene families have been identified [8,20]. However, little information is known about the LAC gene family in the tea plant, which is one of the most important woody cash crops in the world. In this study, 43 candidates of CsLACs were identified via a genome-wide search. Large-scale spatiotemporal expression patterns and the potential CsLAC-mediated defense response were investigated.
LAC, as a class of polyphenol oxidases, is widely distributed across plant species and encoded by multigene families distributed on different chromosomes. To date, LACs have been characterized at the genome-wide level in many plant species, and their numbers differ greatly, e.g., A. thaliana (17 AtLACs on four chromosomes), Gossypium arboreum (44 GaLACs on 10 chromosomes), Citrus sinensis (24 CsLACs on six chromosomes and chromosome Un), O. sativa (30 OsLACs on eight chromosomes), Glycine max (93 GmLACs on 19 chromosomes), and Sorghum bicolor (27 SbLACs on eight chromosomes) [11,26,27,28,29,30]. The 43 CsLACs in tea plant were unevenly distributed across 10 chromosomes and tended to form five clusters on chromosomes 4, 7, 9, and 10 (Figure 2). Compared with the LAC family in other plant species, a moderate expansion of CsLACs was observed, which was likely consistent with a recent study showing that 28.6% of tea genes occurred in tandem duplication, and most of these genes expanded after a recent tetraploidization event in tea plants [21]. Meanwhile, our results are relatively similar to LAC gene-related studies conducted in Glycine max, Setaria viridis and Zea mays [20,31]. These findings indicated that tandem duplication and chromosomal segmental duplication played the determinant roles in LAC expansion, which seems to follow the most commonly evaluated mechanism underlying gene family expansion [32]. Additionally, the exon-intron structure of 43 CsLACs is similar to that of LAC genes in other plant species [20,27], which indicates that the organization of the gene structure of CsLACs was strongly conserved during evolution. But there were significant variations in the pI values of 43 CsLAC proteins, indicating their functional diversity in the developmental processes and stress responses in tea plants.
Phylogenetic analysis suggested that CsLAC members of subclades I, II and III closely clustered with AtLAC17 and AtLAC2, AtLAC4 and AtLAC11, and AtLAC12 in Arabidopsis, respectively. Experimental evidence has demonstrated that these AtLACs were necessary and non-redundant factors for lignification during vascular development [10]. The organ- and development-specific expression patterns of CsLACs were investigated in this study. Generally, most of the CsLACs exhibited leaf- and stem-preferential expression. We therefore speculated that most of the CsLACs of subclades I, II and III were likely involved in constitutive lignin biosynthesis during tea plant growth and development.
During tea leaf development, the lignin content continuously increased, and the genes involved in lignin biosynthesis showed higher expression levels in the leaves with active lignification [29]. Among the selected 23 CsLACs, the regular expression patterns of CsLACs in subclades I, II and IV were observed, whereas CsLACs in subclade III were irregularly expressed on pace with maturity. The expression level of CsLACs in subclades II (from 2nd to 4th leaves) and IV (except for CsLAC4-1) decreased on pace with leaf maturity, and this kind of expression pattern was similar to the previously reported 13 SbLACs in Sorghum bicolor and one SofLAC in sugarcane, which were highly expressed in young internodes but lowly expressed with increasing maturity [26,33]. These observations further reconfirmed the hypothesis proposed previously that this kind of LAC genes may function to polymerize monolignols into oligolignols in early stages of lignification [34]. However, unlike these results, we also found that the expression levels of CsLACs in subclade I (except for CsLAC1-5) increased with leaf maturity and decreased when leaves were fully physiologically mature (in 4th leaves). Thus, CsLACs of subclades II, IV and I exhibited opposite expression patterns, implicating potential functional divergence among these subclades during leaf development. Therefore, our results did not fully support the previous hypothesis that LAC might function during early lignification stages, whereas cell wall peroxidases played a role in the follow-up proceedings of xylem development [34]. Thus, we speculated that most CsLACs of subclades II and IV might function during the early lignification process, whereas CsLACs of subclade I were likely involved in the late lignification of xylem development. Thus, the hypothesis we proposed here is that CsLACs in subclades II, IV and I mediated a trade-off for early- and late-stage preferential expression to balance the available resources and facilitate optimal plant development. It is noteworthy that our results were consistent with a recent study on refined model of secondary cell walls lignification, which indicated that LACs were involved in Arabidopsis stem lignification throughout growth and development processes [35].
In this study, cis-element analysis indicated that many plant hormone, stress and growth responsive elements were identified, which imply that most CsLACs might response to diverse plant hormones and environmental stresses, as well as be involved in diverse processes in plant growth and development. For instance, most CsLACs contained hormone-related elements, including JA and SA signaling elements (Figure 4). JA and SA pathways are thought to play key roles in plant-induced defense responses to biotic and abiotic stresses [36,37,38,39]. Likewise, JA signaling was well-established as the core pathway that regulated tea plant defense against herbivores [24]. Our results showed that JA treatment enhanced the expression of nine CsLACs from subclades II, IV and V, whereas SA treatment decreased the expression of seven CsLACs and enhanced the expression of three CsLACs, suggesting that the JA signaling pathway activated the expression of CsLACs, but the SA signaling pathway was relatively inhibited (Figure 7). This result demonstrates that CsLACs may well be involved in herbivore resistance of tea plants, and CsLAC-based defense is positively regulated by the JA signaling pathway. The findings are consistent with the results from our previous studies, which revealed that JA served as a key defense phytohormone that mediated PPO-based resistance via positive regulation of PPO activity along with transcription of CsPPOs [40,41]. LAC, known as a kind of PPO, is capable of oxidizing a wide spectrum of aromatic compounds by a radical-catalysed reaction mechanism [6]. Although LAC gene structures and functions are quite different from other kinds of PPOs, all of which might play a similar defensive role in plants via several mechanisms: (1) direct toxicity of phenolic oxidation products, (2) alkylation and reduced bioavailability of cellular proteins to decrease nutritional quality, (3) cross-linking of quinones with other phenolics and lignification of cell walls to form physical barriers, and (4) oxidative stress in the gut lumen of insects [40,42]. Moreover, the expression levels of five CsLACs were upregulated under JA treatment and downregulated under SA treatment in accordance with the antagonism between the two signaling pathways [43], further verifying their primary roles in defense against herbivore in tea plants.
It would be a formidable task to determine the exact role of each CsLAC member based on phylogenetic analysis and expression patterns, but, from which many hints can be acquired. LAC is a key enzyme for lignin biosynthesis in the formation of secondary plant cell walls, as it catalyses the final step of monolignol polymerization [26]. In this study, we found that most genes of subclades I, II and III were predominantly expressed in leaves and stems and significantly downregulated by infestation with two important insect pests, E. obliqua and T. aurantii (Figure 5, Figure 6 and Figure 8). The subclades I and II are closely grouped with AtLAC17, AtLAC2 and AtLAC4, AtLAC11, the subclade III clustered together with AtLAC12. These AtLACs have been verified to be necessary for lignification during vascular development [9,10,13]. Hence, we speculate that most members of subclades I, II and III are mainly involved in constitutive lignin biosynthesis during tea plant growth and development.
Correspondingly, the CsLACs of subclades IV, V and VI may be involved in the response to herbivory attack, which has been supported by several lines of evidence. First, the members of subclades IV (except for CsLAC4-6, CsLAC4-8), V and VI expressed constitutively at a very low level in leaves (Figure 5), but the expression of CsLACs (CsLAC4-3, CsLAC4-6, CsLAC5-2, CsLAC5-3, CsLAC6-1) were significantly upregulated in tea leaves attacked by chewing or/and pierce-sucking insect pests (Figure 8). Furthermore, the expression of the subclade IV (CsLAC4-5, CsLAC4-6, CsLAC4-8) and subclade V (CsLAC5-2, CsLAC5-3, CsLAC5-4) were significantly upregulated by exogenous application of JA (Figure 7). These results clearly indicated that some CsLACs in subclades IV, V and VI exhibited stress-inducible expression patterns under biotic and abiotic stresses. Second, CsLAC4-6 and CsLAC4-9 were clustered with Arabidopsis AtLAC14 and AtLAC15, which have been previously reported to take part in the polymerization of phenolic compounds. Furthermore, AtLAC14 and AtLAC15 were phylogenetically related to GhLAC1 and GhLAC15 in upland cotton, which have been demonstrated to be involved in positively regulating defense-induced lignification in the cell wall to enhance the broad-spectrum biotic stress response [12,13]. A recent study demonstrated that the overexpression of poplar PeuLAC2, which phylogenetically clustered with AtLAC14 and AtLAC15 of Arabidopsis, altered the xylem structure of plants, including thickening the secondary cell wall (SCW) and increasing the fiber cell length and stem tensile strength, and thereby mediated stronger antioxidant response and greater drought tolerance [44]. In our study, most CsLACs in subclade IV were remarkably upregulated in E. obliqua simulated feeding leaves. Third, CsLACs of subclade V clustered together with wound-induced AtLAC8 and AtLAC9 (Figure 1), implying that they may have similar functions in response to mechanical damage [8]. There have been few reports of the defense functions of LACs in subclade V under herbivory infestation in other plant species thus far. Our finding has obviously shown that CsLACs of subclade V are potentially crucial for the herbivore resistance of tea plants, but the in-depth mechanism by which members contribute to tea plant resistance needs to be fully clarified. Based on these results, the study showed that the some CsLACs of subclades IV, V and VI were stress-induced and likely involved in induced lignin biosynthesis to facilitate tea plant resistance against herbivory.
In this study, herbivore attack enhanced the expression of most CsLACs in subclades IV, V and VI, but suppressed most genes in subclades I, II and III, which exhibited an opposite expression pattern. These results seems to obey the law that plants suffer limited resource reallocation to facilitate the prioritization of defense towards growth to survive during herbivore attack [4,45], and may account for the incompatibility between growth and defense programmes, known as the “growth-defense trade-off” phenomenon established upon plant-herbivore interactions [5,46,47,48,49,50]. Altogether, our findings provide one CsLACs-based explanation for the growth-defense trade-off in tea plant at the molecular level.
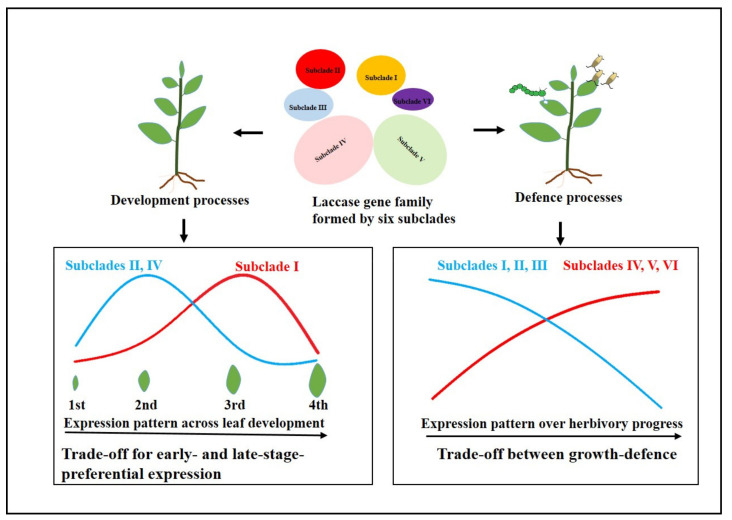
In summary, 43 CsLACs were investigated through genome-wide analysis of C. sinensis. Considering these findings, we shed new light on this phenomenon and propose a simplified model in Figure 9. A spatiotemporal expression analysis indicated that most CsLACs displayed tissue-special or stage-preferential expression patterns, which suggesting a trade-off for early- and late-stage-preferential expression patterns during leaf development. According to expression profiles of CsLACs responding to biotic and abiotic stress, a delicate trade-off between growth and induced defense was presented in a manner that prioritizes defense over growth. Additionally, our results provide evidence for the defense function of CsLACs in tea plants, which was positively regulated by the JA signaling pathway. More in-depth investigation to delineate the upstream regulation and downstream targets mediating herbivore defense should be carried out in the future.
Figure 9.
Schematic diagram showing LAC gene family-mediated trade-offs during tea plant development and defense processes. The curve (red) indicates transcripts that increase with leaf development or damages under herbivore attack. The curve (blue) represents transcripts that decrease with the progress of leaf development or herbivory.
4. Materials and Methods
4.1. Plants and Insects
The plant materials were collected from 3-year old tea plants (C. sinensis cv. Longjing 43), growing in a greenhouse programmed at 25 ± 2 °C, 14 h light (L): 10 h dark (D), and 60–80% relative humidity (RH). Healthy plants at same growth stage were chosen for experiments.
The tea looper (Ectropis obliqua Warren) and tea aphid (Toxoptera aurantii Boyer) were collected from the tea plantation of the Tea Research Institute of the Chinese Academy of Agricultural Sciences (TRI, CAAS, N 30° 10′, E 120°5′), Hangzhou, China and fed with fresh tea shoots in the controlled climate room at 26 ± 2 °C, 70 ± 5% RH, and a photoperiod of 14:10 h (L: D). Newly hatched larvae/nymphs were fed in 75 × 75 × 75 cm net cages with fresh tea shoots. Over one generation, three-instar larvae of E. obliqua and mixed age nymphs of T. aurantii were used for plant treatments.
4.2. Regurgitant Collection
Following the method proposed by Yang et al. [51], regurgitant was collected from the oral cavity of E. obliqua with a P200 Pipetteman (Gilson, Middleton, WI, USA) and stored at −80 °C.
4.3. Confirmation of the CsLACs Gene Family
Total RNA extractions were performed with the TRIzol™ kit according to the manufacturer’s instructions (TIANGEN, Beijing, China). RNAs were reverse-transcribed using PrimeScript RT Master Mix (TaKaRa Bio, Dalian, China). The cDNA fragments were obtained by transcriptome. The 5′ and 3′ sequences of CsLACs were acquired by rapid amplification of cDNA ends (RACE) using the manufacturer’s protocol (SMARTer® RACE 50/30Kit, Clontech Lab, Inc., Mountain View, CA, USA). The primers for gene cloning of 30 CsLACs used are listed in Supplementary Table S2. For another 13 candidate CsLACs, BLASTp was performed in the C. sinensis genome using the AtLAC protein sequences as queries and sequences with E-value < 10−20 were selected. All the selected genes were checked against information from the NCBI database. The sequences with essential conserved domains of multicopper oxidase type were deemed as candidate CsLACs.
4.4. Phylogenetic Analysis and Distribution Pattern of CsLACs
A neighbor-joining phylogenetic tree was constructed by MEGA (http://www.megasoftware.net/ accessed on 18 February 2020) using 60 protein sequences from A. thaliana (17) and C. sinensis (43), with bootstrap tests for 1000 replicates. The sequences form A. thaliana used for phylogenetic analysis were listed in Supplementary Table S1. The chromosomal location map of CsLACs was generated using MapInspect software (http://mapinspect.software.informer.com/ accessed on 18 February 2020).
4.5. Analysis of Gene Structure, Conserved Motif and Cis-Elements
The exon-intron structures of CsLACs were determined using Gene Structure Display Server (GSDS v2.0; http://gsds.cbi.pku.edu.cn/ accessed on 28 May 2020). The molecular weight (Mw) and theoretical isoelectric (pI) points of CsLACs were calculated by ExPASy (http://web.expasy.org/compute_pi/ accessed on 28 May 2020). Subcellular localization pattern of CsLACs were predicted using web based tool TargetP 1.1 server. The promoter sequences of CsLACs were investigated for with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 28 May 2020).
4.6. Plant Treatments
4.6.1. Different Tissues and Leaves at Different Stages of Maturity
Various tissues, including leaves, stems, roots, flowers and seeds, were harvested. The 1st, 2nd, 3rd and 4th leaves were collected from the same branch of tea plant. Six replications were collected and all samples were quickly frozen in liquid nitrogen and then stored at −80 °C for further study.
4.6.2. Simulated Feeding of E. obliqua
The mechanical damage was made by a fabric pattern wheel following the method described in the reference of Li et al. [52]. The second fully expanded leaves were used in the following treatments. Each leaf was rolled six times, and 15 μL regurgitant of E. obliqua was painted to the puncture wounds. The second leaves kept intact were used as controls. All the second leaves were harvested at 3 h and 6 h after the start of treatment. Eight replications were carried out.
4.6.3. T. Aurantii Infestation
In this study, the T. aurantii in 3rd, 4th, and 5th instar nymphs were used for plant treatments. Fifty aphids were inoculated on the tender buds and 1st leaves, while the 2nd leaves were covered with fine-mesh sleeves to keep them from infestation and contamination of honeydews. Second leaves covered with fine-mesh sleeves from plants without aphids were used as controls. The 2nd leaves were harvested at 12h and 24h after the start of treatment. Six replications were carried out.
4.6.4. JA and SA Treatment
JA and SA (Sigma Aldrich, Saint Louis, MO, USA) were dissolved in ethanol to make 100 mM stock solutions. The final spray concentrations were 100 µM for JA, and 1 mM for SA, which were diluted in distilled water containing 0.02% (v/v) Tween 20. For the control, 0.02% (v/v) Tween 20 in distilled water containing 0.01% ethanol was applied. The solution was generously sprayed onto the surfaces of the tea leaves until liquid dripped off the leaves. After spraying, the plants were covered with sealed square plastic containers (65 cm × 65 cm × 65 cm). The second leaves were harvested at 6 h and 24 h after the start of treatment. Eight replications were carried out.
4.7. RNA Sequencing and Data Processing
RNA samples of tea leaves were prepared for RNAseq analysis. Paired end of 2 × 150 bp sequencings were carried on Illumina HiSeq platform commercially provided by Metware Biotechnology Co., Ltd. (Wuhan, China). Fastp was used to further improve the quality of sequence data and assess the quality of clean reads [53]. Then the clean reads were mapped to the transcriptome extracted from tea genome data [22,54]. DEGs (differential expressed gene) were identified by DESeq2 with the criteria of absolute value of FDR (false discovery rate) < 0.05 [55].
4.8. qRT-PCR Analysis
The qRT-PCR reactions were performed on a LightCycle® 480 Real-Time PCR System (Roche Diagnostics, Mannheim, Germany) with 10μL reaction mixture. A five-fold dilution series of cDNA was used as a template for each treatment using a linear regression model to create the standard curves. The relative expressions were calculated by the standard curve method. All the primers for CsLACs and the reference genes used are listed in Supplementary Tables S3 and S4.
4.9. Data Analysis
All data were analyzed with SPSS software version 20 (SAS Institute, Inc., Cary, NC, USA, http://www.sas.com/ accessed on 1 January 2021). Differences in data among different treatments were analyzed using one-way ANOVA. If the ANOVA analysis was significant (p < 0.05), Turkey’s honestly significant difference (HSD) post-hoc test was used to detect differences between groups. Student’s t-test was used for comparing the difference between two treatments. When necessary, data were log-transformed to meet requirements for the homogeneity of variance.
Acknowledgments
We thank all the lab members for their useful suggestions, support, and encouragement.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222212554/s1, Figure S1: Multiple sequence alignment of subgroup 1-6 of CsLAC gene family, Table S1: The gene ID used for phylogenetic analysis, Table S2: Primers used for cloning laccase genes in Camellia sinensis, Table S3: Primers used for qRT-PCR, Table S4: Sequence information of the reference genes used under different treatments.
Author Contributions
Conceptualization, X.S., J.Z., Y.Y. and Y.X.; Funding acquisition, X.S. and J.Z.; Investigation, Y.Y., Y.X., F.L., X.Z. and X.L.; Methodology, X.S., J.Z., Y.Y., Y.X., F.L., X.Z. and X.L.; Project administration, X.S. and J.Z.; Supervision, X.S. and J.Z.; Visualization, Y.Y. and Y.X.; Writing—original draft, Y.Y and Y.X.; Writing—review & editing, X.S., J.Z., F.L., X.Z. and X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31972280, 31901962).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Erb M., Reymond P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019;70:527–557. doi: 10.1146/annurev-arplant-050718-095910. [DOI] [PubMed] [Google Scholar]
- 2.Heil M., Baldwin I.T. Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7:61–67. doi: 10.1016/S1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang W., Zhou P., Mo X., Hu L., Jin N., Chen X., Yu Z., Meng J., Erb M., Shang Z., et al. Induction of defense in cereals by 4-fluorophenoxyacetic acid suppresses insect pest populations and increases crop yields in the field. Proc. Natl. Acad. Sci. USA. 2020;117:12017–12028. doi: 10.1073/pnas.2003742117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia A., Martinez M., Diaz I., Santamaria M.E. The price of the induced defense against pests: A meta-analysis. Front. Plant Sci. 2021;11:615122. doi: 10.3389/fpls.2020.615122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueroa-Macías J.P., García Y.C., Núñez M., Díaz K., Olea A.F., Espinoza L. Plant growth-defense trade-offs: Molecular processes leading to physiological changes. Int. J. Mol. Sci. 2021;22:693. doi: 10.3390/ijms22020693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mot A.C., Silaghi-Dumitrescu R. Laccases: Complex architectures for one-electron oxidations. Biochemistry. 2012;77:1395–1407. doi: 10.1134/S0006297912120085. [DOI] [PubMed] [Google Scholar]
- 7.Arregui L., Ayala M., Gómez-Gil X., Gutiérrez-Soto G., Hernández-Luna C.E., Santos M.H.D.L., Levin L., Rojo-Domínguez A., Romero-Martínez D., Saparrat M.C.N., et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Factories. 2019;18:200. doi: 10.1186/s12934-019-1248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turlapati P.V., Kim K.-W., Davin L.B., Lewis N.G. The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s) Planta. 2011;233:439–470. doi: 10.1007/s00425-010-1298-3. [DOI] [PubMed] [Google Scholar]
- 9.Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.-Y., Dixon R.A. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q., Luo L., Wang X., Shen Z., Zheng L. Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci. 2017;18:209. doi: 10.3390/ijms18020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Q., Min L., Yang X., Jin S., Zhang L., Li Y., Ma Y., Qi X., Li D., Liu H., et al. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2018;176:1808–1823. doi: 10.1104/pp.17.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Wu L., Wang X., Chen B., Zhao J., Cui J., Li Z., Yang J., Wu L., Wu J., et al. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2019;20:309–322. doi: 10.1111/mpp.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng X., Li G., Ma C., Abdullah M., Zhang J., Zhao H., Jin Q., Cai Y., Lin Y. Comprehensive genome-wide analysis of the pear (Pyrus bretschneideri) laccase gene (PbLAC) family and functional identification of PbLAC1 involved in lignin biosynthesis. PLoS ONE. 2019;14:e0210892. doi: 10.1371/journal.pone.0210892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pourcel L., Routaboul J.-M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I. TRANSPARENT TESTA10 Encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell. 2005;17:2966–2980. doi: 10.1105/tpc.105.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai X., Davis E.J., Ballif J., Liang M., Bushman E., Haroldsen V., Torabinejad J., Wu Y. Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 2006;57:2563–2569. doi: 10.1093/jxb/erl022. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y.C., Yu Y., Wang C.Y., Li Z.Y., Liu Q., Xu J., Liao J., Wang X., Qu L., Chen F., et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013;31:848–852. doi: 10.1038/nbt.2646. [DOI] [PubMed] [Google Scholar]
- 18.Soni N., Hegde N., Dhariwal A., Kushalappa A.C. Role of laccase gene in wheat NILs differing at QTL-Fhb1 for resistance against Fusarium head blight. Plant Sci. 2020;298:110574. doi: 10.1016/j.plantsci.2020.110574. [DOI] [PubMed] [Google Scholar]
- 19.Liang M., Haroldsen V., Cai X., Wu Y. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant Cell Environ. 2006;29:746–753. doi: 10.1111/j.1365-3040.2005.01435.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu M., Dong H., Wang M., Liu Q. Evolutionary divergence of function and expression of laccase genes in plants. J. Genet. 2020;99:23. doi: 10.1007/s12041-020-1184-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen J.D., Zheng C., Ma J.Q., Jiang C.K., Ercisli S., Yao M.Z., Chen L. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic. Res. 2020;7:63. doi: 10.1038/s41438-020-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia E., Tong W., Hou Y., An Y., Chen L., Wu Q., Liu Y., Yu J., Li F., Li R., et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant. 2020;13:1013–1026. doi: 10.1016/j.molp.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Liao Y., Yu Z., Liu X., Zeng L., Cheng S., Li J., Tang J., Yang Z. Effect of major tea insect attack on formation of quality-related nonvolatile specialized metabolites in tea (Camellia sinensis) leaves. J. Agric. Food Chem. 2019;67:6716–6724. doi: 10.1021/acs.jafc.9b01854. [DOI] [PubMed] [Google Scholar]
- 24.Yang H., Wang Y., Li L., Li F., He Y., Wu J., Wei C. Transcriptomic and phytochemical analyses reveal root-mediated resource-based defense response to leaf herbivory by Ectropis oblique in tea plant (Camellia sinensis) J. Agric. Food Chem. 2019;67:5465–5476. doi: 10.1021/acs.jafc.9b00195. [DOI] [PubMed] [Google Scholar]
- 25.War A.R., Paulraj M.G., Ahmad T., Buhroo A.A., Hussain B., Ignacimuthu S., Sharma H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012;7:1306–1320. doi: 10.4161/psb.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Feng J., Jia W., Fan P., Bao H., Li S., Li Y. Genome-wide identification of Sorghum bicolor laccases reveals potential targets for lignin modification. Front. Plant Sci. 2017;8:714. doi: 10.3389/fpls.2017.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaig B.C., Meagher R.B., Dean J.F.D. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta. 2005;221:619–636. doi: 10.1007/s00425-004-1472-6. [DOI] [PubMed] [Google Scholar]
- 28.Balasubramanian V.K., Rai K.M., Thu S.W., Hii M.M., Mendu V. Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development. Sci. Rep. 2016;6:34309. doi: 10.1038/srep34309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X., Zhou Y., Wang B., Ding L., Wang Y., Luo L., Zhang Y., Kong W. Genome-wide identification and characterization of laccase gene family in Citrus sinensis. Gene. 2018;689:114–123. doi: 10.1016/j.gene.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q., Li G., Zheng K., Zhu X., Ma J., Wang D., Tang K., Feng X., Leng J., Yu H., et al. The soybean laccase gene family: Evolution and possible roles in plant defense and stem strength selection. Genes. 2019;10:701. doi: 10.3390/genes10090701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simões M.S., Carvalho G.G., Ferreira S., Hernandes-Lopes J., De Setta N., Cesarino I. Genome-wide characterization of the laccase gene family in Setaria viridis reveals members potentially involved in lignification. Planta. 2020;251:46. doi: 10.1007/s00425-020-03337-x. [DOI] [PubMed] [Google Scholar]
- 32.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cesarino I., Araújo P., Mayer J.L.S., Vicentini R., Berthet S., Demedts B., Vanholme B., Boerjan W., Mazzafera P. Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J. Exp. Bot. 2013;64:1769–1781. doi: 10.1093/jxb/ert045. [DOI] [PubMed] [Google Scholar]
- 34.Sterjiades R., Dean J., Gamble G., Himmelsbach D., Eriksson K.-E. Extracellular laccases and peroxidases from sycamore maple (Acer pseudoplatanus) cell-suspension cultures. Planta. 1993;190:75–87. doi: 10.1007/BF00195678. [DOI] [Google Scholar]
- 35.Hoffmann N., Benske A., Betz H., Schuetz M., Samuels A.L. Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 2020;184:806–822. doi: 10.1104/pp.20.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bürger M., Chory J. Stressed out about hormones: How plants orchestrate immunity. Cell Host Microbe. 2019;26:163–172. doi: 10.1016/j.chom.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye M., Liu M., Erb M., Glauser G., Zhang J., Li X., Sun X. Indole primes defence signalling and increases herbivore resistance in tea plants. Plant Cell Environ. 2021;44:1165–1177. doi: 10.1111/pce.13897. [DOI] [PubMed] [Google Scholar]
- 38.Ullah C., Tsai C., Unsicker S., Xue L., Reichelt M., Gershenzon J., Hammerbacher A. Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol. 2019;221:960–975. doi: 10.1111/nph.15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu W., Jin G., Jimenez-Aleman G.H., Wang X., Song J., Li S., Lou Y., Li R. The jasmonic acid-amino acid conjugates JA-Val and JA-Leu are involved in rice resistance to herbivores. Plant Cell Environ. 2021 doi: 10.1111/pce.14202. [DOI] [PubMed] [Google Scholar]
- 40.Constabel C.P., Barbehenn R. Defensive roles of polyphenol oxidase in plants. In: Schaller A., editor. Induced Plant Resistance to Herbivory. Springer; Dordrecht, The Netherlands: 2008. pp. 253–270. [Google Scholar]
- 41.Zhang J., Zhang X., Ye M., Li X.W., Lin S.B., Sun X.L. The jasmonic acid pathway positively regulates the polyphenol oxidase-based defense against tea geometrid caterpillars in the tea plant (Camellia sinensis) J. Chem. Ecol. 2020;46:308–316. doi: 10.1007/s10886-020-01158-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J., Sun X. Recent advances in polyphenol oxidase-mediated plant stress responses. Phytochemistry. 2021;181:112588. doi: 10.1016/j.phytochem.2020.112588. [DOI] [PubMed] [Google Scholar]
- 43.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 44.Niu Z., Li G., Hu H., Lv J., Zheng Q., Liu J., Wan D. A gene that underwent adaptive evolution, LAC2 (LACCASE), in Populus euphratica improves drought tolerance by improving water transport capacity. Hortic. Res. 2021;8:88. doi: 10.1038/s41438-021-00518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan Z., Krosse S., Achard P., van Dam N., Bede J.C. DELLA proteins modulateArabidopsisdefences induced in response to caterpillar herbivory. J. Exp. Bot. 2014;65:571–583. doi: 10.1093/jxb/ert420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li R., Zhang J., Li J., Zhou G., Wang Q., Bian W., Erb M., Lou Y. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife. 2015;4:e04805. doi: 10.7554/eLife.04805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J., Luo T., Wang W., Cao T., Li R., Lou Y. Silencing OsSLR1enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Plant Cell Environ. 2017;40:2147–2159. doi: 10.1111/pce.13012. [DOI] [PubMed] [Google Scholar]
- 48.Panda S., Kazachkova Y., Aharoni A. Catch-22 in specialized metabolism: Balancing defense and growth. J. Exp. Bot. 2021;72:6027–6041. doi: 10.1093/jxb/erab348. [DOI] [PubMed] [Google Scholar]
- 49.Chen D., Li J., Jiao F., Wang Q., Li J., Pei Y., Zhao M., Song X., Guo X. ZmACY-1 antagonistically regulates growth and stress responses in Nicotiana benthamiana. Front. Plant Sci. 2021;12:593001. doi: 10.3389/fpls.2021.593001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai W., Yang S., Wu R., Cao J., Shen L., Guan D., Shuilin H. Pepper NAC-type transcription factor NAC2c balances the trade-off between growth and defense responses. Plant Physiol. 2021;186:2169–2189. doi: 10.1093/plphys/kiab190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z.W., Duan X.N., Jin S., Li X.W., Chen Z.M., Ren B.Z., Sun X.L. Regurgitant derived from the tea geometrid Ectropis obliqua suppresses wound-induced polyphenol oxidases activity in tea plants. J. Chem. Ecol. 2013;39:744–751. doi: 10.1007/s10886-013-0296-x. [DOI] [PubMed] [Google Scholar]
- 52.Li X., Zhang J., Lin S., Xing Y., Zhang X., Ye M., Chang Y., Guo H., Sun X. (+)-Catechin, epicatechin and epigallocatechin are important inducible defensive compounds against Ectropis grisescens in tea plants. Plant Cell Environ. 2021 doi: 10.1111/pce.14216. [DOI] [PubMed] [Google Scholar]
- 53.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:884–890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.