Abstract
Background: Trimethylaminuria (TMAU) is a rare genetic disease characterized by the accumulation of trimethylamine (TMA) and its subsequent excretion trough main body fluids, determining the characteristic fish odour in affected patients. We realized an experimental study to investigate the role of several coding variants in the causative gene FMO3, that were only considered as polymorphic or benign, even if the available literature on them did not functionally explain their ineffectiveness on the encoded enzyme. Methods: Mutational analysis of 26 TMAU patients was realized by Sanger sequencing. Detected variants were, subsequently, deeply statistically and in silico characterized to determine their possible effects on the enzyme activity. To achieve this goal, a docking prediction for TMA/FMO3 and an unbinding pathway study were performed. Finally, a TMAO/TMA urine quantification by 1H-NMR spectroscopy was performed to support modelling results. Results: The FMO3 screening of all patients highlighted the presence of 17 variants distributed in 26 different haplotypes. Both non-sense and missense considered variants might impair the enzymatic kinetics of FMO3, probably reducing the interaction time between the protein catalytic site and TMA, or losing the wild-type binding site. Conclusions: Even if further functional assays will confirm our predictive results, considering the possible role of FMO3 variants with still uncertain effects, might be a relevant step towards the detection of novel scenarios in TMAU etiopathogenesis.
Keywords: FMO3, TMAU, in silico, proteomics, genetic variants
1. Introduction
Trimethylaminuria (TMAU) is a rare genetic disease characterized by the accumulation of trimethylamine (TMA) and its subsequent excretion trough breath sweat, saliva, vaginal secretions, and urine, determining a characteristic fish odour in affected patients [1]. TMA is a volatile tertiary amine, synthesized by gut microbiota. Choline, betaine, lecithin, and L-carnitine introduced by diet are TMA main precursors [2]. In a non-pathological condition, TMA is oxidized in an in-odorous molecule, the trimethylamine n-oxide (TMAO), by a hepatic microsomal enzyme known as Flavin-containing monooxygenase 3 (FMO3) [3].
TMA accumulation can be caused by both genetic and environmental factors, leading to TMAU different forms. To date, the primary form (TMAU1) is known to be determined by the FMO3 enzyme deficiency or functional impairment, as consequence of the FMO3 gene mutation induced effects [4]. The secondary form (TMAU2) is mainly caused by a gut dysbiosis condition. A higher relative abundance of some bacterial families could determine a TMA overproduction. Lastly, the transient form could be induced by excessive sweating due to intense physical effort, stress, mood disturbances, or influence of steroid hormones during the peri-menstrual period [5].
The absence of physical anomalies has brought this syndrome little medical interest, underestimated in its most purely psycho-affective aspects. Although this metabolic disorder is not physically disabling, it has a strong impact on the patient’s socio-psychic sphere. Social isolation, behaviour disorders, and depression are the main psychic manifestations and, in the most severe cases, suicide can also occur [6]. TMAU is particularly severe in children and adolescents due to the negative impact within the school context associated with humiliation experiences [7]. At this age, such social difficulties, in addition to the bad smell, lead to the consultation of a medical specialist. Unfortunately, there is no definitive therapy to date, only a combination of different approaches to reduce, temporarily, the bad smell [8]. Products such as chlorophyllin and activated charcoal are used to sequester the trimethylamine produced in the gut, riboflavin supplements are given to enhance residual FMO3 enzyme activity, while antibiotics (metronidazole, amoxicillin, and neomycin) are used to reduce the levels of TMA-producing gut bacteria [9,10]. In addition, it is advisable to avoid foods such as shellfish, eggs, meat, legumes, and mushrooms, which contain TMA precursors (choline, carnitine, and betaine) [11].
Recently, a strong interest has arisen on the study of the secondary form of trimethylaminuria (TMAU2), but still little is known about the primary form (TMAU1), especially about the exact physiological and pathophysiological activity of the FMO3 [12].
The FMO3 enzyme catalyses the NADPH-dependent oxidative metabolism of a wide range of exogenous chemicals, including drugs, dietary-derived compounds, and environmental pollutants [13]. As already cited, it is responsible for the conversion of TMA, synthesized from precursors introduced with diet, in its oxidized and odourless form TMAO. Genetic variants in the FMO3 gene, inherited in an autosomal recessive manner, could cause an enzymatic activity deficit and, consequently, a TMA accumulation [14,15,16].
To date, 91 FMO3 variants were identified, and more than half of them are responsible for the TMAU phenotype (http://www.hgmd.cf.ac.uk/ac/index.php, accessed on 19 October 2021). Among the remaining variants, some are of uncertain clinical significance, while others are classified as “benign” and, therefore, should not affect the FMO3 enzyme activity [17,18,19].
However, as such variants have a high frequency in TMAU patients, frequently without the contemporary presence of causative mutations, we hypothesized that their haplotypes could play a significant role in FMO3 activity reduction or alteration. To confirm this hypothesis, we performed a proteomic in silico analysis, using different platforms and software, with the final aim of revealing how these variant combinations could influence the enzyme folding, also simulating its dynamic behaviour with the TMA substrate.
2. Results
2.1. Genotyping of TMAU Patients Revealed Different Haplotypes Made of Causative or Associated Variants Carried by FMO3
We screened the whole cohort of patients for the FMO3 gene, founding 17 variants distributed in 26 different haplotypes. Among them, 11 haplotypes were made of missense or non-sense variants only (2, 4, 5, 6, 7, 8, 9, 10, 11, 12, and 13). Four patients (12, 17, 20, and 21) showed variants in homozygous condition, with a missense common to three patients (c. G472A p.E158K) and a splicing one common to two patients (c. 627 + 10 C > G). All known TMAU causative mutations were detected in heterozygous condition only. In detail, such variants were carried by patients 2 and 5 (c. 458C > T p.P153L), 9 (c. 1474C > T p.R492W), 10 (c. 713G > A p.R238Q and c. 713G > C p.R238P), 11 (c. 1424G > A G475D) and 13 (c. 1424G > A G475D and c. 713G > C p.R238P). The most complex haplotypes were exhibited by patients 5 and 8, which carried four common variants (c. G472A p.E158K, c. 485-22G > A, c. 1184-32_1184-31insT, and c. 923A > G p.E308G) and, respectively, c. 458C > T (p.P153L) and c. 441C > T (p.S147=). Finally, the most frequent variants resulted the c. G472A (p.E158K), carried by 18 TMAU patients. Further details are available in Table 1.
Table 1.
FMO3 detected variants genotyping and distribution through the cohort of patients. Seventeen variants were identified during FMO3 genotyping of all twenty-six considered patients. DM = disease mutation (HGMD). DM? = disease mutation without certain evidence (HGMD). DFP = disease-associated polymorphism with additional supporting functional evidence (HGMD). DP = disease-associated polymorphism (HGMD). FP = in vitro/laboratory or in vivo functional polymorphism (HGMD). Variants without HGMD acronym are not yet fully evaluated. HET = heterozygous variant. HOM = homozygous variant.
| ID | FMO3 Variants | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c. G472A (p.E158K) (DFP) | c.627+10 C>G (DM?) | c.485-22G>A | c.1184-32_1184-31insT | c.923A>G (p.E308G) (DFP) | c.769G>A (p.V257M) (FP) | c.458C>T (p.P153L) (DM) | c.441C>T (p.S147=) (DP) | c.993_994delTA (p.Tyr331Stop) | c.1474C>Tp.R492W (DM) | c.1139_1140del (p.Pro380fs) | c.713G>A (p.R238Q) (DM) | c.1424G>A (G475D) (DM) | c.713G>C (p.R238P) (DM) | c.855C>T (Asn285=) | c.422A>T (Asp141Val) | c.539G>T (Gly180Val) | |
| 1 | HET | ||||||||||||||||
| 2 | HET | HET | HET | ||||||||||||||
| 3 | HET | ||||||||||||||||
| 4 | HET | HET | |||||||||||||||
| 5 | HET | HET | HET | HET | HET | ||||||||||||
| 6 | HET | HET | |||||||||||||||
| 7 | HET | HET | HET | ||||||||||||||
| 8 | HET | HET | HET | HET | HET | ||||||||||||
| 9 | HET | HET | HET | ||||||||||||||
| 10 | HET | HET | HET | ||||||||||||||
| 11 | HET | HET | |||||||||||||||
| 12 | HOM | HOM | HOM | HOM | |||||||||||||
| 13 | HET | HET | |||||||||||||||
| 14 | HET | HET | HET | ||||||||||||||
| 15 | HET | HET | HET | ||||||||||||||
| 16 | HET | HET | |||||||||||||||
| 17 | HOM | HOM | HOM | ||||||||||||||
| 18 | HET | HET | HET | ||||||||||||||
| 19 | HET | HET | |||||||||||||||
| 20 | HOM | HOM | |||||||||||||||
| 21 | HOM | HET | HET | ||||||||||||||
| 22 | HET | HET | HET | ||||||||||||||
| 23 | HET | ||||||||||||||||
| 24 | HET | ||||||||||||||||
| 25 | HET | HET | |||||||||||||||
| 26 | HET | HET | HET | HET | |||||||||||||
2.2. Primary Structural and Biochemical Analyses Highlighted Possible Altered Chemical-Physical Features in Mutated FMO3
The physicochemical properties of each mutated FMO3 protein computed by ProtParam are shown in Table 2. The molecular weight ranged from 36,917 g/mol of Y331Stop to 60,090 g/mol of E158K_G475D. Theoretical isoelectric points (pI) for these proteins ranged from 6.26 of Y331Stop to 8.47 of E158K_E308G. The theoretical pI represents the pH at which a particular molecule or surface carries no net electrical charge, and it could help to understand the protein charge stability [20]. The mutated FMO3 proteins of this study had GRAVY indexes ranging from −0.06 of D141V_G180V to −0.21 of Y331Stop. The GRAVY value for a peptide or protein is calculated as the sum of hydropathy values of all of the amino acids, divided by the number of residues in the sequence [21]. Thus, these low GRAVY ranges indicate the possibility of being a globular (hydrophilic) protein rather than membranous (hydrophobic). This information might be useful for localizing these proteins. The aliphatic index for all FMO3 proteins involved in this study ranged from 73.21 of Y331Stop to 83.16 of D141V_G180V. A high aliphatic index indicates that a protein is thermo-stable over a wide temperature range [22]. The aliphatic index of a protein is defined as the relative volume occupied by aliphatic side chains (alanine (Ala), valine (Val), isoleucine (Ile), and leucine (Leu)). It may be regarded as a positive factor for the increase in thermo-stability of globular proteins [22]. All proteins were stable (a protein whose instability index is <40 is predicted as stable [22]), with the instability index values for our hypothetical and conserved proteins range from 33.02 of Y331Stop to 35.12 of V267M. Further details on biochemical properties of wild-type and mutated FMO3 proteins are available in Table S1.
Table 2.
Biochemical and physical changes prediction between wild-type and mutated FMO3. The ProtParam computed features included the molecular weight, theoretical pI, amino acid composition, atomic composition, instability index, aliphatic index, and grand average of hydropathicity (GRAVY). Analyses of such biochemical and physical parameters showed a decreased global instability from wild-type to mutated FMO3 protein encoded by FMO3 gene carrying SNVs only.
| WILD | P153L_E158K | V267M | E158K | E158K_E308G | E158K_R492W | E158K_R238Q | E158K_G475D | D141V_G180V | Y331Stop | R238P_G475D | P380fs_P153L_E158K | P380fs | P380fs_E308G | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residues | 532 | 532 | 532 | 532 | 532 | 532 | 532 | 532 | 532 | 330 | 532 | 381 | 381 | 381 |
| MW (g/mol) | 60,033 | 60,048 | 60,065 | 60,032 | 59,960 | 60,062 | 60,004 | 60,090 | 60,059 | 36,917 | 60,032 | 42,556 | 42,541 | 42,469 |
| 1 μg to pmol | 16.6 | 16.65 | 16.65 | 16.66 | 16.68 | 16.65 | 16.67 | 16.64 | 16.65 | 27.09 | 16.66 | 23.5 | 23.51 | 23.55 |
| Net charge (pH = 7) | 2.17 | 4.17 | 2.17 | 4.17 | 5.17 | 3.17 | 3.17 | 3.17 | 3.17 | −2.74 | 0.17 | −0.65 | −2.65 | −1.65 |
| pI | 7.9 | 8.33 | 7.9 | 8.33 | 8.47 | 8.15 | 8.15 | 8.15 | 8.16 | 6.26 | 7.09 | 6.76 | 6.33 | 6.52 |
| Avg. hydropathy (GRAVY) | −0.08 | −0.07 | −0.08 | −0.08 | −0.07 | −0.07 | −0.08 | −0.09 | −0.06 | −0.21 | −0.08 | −0.10 | −0.11 | −0.10 |
| Aliphatic index | 82.07 | 82.8 | 81.52 | 82.07 | 82.07 | 82.07 | 82.07 | 82.07 | 83.16 | 73.21 | 82.07 | 80.05 | 79.03 | 79.03 |
| Instability index | 34.24 | 33.64 | 35.12 | 33.87 | 33.19 | 33.19 | 33.59 | 33.75 | 34.37 | 33.02 | 34.48 | 33.48 | 34.31 | 33.36 |
| 1 mg/mL to A₂₈₀ | 1.46 | 1.46 | 1.46 | 1.46 | 1.46 | 1.55 | 1.46 | 1.46 | 1.46 | 1.23 | 1.46 | 1.14 | 1.14 | 1.14 |
| ε₂₈₀ (M⁻¹ cm⁻¹) | 87,485 | 87,485 | 87,485 | 87,485 | 87,485 | 92,85 | 87,485 | 87,485 | 87,485 | 45,420 | 87,485 | 48,400 | 48,400 | 48,400 |
| 1 A₂₈₀ to mg/mL | 0.69 | 0.69 | 0.69 | 0.69 | 0.69 | 0.65 | 0.69 | 0.69 | 0.69 | 0.81 | 0.69 | 0.88 | 0.88 | 0.88 |
| 1 mg/mL to A₂₈₀ (red.) | 1.45 | 1.45 | 1.45 | 1.45 | 1.45 | 1.54 | 1.45 | 1.45 | 1.45 | 1.22 | 1.45 | 1.13 | 1.13 | 1.13 |
| ε₂₈₀ (M⁻¹ cm⁻¹) (red.) | 86,860 | 86,860 | 86,860 | 86,860 | 86,860 | 92,360 | 86,860 | 86,860 | 86,860 | 44,920 | 86,860 | 47,900 | 47,900 | 47,900 |
| 1 A₂₈₀ to mg/mL (red.) | 0.69 | 0.69 | 0.69 | 0.69 | 0.69 | 0.65 | 0.69 | 0.69 | 0.69 | 0.82 | 0.69 | 0.89 | 0.89 | 0.89 |
| Features | NAP, FAD, WP5, OXY, ADP, IND, NA, 23 helices, 22 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, OXY, MG, IND, MMZ, CYH, 11 helices, 20 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 46 helices, 44 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 28 helices, 52 strands |
NAP, FAD, OXY, IND, MMZ, CYH, 14 helices, 26 strands | NAP, FAD, WP5, OXY, ADP, IND, NA, 28 helices, 52 strands |
2.3. Tertiary Structure Prediction of Mutated FMO3 Showed That Mutated Amino Acids Changed Their Distance from Surrounding Amino Acids
Due to the unavailability of FMO3 3D structure in the PDB Molecule Database, the i-TASSER servers were used to predict and model multiple domains of wild-type and mutated proteins with high confidence based on the high scoring template. Prediction by i-TASSER resulted the most confident, showing a normalized Z-score of 8.91 for wild-type structure and a mean value of 8.77 for the mutated ones.
A deep analysis of tertiary structures of FMO3 mutated forms highlighted a global distribution of amino acids involved in missense variants along the outer domains of the enzyme (Figure 1). The truncated forms of FMO3 determined by non-sense variants, instead, seem to lose the binding sites for co-enzymes and co-factors (Figure 2).
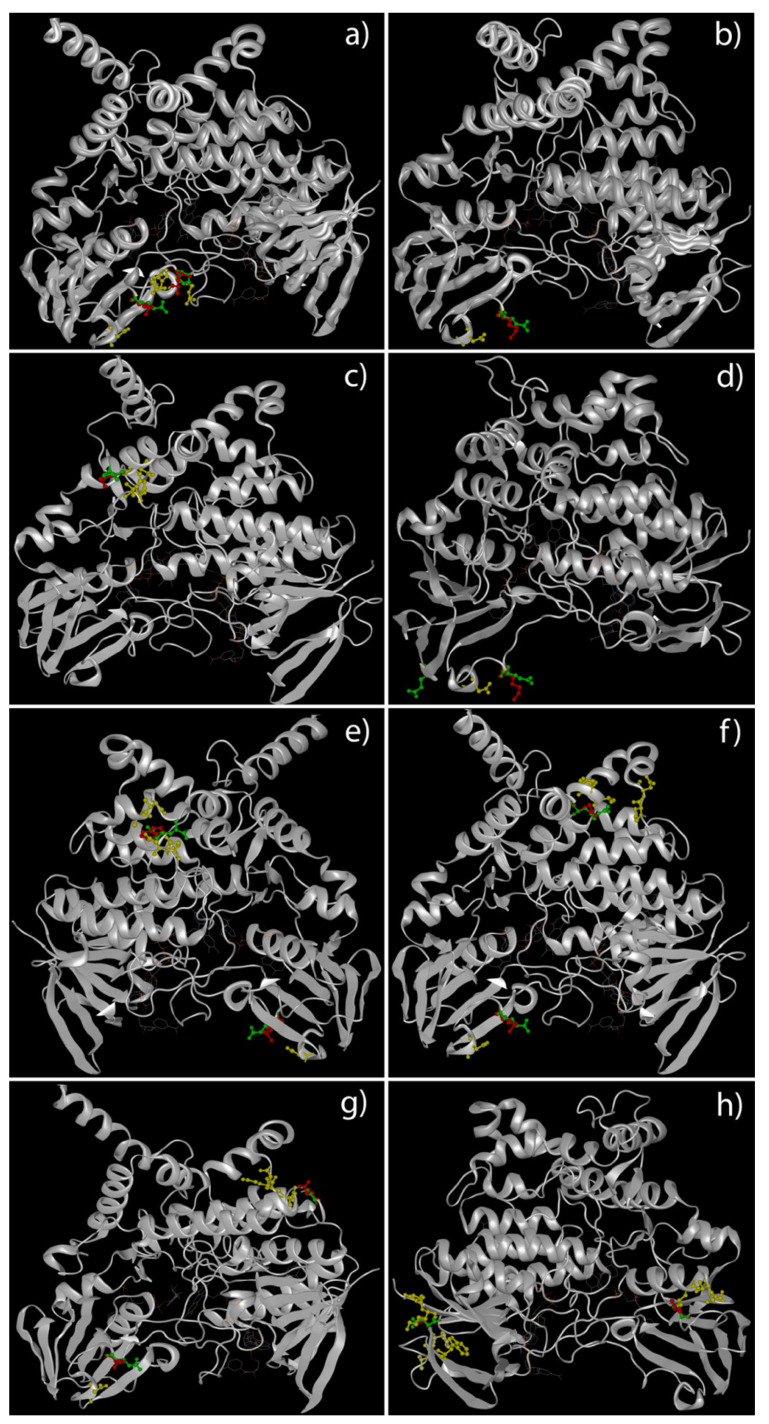
Figure 1.
3D structural models of FMO3 proteins in patients carrying missense variants. This panel highlights the predicted tertiary structure of FMO3 in patients 5 (P153L and E158K) (a), 6 (E158K) (b), 7 (V257M) (c), 8 (E158K and E308G) (d), 9 (E158K and R492W (e), 10 (E158K and R238Q) (f), 11 (E158K and G475D) (g) and 12 (D141V and G180V) (h). Green ball-and-stick aa = wild-type aa. Red ball-and-stick aa = mutated aa. Yellow ball-and-stick aa = aa nearest to aa involved in variant.
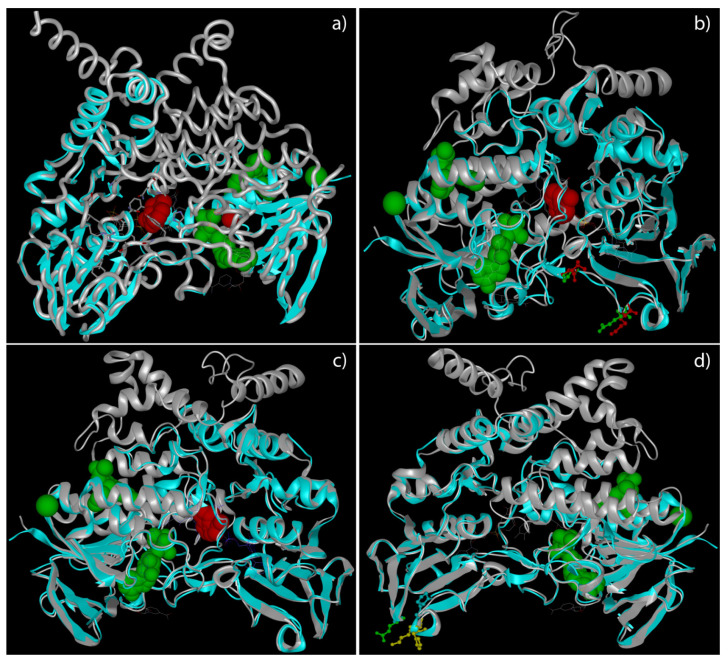
Figure 2.
3D structural models of FMO3 proteins in patients carrying non-sense variants. This panel highlights the predicted tertiary structure of FMO3 in patients 1 (Y331Stop) (a), 2 (P153L, E158K and P380fs) (b), 3 (P380fs) (c), and 4 (E308G and P380fs) (d). The grey tube/ribbons represent the wild-type structures, while the light blue the over imposed mutated ones. The green spheres represent the predicted ligands/coenzymes/cofactors exclusive of wild-type FMO3 (ADP, Na, and TMA), while the red spheres the predicted ones exclusive of different truncated FMO3 (Indole, Mg, and TMA; exceptions are represented by patient 3, who does not present the Mg, and by patient 4, who does not present any exclusive ligand compared to wild-type form of FMO3). Lateral chains of amino acids involved in missense variants are represented in green for the wild-type allele and in red for the mutated one. Yellow chains are characteristics of amino acids nearest to mutated ones.
Looking inside the tridimensional structure and measuring the distance between the amino acid involved in missense variant and the surrounding ones, it was possible to evidence relevant changes in at least eight patients. Two altered forms of FMO3 (E158K + P153L and E158K + G475D) showed a global reduction in distance between mutated amino acid and its nearest ones, while two others (E158K + R492W and E158K + R238Q) evidenced a wider distance between the same considered amino acids (Table 3).
Table 3.
FMO3 missense variants highlighting the highest distance differences between changed amino acid and nearest ones. The wild-type alleles compared to mutated ones showed possible interactions with difference nearest amino acids, also involving different atoms or functional groups (between brackets). Side chains functional groups are reported following the IUPAC-IUB Commission on Biochemical Nomenclature. Wt = wild-type amino acid and atom/functional group involved in a possible interaction with the nearest aa. Mut = mutated amino acid and atom/functional group involved in a possible interaction with the nearest aa.
| ID | Variant | Wt | Nearest a.a. | a.a. Distance (Å) | Mut | Nearest a.a. | a.a. Distance (Å) |
|---|---|---|---|---|---|---|---|
| 2 | P153L | Pro153 (CG) | Arg174 (NE) | 4.33 | Leu153 (CD1) | Arg174 (NE) | 5.20 |
| Pro153 © | Leu155 (N) | 3.20 | Leu153 (CD2) | Leu155 (CD2) | 1.71 | ||
| Pro153 (CB) | His172 (CE1) | 3.32 | Leu153 (CB) | His172 (CG) | 4.00 | ||
| E158K | Glu158 (N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.71 | |
| 4 | E308G | Glu308 | / | / | Gly308 | / | / |
| 5 | P153L | Pro153 (CG) | Arg174 (NE) | 4.33 | Leu153 (CD1) | Arg174 (NE) | 5.20 |
| Pro153 (C) | Leu155 (N) | 3.20 | Leu153 (CD2) | Leu155 (CD2) | 1.71 | ||
| Pro153 (CB) | His172 (CE1) | 3.32 | Leu153 (CB) | His172 (CG) | 4.00 | ||
| E158K | Glu158 (N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.71 | |
| 6 | E158K | Glu158 (N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.71 |
| 7 | V257M | Val257 (N) | Tyr256 (C) | 1.35 | Met257 (N) | Tyr256 (C) | 1.2 |
| Asp253 (O) | 2.97 | Asp253 (O) | 3.13 | ||||
| Val257 (CG2) | Val277 (N) | 4.03 | Met257 (CE) | Val277 (N) | 4.09 | ||
| 8 | E158K | Glu158 (N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.71 |
| E308G | Glu308 | / | / | Gly308 | / | / | |
| 9 | E158K | Glu158 (N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.71 |
| R492W | Arg492 (CD) | Asp76 (OD2) | 3.07 | Tyr492 (NE1) | ASp76 (OD2) | 2.53 | |
| Arg492 (NH2) | Thr488 (OG1) | 2.6 | Trp492 (CH2) | Thr488 (O) | 3.11 | ||
| Arg492 (NH1) | Ala485 (O) | 2.72 | Trp492 (N) | Ala485 (CB) | 4.3 | ||
| Arg492 (NE) | Glu65 (OE2) | 2.68 | Trp492 (CE3) | Glu65 (OE2) | 2.92 | ||
| 10 | E158K | Glu158 (N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.71 |
| R238Q | Arg238 (NH2) | Pro445 (O) | 4.95 | Gln238 (OE1) | Pro445 (O) | 7.59 | |
| Asn446 (N) | 7.15 | Asn446 (N) | 9.81 | ||||
| Arg238 (CG) | Val261 (O) | 3.12 | Gln238 (CB) | Val461 (O) | 3.59 | ||
| Arg238 (CB) | Tyr462 (O) | 3.06 | Tyr462 (O) | 2.59 | |||
| 11 | E158K | Glu158(N) | Asn164 (OD1) | 5.94 | Lys158 (CA) | Asn164 (ND2) | 5.7 |
| G475D | Gly475 (N) | Pro445 (N) | 7.07 | Asp475 (OD2) | Pro445 (N) | 5.90 | |
| Gly475 (CA) | Lys444 (N) | 6.18 | Lys444 (N) | 3.83 | |||
| Ala443 (CA) | 4.30 | Ala443 (CA) | 3.02 | ||||
| Gly442 (O) | 4.52 | Gly442 (O) | 1.41 | ||||
| 12 | D141V | D141 (N) | Lys4 (O) | 2.87 | Val141 (N) | Lys4 (O) | 3.02 |
| Val139 (C) | 4.52 | Val139 (C) | 4.50 | ||||
| Phe140 (C) | 1.34 | Phe140 (C) | 1.25 | ||||
| D141 (OD1) | Lys3 (CA) | 3.52 | Val141 (CG1) | Lys3 (CG) | 3.88 | ||
| D141 (C) | Trp125 (CD1) | 4.59 | Val141 (C) | Trp125 (CD1) | 4.44 | ||
| G180V | Gly180 (N) | Leu203 (CD2) | 6.28 | Val180 (N) | Leu203 (CD2) | 6.22 | |
| Ala207 (N) | 8.42 | Ala207 (N) | 8.28 | ||||
| Thr206 (OG1) | 5.63 | Thr206 (OG1) | 5.49 | ||||
| 13 | R238P | Arg238 (NH1) | Ile447 (CD) | 3.62 | Pro238 (CB) | Ile447 (CD) | 6.78 |
| Arg238 (NH2) | Pro445 (O) | 4.95 | Pro238 (CG) | Pro445 (O) | 9.01 | ||
| Arg238 (N) | Gly464 (O) | 3.44 | Pro238 (CD) | Gly464 (O) | 2.74 | ||
| Arg238 (N) | Cys466 (N) | 4.56 | Pro238 (CD) | Cys466(N) | 3.65 | ||
| G475D | Gly475 (N) | Pro445 (N) | 7.07 | Asp475 (OD2) | Pro445 (N) | 5.90 | |
| Gly475 (CA) | Lys444 (N) | 6.18 | Lys444 (N) | 3.83 | |||
| Ala443 (CA) | 4.30 | Ala443 (CA) | 3.02 | ||||
| Gly442 (O) | 4.52 | Gly442 (O) | 1.41 |
2.4. Docking Analysis of FMO3 Forms Unveiled the Most Probable Binding Sites of TMA and How Considered Variants Could Impair This Binding
The docking analyses calculated a total of 10 modes with affinity (kcal/mol), Ki (µmol), and RMSD lower and upper bound (A) for each protein (Table S2). The mode presenting the lowest values for such variables, considered the most significant, was chosen for next steps (Table 4). The wild-type FMO3 highlighted that the TMA binding site could be made of Ser216, Gly217, Ser218, Trp219, Pro273, Asn275, Gly276, Leu278, Arg279, Lys280, Glu 281, and Pro 282. The same amino acids should constitute the active site of FMO3 in patients 7 only (V257M). It is interesting to notice that patients 2, 10, 11, and 13 share the same aa in their FMO3 active sites, as well as patient 8 with patient 9 and patient 4 with patient 5. On the contrary, little difference was found between active site aa of patient 8 and 9, which evidenced the loss of Lys422 if compared with patients 2, 10, 11, and 13. Moreover, patient 3 presented the loss of two aa (Gly38 and Val151) rather than patients 4 and 5. The absence of Arg417 altered the FMO3 of patient 6 from the enzyme of patients 2, 10, 11, and 13. Finally, the only FMO3 forms which presented unique aa in their active sites were the ones carried by patients 1 and 12. More details about aa of mutated FMO3 active sites are available in Table 5 and Figure 3 and Figure S1.
Table 4.
Best significant modes of TMA/FMO3 docking models. AutoDock Vina permitted to compute 10 different modes for docking analysis between TMA and FMO3. This table shows the most reliable models, based on energetic variables. Ki = inhibition constant. RMSD values are calculated relative to the best mode (the first model) and use only movable heavy atoms (i.e., only ligand atoms, not hydrogen). RMSD upper bound matches each atom in one conformation with itself in the other conformation, ignoring any symmetry. RMSD lower bound matches each atom in one conformation with the closest atom of the same element type in the other conformation.
| Protein | Affinity (kcal/mol) | Ki (µmol) | Lower Bound of the RMSD from This Ligand’s Best Mode (A) | Upper Bound of the RMSD from This Ligand’s Best Mode (A) |
|---|---|---|---|---|
| WILD-TYPE | −2.103 | 28,756.2 | 0.000 | 0.000 |
| D141V_G180V | −2.160 | 26,096.6 | 0.000 | 0.000 |
| E158K_E308G | −2.242 | 22,732.4 | 0.000 | 0.000 |
| E158K_G475D | −2.239 | 22,844.0 | 0.000 | 0.000 |
| E158K_R238Q | −2.240 | 22,789.5 | 0.000 | 0.000 |
| E158K_R492W | −2.242 | 22,739.5 | 0.000 | 0.000 |
| E158K | −2.240 | 22,791.2 | 0.000 | 0.000 |
| P153L_E158K | −2.240 | 22,807.7 | 0.000 | 0.000 |
| P380fs | −2.079 | 29,951.3 | 0.000 | 0.000 |
| P380fs_P153L_E158K | −2.082 | 29,760.4 | 0.000 | 0.000 |
| P380fs_E308G | −2.082 | 29,789.6 | 0.000 | 0.000 |
| R238P_G475D | −2.242 | 22,736.2 | 0.000 | 0.000 |
| V257M | −2.242 | 22,749.4 | 0.000 | 0.000 |
| Y331Stop | −2.033 | 32,327.0 | 0.000 | 0.000 |
Table 5.
Mutated FMO3 showed different amino acids predicted for the TMA binding sites. Different haplotypes present in the mutated forms of FMO3 were predicted to change the aa characteristic of wild-type enzyme catalytic site, probably leading to an impairment of FMO3 activity. The “X” indicates that the specific aa in the related row is an integral part of the FMO3 active site when the variant/s reported in the columns is/are carried by the FMO3 gene.
| FMO3 Haplotypes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active site a.a. Residues | Wild-Type | Y331Stop | P153L + E158K | P380Fs | P380Fs + E308G | P153L + E158K + P380Fs | E158K | V257M | E308G + E158K | R492W + E158K | R238Q + E158K | G475D + E158K | D141V + G180V | R238P + G475D |
| Gly 9 | X | X | X | |||||||||||
| Ala 10 | X | X | X | |||||||||||
| Gly 11 | X | X | X | |||||||||||
| Val 12 | X | X | X | |||||||||||
| Ser 13 | X | X | X | |||||||||||
| Gly 14 | X | X | X | |||||||||||
| Glu 32 | X | X | X | |||||||||||
| Gly 38 | X | X | ||||||||||||
| Gly 39 | X | X | X | |||||||||||
| Leu 40 | X | X | X | |||||||||||
| Ala 52 | X | X | X | |||||||||||
| Cys 146 | X | X | X | |||||||||||
| Ser 147 | X | X | X | |||||||||||
| Gly 148 | X | X | X | |||||||||||
| Hys 149 | X | X | X | |||||||||||
| Val 151 | X | X | ||||||||||||
| Ser 216 | X | X | ||||||||||||
| Gly 217 | X | X | ||||||||||||
| Ser 218 | X | X | ||||||||||||
| Trp 219 | X | X | ||||||||||||
| Gly 240 | X | |||||||||||||
| Leu 243 | X | |||||||||||||
| Lys 244 | X | |||||||||||||
| Leu 247 | X | |||||||||||||
| Ile 251 | X | |||||||||||||
| Ser 252 | X | |||||||||||||
| Asp 253 | X | |||||||||||||
| Leu 255 | X | |||||||||||||
| Tyr 256 | X | |||||||||||||
| Gln 259 | X | |||||||||||||
| Pro 273 | X | X | ||||||||||||
| Asn 275 | X | X | ||||||||||||
| Gly 276 | X | X | ||||||||||||
| Leu 278 | X | X | ||||||||||||
| Arg 279 | X | X | X | X | X | X | X | X | X | X | ||||
| Lys 280 | X | X | ||||||||||||
| Glu 281 | X | X | ||||||||||||
| Pro 282 | X | X | ||||||||||||
| Leu 352 | X | |||||||||||||
| Phe 353 | X | |||||||||||||
| Lys 354 | X | |||||||||||||
| Gly 355 | X | |||||||||||||
| Phe 371 | X | |||||||||||||
| Val 372 | X | |||||||||||||
| Ser 381 | X | X | X | |||||||||||
| Lys 412 | X | X | X | X | X | X | X | X | ||||||
| Met 413 | X | |||||||||||||
| Lys 415 | X | X | X | X | X | X | X | |||||||
| Lys 416 | X | X | X | X | X | X | X | X | ||||||
| Arg 417 | X | X | X | X | X | X | ||||||||
| Trp 419 | X | X | X | X | X | X | X | |||||||
| Phe 420 | X | X | X | X | X | X | X | |||||||
| Lys 422 | X | X | X | X | X | |||||||||
| Thr 425 | X | X | X | X | X | X | X | |||||||
| Ile 426 | X | |||||||||||||
| Gln 427 | X | X | X | X | X | X | X | |||||||
| Thr 428 | X | X | X | X | X | X | X | X | ||||||
| Asp 429 | X | X | X | X | X | X | X | |||||||
| Tyr 433 | X | |||||||||||||
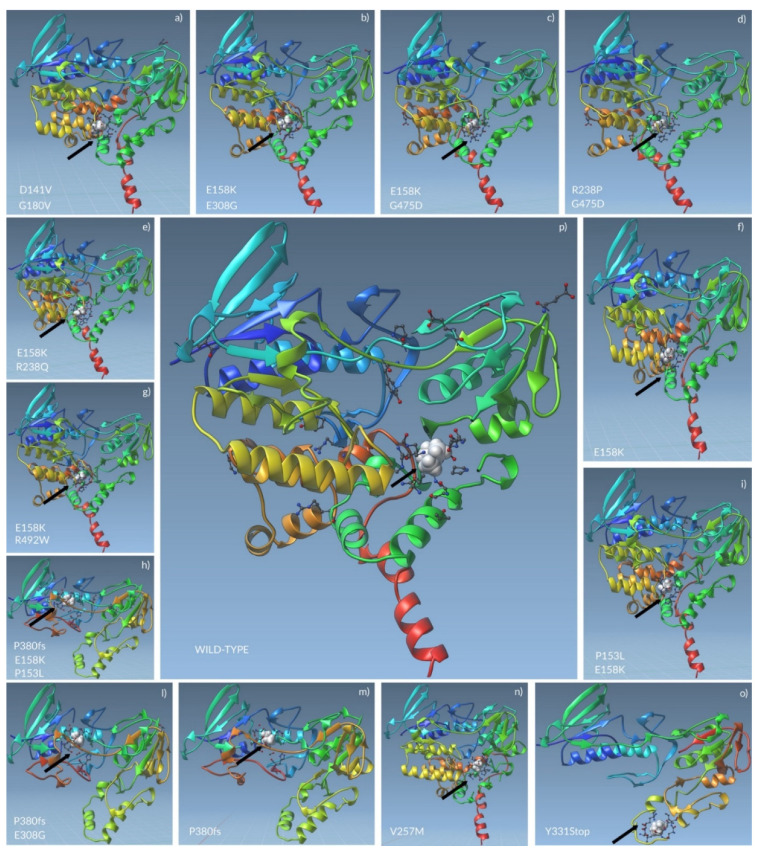
Figure 3.
TMA docking to FMO3 could involve different amino acids in mutated enzymes. The non-sense and missense variants carried by mutated FMO3 (a–o) might shift the TMA binding sites far from the wild-type active site of the enzyme (p). The black arrows indicate the TMA (white spheres) bonded to the active site of FMO3, whose aa are represented as ball-and-stick. The other aa, represented as ball-and-stick, separated from the ones in the catalytic site, are the aa involved in mutations.
2.5. The Unbinding Pathway Analyses of Mutated FMO3 Revealed an Impaired Interaction between TMA and Enzyme Active Site
The unbinding pathways emerged from Ligand Path Finder analyses showed the TMA could follow the identical route in patients 6, 8, 10, and 11, as well as in patients 9 and 13. Only patient 7 (carrying V257M) shares a pathway common to the wild-type FMO3.
In the wild-type FMO3, the TMA mainly interacted with the aa Ser216, Trp219, Leu278, and Lys 280. It went through the entire protein, also interacting with the Pro282 and the Glu 281, passing along the area containing the binding sites for FAD and NADP+.
The TMA unbinding pathway through the mutated FMO3 of patients 6, 8, 10, and 11 was the exact opposite of the wild-type. Here, the TMA firstly interacted with the Trp419 (near to the FAD binding site), then bounced on the Arg279, the Asn245, and the Asn246.
A similar pathway was shown for patient 9, with the difference made of the interaction with the Leu278 after Trp419.
The FMO3 carrying the V257M highlighted a TMA which interacted with Lys280, Pro 273, Asn275 and, at the end, with Ser216. During its exit path, the TMA seemed to be able to interact with NADP+. Even if such an exit way was opposite if compared to the wild-type, the TMA seemed to bind several aa of wild-type FMO3 active site.
An interesting case is the one of patient 2, wherein the TMA interacted with Trp419 first, then with the Leu278 and the Arg279. After these interactions, due to the FMO3 folding, Lys280 and Glu281 changed their conformation.
All the truncated forms of FMO3 showed the loss of FAD binding sites.
In patient 1, the TMA interacted with Leu235 before exiting in a way similar to the wild-type (from top to bottom).
In patient 3, the TMA interacted with Ala377, Ala378, Ile379, Glu9, Ala10, Gly11, and, finally, with Glu281, before exiting as before.
The FMO3 of patient 4 foresaw the interaction between TMA and Glu281 before Lys 280, preceding the exit in a way opposite to the wild-type FMO3 (from the bottom to the top).
Finally, in patient 5, the TMA interacted with Ile87, Glu39, Gly38, Gly11, Ser85, and Asn84, tracing a route opposite to the wild-type FMO3, completely away from the enzyme active site.
A graphical representation of unbinding pathways for wild-type and all mutated FMO3 enzymes is shown in Figure 4, while detailed routes for all proteins are shown in Videos S1–S14.
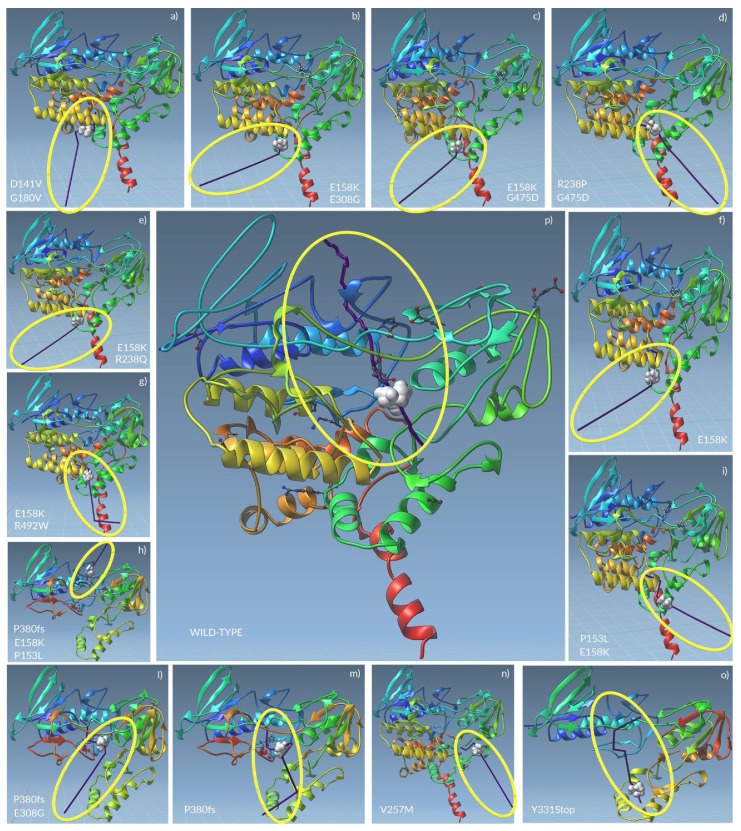
Figure 4.
The unbinding pathways analyses showed that FMO3 catalytic activity might not be fully performed. The TMA unbinding pathways through the whole FMO3 mutated proteins (a–o) seemed to impair the enzyme functionality when compared to the wild-type one (p). The yellow circles indicate the unbinding pathway produced by pathlines (purple line) of TMA (white spheres) through the FMO3.
2.6. NMR Spectra Functionally Confirmed an Altered Catalytic Activity of Mutated FMO3
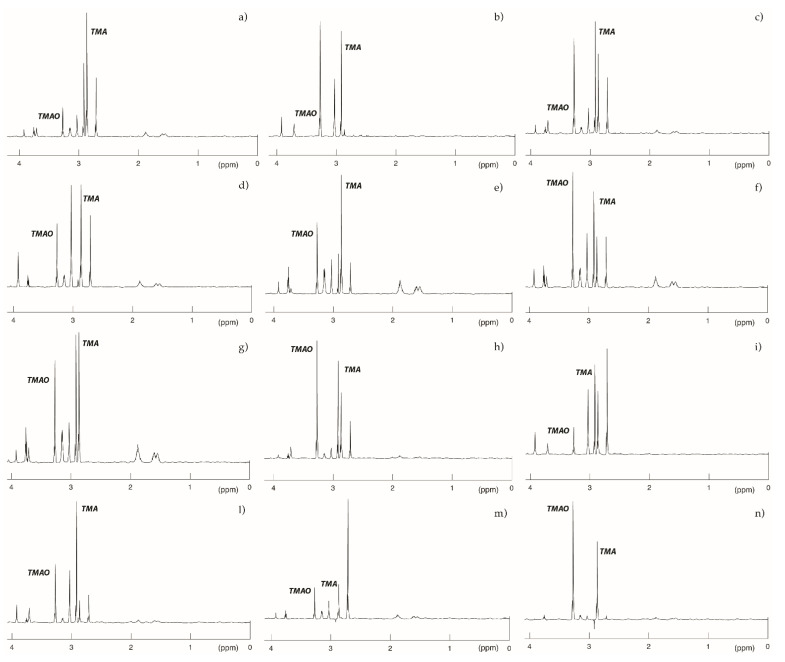
The 1H-NMR spectra of urine samples from TMAU patients with unique haplotype mostly reflected what we already predicted with genotyping and modelling analyses (Figure 5).
Figure 5.
500 MHz 1H spectra of patients included in the study. Here, are represented the spectra resulting from 1H NMR spectroscopy of the urine collected from patients showing a unique FMO3 variant haplotype. A healthy subject should present no peak for TMA and an intermediate peak for TMAO (not shown). Obtained results mostly confirmed genotyping and modelling analyses. (a) Patient 3; (b) Patient 10; (c) Patient 2; (d) Patient 9; (e) Patient 11; (f) Patient 6; (g) Patient 1; (h) Patient 8; (i) Patient 2; (l) Patient 4; (m) Patient 12; and (n) Patient 7. Metabolites peaks are assigned as follows: Trimethylamine-N-oxide (TMAO): 3.27 ppm; Creatinine (Cr): 3.06 ppm; Creatine (Cn): 3.04 ppm; Trimethylamine (TMA): 2.92 ppm; Dimethylamine (DMA): 2.73 ppm; and Citrulline (cit): 1.62 and 1.92 ppm.
The TMA peak at 2.92 ppm was much more intense in the urine samples from patients 1 (Figure 5a), 2 (Figure 5i), and 4 (Figure 5l), whose FMO3 genotype was characterized by nonsense variants. Conversely, patients 6 (Figure 5f), 7 (Figure 5n), 8 (Figure 5h), and 10 (Figure 5b) showed a higher peak for TMAO, suggesting that FMO3 variants carried by these subjects might only slightly alter the catalytic activity of the enzyme.
3. Discussion
The primary form of TMAU represent the genetic form of this social-impacting disease, and the one characterized by the most certain diagnosis. The latter consists of the screening of the FMO3 gene for causative mutations [23]. However, despite this, many patients suffering of TMAU symptomatology did not carry causative mutations in the flavin containing monooxygenase 3 encoding gene. One hypothesis developed to explain such phenomenon relies on the possibility that other genes could be involved in TMAU etiopathogenesis. In the meantime, the role of many polymorphic variants able to impact on FMO3 folding and activity is still unexplored, as well as the involvement of haplotypes in the onset and progression of trimethylaminuria [24].
It has been widely acknowledged that haplotype analysis in association studies can provide much more useful information than the information derived from single polymorphisms analysis [25]. The primary reason for considering the haplotype organization of variation resides in the fact that the folding kinetics, stability, and other physical features of a protein may depend on interactions between pairs or higher-order combination of aminoacidic sites; if such interactions are relevant, then haplotypes are of direct biological importance.
Thus, in our study we investigated 26 patients affected by TMAU, identifying 17 coding-sequence variants making 26 different haplotypes. Such variants were predicted to determine relevant conformational and biochemical changes in FMO3 mutated structures, leading to possible impairment of its activity. By docking and unbinding analyses, we shed light on possible alterations involving the enzymatic role of FMO3 on TMA.
Thanks to AutoDock Vina it was possible to determine the FMO3 active site aminoacidic residues, so far unknown. Today, indeed, only isolated domains were homologically modelled on the basis of the bacterial FMO3 [26].
The unbinding pathway analysis showed that TMA mainly interacts with Ser216, Gly217, Ser218, Trp219, Val220, Pro273, Asn 275, Gly 276, Leu278, Lys280, Glu281, and Pro282 amino acids of FMO3, suggesting that these aa might play an important role in the TMA oxidation process.
The single variant and haplotypic analyses results of the mutated FMO3 revealed that V257M was the only one that does not appear to cause a change of the enzyme active site. Furthermore, from the ligand pathfinder analyses, it emerged that the TMA unbinding path routed in the FMO3 carrying this variant, presented an opposite trajectory compared to that in wild-type enzyme. Probably, this variant might induce a protein folding change obstructing the TMA transit into the enzymatic pocket. Nevertheless, TMA seems to interact with most of the active site amino acids.
The situation resulted completely different for FMO3 carrying Y331stop and P380Fs variants in haplotypes (P380Fs + E308G and P380Fs + P153L + E158K, respectively). It is already known from the literature that both variants lead to a truncated protein whose enzymatic activity is reduced or totally absent, depending on whether this variant is in a heterozygous or a homozygous condition [27]. However, the molecular mechanisms underlying this deficit are not yet fully known. As shown in Figure 2, the stop variants resulted in the loss of the binding site for the FAD prosthetic group, therefore blocking the TMA oxidation process.
Comparing results obtained from both docking and unbinding pathway analyses, it was clear that both the single E158K and this variant in combination with P153L, R238Q, G475D, E308G, and R492W, as well as the R238P + G475D haplotype might influence protein folding, causing a lower TMA-active site binding stability. Such hypothesis is supported by the fact that the TMA would seem to form more stable bonds with different amino acid residues than those present in the active site.
Furthermore, in all haplotypes, except for the D141V + G180V, Arg279 is the only active site amino acid with whom the TMA would appear to form a stable interaction.
All previously obtained results were mostly confirmed by 1H-NMR analysis, which underlined the role of nonsense variants, especially if associated with other missense ones, in the alteration of FMO3 catalytic activity.
Thus, it seems to be clear that TMA might take a different path through the mutated FMO3 if compared to the wild-type enzyme. Both trajectory and direction are different.
Therefore, the described haplotypes might cause an enzymatic pocket narrowing, altering the TMA transit through the FMO3. Furthermore, such impairments could determine not only a different route for the TMA, but also a reduced interaction time of the ammine with the catalytic site of the enzyme. The final result of this altered enzymatic kinetics might be the reduced levels of TMAO and an accumulation of TMA, leading to the characteristic TMAU phenotype, whose different expression levels could depend on the particular haplotype carried by the patient.
4. Methods
4.1. Clinical Data
A cohort of 26 patients, with a mean age of 38 years and with a predominance of female sex, presented to our molecular genetic laboratories with a clinical diagnosis of suspected TMAU. All patients were subjected to a deep smell analysis, trying to classify the exact emitted odour, which is characteristic of TMAU phenotype. Then, several gastro-intestinal exams were performed, such as abdominal ultrasounds and microbiome analyses. Finally, a specific anamnestic study was realized in order to support the clinical diagnosis. Among personal details reported by patients, the intake of drugs and/or probiotics was carefully investigated, in order to rule out any confounding factors. A detailed list of patients’ clinical features is available in Table 6.
Table 6.
Clinical features of the 26 TMAU patients involved in this study. Signs, symptoms, and pathological features listed are those most frequently observed in TMAU affected individuals, as well as several ones specific of particular cases.
| ID | Age | Smell Description | Intestinal Disorders | Liver Diseases | Age of Onset | Drugs | Supplements/Probiotics |
|---|---|---|---|---|---|---|---|
| 1 | 9 | Rotten fish | / | / | 6 months | / | / |
| 2 | 42 | Rotten fish | Colitis | no | 7/8 years | no | no |
| 3 | 48 | Genital fishy odour, body garbage odour, and scalp acid/sulphur odour | Irritable colon | no | 10 years | Eutirox, Prisma 50 | no |
| 4 | Rotten fish | ||||||
| 5 | 36 | Rotten fish | Moderate degree of dysbiosis | no | 2/3 years | ||
| 6 | 71 | Rotten fish | Megacolon, anorectal stenosis, constipation, and congenital sacro-coccygeal malformation with fistulas | / | 2/3 years | / | / |
| 7 | 38 | Rotten fish | / | / | 37 years | Atazanavir, Abacavoir, and Lamivudin | Riboflavin |
| 8 | 45 | Rotten fish | / | / | 38 years | Oral contraceptives | / |
| 9 | 21 | Rotten fish | no | no | 6/7 years | no | no |
| 10 | 73 | Rotten fish | no | no | 63 years | / | no |
| 11 | 51 | Fish | no | no | 2/3 years | no | no |
| 12 | 20 | Rotten fish | no | no | 6 months | no | no |
| 13 | 28 | Fish | no | no | After weaning | no | no |
| 14 | 12 | Rotten fish | / | / | 5 years | / | / |
| 15 | 31 | Rotten fish, garbage, and garlic | Moderate dysbiosis | no | 17 years | / | / |
| 16 | 51 | Rotten fish | / | / | 44 years | Medicinal herbs | / |
| 17 | 26 | Rotten fish | no | no | 14 years | Psychotropic drugs and tranquilizers | |
| 18 | 48 | Rotten fish, faecal odour | Mild dysbiosis, slow oro-caecal transit | no | / | / | |
| 19 | 52 | Rotten fish | Mild dysbiosis | no | 6 years | / | Flebinic (carnitine-based supplement) |
| 20 | 24 | Rotten fish | no | no | After weaning | no | Activated carbon |
| 21 | 38 | Rotten fish | no | no | 34 years | no | no |
| 22 | 13 | Rotten fish | no | no | 10 years | no | no |
| 23 | 28 | Rotten fish | no | no | 28 years | no | no |
| 24 | 27 | Rotten fish, faecal odour, rot, and sulphur | Mild dysbiosis | no | 4 years | no | no |
| 25 | 47 | Rotten fish | / | / | 39 years | / | / |
| 26 | 71 | Rotten fish | / | / | 2/3 years | / | Assumption of a.a. |
4.2. Mutational Analysis of FMO3 Gene
DNA was extracted from peripheral blood by using a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The FMO3 (9 coding exons) gene was amplified using primers designed according to the genes published nucleotide sequence of GenBank (accessed on 18 July 2021). Sequences of such primers are available upon request.
PCR amplifications were carried out in a 50 µL solution containing 2 µL of each primer (10 µM), 0.8 µg of genomic, and 1.5 U MyTaq DNA Polymerase of (Bioline, London, UK). After an initial denaturation step at 95 °C for 1 min, the samples were subjected to 35 cycles of amplification consisting of 15 s of denaturation at 95 °C and 10 s of annealing. The annealing temperature was optimized for each primer set. Following PCR, 5 µL of amplified product was examined by electrophoresis on a 1% agarose gel. Direct sequencing was, then, performed using BigDye Terminator v.3.1 chemistry on a 3500 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
Informed consent was obtained from all subjects involved in the study.
4.3. In Silico Structural Analyses of Wild-Type and Muted Forms of FMO3
In order to investigate the possible effects of found variants on the coding sequence and their impact on FMO3 enzyme, a proteomic in silico approach was exploited.
ProtParam [28] permitted us to compare various physical and chemical parameters between the wild-type and the mutated FMO3 protein sequences. The computed features included the molecular weight, theoretical pI, amino acid composition, atomic composition, instability index, aliphatic index, and grand average of hydropathicity (GRAVY).
The prediction of tertiary structure has been realized thanks to Iterative Threading ASSEmbly Refinement (i-TASSER) hierarchical approach [29]. 3D structures of wild-type and mutated FMO3 were, then, analysed by UCSF ChimeraX [30], to visualize conformational changes and variations in intramolecular contacts.
4.4. Molecular Dynamics Analyses of FMO3/TMA Complex
The use of an unbinding pathway analysis tool is fundamental for the dynamical study of ligand-protein interaction, as the ligand binding might be simply treated as a two-state process between the unbound and bound states that form the stable basins of attraction. While the relative free energy difference between the two states controls the binding affinity, the kinetics of the binding is theoretically determined by the height of the relevant free energy barrier. Thus, the binding affinity can be exclusively governed by the endpoint states, while the binding kinetics is influenced by the detailed pathway connecting the endpoints.
In order to evaluate the possible docking between TMA and FMO3 and the dynamical molecular modelling of their complex, the SAMSON-Connect platform for integrated molecular design (https://www.samson-connect.net, accessed on 19 October 2021), along with its extensions AutoDock Vina Extended [31], FIRE state updater [32], GROMACS Model Generator [33], and Ligand Path Finder [34] was exploited.
Before using the extensions described, each structural model was validated to find small free molecules and clashes (bound ligands which are superposed and covalently docked with each other and with the protein), check the bond length, and check for alternate locations which must be removed.
In detail, the AutoDock Vina Extended permitted a highly accurate docking between ligand and enzyme, reducing computation time with a multithreaded algorithm, and mixing a “physical” scoring with a machine learning one. The analyses were performed setting minimization to 5000 steps and docking exhaustiveness to 8. The analyses calculated a total of 10 modes with affinity (kcal/mol), Ki (µmol), and RMSD lower and upper bound (A).
The Ligand Path Finder permitted to validate the predicted complexes from the previous docking analysis and allowed the prediction of kinetics properties such as binding affinity, binding/unbinding rates, and binding potency. Moreover, the tool helped to identify protein sites which hinder/facilitate the ligand entry/exit or whether certain entry/exit routes are more favourable. Mode 1 was chosen for each protein–ligand complex to find unbinding paths with Ligand Path Finder. Before applying Ligand Path Finder, each protein model was minimized without changing its backbone conformation and without changing the ligand’s position. Next, another minimization was applied to refine the systems, using Universal Force Field (UFF) as the interaction model and FIRE as the state updater. Geometry optimization is a fundamental step in many molecular modelling applications, used to produce stable, realistic structures which correspond to energy minima, so FIRE (Fast Inertial Relaxation Engine) is an efficient optimizer for molecular structures that implements the FIRE minimizer described by Bitzek et al. [32]. In the FIRE properties, the following parameters have been set: step size (the initial step size given to the algorithm) = 1.00 fs, steps (frequency of the viewport update) = 100, and fixed (forces the time step to be constant).
After the minimization, the parameters were set in Ligand Path Finder as follows: use seed = 1234, ARAP-modelling iterations = 20, initial temperature (T) = 0.0010 K, temperature factor = 2.00, RRT extension step size = 1.0000 A, runs = 2, minimization iterations = 10, failures before T increases = 1, and max. ligand displacement = 40.00 A. The sampling region was set according to the region of interest for the analysis. A single atom in the ligand was set as active atoms (nitrogen) to control the movement of the ligand and extract it using the active atom from the receptor. The choice of fixed atoms may influence the resulting unbinding pathways and ensure that the protein does not drift along with the ligand, so a single atom in the centre of the protein from one of its backbones away from the binding site was set as the fixed one (CA from SER 195 Backbone). The maximum time per run was set to 4 h and 2 paths were found per model.
The Pathlines extension was used to create a visual model that represent the trajectory of the centre of mass of selected atoms along selected path and to understand the motion of atoms along a path.
4.5. Urine Sample Preparation and 1H NMR Spectroscopy
Once collected, the patients’ urine samples were immediately frozen at −20 °C and thawed at room temperature only before analysis. In detail, 600 μL aliquots were used directly for 1H-NMR analysis in 5 mm diameter NMR tubes, together with 100 μL of deuterium oxide 99.96% (Eurisotop) as an internal field-frequency lock. Dietary overload (a 3 day, amine-rich diet) was performed by evaluated patients. Fish, cabbage, eggs, and crustaceans were recommended. The urine sample was collected on the fourth day.
NMR experiments were performed in a Bruker AVANCE III spectrometer (Bruker Biospin) operating at 500 MHz, set with a 5 mm gradient indirect detection probe and a probe temperature of 300 K. The one-dimensional proton spectra were acquired with 64 scans, 32 K data points, and a spectral width of 5000 Hz. A conventional proton 90° pulse with a relaxation delay of 2 s was applied. The water signal was suppressed by irradiation at the water resonance frequency (i.e., with a presaturation sequence). Resonances were assigned by reference to a spectral database of standard chemical shifts [35]. The Cr resonance at 3.05 ppm was used as an internal chemical shift reference. In these conditions, TMA resonance was detected at 2.92 ± 0.02 ppm, while TMAO was detected at approximately 3.27 ± 0.03 ppm, depending on the urine’s pH. The metabolite peaks for Cr, TMA, and TMAO were quantified by integration. The total amounts of TMA and TMAO excreted were normalized against the amount of Cr eliminated, in order to estimate the quantity considering the glomerular filtration. The values considered for healthy subjects were TMA/Cr < 10, TMAO/Cr ranging from 50 to 1000, and TMA/TMAO < 0.1 [36].
5. Conclusions
We realized an experimental study to investigate the role of several coding variants carried by the FMO3 gene in a cohort of patients affected by trimethylaminuria. Evaluated variants were only considered as polymorphic or benign, but the available literature on them has never been able to functionally explain their ineffectiveness on the encoded enzyme.
Both molecular modelling and 1H-NMR spectroscopy suggested that both non-sense and missense considered variants might impair the enzymatic kinetics of FMO3 with different severities, probably reducing the interaction time between the protein catalytic site and TMA, or losing the wild-type binding site.
However, we cannot assert with certainty that the same effect, in vivo, is limited to their presence, as there will be other factors involved into FMO3 catalysis alterations, e.g., variant impact on co-enzymes and co-factors binding sites and activity.
Despite this, considering the possible role of FMO3 variants with still uncertain effects, might be a relevant step towards the detection of novel scenarios in TMAU etiopathogenesis.
Acknowledgments
Not applicable.
Supplementary Materials
Figure S1: TMA docking to FMO3 could involve different amino acids in mutated enzymes (details). Table S1: Secondary structure and biochemical comparisons between wild-type and mutated FMO3 by different algorithms. Table S2: AutoDock Vina Extended docking analysis of wild-type and mutated FMO3 proteins. Videos S1–S14: Unbinding pathways of wild-type and mutated FMO3 enzymes.
Author Contributions
Conceptualization, L.D.; methodology, L.D.; software, L.D. and C.S.; validation, C.S. and S.A.; formal analysis, L.D. and F.N.; investigation, L.D.; resources, F.N.; data curation, L.D.; writing—original draft preparation, L.D. and S.A.; writing—review and editing, C.S., R.D. and C.R.; visualization, S.A. and R.D.; supervision, A.S.; and project administration, A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the “Azienda Policlinico Universitario di Messina” (protocol code 54-20 of 24 June 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent for publication was obtained from each member of screened family, and for children (under 16 years old) participating in the study the written informed consent for publication was obtained from their parents.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Doyle S., O’Byrne J.J., Nesbitt M., Murphy D.N., Abidin Z., Byrne N., Pastores G., Kirk R., Treacy E.P. The genetic and biochemical basis of trimethylaminuria in an Irish cohort. JIMD Rep. 2019;47:35–40. doi: 10.1002/jmd2.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller N.B., Beigelman A., Utterson E., Shinawi M. Transient massive trimethylaminuria associated with food protein-induced enterocolitis syndrome. JIMD Rep. 2014;12:11–15. doi: 10.1007/8904_2013_238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chhibber-Goel J., Gaur A., Singhal V., Parakh N., Bhargava B., Sharma A. The complex metabolism of trimethylamine in humans: Endogenous and exogenous sources. Expert Rev. Mol. Med. 2016;18:e8. doi: 10.1017/erm.2016.6. [DOI] [PubMed] [Google Scholar]
- 4.Phillips I.R., Shephard E.A. Primary Trimethylaminuria. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Mirzaa G., Amemiya A., editors. GeneReviews((R)) University of Washington; Seattle, WA, USA: 1993. [Google Scholar]
- 5.Donato L., Alibrandi S., Scimone C., Castagnetti A., Rao G., Sidoti A., D’Angelo R. Gut-Brain Axis Cross-Talk and Limbic Disorders as Biological Basis of Secondary TMAU. J. Pers. Med. 2021;11:87. doi: 10.3390/jpm11020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roddy D., McCarthy P., Nerney D., Mulligan-Rabbitt J., Smith E., Treacy E.P. Impact of trimethylaminuria on daily psychosocial functioning. JIMD Rep. 2021;57:67–75. doi: 10.1002/jmd2.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramos N., Wystrach C., Bolton M., Shaywitz J., IsHak W.W. Delusional disorder, somatic type: Olfactory reference syndrome in a patient with delusional trimethylaminuria. J. Nerv. Ment. Dis. 2013;201:537–538. doi: 10.1097/NMD.0b013e31829482fd. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt A.C., Leroux J.C. Treatments of trimethylaminuria: Where we are and where we might be heading. Drug Discov. Today. 2020;25:1710–1717. doi: 10.1016/j.drudis.2020.06.026. [DOI] [PubMed] [Google Scholar]
- 9.Manning N.J., Allen E.K., Kirk R.J., Sharrard M.J., Smith E.J. Riboflavin-responsive trimethylaminuria in a patient with homocystinuria on betaine therapy. JIMD Rep. 2012;5:71–75. doi: 10.1007/8904_2011_99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamazaki H., Fujieda M., Togashi M., Saito T., Preti G., Cashman J.R., Kamataki T. Effects of the dietary supplements, activated charcoal and copper chlorophyllin, on urinary excretion of trimethylamine in Japanese trimethylaminuria patients. Life Sci. 2004;74:2739–2747. doi: 10.1016/j.lfs.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., Bergeron N., Levison B.S., Li X.S., Chiu S., Jia X., Koeth R.A., Li L., Wu Y., Tang W.H.W., et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur. Heart J. 2019;40:583–594. doi: 10.1093/eurheartj/ehy799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fennema D., Phillips I.R., Shephard E.A. Trimethylamine and Trimethylamine N-Oxide, a Flavin-Containing Monooxygenase 3 (FMO3)-Mediated Host-Microbiome Metabolic Axis Implicated in Health and Disease. Drug Metab. Dispos. 2016;44:1839–1850. doi: 10.1124/dmd.116.070615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scimone C., Alibrandi S., Donato L., Giofre S.V., Rao G., Sidoti A., D’Angelo R. Antiretroviral treatment leading to secondary trimethylaminuria: Genetic associations and successful management with riboflavin. J. Clin. Pharm. Ther. 2021;46:304–309. doi: 10.1111/jcpt.13315. [DOI] [PubMed] [Google Scholar]
- 14.D’Angelo R., Scimone C., Esposito T., Bruschetta D., Rinaldi C., Ruggeri A., Sidoti A. Fish odor syndrome (trimethylaminuria) supporting the possible FMO3 down expression in childhood: A case report. J. Med. Case Rep. 2014;8:328. doi: 10.1186/1752-1947-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esposito T., Varriale B., D’Angelo R., Amato A., Sidoti A. Regulation of flavin-containing mono-oxygenase (Fmo3) gene expression by steroids in mice and humans. Horm. Mol. Biol. Clin. Investig. 2014;20:99–109. doi: 10.1515/hmbci-2014-0012. [DOI] [PubMed] [Google Scholar]
- 16.Scimone C., Donato L., Rinaldi C., Sidoti A., D’Angelo R. First case of Currarino syndrome and trimethylaminuria: Two rare diseases for a complex clinical presentation. J. Dig. Dis. 2016;17:628–632. doi: 10.1111/1751-2980.12373. [DOI] [PubMed] [Google Scholar]
- 17.D’Angelo R., Esposito T., Calabro M., Rinaldi C., Robledo R., Varriale B., Sidoti A. FMO3 allelic variants in Sicilian and Sardinian populations: Trimethylaminuria and absence of fish-like body odor. Gene. 2013;515:410–415. doi: 10.1016/j.gene.2012.12.047. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J., Shephard E.A. Mutation, polymorphism and perspectives for the future of human flavin-containing monooxygenase 3. Mutat. Res. 2006;612:165–171. doi: 10.1016/j.mrrev.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Krueger S.K., Williams D.E., Yueh M.F., Martin S.R., Hines R.N., Raucy J.L., Dolphin C.T., Shephard E.A., Phillips I.R. Genetic polymorphisms of flavin-containing monooxygenase (FMO) Drug Metab. Rev. 2002;34:523–532. doi: 10.1081/DMR-120005653. [DOI] [PubMed] [Google Scholar]
- 20.Razmazma H., Ebrahimi A. The effects of cation-pi and anion-pi interactions on halogen bonds in the [Ncdots, three dots, centeredXcdots, three dots, centeredN](+) complexes: A comprehensive theoretical study. J. Mol. Graph. Model. 2018;84:134–144. doi: 10.1016/j.jmgm.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Chang K.Y., Yang J.R. Analysis and prediction of highly effective antiviral peptides based on random forests. PLoS ONE. 2013;8:e70166. doi: 10.1371/journal.pone.0070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- 23.Zhang J., Tran Q., Lattard V., Cashman J.R. Deleterious mutations in the flavin-containing monooxygenase 3 (FMO3) gene causing trimethylaminuria. Pharmacogenetics. 2003;13:495–500. doi: 10.1097/00008571-200308000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Lambert D.M., Mamer O.A., Akerman B.R., Choiniere L., Gaudet D., Hamet P., Treacy E.P. In vivo variability of TMA oxidation is partially mediated by polymorphisms of the FMO3 gene. Mol. Genet. Metab. 2001;73:224–229. doi: 10.1006/mgme.2001.3189. [DOI] [PubMed] [Google Scholar]
- 25.Donato L., Scimone C., Nicocia G., Denaro L., Robledo R., Sidoti A., D’Angelo R. GLO1 gene polymorphisms and their association with retinitis pigmentosa: A case-control study in a Sicilian population. Mol. Biol. Rep. 2018;45:1349–1355. doi: 10.1007/s11033-018-4295-4. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y., Patel N.A., Crombie A., Scrivens J.H., Murrell J.C. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. USA. 2011;108:17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bortolussi S., Catucci G., Gilardi G., Sadeghi S.J. N- and S-oxygenation activity of truncated human flavin-containing monooxygenase 3 and its common polymorphic variants. Arch. Biochem. Biophys. 2021;697:108663. doi: 10.1016/j.abb.2020.108663. [DOI] [PubMed] [Google Scholar]
- 28.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 29.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitzek E., Koskinen P., Gähler F., Moseler M., Gumbsch P. Structural Relaxation Made Simple. Phys. Rev. Lett. 2006;97:170201. doi: 10.1103/PhysRevLett.97.170201. [DOI] [PubMed] [Google Scholar]
- 33.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen M.K., Jaillet L., Redon S. ART-RRT: As-Rigid-As-Possible search for protein conformational transition paths. J. Comput. Aided Mol. Des. 2019;33:705–727. doi: 10.1007/s10822-019-00216-w. [DOI] [PubMed] [Google Scholar]
- 35.Zuppi C., Messana I., Forni F., Rossi C., Pennacchietti L., Ferrari F., Giardina B. 1H NMR spectra of normal urines: Reference ranges of the major metabolites. Clin. Chim. Acta. 1997;265:85–97. doi: 10.1016/S0009-8981(97)00110-1. [DOI] [PubMed] [Google Scholar]
- 36.Chalmers R.A., Bain M.D., Michelakakis H., Zschocke J., Iles R.A. Diagnosis and management of trimethylaminuria (FMO3 deficiency) in children. J. Inherit. Metab. Dis. 2006;29:162–172. doi: 10.1007/s10545-006-0158-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.