Abstract
The potential presence of spore-forming bacteria related to the Bacillus cereus group in Mexican chili powder elaborated from Capsicum annuum L. is of commercial and clinical interest, because chili powder is an essential spice in the Mexican diet and in diets around the globe. To facilitate detection and isolation of members of this group of spore-forming bacteria from Mexican chili powder samples, we identified colonies that grew on agar medium selective for Bacillus cereus sensu lato, supplemented with polymyxin B (10 µg/mL) and ampicillin (10 to 100 µg/mL). The presumptive B. cereus (s.l.) isolates were tested using a tRNACys-PCR-based approach and the results identified species related phylogenetically to B. cereus, B. thuringiensis, and B. toyonensis. Their toxigenic potential was assessed by serological tests to detect enterotoxins (Nhe and Hbl) and by PCR targeting the hemolysin BL (hbl) component C (hblC) and non-hemolytic enterotoxin component A (nheA). The antibiotic profiles of the isolates showed a high resistance to β-lactams (100% of the isolates), trimethoprim-sulfamethoxazole (100%), tetracycline (90%), erythromycin (77%), clindamycin (74%), and chloramphenicol (42%). Our results indicate the presence of B. cereus s.l. with toxigenic characteristics in Mexican chili powder. Because of the potential for these organisms to cause disease through their production of various toxins, and resistance to antibiotics, we recommend that a microbiological risk assessment must be considered in the Mexican regulatory requirements.
Keywords: Bacillus cereus sensu lato, tRNACys-PCR, antibiotics resistance, Mexican chili powder
1. Introduction
Within the Solanaceae family, pepper (Capsicum annuum L.) is one of the most economical and agriculturally important plants cultivated all over the world [1,2]. After China, with 17.5 million tons, Mexico is the second largest producer of fresh pepper with 2.7 million tons at 150,000 hectares cultivated annually [2]. Its importance is based on its nutritional content, diverse bioactive compounds, pungency, aroma, and health benefits for the consumers [1,3]. Additionally, diverse genetic lines of Capsicum annuum L. have been developed to produce carotenoids with high commercial value in powdered form as spices or as colorants on agro-food, cosmetics, and products from pharma industries [1,2,3,4,5].
In Mexico, chili powder is used as a spice in diverse seasoned foods, fast food, beverages, snacks, fruits, grains, regional spicy candies, and diverse sauces. Thus, chili powder is an important spice in the Mexican diet [2,6]. However, chili powder is produced from sun-dried peppers, and this production process increases the risk of microbial contamination of the product [2]. In some cases, industrial dryers are used to reduce drying time [2]; however, retention of important traits in chili powder is dependent on the drying procedure. For example, color deterioration in chili powder is greatly influenced by moisture content, storage, temperature, atmospheric conditions, and light [1,7]. Thus, drying chili at high temperatures can reduce the volatile compounds, nutrients, and color content in chili powder. All of these conditions during chili powder production are favorable for microbial contamination and demand a comprehensive microbiological risk assessment, with a focus on potential pathogens.
Among the approaches used for the characterization of microbial load in chili powder from various countries are the determination of total aerobic mesophilic bacteria, aerobic spore-forming bacteria, B. cereus detection, and determination of members of the Enterobacteriaceae, yeast, and molds [8,9,10,11,12,13,14,15]. Other studies have been conducted for pathogenic species detection, such as Bacillus cereus, Salmonella spp., Clostridium perfringens, or Escherichia coli in paprika powder [8,9,10,11,12,13,14,15]. A documented outbreak of human salmonellosis was traced to paprika powdered potato chips as the main vehicle of transmission in Germany when paprika powder imported from South America was used to flavor the product [16]. It is noteworthy that a molecular approach using 16S rRNA gene sequencing of bacteria, isolated from paprika powder, produced in different countries, identified spore-forming bacteria, facilitating the association of a particular species with its geographical origin. This study was limited, however, by the number of bacterial isolates that were examined [12].
In other studies, several B. cereus sensu lato (s.l.) strains have been identified as opportunistic pathogens in chili powder, paprika, and other spices of different geographical origins, and several different toxins have been associated with these strains, viz. cereulide, cytotoxin K, hemolysin BL (HBL), and non-hemolytic enterotoxin (NHE) [14,15]. Toxins from these bacteria have been associated with the diarrheal type of B. cereus food poisoning, which is typically characterized by abdominal pain and watery diarrhea [17,18,19,20,21]. In addition to these toxigenic characteristics, B. cereus s.l. strains are naturally resistant to penicillin and other β-lactam antibiotics because of their content of β-lactamases [13,14,15,22,23], and some studies reveal that resistance may be extended to other commonly used antibiotics, such as chloramphenicol, gentamicin, imipenem, erythromycin, tetracycline, and the trimethoprim/sulfamethoxazole combination [13,14,15,23]. With these genetic characteristics present in B. cereus s.l., its detection is essential in controlling the spread of potential pathogens present on Mexican chili powder.
Unfortunately, knowledge about the presence of the pathogen B. cereus in Mexican chili powder is limited and of equal or greater concern is the lack of phylogenetic information on the microbes that may be present as contaminants in the chili powder. To address these deficiencies in our knowledge, we used standard and molecular microbial procedures on Mexican chili powder samples elaborated from Capsicum annuum L., and extended our tRNACys-PCR method previously reported [24] to detect the presence of antibiotic-resistant B. cereus s.l. isolates. Further, we examined the capability of the identified microbes to produce enterotoxins (Nhe and Hbl).
2. Materials and Methods
2.1. Ampicillin-Resistant Detection of B. cereus s.l.
In a first effort to obtain knowledge about the presence of B. cereus s.l. in chili powder elaborated from Capsicum annuum L., we selected four chili powder varieties, commercially available and economically competitive in a local market in Queretaro, Mexico. Sample A was a red-hot chili powder used as spice principally for fruit, grains, and beverages. Sample B was a red chili powder used principally as a spice for fast food. Sample C was a red extra hot chili powder used as a spice principally for fruit and grains, and sample D was a red chili powder used for its color content in snacks. Each replicate of 10 g of a sample was homogenized in 90 mL of peptone (0.1% w/v; Difco Laboratories; Detroit, MI, USA) and treated at 80 °C for 10 min to select endospore-forming bacteria. Serial dilutions were then inoculated into triplicated tryptic soy agar (TSA) medium (Difco Laboratories; Detroit, MI, USA) and incubated at 37 °C during 24 h. Numbers of spore-forming mesophilic bacteria (SMB) from each sample were determined. For the isolation of B. cereus s.l., 0.1 mL of the first dilution was inoculated into triplicated agar plates containing B. cereus agar base (Sigma-Aldrich, CDMX, México) supplemented with 100 mL/L of Egg Yolk Emulsion and 10 µg/mL of Polymyxin B. To select colonies of B. cereus s.l. with ampicillin-resistance, the medium was supplemented at concentrations of 5, 10, 15, 20, 25, 50, 75, and 100 µg/ mL of ampicillin. Plates were incubated at 37 °C for 24 h and observed for growth. The theoretical limit of detection (LOD) was therefore 100 cfu/g. Numbers below this limit (˂100 cfu/g) in our results mean that bacterial growth was not detected in these conditions. Suspected B. cereus s.l. colonies that were typically mannitol-negative and lecithinase-positive (zone of precipitation around colonies) were selected, and their identities were confirmed in the same culture medium. B. cereus ATCC 10876 was used as reference strains for phenotypic tests and phylogenetic analysis.
2.2. tRNACys Region Amplification Conditions
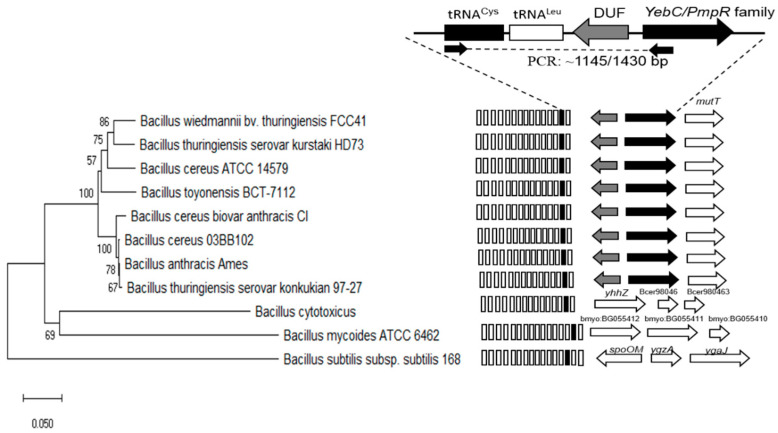
To facilitate detection of suspected B. cereus s.l. colonies and B. cereus ATCC 10876 as control, we used our tRNACys-PCR method previously reported [24]. PCR primers were designed to amplify the tRNACys region in the B. anthracis/cereus/thuringiensis/toyonensis/ wiedmannii group. Primer 1517 (5′-GGCGGCATAGCCAAGTGGTAAGGC-3′) was designed to target the tRNACys gene, while primers 1518 (5′-GCTGCCACATAAATTTCACGCCC-3′) and 1520 (5′-GCTACAGAACCGTTCACACCC-3′) were designed to target the yebC/pmpR-like gene. Primers were synthesized by T4OLIGO, Guanajuato, Mexico. The primers were predicted to yield products of 1145 and 1430 bp, respectively (see Figure 1). Amplicons were sequenced using the platform at Macrogen Inc. (Seoul, Korea). Gene sequences were analyzed using MEGA X using the neighbor-joining method [25,26,27,28] and compared with sequences representative of the B. cereus group, by a BLAST search, using the GenBank database (http://www.ncbi.nlm.nih.gov, accessed on 16 august 2021).
Figure 1.
Schematic representation of tRNACys-PCR strategy for the B. cereus group. The cluster of 15 to 17 tRNA genes and the orientation of three specific genes located downstream of tRNALeu is shown. The primers indicated by the arrows were predicted to yield products of 1145 and 1430 bp, respectively (see Methodology). Phylogenetic analysis from these regions were obtained in MEGA X, using the neighbor-joining method. B. subtilis subsp. subtilis 168 was designated as the outgroup taxon.
2.3. Duopath® Test for Detection of Nhe and Hbl Bacterial TOXIN Formation
We used the Duopath® Cereus Enterotoxins test (Merck) to detect the non-hemolytic enterotoxin (Nhe) and hemolysin BL (HBL) [29] and tests were performed according to the manufacturer’s specifications. The ampicillin-resistant isolates were streaked onto agar plates containing B. cereus agar base (Sigma-Aldrich) supplemented with 100 mL/L of egg yolk emulsion and 100 µg/ mL of polymyxin B. After incubation at 37 °C for 24 h, the colonies were picked and suspended in 1 mL of casein hydrolysate-glucose-yeast extract broth with the following composition (g/L): casein hydrolysate 20.0 (MCD Lab, Mexico), yeast extract 6.0 (Difco, CDMX, México), ammonium sulfate 2.0 (J.T.Baker, CDMX, México), tri-sodium citrate 1.0 (USB Corporation USA), di-potassium hydrogen phosphate 14.0 (J.T.Baker), potassium dihydrogen phosphate 6.0 (J.T.Baker), magnesium sulfate 2.0 (J.T.Baker), and supplemented with 1% glucose (MEYER Lab, CDMX, México). After 5 h incubation at 37 °C, and cooling to room temperature, a 150 μL aliquot of the enrichment was placed into the circular sample port on the Duopath® Cereus Enterotoxins test (Merck) and incubated at room temperature and the interpretation was following manufacturer´s specifications. All strains were evaluated for the presence of representative enterotoxin genes (nheA and hblC) with primers designed, as well as PCR conditions reported previously [24,30].
2.4. Antibiotic Susceptibility Testing
The antibiotic susceptibility of each strain was tested by triplicate using the disc diffusion method determined by the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines; criteria were adopted from Staphylococcus spp. and Bacillus spp. [31]. The bacterial cultures were incubated in liquid soya medium with shaking at 37 °C until the suspension reached an optical density (OD) between 0.4 and 0.5 at 600 nm. A total volume of 100 µL of each strain was spread on Mueller Hinton agar (Bioxon). A total of 18 different antibiotic discs (Oxoid) containing the antibiotics amikacin (AMK 30 μg), ampicillin (AMP 10 μg), carbenicillin (CAB 100 µg), cefalotin (CFT 30 µg), cefotaxime (CTX 30 μg), chloramphenicol (CHL 30 μg), ciprofloxacin (CIP 5 μg), clindamycin (CDM 30 µg), dicloxacillin (DCX 1 µg), erythromycin (ERY 15 μg), gentamicin (GEN 10 μg), netilmicin (NET 30 µg), nitrofurantoin (NTF 300 µg), norfloxacin (NFX 10 µg), penicillin (PEN 10 μg), tetracycline (TET 30 μg), trimethoprim-sulfamethoxazole (SXT 25 μg), and vancomycin (VCM 30 µg) were used for susceptibility testing. After overnight incubation, the mean of diameter in mm of the inhibitory or clear zones around the disc was recorded, and these were interpreted as susceptible, susceptible increased exposure or resistant, according to EUCAST guidelines.
3. Results
3.1. SMB and B. cereus s.l. Determination
The content of spore-forming mesophilic bacteria (SMB) and presumptive B. cereus s.l. counts from four chili powder samples were determined, as described in Methodology. As shown in Table 1, sample B showed the lowest SMB content with 8.0 × 102 cfu/g, followed by sample A with 1.73 × 103 cfu/g, while sample C and D counts were 2.92 × 105 and 3.24 × 105 cfu/g, respectively. These results document the presence of spore-forming bacteria in the tested chili powder samples, validating the need for the next step, detecting the presence of B. cereus s.l. The literature indicates that B. cereus s.l. is typically resistant to β-lactam antibiotics [13,14,15,22,23]. Accordingly, we used various concentrations of ampicillin in the culture medium during selection with two purposes, first to eliminate ampicillin-susceptible bacteria and second, to facilitate detection and isolation of ampicillin-resistant B. cereus s.l. Table 1 shows the presumptive B. cereus s.l. counts according with the phenotypical characteristics mentioned in Methodology. The lowest presumptive B. cereus s.l. counts were observed in sample B, with the limit of detection (LOD) of 100 cfu/g in absence of ampicillin, while in the presence of different concentrations of ampicillin, no presumptive B. cereus s.l. could be detected (LOD = 100 cfu/g). Sample A had 200 cfu/g in the absence of ampicillin, while in the presence of ampicillin (5 to 50 µg/mL), 100 cfu/g was detected. At 75 and 100 µg/mL of ampicillin, no presumptive B. cereus s.l. could be detected (LOD = 100 cfu/g). In samples C and D, the counts were 600 and 700 cfu/g, respectively, in the absence of ampicillin. In the presence of ampicillin, presumptive B. cereus s.l. in sample C could be detected at a concentration of 5 to 25 µg/mL of ampicillin, with an average of 580 cfu/g. In Sample D, presumptive B. cereus s.l. counts averaged 470 cfu/g at concentrations of 5 to 75 µg/mL of ampicillin. From these results, a total of 30 presumptive ampicillin-resistant B. cereus s.l. colonies from all chili powder samples were selected, and the properties described above were confirmed in the same culture medium.
Table 1.
Spore-forming mesophilic bacteria (SMB) and presumptive B. cereus s.l. counts in Mexican chili powder samples.
| Sample | SMB a | BC b (0) | BC (5) | BC (10) | BC (15) | BC (20) | BC (25) | BC (50) | BC (75) | BC (100) |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 1.73 × 103 | 200 | 100 | 100 | 100 | 100 | 100 | 100 | ˂100 | ˂100 |
| B | 8.0 × 102 | 100 | ˂100 | ˂100 | ˂100 | ˂100 | ˂100 | ˂100 | ˂100 | ˂100 |
| C | 2.92 × 105 | 600 | 633 | 600 | 666 | 566 | 433 | ˂100 | ˂100 | ˂100 |
| D | 3.24 × 105 | 700 | 600 | 433 | 633 | 533 | 533 | 333 | 233 | ˂100 |
a Spore-forming mesophilic bacteria (SMB) counts. b Presumptive Bacillus cereus s.l. (BC) counts. To select colonies with ampicillin-resistance, the medium was supplemented at concentrations of 5, 10, 15, 20, 25, 50, 75, and 100 µg/ mL of ampicillin, indicated by numbers in parentheses. The theoretical limit of detection (LOD) was therefore 100 cfu/g. Numbers below this limit (˂100 cfu/g) in our results mean that bacterial growth was not detected in these conditions.
3.2. Phylogenetic Analysis
For the confirmation of the identities of these ampicillin-resistant presumptive B. cereus s.l. colonies, we used the tRNACys-PCR strategy previously published [24]. In Figure 1, we show that the tRNACys gene in B. cereus s.l. is part of cluster of 15 to 17 tRNA genes localized downstream of a ribosomal RNA operon. For the diagnostic and phylogenetic analysis we used the specific DNA region between tRNACys and yebC/pmpR-like gene (which encodes a probable transcriptional regulatory protein) located downstream of a gene sequence of unknown function (DUF gene), indicated in Figure 1. The phylogenetic analysis show that this DNA region is specific for B. anthracis, B. cereus s.s., B thuringiensis, B. toyonensis, and B. wiedmannii related species, while B. mycoides and B. cytotoxicus show a different gene organization. Therefore, our PCR test is a suitable approach for verification of the identities of the chili powder isolates.
From a total of 30 presumptive ampicillin-resistant B. cereus s.l. colonies selected (6 colonies from sample A, 3 colonies from sample B, 4 colonies from sample C, and 17 colonies from sample D), 25 (83 %) were positive for the tRNACys-PCR product (data not shown), when the primers 1517 and 1518 were used as the first option (~1145 bp PCR product). A negative result for tRNACys-PCR was obtained for five colonies from sample D, and could be due to a small nucleotide variation in the yebC/pmpR-like gene region, from which the 1518 primer was designed. Therefore, a new primer, designated 1520, was designed, beginning 285 nucleotides downstream of the DNA region, targeted by the 1518 primer. The tRNACys-PCR results (~1430 bp PCR product) were positive (data not shown) with this last primer designed for the rest of presumptive ampicillin-resistant B. cereus s.l. colonies (17%).
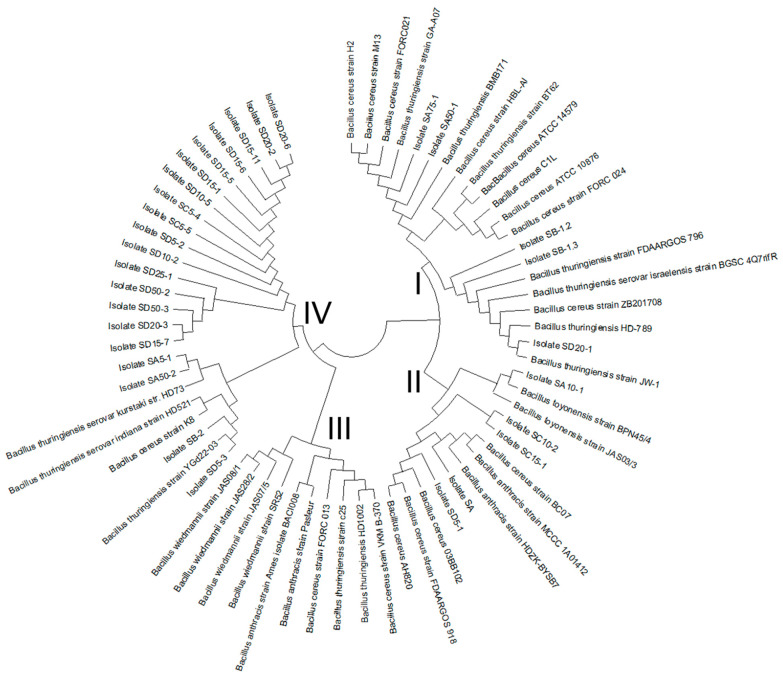
Our phylogenetic analysis confirmed the results obtained by tRNACys-PCR; the presumptive ampicillin-resistant B. cereus s.l. colonies are members of the B. cereus group. Clustering of the tRNACys- yebC/pmpR genes sequences revealed four major species groups (Figure 2). The species isolated from chili powder samples were clustered in Groups I, II, and IV, with the majority of isolates related phylogenetically to B. cereus and B. thuringiensis, and the isolate SA10-1 closely related to B. toyonensis strains from various environmental sources.
Figure 2.
Phylogenetic analysis for tRNACys-yebC/pmpR region. The sequences of the isolates obtained from chili powder samples were compared with sequences of representative strains of B. cereus group. The isolated colonies obtained clustered within groups I, II, and IV, respectively (see Table 2 for correspondence of each isolate).
3.3. Toxigenic Characteristics
The toxigenic potential for the isolates was investigated by PCR for hblC and nheA, as representative genes. Figure 3, panel A shows a representative PCR for hblC and nheA genes. Lane 1 shows the negative PCR for the strain SA5-1, while lanes 2 and 3 are PCR multiplex positive for the strain SA and SB-2. Lanes 4, 5, and 6 show the PCR positive control using the strain B. cereus ATCC 10876 with single or multiplex PCR, respectively. The PCR results are summarized in Table 2. With the exception of one strain (SA5-1) all isolates were positive for at least two toxin genes (97%). The Duopath test for detection of toxin production showed a positive result for the Hbl toxin (Table 2) in 28 strains (90%), including the B. cereus ATCC 10876, while the negative strains were SA, SC10-2, and SD20-1. The Nhe toxin was detected in all strains using the Duopath test, including strain SA5-1, even though the PCR result for this strain was negative. In Figure 3B, we show a representative Duopath test for detection of toxin production (NHE and HBL), the results show from left to right production of both toxins for the strain B. cereus ATCC 10876 and SA75-1, while for the strain SD20-1, only the NHE toxin was produced. The toxin production capabilities reflected the potential health risk of B. cereus s.l. present in Mexican chili powder samples.
Figure 3.
Representative results for toxigenic characteristics. Panel (A) shows a representative electrophoresis gel for multiplex PCR. Lane M contains a Thermo Scientific GeneRuler 1 Kb DNA ladder, lane 1; strain SA5-1, lane 2; strain SA, lane 3; strain SB-2, lane 4, 5, and 6 is the control (B. cereus ATCC 10876) with single or multiplex PCR respectively. The arrows show the PCR amplification product for hblC (750 bp) and nheA (500 bp) toxin genes. Panel (B) shows a representative Duopath test for detection of toxin production indicated by a red line in both the test (Nhe and/or Hbl) and control zones (C). From left to right; B. cereus ATCC 10876 (1), SA75-1 (2), and SD20-1 (9).
Table 2.
Toxigenic and antibiotics characteristics for B. cereus s.l isolates from Mexican chili powder samples. Presence (+) or Absence (−) for toxin production and toxin genes.
| Strain | Chili Powder Sample | Toxin Production | Toxin Genes | Antibiotics | Susceptible Increased Exposure | |||
| Hbl | Nhe | hblC | nheA | Resistance | Susceptible | |||
| B. cereus ATCC 10876 | − | + | + | + | + | AMP, CAB, CFT, CTX, DCX, PEN, SXT, CDM | AMK, CHL, CIP, GEN, NET, NTF, NFX, TET, VCM | ERY |
| SA | A | − | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT, TET | AMK, CHL, CIP, CDM, GEN, NET, NTF, NFX, VCM | |
| SA5-1 | A | + | + | − | − | AMP, CAB, CFT, CTX, DCX, PEN, SXT, CHL, TET | AMK, CIP, GEN, NET, NTF, NFX, VCM | ERY, CDM |
| SA10-1 | A | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT | AMK, CHL, CIP, CDM, GEN, NET, NTF, NFX, TET, VCM | |
| SA50-1 | A | + | + | + | + | AMP, CAB, CFT, CTX, DCX, PEN, TET, SXT | AMK, CHL, CIP, ERY, GEN, NET, NTF, NFX, VCM | CDM |
| SA50-2 | A | + | + | + | + | AMP, CAB, CFT, CTX, DCX, PEN, TET, SXT, CHL | AMK, ERY, GEN, NET, NTF, NFX, VCM | CIP, CDM |
| SA75-1 | A | + | + | + | + | AMP, CAB, CFT, CTX, DCX, NTF, PEN, TET, SXT | AMK, CHL, CIP, CDM, ERY, GEN, NET, NFX, VCM | |
| SB-2 | B | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT, TET, CDM | AMK, CHL, NET, NTF, NFX, VCM | CIP, GEN |
| SB-1.2 | B | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT, TET, CDM | AMK, CHL, NET, NTF, NFX, VCM | CIP, GEN |
| SB-1.3 | B | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT, TET, CDM | AMK, CHL, NET, NTF, NFX, VCM | CIP, GEN |
| SC5-4 | C | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CDM | AMK, CHL, CIP, NET, NTF, NFX, VCM | GEN |
| SC5-5 | C | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CDM | AMK, CHL, CIP, NET, NTF, NFX, VCM | GEN |
| SC10-2 | C | − | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT, TET, CDM | AMK, CHL, NET, NTF, NFX, VCM | CIP, GEN |
| SC15-1 | C | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, SXT, CDM | AMK, CHL, CIP, GEN, NET, NTF, NFX, TET, VCM | |
| SD5-1 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CDM | AMK, CHL, CIP, NET, NTF, NFX, VCM | GEN |
| SD5-2 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, CIP, NET, NTF, NFX, VCM | GEN |
| SD5-3 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, PEN, SXT, CHL, TET, CDM | AMK, CIP, NET, NTF, NFX, VCM | GEN, ERY |
| SD10-2 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, CIP, GEN, NET, NTF, NFX, VCM | |
| SD10-5 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, GEN, NET, NTF, NFX, VCM | CIP |
| SD15-1 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL | AMK, CIP, CDM, GEN, NET, NTF, NFX, VCM | |
| SD15-5 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, CIP, GEN, NET, NTF, NFX, VCM | |
| SD15-6 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CDM | AMK, CHL, CIP, NET, NTF, NFX, VCM | GEN |
| SD15-7 | D | + | + | + | + | AMK, AMP, CAB, CFT, CTX, DCX, ERY, NET, PEN, TET, SXT, CHL, GEN, CDM | CIP, NTF, NFX, VCM | |
| SD15-11 | D | + | + | + | + | AMK, AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CDM | CHL, GEN, NET, NTF, NFX, VCM | CIP |
| SD20-1 | D | − | + | + | + | AMP, CAB, CFT, CTX, DCX, PEN, TET, SXT, CDM | AMK, CHL, CIP, GEN, NET, NTF, NFX, VCM | ERY |
| SD20-2 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, NET, NTF, NFX, VCM, GEN | CIP |
| SD20-3 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, NET, NTF, NFX, VCM | CIP, GEN |
| SD20-6 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT | AMK, CHL, CIP, GEN, NET, NTF, NFX, VCM | CDM |
| SD25-1 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, GEN, CDM | AMK, CHL, NET, NTF, NFX, VCM | CIP, |
| SD50-2 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, CIP, NET, NTF, NFX, VCM | GEN |
| SD50-3 | D | + | + | + | + | AMP, CAB, CFT, CTX, DCX, ERY, PEN, TET, SXT, CHL, CDM | AMK, CIP, NET, NTF, NFX, VCM, GEN | |
3.4. Antibiotic Resistance
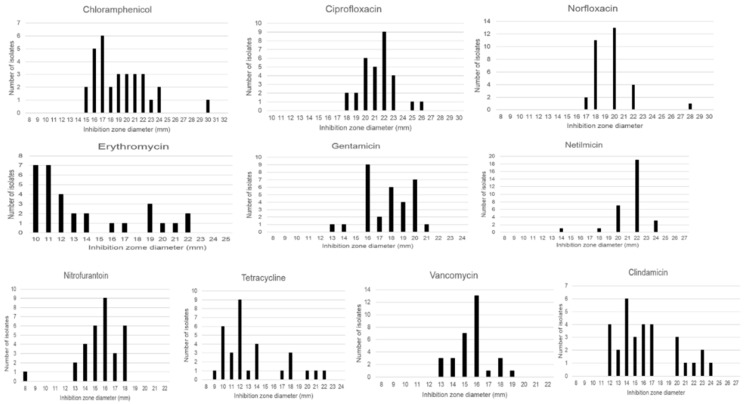
The detection and isolation of ampicillin-resistant B. cereus s.l. colonies confirmed the following results. All B. cereus s.l. isolates, including the B. cereus ATCC 10876, were significantly resistant to the tested concentrations of the β-lactam antibiotics penicillin G (PEN), ampicillin (AMP), carbenicillin (CAB), cefalotin (CFT), cefotaxime (CTX), and dicloxacillin (DCX) (Table 2, and see Methodology). All strains were also significantly resistant to the tested concentrations of trimethoprim-sulfamethoxazole (SXT). Our interpretation based on the EUCAST breakpoints, the results in Table 2 and Figure 4 show that the B. cereus s.l. isolates were resistant to tetracycline (TET) (90% of the isolates), erythromycin (ERY) (77%), clindamycin (CDM) (74%), and chloramphenicol (CHL) (42%). A lower percentage of strains showed resistance to amikacin (AMK) (6%), gentamicin (GEN) (6%), netilmicin (NET) (3%), and nitrofurantoin (NTF) (3%). On the other hand, B. cereus s.l. isolates were susceptible to ciprofloxacin (CIP) (100%), norfloxacin (NFX) (100%), vancomycin (VCM) (100%), netilmicin (NET) (97%), nitrofurantoin (NTF) (97%), amikacin (AMK) (94%), gentamicin (GEN) (94%), chloramphenicol (CHL) (58%), clindamycin (CDM) (26%), erythromycin (ERY) (23%), and tetracycline (TET) (10%).
Figure 4.
Determination of inhibition zone diameter in the antibiotic disc diffusion tests. Interpretation is based on the EUCAST breakpoints; isolates categorized as susceptible to norfloxacin (S ≥ 17 mm) can be reported as “susceptible increased exposure” to ciprofloxacin (R ˂ 21 mm, S ≥ 50 mm) (see Table 2), vancomycin (R ˂ 10 mm, S ≥ 10 mm), netilmicin (R ˂ 18 mm, S ≥ 18 mm), nitrofurantoin (R ˂ 13 mm, S ≥ 13 mm), gentamicin (R ˂ 18 mm, S ≥ 18 mm), chloramphenicol (R ˂ 18 mm, S ≥ 18 mm), clindamycin (R ˂ 19 mm, S ≥ 22 mm), erythromycin (R ˂ 18 mm, S ≥ 21 mm), and tetracycline (R ˂ 19 mm, S ≥ 22 mm).
4. Discussion
In Mexico, the cultivated pepper (Capsicum spp.) is a phenotypically diverse species grown throughout the country, in different geographical areas, and using various cultivation methods [2,6]. The peppers are used to produce a wide range of chili powders that are subsequently used in a variety of foodstuffs. One potential issue, from a biosecurity perspective, is the possibility that chili powder might be exposed to microbial contamination during its production, storage, commercialization, or when combined with other ingredients to prepare seasoned foodstuffs. Therefore, quality and safety must be important considerations in certifying chili powder for use, with the specific focus on detection of potential pathogens to avoid the spread of food-borne illnesses in consumers.
With these considerations in mind, the studies presented here demonstrate the presence in chili powder samples of levels of aerobic-mesophilic bacterial spores of 8.0 × 102 to 3.24 × 105 cfu/g and bacterial species closely related phylogenetically to the B. cereus s.l. group, and in low quantities (˂700 cfu/g). With regard to this results, in some reports, B. cereus s.l. levels of ˂104 cfu/g have been found in red chili powder from India [32], in paprika and pepper (˂80 cfu/g) in Germany [14], and in chili powder and organic paprika (˂23 cfu/g) in the USA [15]. The European Food Safety Authority (EFSA) has noted that some reports of food-borne disease outbreaks caused by diverse B. cereus isolates have been associated with bacterial concentrations above 105 cfu/g foodstuff, but in a few reports, concentrations of 103 cfu/g [33] were involved in reported outbreaks.
The studies reported are also significant in that they demonstrate the presence of enterotoxin genes (hblC and nheA) in the B. cereus s.l. isolates. The PCR results were confirmed and extended by the Duopath test for detection of toxin production, which showed the presence of Hbl and Nhe toxins in 90% and 100% of isolates, respectively. The presence of these toxins increases the potential for these strains to cause food-borne illnesses in the consumers of chili powder. Other reports on studies involving paprika, chili powder, and other seasonings have also shown that the nhe and hbl gene sequences and toxin production are detected at high frequency [14,15]. Taking into account the available information and in agreement with our results, the levels of B. cereus s.l. found in Mexican chili powder are low and probably not represent a health risk according to EFSA. However, the results are a minimum representative sample from a wide chili powder diversity in Mexico; thus, requiring additional efforts to extend similar microbial analysis throughout the country, where chili powder is produced.
Generally, chili powder is a ready-to-eat spice in different foodstuffs in Mexico, and for the consumers, the inherent health risk occasioned by the presence of B. cereus s.l. could be increased due to the antibiotic resistance properties of these organisms. That resistance could easily compromise treatment procedures in cases of infections caused by these toxigenic bacteria after consumption of chili powder. Thus, obtaining the B. cereus antibiotic resistance profile is highly relevant to public health. Our results in this context are in concordance with other reports where B. cereus s.l. has been shown to be resistant to β-lactam antibiotics resulting from the presence in their genomes of genes coding for β-lactamase enzymes [13,14,15,22,23]. It is worth mentioning that B. cereus s.l. isolates were also significantly resistant to trimethoprim-sulfamethoxazole, tetracycline, erythromycin, clindamycin, and chloramphenicol. Similar antibiotic resistances have been reported in the USA, Europe, and China [14,23,34]. Similar to the results of other reports, our B. cereus s.l. isolates were generally susceptible to other antibiotics, including ciprofloxacin, norfloxacin, vancomycin, netilmicin, nitrofurantoin, amikacin, and gentamicin [14,23,34]. A few B. cereus s.l. isolates, however, showed reduced susceptibility to chloramphenicol, clindamycin, erythromycin, and tetracycline in our studies. Although the number is limited, there are studies that indicate that vancomycin is the antibiotic of choice for B. cereus infections [23,34,35]. The results obtained in this study confirm the need for further studies of such resistance breakpoints.
Additional insight into the taxonomic and toxigenic relationship of B. cereus s.l. isolates obtained in this study is necessary by comparative genomics, as well as by other methods of molecular and biochemical discrimination analyses necessary to understand its metabolic capabilities and close relationship among members of the B. cereus group.
5. Conclusions
The results obtained in this study show that Mexican chili powder can be contaminated with B. cereus s.l. strains, which represent a potential health risk because of their antibiotic resistance and their production of biological toxins. Furthermore, chili powder has a long shelf life and the spore-forming ability of B. cereus s.l. allows it to persist for long periods. All of these considerations would seem to support the development of appropriate monitoring programs for microbial quality to assess the distribution of microorganisms in seasonings, such as chili powder, and the potential risk to public health.
Author Contributions
Conceptualization, J.C.G. and J.L.H.F.; methodology and software, A.G.C.H., V.G.O., E.Á.H., J.N.R., and F.J.F.G.; writing—original draft preparation, J.C.G., J.L.H.F., J.L.A.G., M.Á.R.L., J.A.R.M., A.F.M., and G.H.J.; writing—review and editing, J.C.G., G.H.J., J.L.H.F., M.C.G.G., and S.R.G.; secured funding for the project and the discussion of the final results, conclusions, and project administration, J.C.G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially financed by the Universidad Autónoma de Querétaro (FONDEC-UAQ-2019-FCQ202001 and FONDO “QUÍMICA SOMOS TODOS” 2020).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baenas N., Belović M., Ilic N., Moreno D.A., García-Viguera C. Industrial use of pepper (Capsicum annum L.) derived products: Technological benefits and biological advantages. Food Chem. 2019;274:872–885. doi: 10.1016/j.foodchem.2018.09.047. [DOI] [PubMed] [Google Scholar]
- 2.Aguilar-Rincón V.H., Corona Torres P.T., López López L., Latournerie Moreno M., Ramírez Meraz H., Villalón Mendoza Y.J., Aguilar Castillo A. Los chiles de México y su distribución. Rev Fitotecnia Mex SINAREFI, Colegio de Postgraduados. INIFAP, ITConkal, UANL, UAN; Montecillo, Texcoco, Estado de México, Mexico: 2010. p. 114. [Google Scholar]
- 3.Padilha H.K.M., Pereira E.D.S., Munhoz P.C., Vizzotto M., Valgas R.A., Barbieri R.L. Genetic variability for synthesis of bioactive compounds in peppers (Capsicum annuum) from Brazil. Food Sci. Technol. 2015;35:516–523. doi: 10.1590/1678-457X.6740. [DOI] [Google Scholar]
- 4.De Azevedo-Meleiro C.H., Rodriguez-Amaya D.B. Qualitative and quantitative differences in the carotenoid composition of yellow and red peppers determined by HPLC-DAD-MS. J. Sep. Sci. 2009;32:3652–3658. doi: 10.1002/jssc.200900311. [DOI] [PubMed] [Google Scholar]
- 5.Eggersdorfer M., Wyss A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018;652:18–26. doi: 10.1016/j.abb.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Gaytan V., Gómez-Merino F.C., Trejo-Téllez L.I., Baca-Castillo G.A., García-Morales S. The Chilhuacle Chili (Capsicum annuum L.) in Mexico: Description of the Variety, Its Cultivation, and Uses. Int. J. Agron. 2017;2017:1–13. doi: 10.1155/2017/5641680. [DOI] [Google Scholar]
- 7.Molnár H., Bata-Vidács I., Baka E., Cserhalmi Z., Ferenczi S., Tömösközi-Farkas R., Adányi N., Székács A. The effect of different decontamination methods on the microbial load, bioactive components, aroma and colour of spice paprika. Food Control. 2018;83:131–140. doi: 10.1016/j.foodcont.2017.04.032. [DOI] [Google Scholar]
- 8.Feroz F., Shimizu H., Nishioka T., Mori M., Sakagami Y. Bacterial and Fungal Counts of Dried and Semi-Dried Foods Collected from Dhaka, Bangladesh, and Their Reduction Methods. Biocontrol Sci. 2016;21:243–251. doi: 10.4265/bio.21.243. [DOI] [PubMed] [Google Scholar]
- 9.Van Doren J.M., Neil K.P., Parish M., Gieraltowski L., Gould L.H., Gombas K.L. Foodborne illness outbreaks from microbial contaminants in spices, 1973–2010. Food Microbiol. 2013;36:456–464. doi: 10.1016/j.fm.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 10.González M.G.M., Romero S.M., Arjona M., Larumbe A.G., Vaamonde G. Microbiological quality of Argentinian paprika. Rev. Argent. Microbiol. 2017;49:339–346. doi: 10.1016/j.ram.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Mamun A.A., Masuma A., Majumder D., Ali M., Hossen M., Maruf K. Quality assessment of selected commercial brand of chilli powder in Bangladesh. MOJ Food Process Tech. 2016;3:70–73. doi: 10.15406/mojfpt.2016.03.00070. [DOI] [Google Scholar]
- 12.Bata-Vidács I., Baka E., Tóth Á., Csernus O., Luzics S., Adányi N., Székács A., Kukolya J. Investigation of regional differences of the dominant microflora of spice paprika by molecular methods. Food Control. 2018;83:109–117. doi: 10.1016/j.foodcont.2017.04.030. [DOI] [Google Scholar]
- 13.György É., Laslo É., Antal M., András C.D. Antibiotic resistance pattern of the allochthonous bacteria isolated from commercially available spices. Food Sci. Nutr. 2021;9:4550–4560. doi: 10.1002/fsn3.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frentzel H., Kraushaar B., Krause G., Bodi D., Wichmann-Schauer H., Appel B., Mader A. Phylogenetic and toxinogenic characteristics of Bacillus cereus group members isolated from spices and herbs. Food Control. 2018;83:90–98. doi: 10.1016/j.foodcont.2016.12.022. [DOI] [Google Scholar]
- 15.Hariram U., Labbé R. Spore Prevalence and Toxigenicity of Bacillus cereus and Bacillus thuringiensis Isolates from U.S. Retail Spices. J. Food Prot. 2015;78:590–596. doi: 10.4315/0362-028X.JFP-14-380. [DOI] [PubMed] [Google Scholar]
- 16.Lehmacher A., Bockemühl J., Aleksic S. Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 1995;115:501–511. doi: 10.1017/S0950268800058660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehling-Schulz M., Lereclus D., Koehler T.M. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec.gpp3-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guinebretière M.H., Auger S., Galleron N., Contzen M., De Sarrau B., De Buyser M.L., Lamberet G., Fagerlund A., Granum P.E., Lereclus D., et al. Bacillus cytotoxicus sp. nov. is a new thermotolerant species of the Bacillus cereus Group occasionally associated with food poisoning. Int. J. Syst. Evol. Microbiol. 2012;63:31–40. doi: 10.1099/ijs.0.030627-0. [DOI] [PubMed] [Google Scholar]
- 19.Chang T., Rosch J.W., Gu Z., Hakim H., Hewitt C., Gaur A., Wu G., Hayden R.T. Whole-Genome Characterization of Bacillus cereus Associated with Specific Disease Manifestations. Infect. Immun. 2018;86 doi: 10.1128/IAI.00574-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapidus A., Goltsman E., Auger S., Galleron N., Ségurens B., Dossat C., Land M.L., Broussolle V., Brillard J., Guinebretiere M.-H., et al. Extending the Bacillus cereus group genomics to putative food-borne pathogens of different toxicity. Chem. Interact. 2008;171:236–249. doi: 10.1016/j.cbi.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Klee S.R., Brzuszkiewicz E.B., Nattermann H., Brüggemann H., Dupke S., Wollherr A., Franz T., Pauli G., Appel B., Liebl W., et al. The Genome of a Bacillus Isolate Causing Anthrax in Chimpanzees Combines Chromosomal Properties of B. cereus with B. anthracis Virulence Plasmids. PLoS ONE. 2010;5:e10986. doi: 10.1371/journal.pone.0010986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson B.D., Elkins C., Mullis L.B., Heinze T.M., Wagner R.D., Cerniglia C.E. A metallo-β-lactamase is responsible for the degradation of ceftiofur by the bovine intestinal bacterium Bacillus cereus P41. Vet. Microbiol. 2014;172:499–504. doi: 10.1016/j.vetmic.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Luna V.A., King D.S., Gulledge J., Cannons A.C., Amuso P.T., Cattani J. Susceptibility of Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides and Bacillus thuringiensis to 24 antimicrobials using Sensititre (R) automated microbroth dilution and Etest (R) agar gradient diffusion methods. J. Antimicrob. Chemother. 2007;60:555–567. doi: 10.1093/jac/dkm213. [DOI] [PubMed] [Google Scholar]
- 24.Hernández Flores J.L., Salinas Landaverde D., Pacheco Huerta Y., Guerra Castillo V.L., Barrios Sánchez M.D.L.Á., Arvizu Hernández I., Ramos López M.Á., Álvarez Hidalgo E., Jones G.H., Campos Guillén J. Phylogenetic Analysis of Bacillus cereus sensu lato Isolates from Commercial Bee Pollen Using tRNACys-PCR. Microorganisms. 2020;8:524. doi: 10.3390/microorganisms8040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 27.Jukes T.H., Cantor C.R. Evolution of protein molecules. In: Munro H.N., editor. Mammalian Protein Metabolism. Academic Press; Cambridge, MA, USA: 1969. pp. 21–132. [Google Scholar]
- 28.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause N., Moravek M., Dietrich R., Wehrle E., Slaghuis J., Märtlbauer E. Performance characteristics of the Duopath® Cereus Enterotoxins assay for rapid detection of enterotoxinogenic Bacillus cereus strains. Int. J. Food Microbiol. 2010;144:322–326. doi: 10.1016/j.ijfoodmicro.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Hansen B.M., Hendriksen N.B. Detection of Enterotoxic Bacillus cereus and Bacillus thuringiensis Strains by PCR Analysis. Appl. Environ. Microbiol. 2001;67:185–189. doi: 10.1128/AEM.67.1.185-189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Testing E.T. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. 2021. [(accessed on 5 July 2021)]. Available online: http://www.eucast.org.
- 32.Banerjee M., Sarkar P.K. Microbiological quality of some retail spices in India. Food Res. Int. 2003;36:469–474. doi: 10.1016/S0963-9969(02)00194-1. [DOI] [Google Scholar]
- 33.EFSA Panel on Biological Hazards (BIOHAZ) Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J. 2016;14:e04524. [Google Scholar]
- 34.Gao T., Ding Y., Wu Q., Wang J., Zhang J., Yu S., Yu P., Liu C., Kong L., Feng Z., et al. Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Bacillus cereus Isolated From Pasteurized Milk in China. Front. Microbiol. 2018;9:533. doi: 10.3389/fmicb.2018.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godič Torkar K., Bedenić B., Plecko V. Antimicrobial susceptibility and the in vitro postantibiotic effects of vancomycin and ciprofloxacin against Bacillus cereus isolates. J. Chemother. 2015;28:1973947815Y0000000069. doi: 10.1179/1973947815Y.0000000069. [DOI] [PubMed] [Google Scholar]