Abstract
Ascomycetous yeast species in the genus Wickerhamomyces (Saccharomycetales, Wickerhamomycetaceae) are isolated from various habitats and distributed throughout the world. Prior to this study, 35 species had been validly published and accepted into this genus. Beneficially, Wickerhamomyces species have been used in a number of biotechnologically applications of environment, food, beverage industries, biofuel, medicine and agriculture. However, in some studies, Wickerhamomyces species have been identified as an opportunistic human pathogen. Through an overview of diversity, taxonomy and recently published literature, we have updated a brief review of Wickerhamomyces. Moreover, two new Wickerhamomyces species were isolated from the soil samples of Assam tea (Camellia sinensis var. assamica) that were collected from plantations in northern Thailand. Herein, we have identified these species as W. lannaensis and W. nanensis. The identification of these species was based on phenotypic (morphological, biochemical and physiological characteristics) and molecular analyses. Phylogenetic analyses of a combination of the internal transcribed spacer (ITS) region and the D1/D2 domains of the large subunit (LSU) of ribosomal DNA genes support that W. lannaensis and W. nanensis are distinct from other species within the genus Wickerhamomyces. A full description, illustrations and a phylogenetic tree showing the position of both new species have been provided. Accordingly, a new combination species, W. myanmarensis has been proposed based on the phylogenetic results. A new key for species identification is provided.
Keywords: ascomycetous yeast, distribution, new species, phylogeny, taxonomy, Wickerhamomyces
1. Introduction
The genus Wickerhamomyces was first proposed by Kurtzman et al. [1] in 2008 with W. canadensis (basionym Hansenula canadensis) as the type species. This genus belongs to the family Wickerhamomycetaceae of the order Saccharomycetales [1]. Wickerhamomyces species can reproduce both asexually and sexually. Through asexual reproduction, the species reproduce by budding and some species produce pseudohyphae and/or true hyphae. Alternatively, in sexual reproduction they produce hat-shaped or spherical ascospores with an equatorial ledge for sexual reproduction [1,2]. Most of the known Wickerhamomyces species can utilize various carbon sources, but not methanol or hexadecane. Nitrate utilization was observed in some species, while the diazonium blue B reaction was negative for all species. The predominant ubiquinone in the Wickerhamomyces species is CoQ-7 [1,3].
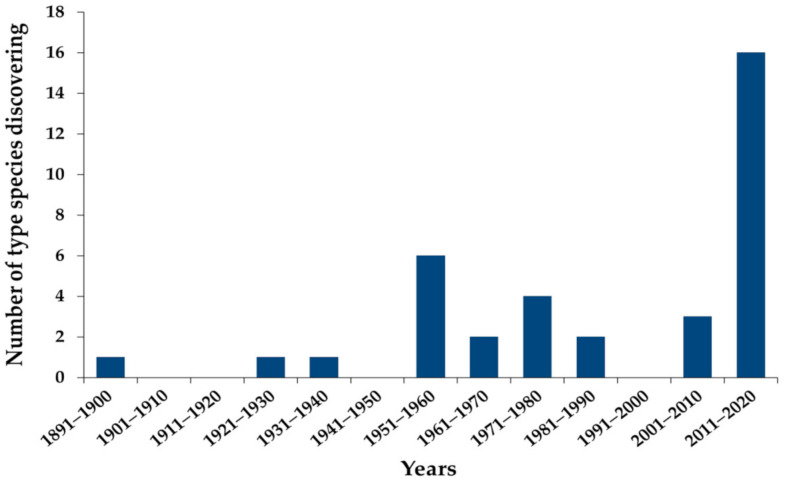
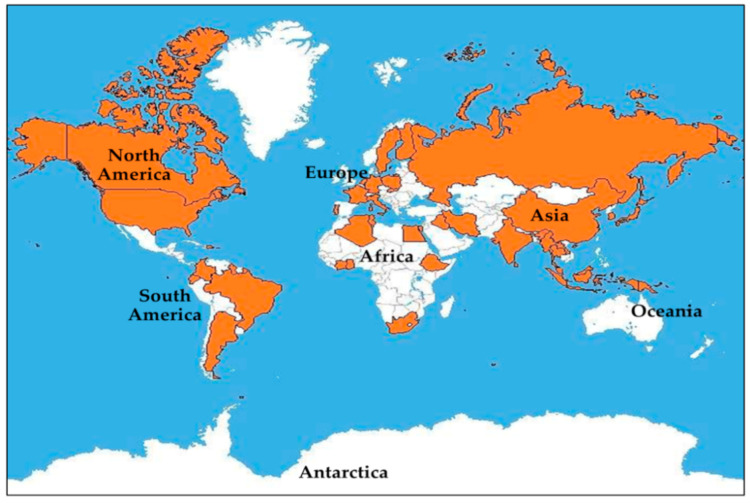
Most species of the genus Wickerhamomyces have been transferred from the genera Candida, Hansenula, Pichia and Williopsis based on phylogenetic analyses [1,2,4,5,6]. Currently, a total of 35 species have been accepted and recorded in the Index Fungorum [7]. A phylogenetic study of Arastehfar et al. [8] has suggested that Pichai myanmarensis [9] should be transferred to the genus Wickerhamomyces, but W. myanmarensis has remained invalidly published. Therefore, in this study, we have proposed the validation of this name for a new combination species. In this case, 35 type species of Wickerhamomyces and P. myanmarensis have been reported since 1891. It has been revealed that the highest number of the Wickerhamomyces species were discovered during the period of 2011–2020, followed by the period of 1951–1960 (6 species), the period of 1971–1980 (4 species) and the period of 2001–2010 (3 species) (Figure 1). An increasing trend with regard to the discovery of new species of Wickerhamomyces is expected to continue in the future. Wickerhamomyces species are widely distributed in tropical, subtropical, temperate and subpolar areas throughout the world (Figure 2). It has been reported that the highest number of Wickerhamomyces species were found in Asia (18 species), followed by Europe (12 species), South America (8 species), Africa (7 species), North America (3 species), Oceania (1 species) and Antartica (1 species) [7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82] (Table 1). Both W. anomalus and W. onychis are known to be from Asia, Africa, Europe and South America [13−35,54−59]. Moreover, W. anomalus and W. rabaulenis have been discovered in Antarctica (King George Island) [17] and Oceania (Papua New Guinea) [67], respectively. Recently, W. psychrolipolyticus has been discovered from Japan [65]. Consequently, Wickerhamomyces species have been successfully isolated from various habitats, as has been summarized in Table 1.
Figure 1.
The discovering of Wickerhamomyces type species since 1891 till the present time.
Figure 2.
Global distribution of Wickerhamomyces species. The countries where Wickerhamomyces species have been discovered are indicated in orange color.
Table 1.
Global distribution and isolation sources of Wickerhamomyces species.
| No. | Spices | Known Distribution | Isolation Source | Reference |
|---|---|---|---|---|
| 1 | Wickerhamomyces alni (Phaff, M.W. Mill. and M. Miranda) Kurtzman, Robnett and Bas.-Powers | Canada | Exudate of Alnus rubra | [12] |
| 2 | Wickerhamomyces anomalus (E.C. Hansen) Kurtzman, Robnett and Bas.-Powers | Algeria, Brazil, China, Colombia, Ethiopia, India, Iraq, King George Island, Lao, Russia, Slovakia Sweden and Thailand |
Process of beer and wine, phylloplane, soil, water, coral reefs, Thai traditional alcoholic starter, mangrove forest, fermented food, flowers, fruits, fermented grains, coffee processing, wastewater treatment plant, Colombian fermented beans and Brazilian spirit | [13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35] |
| 3 | Wickerhamomyces arborarius S.A. James, E.J. Carvajal, Barahona, T.C. Harr., C.F. Lee, C.J. Bond and I.N. Roberts | Ecuador | Flower | [36] |
| 4 | Wickerhamomyces bisporus (O. Beck) Kurtzman, Robnett and Bas.-Powers | Finland, France and USA | Platypus compositus, phoretic mites on Ips typographus and bark beetles (Dendroctonus) | [37,38,39] |
| 5 | Wickerhamomyces bovis (Uden and Carmo Souza) Kurtzman, Robnett and Bas.-Powers | Portugal | Caecum of feral cattle (Bos taurus) | [40] |
| 6 | Wickerhamomyces canadensis (Wick.) Kurtzman, Robnett and Bas.-Powers | Canada | Beetle frass from Pinus resinosa | [41] |
| 7 | Wickerhamomyces chambardii (C. Ramírez and Boidin) Kurtzman, Robnett and Bas.-Powers | France | Chestnut | [42] |
| 8 | Wickerhamomyces chaumierensis M. Groenew., V. Robert and M.T. Sm. | Guyana | Surface of flower | [43] |
| 9 | Wickerhamomyces ciferrii (Lodder) Kurtzman, Robnett and Bas.-Powers | Dominican Republic, USA and Thailand | Fruit of Dipteryx odorata and male olive fruit fly (Bactrocera oleae) | [44,45,46] |
| 10 | Wickerhamomyces edaphicus Limtong, Yongman., H. Kawas. and Fujiyama | India and Thailand | Forest and mangrove soils | [4,47] |
| 11 | Wickerhamomyces hampshirensis (Kurtzman) Kurtzman, Robnett and Bas.-Powers | USA | Frass of cut and dead of Quercus and beetle (Xyloterinus politus) | [48,49] |
| 12 | Wickerhamomyces kurtzmanii A.H. Li, Y.G. Zhou and Q.M. Wang | China | Crater lake water | [6] |
| 13 | Wickerhamomyces lynferdii (Van der Walt and Johannsen) Kurtzman, Robnett and Bas.-Powers | South Africa | Soil | [50] |
| 14 | Wickerhamomyces menglaensis F.L. Hui and L.N. Huang | China | Rotting wood | [51] |
| 15 | Wickerhamomyces mori F.L. Hui, Liang Chen, X.Y. Chu, Niu and T. Ke | China | Gut of larvae of wood-boring insect on trunk of Morus alba | [52] |
| 16 | Wickerhamomyces mucosus (Wick. and Kurtzman) Kurtzman, Robnett and Bas.-Powers | USA | Soil | [53] |
| 17 | Wickerhamomyces myanmarensis (Nagats., H. Kawas. and T. Seki) J. Kumla, N. Suwannarach and S. Lumyong | Iran and Myanmar | Palm sugar in rum distiller, and blood and central venous catheter of patients | [8,9] |
| 18 | Wickerhamomyces ochangensis K.S. Shin | South Korea | Soil of potato field | [11] |
| 19 | Wickerhamomyces onychis (Yarrow) Kurtzman, Robnett and Bas.-Powers | Brazil, Ethiopia, Iraq, Malaysia, Netherlands, Poland and Tunisia | Nail infection of Homo sapiens, fermented food, cocoa beans, grape and tomato during spontaneous fermentation, and soil | [23,54,55,56,57,58,59] |
| 20 | Wickerhamomyces orientalis Sipiczki, S. Nasr, H.D.T. Nguyen and Soudi | Iran and Sri Lanka | Fruits and rhizosphere soil | [27,60] |
| 21 | Wickerhamomyces patagonicus V. de García, Brizzio, C.A. Rosa, Libkind and Van Broock | Argentina | Sap exudate on cut branches of Nothofagus dombeyi and glacier meltwater river | [61] |
| 22 | Wickerhamomyces pijperi (Van der Walt & Tscheuschner) Kurtzman, Robnett and Bas.-Powers | Egypt, Ghana and South Africa | Buttermilk, cocoa fermentation and orange juice | [62,63,64] |
| 23 | Wickerhamomyces psychrolipolyticus Y. Shimizu and K. Konno | Japan | Soil | [65] |
| 24 | Wickerhamomyces queroliae C.A. Rosa, P.B. Morais, Lachance and Pimenta | Brazil | Larva of Anastrepha mucronata from fruit of Peritassa campestris | [66] |
| 25 | Wickerhamomyces rabaulensis (Soneda and S. Uchida) Kurtzman, Robnett and Bas.-Powers | Ethiopia, Papua New Guinea and Thailand | Excreta of snail, soils, decaying agricultural residues, decaying leaves and tree bark, and fermented food |
[23,67,68] |
| 26 | Wickerhamomyces scolytoplatypi Ninomiya | Japan | Gallery of beetles (Scolytoplatypus shogun) in Fagus crenata | [69] |
| 27 | Wickerhamomyces siamensis Kaewwich., Yongman., H. Kawas. and Limtong | Thailand | Phylloplane of Saccharum officinarum | [70] |
| 28 | Wickerhamomyces silvicola (Wick.) Kurtzman, Robnett and Bas.-Powers | Germany, South Korea and USA |
Flowers, gum of Prunus serotina and Prunus wood | [41,71,72] |
| 29 | Wickerhamomyces spegazzinii Masiulionis and Pagnocca | Argentina | The fungus garden of an attine ant nest (Acromyrmex lundii) | [73] |
| 30 | Wickerhamomyces strasburgensis (C. Ramírez and Boidin) Kurtzman, Robnett and Bas.-Powers | France | On leather tanned by vegetable means | [74] |
| 31 | Wickerhamomyces subpelliculosus (Kurtzman) Kurtzman, Robnett and Bas.-Powers | Egypt and USA | Fermenting cucumber brines, gut of honey bee and molasses | [75,76] |
| 32 | Wickerhamomyces sydowiorum (D.B. Scott and Van der Walt) Kurtzman, Robnett and Bas.-Powers | Brazil, Ivory Coast, South Africa and Thailand |
Frass of Sinoxylon ruficorne in dead Combretum apiculatum, decayed plant leaf, fermented cocoa, honey, sand and water | [59,77,78,79,80] |
| 33 | Wickerhamomyces sylviae Moschetti and J.P. Samp. | Italy | Cloaca of migratory birds (Sylvia communis) |
[81] |
| 34 | Wickerhamomyces tratensis Nakase, Jindam., Am-In, Ninomiya and H. Kawas. | Thailand | Flower of mangrove apple (Sonneratia caseolaris) |
[82] |
| 35 | Wickerhamomyces xylosicus Limtong, Nitiyon, Kaewwich., Jindam., Am-In and Yongman | Thailand | Soil | [5] |
| 36 | Wickerhamomyces xylosivorus R. Kobay., A. Kanti and H. Kawas. | Indonesia | Decayed wood | [4] |
Many species of Wickerhamomyces have been used in a variety of industries including the medicinal, agricultural, biofuel, food and beverage industries, and a number of others [64]. Most previous studies have focused on different strains of W. anomalus for biotechnological applications. For example, the W. anomalus strains CBS261, HN006 and HN010 are capable of excessively producing ethyl acetate. As a result, this species has been used in the brewing of Baijiu (Chinese liquor) and in winemaking to improve the aroma and quality of the finished product [83,84,85].
Wickerhamomyces anomalus strains BS91 and DMKU-RP04 could effectively inhibit plant pathogenic fungi and have been used as a biological control agent in agriculture [86,87,88]. Notably, W. anomalus strains SDBR-CMU-S1-06 and Wa-32 have exhibited plant growth promotion potential by solubilizing insoluble minerals, producing indole-3-acitic acid (IAA) and siderophores, and by secreting various extracellular enzymes [16,89]. Moreover, most strains of W. anomalus are known to produce killer toxins that possess antimicrobial and larvicidal activities [90,91]. Wickerhamomyces bovis and W. silvicola have been observed to produce mycocin, which exhibited fungicidal activity [92,93]. In addition, W. lynferdii and W. sydowiorum have been recognized as relevant yeast species for the improvement of the fermentation processes for coffee cherries and cocoa, respectively [79,94]. Furthermore, W. subpelliculosus has been used as an alternative to baker’s yeast [95], while W. chambardii could produce amylase and cellulase enzymes that could be used to produce bioethanol from corn straw [96,97]. Previous studies have found that the biosurfactants produced by W. anomalus and W. edaphicus [47,98,99], the saturated fatty acids produced by W. siamensis [100], xylitol produced by W. rabaulensis [101], cellulase enzymes produced by W. psychrolipolyticus [65] and the extracellular polysaccharide produced by W. mucosus [102] could be applied in the bioremediation, biotechnological and cosmetic industries. Furthermore, these substances could also be employed in the production of detergents, food and various pharmaceuticals, as well as in the process of oil recovery enhancement.
On the other hand, some Wickerhamomyces species (e.g., W. anomalus and W. lynferdii) have been responsible for the spoilage of beer and bakery products [103,104,105,106]. Some cases of human infection caused by W. anomalus, W. myanmarensis and W. onychis have also been reported, but only with patients with serious illness [8,107,108,109,110,111]. Based on this evidence, W. anomalus has been labeled a biosafety level 1 organism by the European Food Safety Authority [112] and is considered safe for consumption by healthy individuals.
Currently, only eight Wickerhamomyces species, namely W. anomalus, W. ciferrii, W. edaphicus, W. rabaulensis, W. siamensis, W. sydowiorum, W. tratensis and W. xylosicus, have been reported in Thailand [4,8,16,17,19,20,46,70,78]. Accordingly, Thailand has been identified as a hotspot for unexpected novel species and the newly recorded discovery of many microorganisms [113,114]. In our previous investigation on yeasts in soil samples collected from Assam tea (Camellia sinensis var. assamica) plantations in northern Thailand [16], we obtained five yeast strains belonged to the genus Wickerhamomyces that represent potentially new species. In our present study, we have described them into two novel species. These two novel species are introduced based on their phenotypic (morphological, biochemical and physiological data) and molecular characteristics. To confirm their taxonomic status, phylogenetic relationship was determined by analysis of the combined sequence dataset of the D1/D2 domains of LSU and ITS sequences.
2. Materials and Methods
2.1. Yeast Strain
Five yeasts strains (SDBR-CMU-S2-02, SDBR-CMU-S2-06, SDBR-CMU-S2-14, SDBR-CMU-S2-17 and CMU-S3-15) isolated from soils of Assam tea (C. sinensis var. assamica) plantations in Thep Sadej, Doi Saket District, Chiang Mai Province and Sri Na Pan, Muang District, Nan Province, northern Thailand [16] were selected for this present study. All strains were deposited in the culture collection of the Sustainable Development of Biological Resources, Faculty of Science, Chiang Mai University (SDBR-CMU), Chiang Mai Province and Thailand Bioresource Research Center (TBRC), Pathum Thani Province, Thailand.
2.2. Yeast Identification
2.2.1. Morphological Study
The morphological characteristics of yeast strains were determined according to established methods by Kurtzman et al. [2], Yarrow [3] and Limtong et al. [10]. Colony characters were observed on yeast extract-malt extract agar (YMA) after two days of incubation in darkness at 30 °C. Ascospore formation was investigated on YMA, 5% malt extract agar (MEA), potato dextrose agar (PDA) and V8 agar after incubation at 25 °C in the dark for four weeks. Micromorphological characteristics were examined under a light microscope (Nikon Eclipse Ni U, Tokyo, Japan). Size data of the anatomical structure (e.g., cells, pseudohyphae, asci and ascospores) were based on at least 50 measurements of each structure using the Tarosoft (R) Image Frame Work program.
2.2.2. Biochemical and Physiological Studies
Biochemical and physiological characterizations of yeast strains was followed the previous studies [2,3,115]. Fermentation of carbohydrates including glucose, galactose, maltose, sucrose, trehalose, melibiose, lactose, raffinose, and xylose were performed. Additionally, assimilation tests for carbon (D-glucose, D-galactose, L-sorbose, N-acetyl glucosamine, D-ribose, D-xylose, L-arabinose, D-arabinose, rhamnose, sucrose, maltose, α,α-trehalose, α-methyl-D-glucoside, cellobiose, salicin, melibiose, lactose, raffinose, melezitose, inulin, soluble starch, glycerol, erythritol, ribitol, D-glucitol, D-mannitol, galactitol, myo-inositol, D-glucono-1,5-lactone, 2-ketogluconic acid, 5-ketogluconic acid, D-gluconate, D-glucuronate, D-galacturonic acid, DL-lactate, succinate, citrate, methanol, ethanol, and xylitol) and nitrogen compounds (ammonium sulfate, potassium nitrate, sodium nitrite, ethylamine hydrochloride, L-lysine, cadaverine, and creatine) were determined. Moreover, the effects of temperature on growth were examined by cultivation on YMA at temperature ranging from 15–45 °C and diazonium blue B reactions were tested [116].
2.2.3. Molecular Study
Each yeast strain was grown in 5 mL of yeast extract-malt extract broth in 18 × 180 mm test tubes with shaking at 150 rpm on an orbital shaker in the dark for two days. Yeast cells were harvested by centrifugation at 11,000 rpm and washed three times with sterile distilled water. Genomic DNA was extracted from yeast cells using DNA Extraction Mini Kit (FAVORGEN, Taiwan) following the manufacturer’s protocol. The ITS region and D1/D2 domains of LSU gene were amplified by polymerase chain reactions (PCR) using ITS1/ITS4 primers [117] and NL1/NL4 primers [118], respectively. The amplification of both D1/D2 domains and ITS region process consisted of an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 45 s, an extension at 72 °C for 1 min and 72 °C for 10 min on a peqSTAR thermal cycler (PEQLAB Ltd., UK). PCR products were checked and purified by a PCR clean up Gel Extraction NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel, Germany). Final PCR products were sent to 1st Base Company Co., Ltd., (Kembangan, Malaysia) for sequencing. The obtained sequences were used to query GenBank via BLAST (http://blast.ddbj.nig.ac.jp/top-e.html, accessed on 25 August 2021).
Phylogenetic analysis was carried out based on the combined dataset of ITS and D1/D2 domains of LSU sequences. Sequences from this study along with those obtained from previous studies and the GenBank database were selected and provided in Table 2. Multiple sequence alignment was performed using MUSCLE [119]. A combination of D1/D2 domains of LSU and ITS alignment was deposited in TreeBASE under the study ID number 28785. A phylogenetic tree was constructed under maximum likelihood (ML) and Bayesian inference (BI) methods. The ML analysis was carried out using RAxML-HPC2 on XSEDE (8.2.10) in CIPRES Science Gateway V. 3.3 [120] using GTRCAT model with 25 categories and 1000 bootstrap (BS) replications. The optimum nucleotide substitution model was obtained using jModeltest v.2.3 [121] under the Akaike information criterion (AIC) method. The BI analysis was performed using MrBayes 3.2.6 software for Windows [122]. The selected optimal model of each gene is similar as GTR + I + G model. Six simultaneous Markov chains were run with one million generations and starting from random trees and keeping one tree every 100th generation until the average standard deviation of split frequencies was below 0.01. The value of burn-in was set to discard 25% of trees when calculating the posterior probabilities. Bayesian posterior probabilities (PP) were obtained from the 50% majority rule consensus of the trees kept. The tree topologies were visualized in FigTree v1.4.0 [123].
Table 2.
DNA sequences used in the molecular phylogenetic analysis. Type strains indicated by “T”.
| Yeast Species | Strain | GenBank Acession Number | Reference | |
|---|---|---|---|---|
| ITS | D1/D2 | |||
| Wickerhamomyces alni | NRRL Y-11625T | ‒ | EF550294 | [1] |
| CBS 6986 | NR154966 | KY110065 | [124] | |
| Wickerhamomyces anomalus | NRRL Y-366T | ‒ | EF550341 | [1] |
| H1Wh | JQ857021 | JQ856997 | [17] | |
| Wickerhamomyces arborarius | Bq 164T | NR_55000 | FN908198 | [36] |
| Wickerhamomyces bisporus | NRRL Y-1482T | ‒ | EF550296 | [1] |
| Wickerhamomyces bovis | NRRL YB-4184T | ‒ | EF550298 | [1] |
| CBS 2616 | NR154968 | KY110109 | [124] | |
| Wickerhamomyces canadensis | NRRL Y-1888T | ‒ | EF550300 | [1] |
| GoToruMP327 | EF093299 | EF016107 | [125] | |
| Wickerhamomyces chambardii | NRRL Y-2378T | ‒ | EF550344 | [1] |
| CBS 1900 | NR154969 | KY110114 | [124] | |
| Wickerhamomyces chaumierensis | CBS 8565T | HM156503 | HM156533 | [43] |
| Wickerhamomyces ciferrii | NRRL Y-1031T | ‒ | EF550339 | [1] |
| UCDFST 83-22 | MH153583 | MH130275 | [126] | |
| Wickerhamomyces edaphicus | S-29T | AB436771 | AB436763 | [10] |
| CBS 10408 | KY105904 | KY110120 | [124] | |
| Wickerhamomyces hampshirensis | NRRL YB-4128T | ‒ | EF550334 | [1] |
| Wickerhamomyces kurtzmanii | TF5-16-2T | MK573939 | MK573939 | [6] |
| Wickerhamomyces lannaensis | SDBR-CMU-S3-15T | OK135750 | MT639220 | This study, [16] |
| SDBR-CMU-S2-02 | OK135752 | MT623569 | This study, [16] | |
| SDBR-CMU-S2-06 | OK135753 | MT613722 | This study, [16] | |
| Wickerhamomyces lynferdii | NRRL Y-7723T | EF550342 | EF550342 | [1] |
| BCRC 22676 | NR111798 | ‒ | [127] | |
| Wickerhamomyces menglaensis | NYNU 1673T | KY213818 | KY213812 | [51] |
| Wickerhamomyces mori | NYNU 1216T | JX204288 | JX204287 | [52] |
| NYNU 1204 | JX292100 | JX292099 | [52] | |
| Wickerhamomyces mucosus | NRRL YB-1344T | ‒ | EF550337 | [1] |
| CBS 6341 | Z93877 | KY110124 | [124] | |
| Wickerhamomyces myanmarensis | CBS 9786T | ‒ | AB126678 | [9] |
| SU-263 | MH236221 | MH236219 | [8] | |
| Wickerhamomyces nanensis | SDBR-CMU-S2-17T | OK143510 | MT613875 | This study, [16] |
| SDBR-CMU-S2-14 | OK143511 | MT623571 | This study, [16] | |
| Wickerhamomyces ochangensis | N7a-Y2T | NR154971 | HM485464 | [11] |
| CBS 11843 | KY105909 | ‒ | [124] | |
| Wickerhamomyces onychis | NRRL Y-7123T | ‒ | EF550279 | [1] |
| CBS 5587 | KY105910 | KY110125 | [124] | |
| Wickerhamomyces orientalis | KH-D1T | KF938677 | KF938676 | [60] |
| 12-101 | KU253704 | KU253703 | [60] | |
| Wickerhamomyces patagonicus | CRUB 1724T | FJ793131 | FJ666399 | [61] |
| CBS 11398 | NG057185 | KY110126 | [124] | |
| Wickerhamomyces pijperi | NRRL YB-4309T | ‒ | EF550335 | [1] |
| CBS 2887 | HM156502 | KY110127 | [124] | |
| Wickerhamomyces psychrolipolyticus | Y08-202-2T | ‒ | LC333101 | [65] |
| Y08-202-2 | ‒ | LC333102 | [65] | |
| Wickerhamomyces queroliae | UFMG-T05-200T | EU580140 | EU580140 | [66] |
| Wickerhamomyces rabaulensis | NRRL Y-7945T | ‒ | EF550303 | [1] |
| CBS 6797 | KY105914 | KY110128 | [124] | |
| Wickerhamomyces scolytoplatypi | NBRC 11029T | ‒ | AB534166 | [69] |
| CBS 12186 | KY105915 | KY110130 | [124] | |
| Wickerhamomyces siamensis | DMKU-RK359T | NR111029 | AB714248 | [70] |
| CBS 12570 | KY105916 | KY110131 | [124] | |
| Wickerhamomyces silvicola | NRRL Y-1678T | ‒ | EF550302 | [1] |
| GLMC 1708 | MT156140 | MT156324 | [71] | |
| Wickerhamomyces spegazzinii | JLU025T | KJ832072 | KJ832071 | [73] |
| Wickerhamomyces strasburgensis | NRRL Y-2383T | ‒ | EF550333 | [1] |
| Wickerhamomyces subpelliculosus | NRRL Y-1683T | NR111336 | EF550340 | [1,127] |
| Wickerhamomyces sydowiorum | NRRL Y-7130T | NR138219 | EF550343 | [1,36] |
| NRRL Y-10996 | FR690145 | FR690073 | [36] | |
| Wickerhamomyces sylviae | PYCC6345T | ‒ | KF240728 | [81] |
| U92A1 | ‒ | KF240729 | [81] | |
| Wickerhamomyces tratensis | NBRC 107799T | AB607029 | AB607028 | [82] |
| CBS 12176 | KY105935 | KY110150 | [124] | |
| Wickerhamomyces xylosica | CBS 12320T | NR160310 | AB557867 | [5] |
| NT31 | AB704715 | NG064304 | [5] | |
| Wickerhamomyces xylosivorus | NBRC 111553T | NR155013 | LC202858 | [4] |
| 14Y125 | ‒ | NG057186 | [4] | |
| Saccharomyces cerevisiae | NRRL Y-12632T | AY046146 | JQ689017 | [128] |
| Spathaspora allomyrinae | CBS 13924T | KP054268 | KP054267 | [129] |
Note: sepecies obtained in this study are in bold.
3. Results
3.1. Phylogenetic Results
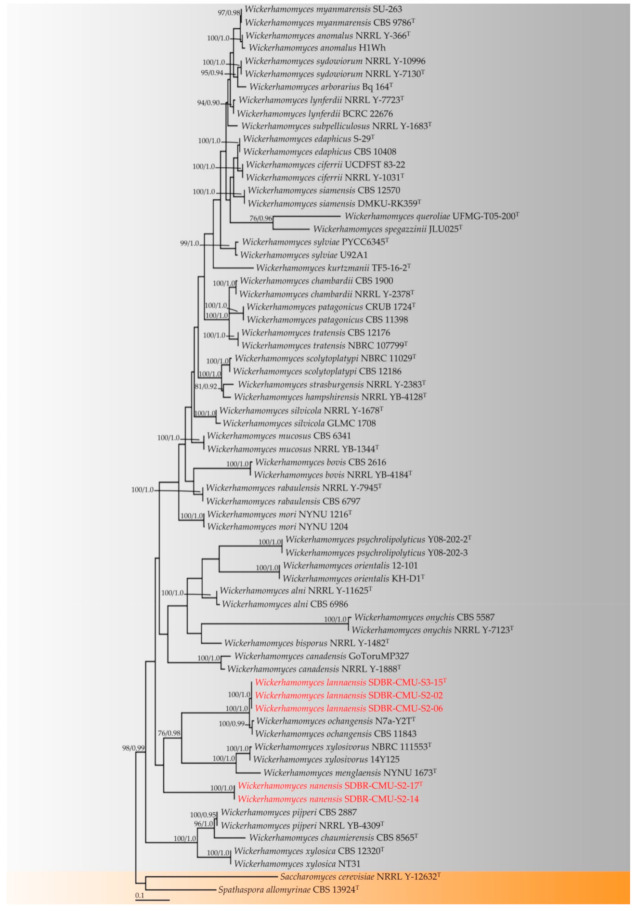
The sequences of five yeast strains were deposited in the GenBank database (Table 2). The alignment of a combination of ITS and D1/D2 domains of the LSU genes contained 1544 characters including gaps (ITS: 1−823 and D1/D2 domains of LSU: 824−1544). RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −12,120.4323. The matrix contained 776 distinct alignment patterns with 42.33% undetermined characters or gaps. Estimated base frequencies were recorded as follows: A = 0.2730, C = 0.1821, G = 0.2603, T = 0.2844; substitution rates AC = 1.0574, AG = 2.0209, AT = 1.4684, CG = 0.6712, CT = 4.4165, GT = 1.0000. The gamma distribution shape parameter alpha was equal to 0.2698 and the Tree-Length was equal to 4.4075. In addition, the final average standard deviation of the split frequencies at the end of the total MCMC generations was calculated as 0.00638 through BI analysis. Phylograms of the ML and BI analyses were similar in terms of topology (data not shown). Therefore, the phylogram obtained from the ML analysis was selected and presented for this study. The phylogram was comprised of 67 sequences of Wickerhamomyces strains (including 37 type strains obtained from either previous studies or the present study) and two sequences (Saccharomyces cerevisiae NRRL 12632 and Spathaspora allomyrinae CBS 13924) of the outgroup (Figure 3). Our phylogenetic analysis separated Wickerhamomyces by different species based on different topologies. Our analysis confirmed that W. myanmarensis (previously known as P. myanmarensis) belonged to the genus Wickerhamomyces according to the phylogenetic results of Arastehfar et al. [8] and Shimizu et al. [65]. Moreover, a phylogram clearly separated our yeast strains into two monophyletic clades with high support values (BS = 100% and PP = 1.0). The results indicated that our two yeast strains, SDBR-CMU-S2-17 and SDBR-CMU-S2-14 (introduced as W. nanensis), were clearly distinguished from the previously known species of Wickerhamomyces. Moreover, three yeast strains in this study, SDBR-CMU-S2-02, SDBR-CMU-S2-15, and CMU-S3-06 (described here as W. lannaensis) formed a sister clade to W. ochangensis with high support (BS = 100% and PP = 1.0).
Figure 3.
Phylogram derived from maximum likelihood analysis of 69 sequences of the combined ITS and D1/D2 sequences. Saccharomyces cerevisiae NRRL 12632 and Spathaspora allomyrinae CBS 13924 were used as the outgroup. The numbers above branches represent bootstrap percentages (left) and Bayesian posterior probabilities (right). Bootstrap values ≥ 75% and Bayesian posterior probabilities ≥ 0.90 are shown. Sequences obtained in this study are in red. Superscription “T” means the type strains.
3.2. Taxonomic Description of New Species
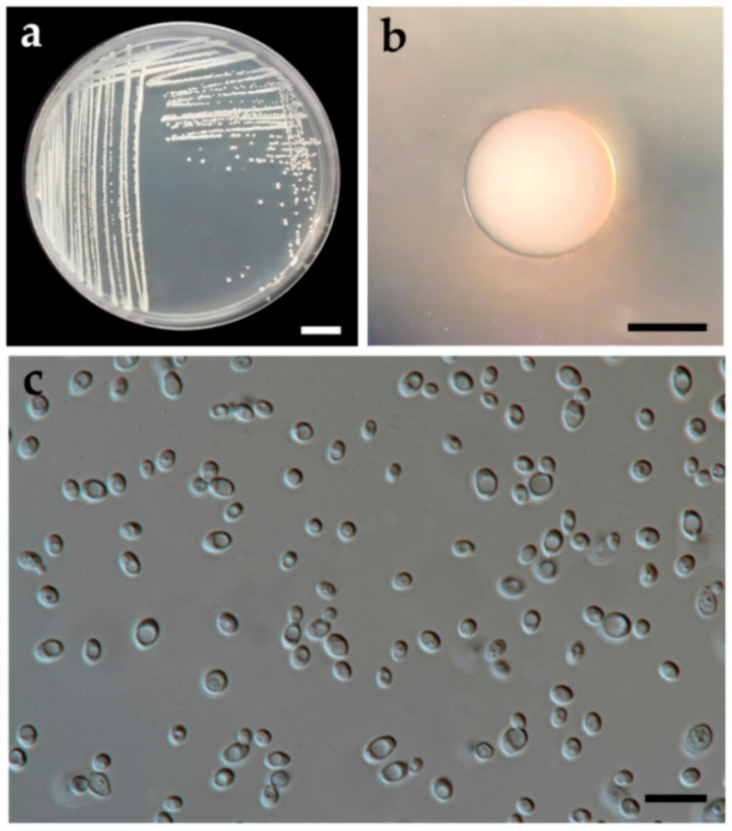
3.2.1. Wickerhamomyces lannaensis S. Nundaeng, J. Kumla, N. Suwannarach and S. Lumyong, sp. nov. (Figure 4)
Figure 4.
Wickerhamomyces lannaensis (holotype SDBR-CMU-S3-15). Culture (a), single colony (b) and budding cells (c) on YMA after two days at 30 °C. Scale bar a and b = 10 cm, c = 10 µm.
Mycobank No.: 841356
Etymology: “lannaensis” refers to Lanna kingdom the historic name of northern Thailand, the collection locality of the type strain of the species.
Holotype: Thailand, Chiang Mai Province, Thep Sadej, Doi Saket District, in soil from Assam tea (C. sinensis var. assamica) plantation, May 2017, J. Kumla and N. Suwannarach, (holotype SDBR-CMU-S3-15T, culture ex-type TBRC 15533)
Description: The streak culture on YMA after two days at 30 °C is circular from (1–2 mm in diameter), white to cream color, smooth surface, dull-shining, entire margin, and raised elevation. After growth on YMA at 30 °C for two days, the cells are spheroidal to short ovoidal (3.6–3.8 × 2.4–2.6 µm), occur singly or in budding pairs. Pseudohyphae and true hyphae were absent. Ascospores were not obtained for individual strains and strain pairs on YMA, 5% MEA, PDA and V8 agar after incubation at 30 °C for one month. Urea hydrolysis and diazonium blue B reactions are negative. Fermentation tests, glucose is delayed positive, but galactose, maltose, sucrose, trehalose, melibiose, lactose, raffinose, and xylose are negative. D-glucose, D-xylose, rhamnose, cellobiose, salicin, inulin (weak), glycerol, D-glucitol, D-mannitol, D-glucono-1,5-lactone, D-gluconate, DL-lactate (weak), succinate, and ethanol are assimilated. No growth was observed in L-sorbose, N-acetyl glucosamine, D-ribose, L-arabinose, D-arabinose, sucrose, maltose, α,α-trehalose, α-methyl-D-glucoside, melibiose, lactose, raffinose, melezitose, soluble starch, erythritol, ribitol, galactitol, myo-inositol, 2-ketogluconic acid, 5-ketogluconic acid, D-glucuronate, D-galacturonic acid, citrate, methanol, and xylitol. For the assimilation of nitrogen compounds, growth on ammonium sulfate, potassium nitrate, sodium nitrite, ethylamine HCl, cadaverine, and creatine (weak) are positive and on L-lysine is latent positive.
Growth in the vitamin-free medium is weak positive. Growth was observed at 15 °C and 30 °C, but not at 35, 37, 40, 42 and 45 °C. Growth in the presence of 50% glucose is positive, but growth in the presence of 0.01% cycloheximide, 0.1% cycloheximide, 60% glucose, 10% NaCl with 5% glucose and 15% NaCl with 5% glucose are negative. Acid formation is negative.
Additional strains examined: Thailand, Nan Province, Muang District, Sri Na Pan, in soil from Assam tea (C. sinensis var. assamica) plantation, September 2016, J. Kumla and N. Suwannarach, SDBR-CMU-S2-02, SDBR-CMU-S2-06.
GenBank accession numbers: holotype SDBR-CMU-S3-15 (D1/D2: MT639220, ITS: OK135750); additional strains SDBR-CMU-S2-02 (D1/D2: MT623569, ITS: OK135752) and SDBR-CMU-S2-06 (D1/D2: MT613722, ITS: OK135753).
Note: Based on phylogenetic analyses, W. lannaensis formed a monophyletic clade in a well-supported clade and was found to be closely related to W. ochangensis (Figure 3). Wickerhamomyces lannaensis can be distinguished from W. ochangensis by its ability to assimilate inulin and creatine and its growth in 50% glucose medium [11]. Additionally, W. ochangensis was able to grow at a temperature of 37 °C, while W. lannaensis could not grow at 37 °C [11].
3.2.2. Wickerhamomyces nanensis J. Kumla, S. Nundaeng, N. Suwannarach and S. Lumyong, sp. nov. (Figure 5)
Figure 5.
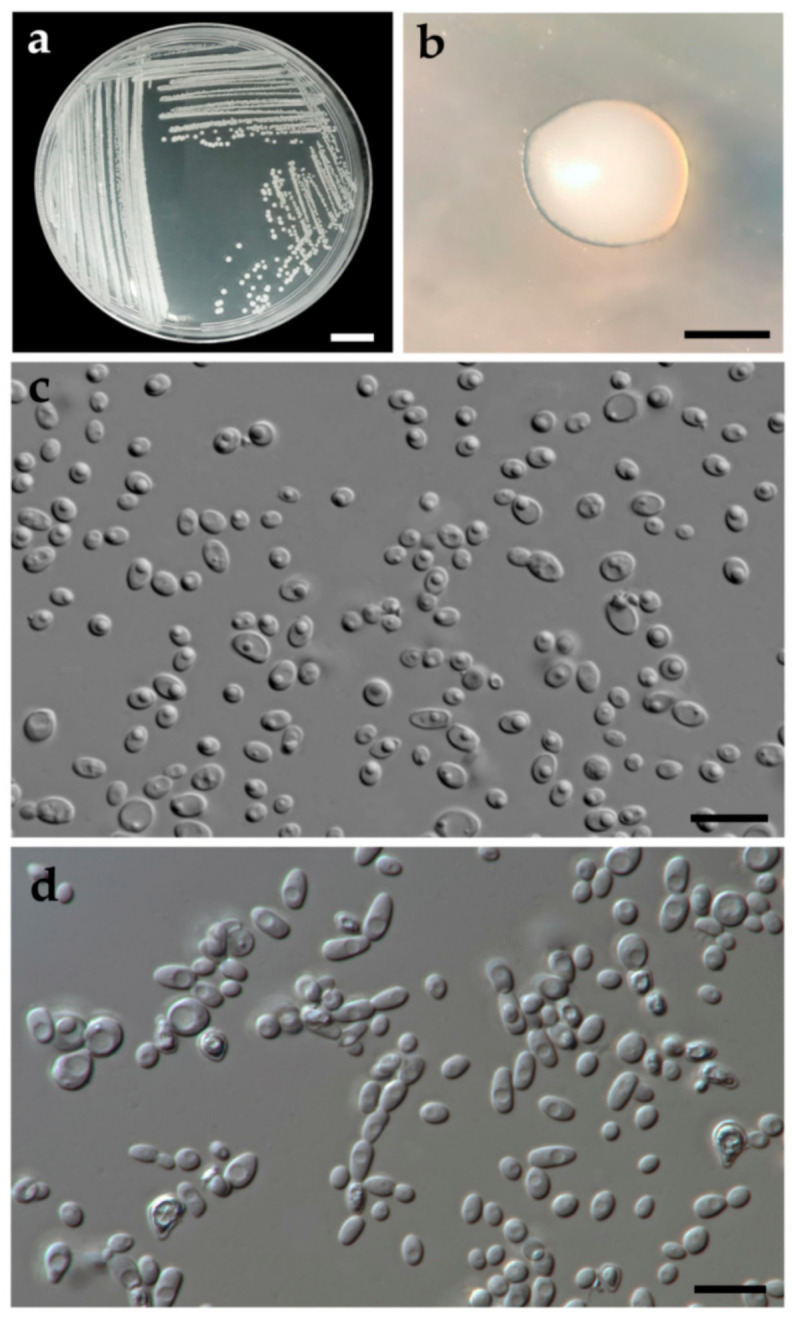
Wickerhamomyces nanensis (holotype SDBR-CMU-S3-15). Culture (a), single colony (b) and budding cells (c) on YMA after two days at 30 °C. Pseudohypahae (d) on 5% MAE agar after 7 days at 25 °C. Scale bar a and b = 10 cm, c and d = 10 µm.
Mycobank No.: 841357
Etymology: “nanensis” refers to Nan Province of Thailand, the collection locality of the type strain of the species.
Holotype: Thailand, Nan Province, Muang District, Sri Na Pan, in soil from Assam tea (C. sinensis var. assamica) plantation, September 2016, J. Kumla, N. Suwannarach and S. Khuna, (holotype SDBR-CMU-S2-17T, culture ex-type TBRC 15534)
Description: The streak culture on YMA after two days at 30 °C is circular from (1–2 mm in diameter), white to cream color, smooth surface, dull-shining, entire margin, and raised elevation. After growth on YMA at 30 °C for two days, the cells are spheroidal to short ovoidal (3.8–4.0 × 2.4–2.5 µm), occur singly or in budding pairs. Pseudohyphae (4.8–6.9 × 2.2–2.9 µm) were produced in Dalmau plate culture on 5% MEA and PDA after 7 days at 25 °C, but true hyphae are not obtained. Ascospores were not observed for individual strains and strain pairs on YMA, 5% MEA, PDA and V8 agar after incubation at 30 °C for one month. Urea hydrolysis and diazonium blue B reactions are negative. Fermentation test, glucose is delayed positive, but galactose, maltose, sucrose, trehalose, melibiose, lactose, raffinose, and xylose are not positive. D-glucose, D-galactose, cellobiose, salicin, glycerol, D-mannitol, D-glucono-1,5-lactone, DL-lactate (weak), succinate, citrate, and ethanol are assimilated. No growth was observed in L-sorbose, N-acetyl glucosamine, D-ribose, D-xylose, L-arabinose, D-arabinose, rhamnose, sucrose, maltose, α,α-trehalose, α-methyl-D-glucoside, melibiose, lactose, raffinose, melezitose, inulin, soluble starch, erythritol, ribitol, D-glucitol, galactitol, myo-inositol, 2-ketogluconic acid, 5-ketogluconic acid, D-gluconate, D-glucuronate, D-galacturonic acid, methanol, and xylitol. For the assimilation of nitrogen compounds, growth on ammonium sulfate, potassium nitrate (weak), sodium nitrite (weak), ethylamine HCl, l-lysine, and creatine (slow) are positive, but cadaverine is not. Growth in the vitamin-free medium is weak. Growth was observed at 15 °C and 30 °C, but not at 35, 37, 40, 42 and 45 °C. Growth in the presence of 50% glucose and acid formation are positive, but growth in the presence of 0.01% cycloheximide, 0.1% cycloheximide, 60% glucose, 10% NaCl with 5% glucose, and 15% NaCl with 5% glucose are negative.
Additional strain examined: Thailand, Nan Province, Muang District, Sri Na Pan, in soil from Assam tea (C. sinensis var. assamica) plantation, September 2016, J. Kumla and N. Suwannarach, SDBR-CMU-S2-14.
GenBank accession numbers: holotype SDBR-CMU-S2-17 (D1/D2: MT613875, ITS: OK143510); additional strain SDBR-CMU-S2-14 (D1/D2: MT623569, ITS: OK143511).
Note: Several morphological and biochemical characteristics of W. nanensis were similar to W. chambardii. However, W. chambardii differed from W. nanensis by its ascospore formation and could not assimilate D-mannitol [2]. Phylogenetic analyses clearly separated W. nanensis and W. chambardii as different species. Moreover, W. nanensis formed a monophyletic clade in a well-supported clade and was separated from other Wickerhamomyces species (Figure 3).
3.3. New Combination
Wickerhamomyces myanmarensis (Nagats., H. Kawas. and T. Seki) J. Kumla, N. Suwannarach and S. Lumyong, comb. nov.
Mycobank No.: 841356
Basionym: Pichia myanmarensis Nagats., H. Kawas. and T. Seki, Int. J. Syst. Evol. Microbiol. 55: 1381, 2005.
Note: The combined ITS and D1/D2 phylogenetic analyses indicated that the type species, P. myanmarensis, belongs to the genus Wickerhamomyces and has a close phylogenetic relationship with W. anomalus (Figure 3). Accordingly, the phylogenetic results of Arastehfar et al. [8] and Shimizu et al. [65] found that P. myanmarensis was placed within the genus Wickerhamomyces.
3.4. Key to Species of Wickerhamomyces
A key to the identification of the Wickerhamomyces species introduced in the present study was derived from the key described by Kurtzman et al. [2]. Key characteristics are shown in Table 3.
Table 3.
Key characteristics of species assigned to the genus Wickerhamomyces.
| Species | Growth in/at * | Ascospores on 5% MEA |
True Hyphae | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ga | Sor | DXy | LAr | DAr | Rh | Su | Cel | Mlb | Raf | St | Rbl | DGlu | Man | Glt | 2-ket | Cit | NO3 | ‒V | 37 °C | |||
| W. alni | ‒ | ‒ | + | ‒ | ‒ | + | + | + | ‒ | ‒ | ‒ | v | + | + | ‒ | ‒ | + | + | ‒ | ‒ | + | ‒ |
| W. anomalus | v | ‒ | v | v | ‒ | ‒ | + | + | ‒ | + | + | v | + | + | ‒ | ‒ | + | + | + | v | + | ‒ |
| W. arborarius | + | l/‒ | + | v | v | l/+ | + | v | + | + | + | + | + | + | ‒ | n | + | + | n | ‒ | ‒ | ‒ |
| W. bisporus | ‒ | ‒ | + | w/‒ | ‒ | + | + | + | ‒ | ‒ | ‒ | v | w/+ | v | ‒ | ‒ | + | + | ‒ | ‒ | n | + |
| W. bovis | ‒ | ‒ | + | + | ‒ | v | + | + | ‒ | ‒ | v | ‒ | + | + | ‒ | ‒ | + | ‒ | ‒ | + | + | ‒ |
| W. canadensis | ‒ | ‒ | + | ‒ | ‒ | w/+ | + | + | ‒ | ‒ | ‒ | v | + | w/+ | ‒ | ‒ | + | v | ‒ | + | + | v |
| W. chambardii | + | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | w | ‒ | ‒ | ‒ | + | ‒ |
| W. chaumierensis | ‒ | ‒ | + | ‒ | n | ‒ | + | + | ‒ | ‒ | n | n | ‒ | ‒ | n | ‒ | n | ‒ | n | ‒ | n | ‒ |
| W. ciferrii | + | ‒ | w/+ | w/+ | ‒ | w/+ | + | w/+ | ‒ | + | + | + | + | + | ‒ | ‒ | + | + | + | w/‒ | + | v |
| W. edaphicus | + | ‒ | + | w/‒ | ‒ | + | + | + | + | + | + | + | + | + | l/‒ | ‒ | + | + | + | w | n | ‒ |
| W. hampshirensis | ‒ | ‒ | + | ‒ | ‒ | s | + | + | ‒ | ‒ | ‒ | w/+ | + | v | ‒ | ‒ | s | ‒ | ‒ | ‒ | + | ‒ |
| W. kurtzmanii | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ | w | ‒ | ‒ | ‒ | ‒ | w | ‒ | n | ‒ | + | ‒ | ‒ | ‒ | ‒ |
| W. lynferdii | + | ‒ | ‒ | ‒ | ‒ | ‒ | + | + | ‒ | + | ‒ | + | + | + | ‒ | ‒ | + | + | + | ‒ | + | ‒ |
| W. menglaensis | ‒ | ‒ | w | w | ‒ | w | ‒ | + | ‒ | ‒ | w | ‒ | + | + | ‒ | ‒ | + | + | + | n | ‒ | ‒ |
| W. mori | ‒ | + | ‒ | ‒ | w | ‒ | + | ‒ | ‒ | ‒ | ‒ | ‒ | n | + | ‒ | ‒ | w | ‒ | + | ‒ | ‒ | ‒ |
| W. mucosus | ‒ | + | + | ‒ | v | ‒ | + | + | ‒ | ‒ | + | ‒ | + | w/+ | ‒ | + | ‒ | ‒ | ‒ | ‒ | + | ‒ |
| W. myanmarensis | + | ‒ | s | w | s | ‒ | + | s | ‒ | s | + | + | + | + | ‒ | n | + | + | + | + | + | ‒ |
| W. ochangensis | ‒ | ‒ | + | ‒ | ‒ | + | ‒ | + | ‒ | ‒ | ‒ | ‒ | + | + | ‒ | ‒ | ‒ | + | n | + | n | ‒ |
| W. onychis | ‒ | ‒ | + | v | v | ‒ | + | + | ‒ | + | ‒ | ‒ | + | + | ‒ | ‒ | + | ‒ | ‒ | + | + | ‒ |
| W. orientalis | + | n | w | w | w | w/‒ | + | + | w | w | ‒ | w | n | w | n | n | ‒ | ‒ | ‒ | + | ‒ | + |
| W. patagonicus | w | ‒ | + | ‒ | ‒ | + | n | w | ‒ | w | w | ‒ | w | ‒ | ‒ | n | ‒ | ‒ | + | ‒ | n | ‒ |
| W. pijperi | ‒ | + | + | ‒ | ‒ | ‒ | ‒ | + | ‒ | ‒ | ‒ | ‒ | + | + | ‒ | ‒ | v | ‒ | ‒ | ‒ | + | ‒ |
| W. psychrolipolyticus | ‒ | ‒ | + | ‒ | + | + | + | + | ‒ | + | + | ‒ | + | + | n | n | + | + | n | ‒ | ‒ | ‒ |
| W. queroliae | ‒ | ‒ | + | + | ‒ | + | + | ‒ | ‒ | ‒ | ‒ | + | + | + | ‒ | ‒ | w/s | + | ‒ | + | ‒ | ‒ |
| W. rabaulensis | ‒ | ‒ | + | + | ‒ | v | + | + | ‒ | + | ‒ | + | + | + | ‒ | ‒ | + | ‒ | ‒ | + | + | ‒ |
| W. scolytoplatypi | + | ‒ | s | ‒ | ‒ | s | + | + | ‒ | ‒ | + | s | + | + | ‒ | ‒ | ‒ | + | ‒ | ‒ | + | ‒ |
| W. siamensis | s | ‒ | s | ‒ | ‒ | ‒ | + | w | ‒ | w | w | ‒ | w | w | ‒ | ‒ | ‒ | ‒ | ‒ | + | + | - |
| W. silvicola | + | v | + | + | ‒ | + | v | + | ‒ | ‒ | ‒ | + | + | v | ‒ | ‒ | v | + | ‒ | v | + | v |
| W. spegazzinii | + | ‒ | + | ‒ | ‒ | + | + | + | + | + | s | ‒ | + | + | ‒ | ‒ | w | + | + | + | + | ‒ |
| W. strasburgensis | + | ‒ | + | + | ‒ | + | + | + | ‒ | + | ‒ | + | + | + | ‒ | ‒ | + | ‒ | ‒ | v | + | ‒ |
| W. subpelliculosus | v | ‒ | v | v | v | ‒ | + | v | ‒ | + | v | v | + | + | ‒ | ‒ | + | + | ‒ | v | + | v |
| W. sydowiorum | + | ‒ | v | + | ‒ | + | + | + | + | + | v | + | + | + | ‒ | ‒ | + | + | + | ‒ | + | ‒ |
| W. sylviae | v | ‒ | s/‒ | + | + | +,‒ | w/‒ | s/‒ | ‒ | ‒ | w/+ | s/‒ | ‒ | ‒ | ‒ | ‒ | ‒ | v | + | + | ‒ | ‒ |
| W. tratensis | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | v | ‒ | ‒ | ‒ | ‒ | v | v | ‒ | ‒ | ‒ | n | n | + | n | ‒ |
| W. xylosicus | ‒ | + | + | ‒ | ‒ | ‒ | + | + | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ | w | ‒ | ‒ | n | ‒ | + | ‒ |
| W. xylosivorus | w/‒ | ‒ | + | ‒ | ‒ | + | ‒ | + | ‒ | w | ‒ | ‒ | + | ‒ | n | ‒ | ‒ | + | + | n | ‒ | ‒ |
| W. lannaensis | ‒ | ‒ | + | ‒ | ‒ | + | ‒ | + | ‒ | ‒ | ‒ | ‒ | + | + | ‒ | ‒ | ‒ | + | w | ‒ | ‒ | ‒ |
| W. nanensis | + | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ | ‒ | + | w | w | ‒ | ‒ | ‒ |
* Ga = Galactose, Sor = L-Sorbose, DXy = D-Xylose, LAr = L-Arabinose, DAr = D-Arabinose, Rh = L-Rhamnose, Su = Sucrose, Cel = Cellobiose, Mlb = Melibiose, Raf = Raffinose, St = Soluble starch, Rbl = Ribitol, DGlu = D-glucitol, Man = D-Mannitol, Glt = Galactitol, 2-ket = 2-ketogluconic acid, Cit = Citrate, NO3 = Potassium nitrate, ‒V = vitamin-free medium, 37 °C = Growth at 37 °C. “+” = strong growth or produce, “‒“ = absence of growth or not produce, “w” = weak growth, “v” = strain variable response, “l” = latent positive, “s” = slow positive, and “n” = no data.
-
1.
a. Melibiose is assimilated………………...……………………..…..…..…………………2
-
b. Melibiose is not assimilated………..……………………………..…..….…….….….…7
-
2.
(1) a. Raffinose is assimilated………….…….…..…….….….….….……..………………3
-
b. Raffinose is not assimilated………………….…………………...….W. kurtzmanii
-
3.
(2) a. Citrate is assimilated………………………….……….………………….…………4
-
b. Citrate is not assimilated…………………….…….……………......….W. orientalis
-
4.
(3) a. Ribitol is assimilated………………...………….……………………………………5
-
b. Ribitol is not assimilated………………...………………………..…W. spegazzinii
-
5.
(4) a. Growth at 37 °C……………………………………………….….…….W. edaphicus
-
b. Growth is absent at 37 °C…………………………...…………..…….……………6
-
6.
(5) a. Ascospores observed on 5% MEA……………………………....…W. sydowiorum
-
b. Ascospores not observed on 5% MEA…………….………..…….…W. arborarius
-
7.
(1) a. Raffinose is assimilated…………………………….….……………………………8
-
b. Raffinose is not assimilated………………………………..………..…………….19
-
8.
(7) a. Nitrate is assimilated………………….……………………….………...………….9
-
b. Nitrate is not assimilated…………………………………………………….……15
-
9.
(8) a. L-Rhamnose is assimilated……………………………………………….……….10
-
b. L-Rhamnose is not assimilated………………………….………………………..12
-
10.
(9) a. L-Arabinose is assimilated…………………………..………………...….W. ciferrii
-
b. L-Arabinose is not assimilated…………………………..………………….……11
-
11.
(10) a. Sucrose is assimilated……………………………...……….W. psychrolipolyticus
-
b. Sucrose is not assimilated…………………………….……...…...W. xylosivorus
-
12.
(9) a. Growth in vitamin-free medium…………………………………………………13
-
b. Growth is absent in vitamin-free medium……….…..………..W. subpelliculosus
-
13.
(12) a. Soluble starch is assimilated…………………...……………………………….14
-
b. Soluble starch is not assimilated………………………….………....W. lynferdii
-
14.
(13) a. D-Arabinose is assimilated……………..……..….……………W. myanmarensis
-
b. D-Arabinose is not assimilated…………………..…………………W. anomalus
-
15.
(8) a. Ribitol is assimilated…………………………………………………………….16
-
b. Ribitol is not assimilated…………………………………….…………….……17
-
16.
(15) a. Galactose is assimilated……………………….....…………….W. strasburgensis
-
b. Galactose is not assimilated……………………..………..………W. rabaulensis
-
17.
(15) a. Growth in vitamin-free medium……….………....…………..…W. patagonicus
-
b. Growth is absent in vitamin-free medium…………………….……………...18
-
18.
(17) a. Citrate is assimilated……………………….…………...….….………W. onychis
-
b. Citrate is not assimilated………………………..…………..………W. siamensis
-
19.
(7) a. 2-Keto-D-gluconate is assimilated……………………………………….……….20
-
b. 2-Keto-D-gluconate is not assimilated……………………………...……………21
-
20.
(19) a. D-Glucitol is assimilated…………………………………….……….W. mucosus
-
b. D-Glucitol is not assimilated……………………….……………….W. xylosicus
-
21.
(19) a. D-Arabinose is assimilated………………………………..………………………22
-
b. D-Arabinose is not assimilated…………………………..…………………….23
-
22.
(21) a. Growth at 37 °C……….………………...………………………...…….W. sylviae
-
b. Growth is absent at 37 °C……………….…………….………………….W. mori
-
23.
(21) a. Galactose is assimilated……………………………………..…………...……..24
-
b. Galactose is not assimilated………………...…………………....…………….27
-
24.
(23) a. L-Arabinose is assimilated……………….……………………..…….W. silvicola
-
b. L-Arabinose is not assimilated………………...…………………....…...…….25
-
25.
(24) a. Sucrose is assimilated…………….………………….………….W. scolytoplatypi
-
b. Sucrose is not assimilated…………………….………………..……….………26
-
26.
(24) a. D-Mannitol is assimilated……………………..………….…….……W. nanensis
-
b. D-Mannitol is not assimilated………………..….……….…....….W. chambardii
-
27.
(23) a. L-Sorbose is assimilated………………….……...…..…………..……...W. pijperi
-
b. L- Sorbose is not assimilated……………………….………….….……………28
-
28.
(27) a. D-Xylose is assimilated……………………………………………...………….29
-
b. D-Xylose is not assimilated……………….…………..….………….W. tratensis
-
29.
(28) a. Sucrose is assimilated……………………………...……………………………30
-
b. Sucrose is not assimilated……………………………...……………….………36
-
30.
(29) a. Cellobiose is assimilated……………………………………..………………....31
-
b. Cellobiose is not assimilated…………..…………………………….W. queroliae
-
31.
(30) a. D-Glucitol is assimilated…………………………………………….………….32
-
b. D-Glucitol is not assimilated…………………….…………….W. chaumierensis
-
32.
(31) a. Growth at 37 °C………………………………….……………………………....33
-
b. Growth is absent at 37 °C………………………………...…………….………34
-
33.
(32) a. L-Arabinose is assimilated……………………….....………..………….W. bovis
-
b. L-Arabinose is not assimilated……………..…………….……….W. canadensis
-
34.
(32) a. Nitrate is assimilated……………………………………………...…………….35
-
b. Nitrate is not assimilated…………………………..……...…...W. hampshirensis
-
35.
(34) a. True hyphae are formed……………..……………………………….W. bisporus
-
b. True hyphae are not formed…………………………………...…………W. alni
-
36.
(29) a. Citrate is assimilated……………………………...……..………..W. menglaensis
-
b. Citrate is not assimilated……………………………….………………………37
-
37.
(36) a. Growth at 37 °C……………………..………..……..…..…………W. ochangensis
-
b. Growth is absent at 37 °C………………..………...…..…….…….W. lannaensis
4. Discussion
Traditional methods of identification and characterization for the Wickerhamomyces species are based primarily on phenotypical characteristics. These are further recognized as relevant morphological, biochemical, and physiological characteristics [41,128]. However, identification can be difficult because some species have similar appearances, and some biochemical characteristics are consistent across a number of species. In accordance with this evidence, previous species of Wickerhamomyces were originally classified into various yeast genera [1,2,4,5,50,70,75]. In 2008, the genus Wickerhamomyces was proposed by Kurtzman et al. [1], wherein this genus was clearly separated from other yeast genera based on phylogenetic evidence. Subsequently, some previously identified species were then transferred from the genera Candida, Hansenula, Pichia, and Williopsis [1,4,5,50,54,70]. Therefore, molecular phylogenetic analysis is necessary to concretely identify the Wickerhamomyces species. Species of the genus Wickerhamomyces are known to be widely distributed throughout the world and have been isolated from various habitats as shown in Table 1. Prior to conducting our study, Wickerhamomyces consisted of 35 accepted and published species according to molecular phylogenetic analysis. Our phylogenetic results were similar to those of Arastehfar et al. [8] and Shimizu et al. [65] who indicated that P. myanmarensis should be placed in the genus Wickerhamomyces. Consequently, we have proposed that this yeast species be named W. myanmarensis.
Yeast diversity has been investigated in various habitats throughout different regions of Thailand [5,10,15,16,18,19,46,70,78]. Wickerhamomyces anomalus was first species reported in Thailand in 2002 [20]. In 2009, the first new species, W. edaphicus has been discovered in Thailand [10]. Until now, a total of eight Wickerhamomyces species have been found [5,10,20,46,68,70,78,82]. However, W. siamensis, W. tratensis, and W. xylosicus were only known to be from Thailand [5,70,82]. In this study, two new Wickerhamomyces species, namely W. lannaensis and W. nanensis, that were isolated from soil collected from Assam tea plantations in northern Thailand were proposed based on identification through molecular phylogenetic and phenotypic (morphological, biochemical, and physiological characteristics) analyses. Therefore, effective identification of the Wickerhamomyces species has increased the number of species found in Thailand to 10 species and has led to 38 global species. This present discovery has increased the number of species of yeast known to be from Thailand and is considered important in terms of stimulating deeper investigations of yeast varieties in Thailand. Ultimately, these findings will help researchers gain a better understanding of the distribution and ecology of Wickerhamomyces.
Many species of the genus Wickerhamomyces have been investigated, and some strains have been used in a variety of biotechnology, food, and beverage industries, as well as in medical and agricultural fields [83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102]. Despite the fact that many Wickerhamomyces species can survive in a variety of environments, climate change has had an impact on both the terrestrial biome and the aquatic environment. These environments are known to serve as habitats for a number of microorganisms [130,131,132,133,134] and may have an impact on the global diversity and distribution of Wickerhamomyces. Therefore, in addition to studying the diversity and distribution of newly identified species, future research should focus on the effects of climate change on Wickerhamomyces.
Acknowledgments
The authors are grateful to Russell Kirk Hollis for kind help in the English correction.
Author Contributions
Conceptualization, S.N., J.K., N.S. and S.L. (Saisamorn Lumyong); methodology, S.N., J.K., N.S. and S.L. (Savitree Limtong); software, N.S. and J.K.; validation, N.S., J.K., S.L. (Savitree Limtong) and S.L. (Saisamorn Lumyong); formal analysis, S.N., J.K., N.S.; investigation, S.N., J.K. and N.S.; resources, J.K., N.S. and S.K.; data curation, N.S., J.K. and N.S.; writing—original draft, S.N., J.K. and N.S.; writing—review and editing, J.K., S.N., N.S., S.L. (Savitree Limtong) and S.L. (Saisamorn Lumyong); supervision, S.L. (Saisamorn Lumyong). All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge the financial support provided from Chiang Mai University, Thailand.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA sequence data obtained from this study have been deposited in GenBank under accession numbers; D1/D2 domains (MT639220, MT623569, MT613722, MT613875 and MT623569) and ITS (OK135750, OK135752, OK135753, OK143510 and OK143511).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kurtzman C.P., Robnett C.J., Basehoar-Powers E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS. Yeast. Res. 2008;8:939–954. doi: 10.1111/j.1567-1364.2008.00419.x. [DOI] [PubMed] [Google Scholar]
- 2.Kurtzman C.P. Wickerhamomyces Kurtzman, Robnett & Basehoar-Powers. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The Yeasts. Elsevier; Amsterdam, The Netherlands: 2011. pp. 899–917. [Google Scholar]
- 3.Yarrow D. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman C.P., Fell J.W., editors. The Yeasts. Elsevier; Amsterdam, The Netherlands: 1998. pp. 77–100. [Google Scholar]
- 4.Kobayashi R., Kanti A., Kawasaki H. Three novel species of d-xylose-assimilating yeasts, Barnettozyma xylosiphila sp. nov., Barnettozyma xylosica sp. nov. and Wickerhamomyces xylosivorus f.a., sp. nov. Int. J. Syst. Evol. Microbiol. 2017;67:3971–3976. doi: 10.1099/ijsem.0.002233. [DOI] [PubMed] [Google Scholar]
- 5.Limtong S., Nitiyon S., Kaewwichian R., Jindamorakot S., Am-In S., Yongmanitchai W. Wickerhamomyces xylosica sp. nov. and Candida phayaonensis sp. nov., two xylose-assimilating yeast species from soil. Int. J. Syst. Evol. Microbiol. 2012;62:2786–2792. doi: 10.1099/ijs.0.039818-0. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y., Jia B.S., Han P.J., Wang Q.M., Li A.H., Zhou Y.G. Wickerhamomyces kurtzmanii sp. nov. an ascomycetous yeast isolated from crater lake water, Da Hinggan Ling Mountain, China. Curr. Microbiol. 2019;76:1537–1544. doi: 10.1007/s00284-019-01773-x. [DOI] [PubMed] [Google Scholar]
- 7.Index Fungorum. [(accessed on 25 July 2021)]. Available online: http://www.indexfungorum.org.
- 8.Arastehfar A., Bakhtiari M., Daneshnia F., Fang W., Sadati S.K., Al-Hatmi A.M., Groenewald M., Sharifi-Mehr H., Liao W., Pan W., et al. First fungemia case due to environmental yeast Wickerhamomyces myanmarensis: Detection by multiplex qPCR and antifungal susceptibility. Future Microbiol. 2019;14:267–274. doi: 10.2217/fmb-2018-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagatsuka Y., Kawasaki H., Seki T. Pichia myanmarensis sp. nov., a novel cation-tolerant yeast isolated from palm sugar in Myanmar. Int. J. Syst. Evol. Microbiol. 2005;55:1379–1382. doi: 10.1099/ijs.0.63558-0. [DOI] [PubMed] [Google Scholar]
- 10.Limtong S., Yongmanitchai W., Kawasaki H., Fujiyama K. Wickerhamomyces edaphicus sp. nov. and Pichia jaroonii sp. nov., two ascomycetous yeast species isolated from forest soil in Thailand. FEMS Yeast Res. 2009;9:504–510. doi: 10.1111/j.1567-1364.2009.00488.x. [DOI] [PubMed] [Google Scholar]
- 11.Shin K.S., Bae K.S., Lee K.H., Park D.S., Kwon G.S., Lee J.B. Wickerhamomyces ochangensis sp. nov., an ascomycetous yeast isolated from the soil of a potato field. Int. J. Syst. Evol. Microbiol. 2011;61:2543–2546. doi: 10.1099/ijs.0.026682-0. [DOI] [PubMed] [Google Scholar]
- 12.Phaff H.J., Miller M.W., Miranda M. Hansenula alni, a new heterothallic species of yeast from exudates of alder trees. Int. J. Syst. Evol. 1979;29:60–63. doi: 10.1099/00207713-29-1-60. [DOI] [Google Scholar]
- 13.Hansen E.C. Sur la germination des spores chez les Saccharomyces. Ann. Microgr. 1891;3:449–474. [Google Scholar]
- 14.Sláviková E., Vadkertiová R., Vránová D. Yeasts colonizing the leaf surfaces. J. Basic Microbiol. 2007;47:344–350. doi: 10.1002/jobm.200710310. [DOI] [PubMed] [Google Scholar]
- 15.Into P., Pontes A., Sampaio J.P., Limtong S. Yeast diversity associated with the phylloplane of corn plants cultivated in Thailand. Microorganisms. 2020;8:80. doi: 10.3390/microorganisms8010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumla J., Nundaeng S., Suwannarach N., Lumyong S. Evaluation of multifarious plant growth promoting trials of yeast isolated from the soil of Assam tea (Camellia sinensis var. assamica) plantations in Northern Thailand. Microorganisms. 2020;8:1168. doi: 10.3390/microorganisms8081168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrasco M., Rozas J.M., Barahona S., Alcaíno J., Cifuentes V., Baeza M. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 2012;12:251. doi: 10.1186/1471-2180-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaewkrajay C., Chanmethakul T., Limtong S. Assessment of diversity of culturable marine yeasts associated with corals and zoanthids in the gulf of Thailand, South China Sea. Microorganisms. 2020;8:474. doi: 10.3390/microorganisms8040474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satianpakiranakorn P., Khunnamwong P., Limtong S. Yeast communities of secondary peat swamp forests in Thailand and their antagonistic activities against fungal pathogens cause of plant and postharvest fruit diseases. PLoS ONE. 2020;15:e0230269. doi: 10.1371/journal.pone.0230269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limtong S., Sintara S., Suwannarit P., Lotong N. Yeast diversity in Thai traditional alcoholic starter. Kasetsart J. (Nat. Sci.) 2002;36:149–158. [Google Scholar]
- 21.Hoondee P., Wattanagonniyom T., Weeraphan T., Tanasupawat S., Savarajara A. Occurrence of oleaginous yeast from mangrove forest in Thailand. World J. Microbiol. Biotechnol. 2019;35:108. doi: 10.1007/s11274-019-2680-3. [DOI] [PubMed] [Google Scholar]
- 22.Tepeeva A.N., Glushakova A.M., Kachalkin A.V. Yeast communities of the Moscow city soils. Microbiology. 2018;87:407–415. doi: 10.1134/S0026261718030128. [DOI] [Google Scholar]
- 23.Koricha A.D., Han D.Y., Bacha K., Bai F.Y. Diversity and distribution of yeasts in indigenous fermented foods and beverages of Ethiopia. J. Sci. Food Agric. 2020;100:3630–3638. doi: 10.1002/jsfa.10391. [DOI] [PubMed] [Google Scholar]
- 24.Vadkertiová R., Molnárová J., Vránová D., Sláviková E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can. J. Microbiol. 2012;58:1344–1352. doi: 10.1139/cjm-2012-0468. [DOI] [PubMed] [Google Scholar]
- 25.PrasannaKumar C., Velmurugan S., Subramanian K., Pugazhvendan S.R., Nagaraj D.S., Khan K.F., Sadiappan B., Manokaran S., Hemalatha K.R. DNA barcoding analysis of more than 1000 marine yeast isolates reveals previously unrecorded species. BioRxiv. 2020:1–39. [Google Scholar]
- 26.Abu-Mejdad N.M.J.A., Al-Badran A.I., Al-Saadoon A.H., Minati M.H. A new report on gene expression of three killer toxin genes with antimicrobial activity of two killer toxins in Iraq. Bull. Natl. Res. Cent. 2020;44:162. doi: 10.1186/s42269-020-00418-5. [DOI] [Google Scholar]
- 27.Horváth E., Sipiczki M., Csoma H., Miklós I. Assaying the effect of yeasts on growth of fungi associated with disease. BMC Microbiol. 2020;20:320. doi: 10.1186/s12866-020-01942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamoudi-Belarbi L., Nouri L.H., Belkacemi K. Effectiveness of convective drying to conserve indigenous yeasts with high volatile profile isolated from algerian fermented raw bovine milk (Rayeb) J. Food Sci. Technol. 2016;36:476–484. doi: 10.1590/1678-457X.00416. [DOI] [Google Scholar]
- 29.You L., Wang S., Zhou R., Hu X., Chu Y., Wang T. Characteristics of yeast flora in Chinese strong-flavoured liquor fermentation in the Yibin region of China. J. Inst. Brew. 2016;122:517–523. doi: 10.1002/jib.352. [DOI] [Google Scholar]
- 30.Borling Welin J., Lyberg K., Passoth V., Olstorpe M. Combined moist airtight storage and feed fermentation of barley by the yeast Wickerhamomyces anomalus and a lactic acid bacteria consortium. Front. Plant. Sci. 2015;6:270. doi: 10.3389/fpls.2015.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pires J.F., de Souza Cardoso L., Schwan R.F., Silva C.F. Diversity of microbiota found in coffee processing wastewater treatment plant. World J. Microbiol. Biotechnol. 2017;33:211. doi: 10.1007/s11274-017-2372-9. [DOI] [PubMed] [Google Scholar]
- 32.Delgado-Ospina J., Triboletti S., Alessandria V., Serio A., Sergi M., Paparella A., Rantsiou K., Chaves-López C. Functional biodiversity of yeasts isolated from Colombian fermented and dry cocoa beans. Microorganisms. 2020;8:1086. doi: 10.3390/microorganisms8071086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Conceição L.E.F.R., Saraiva M.A.F., Diniz R.H.S., Oliveira J., Barbosa G.D., Alvarez F., Correa L.F., Mezadri H., Coutrim M.X., Afonso R.J., et al. Biotechnological potential of yeast isolates from cachaça: The Brazilian spirit. J. Ind. Microbiol. Biotechnol. 2015;42:237–246. doi: 10.1007/s10295-014-1528-y. [DOI] [PubMed] [Google Scholar]
- 34.Da Cunha A.C., Gomes L.S., Godoy-Santos F., Faria-Oliveira F., Teixeira J.A., Sampaio G.M.S., Trópia M.J.M., Castro I.M., Lucas C., Brandão R.L. High-affinity transport, cyanide-resistant respiration, and ethanol production under aerobiosis underlying efficient high glycerol consumption by Wickerhamomyces anomalus. J. Ind. Microbiol. Biotechnol. 2019;46:709–723. doi: 10.1007/s10295-018-02119-5. [DOI] [PubMed] [Google Scholar]
- 35.Cunha A.C., Santos R.A., Riaño-Pachon D.M., Squina F.M., Oliveira J.V., Goldman G.H., Souza A.T., Gomes L.S., Godoy-Santos F., Teixeira J.A. Draft genome sequence of Wickerhamomyces anomalus LBCM1105, isolated from cachaça fermentation. Genet. Mol. Biol. 2020;43:1–5. doi: 10.1590/1678-4685-gmb-2019-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James S.A., Barriga E.J.C., Barahona P.P., Harrington T.C., Lee C.F., Bond C.J., Roberts I.N. Wickerhamomyces arborarius fa, sp. nov., an ascomycetous yeast species found in arboreal habitats on three different continents. Int. J. Syst. Evol. 2014;64:1057–1061. doi: 10.1099/ijs.0.059162-0. [DOI] [PubMed] [Google Scholar]
- 37.Beck O. Eine neue Endomyces-Art, Endomyces bisporus. Ann. Mycol. 1922;20:219–227. [Google Scholar]
- 38.Linnakoski R., Lasarov I., Veteli P., Tikkanen O.P., Viiri H., Jyske T., Kasanen R., Duong T.A., Wingfield M.J. Filamentous fungi and yeasts associated with mites phoretic on Ips typographus in eastern Finland. Forests. 2021;12:743. doi: 10.3390/f12060743. [DOI] [Google Scholar]
- 39.Dohet L., Gregoire J.C., Berasategui A., Kaltenpoth M., Biedermann P.H. Bacterial and fungal symbionts of parasitic Dendroctonus bark beetles. FEMS Microbiol. Ecol. 2016;92:fiw129. doi: 10.1093/femsec/fiw129. [DOI] [PubMed] [Google Scholar]
- 40.Uden V.N., Carmo Souza L.D. Yeast from the bovine caecum. J. Gen. Microbiol. 1957;16:385–395. doi: 10.1099/00221287-16-2-385. [DOI] [PubMed] [Google Scholar]
- 41.Wickerham L.J. Taxonomy of Yeasts. U.S. Department of Agriculture; Washington, DC, USA: 1951. pp. 1–56. [Google Scholar]
- 42.Ramirez C. Note sur deux nouvelles espèces de levures isolées de divers milieux. Rev. Mycol. 1954;19:98–102. [Google Scholar]
- 43.Groenewald M., Robert V., Smith M.T. Five novel Wickerhamomyces-and Metschnikowia-related yeast species, Wickerhamomyces chaumierensis sp. nov., Candida pseudoflosculorum sp. nov., Candida danieliae sp. nov., Candida robnettiae sp. nov. and Candida eppingiae sp. nov., isolated from plants. Int. J. Syst. Evol. 2011;61:2015–2022. doi: 10.1099/ijs.0.026062-0. [DOI] [PubMed] [Google Scholar]
- 44.Lodder J. Über einige durch das “Centraalbureau voor Schimmelcultures” neuerworbene sporogene Hefearten. Zentralbl. Bakteriol. Parasitenkd. Abt. II. 1932;86:227–253. [Google Scholar]
- 45.Sitepu I.R., Enriquez L.L., Nguyen V., Doyle C., Simmons B.A., Singer S.W., Fry R., Simmons C.W., Boundy-Mills K. Ethanol production in switchgrass hydrolysate by ionic liquid-tolerant yeasts. Bioresour. Technol. Rep. 2019;7:100275. doi: 10.1016/j.biteb.2019.100275. [DOI] [Google Scholar]
- 46.Limtong S., Kaewwichian R. The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann. Microbiol. 2015;65:667–675. doi: 10.1007/s13213-014-0905-0. [DOI] [PubMed] [Google Scholar]
- 47.Patel K., Patel F.R. Optimization of culture conditions for biosurfactant production by Wickerhamomyces edaphicus isolated from mangrove region of Mundra, Kutch, Gujarat. Indian J. Sci. Technol. 2020;13:1935–1943. doi: 10.17485/IJST/v13i19.301. [DOI] [Google Scholar]
- 48.Kurtzman C.P. Two new species of Pichia from arboreal habitats. Mycologia. 1987;79:410–417. doi: 10.2307/3807464. [DOI] [Google Scholar]
- 49.Suh S.O., Zhou J. Yeasts associated with the curculionid beetle Xyloterinus politus: Candida xyloterini sp. nov., Candida palmyrensis sp. nov. and three common ambrosia yeasts. Int. J. Syst. Evol. Microbiol. 2010;60:1702–1708. doi: 10.1099/ijs.0.016907-0. [DOI] [PubMed] [Google Scholar]
- 50.van der Walt J.P., Johannsen E. Hansenula lynferdii sp. nov. Antonie Van Leeuwenhoek. 1975;41:13–16. doi: 10.1007/BF02565032. [DOI] [PubMed] [Google Scholar]
- 51.Chai C.Y., Huang L.N., Cheng H., Liu W.J., Hui F.L. Wickerhamomyces menglaensis fa, sp. nov., a yeast species isolated from rotten wood. Int. J. Syst. Evol. Microbiol. 2019;69:1509–1514. doi: 10.1099/ijsem.0.003350. [DOI] [PubMed] [Google Scholar]
- 52.Hui F.L., Chen L., Chu X.Y., Niu Q.H., Ke T. Wickerhamomyces mori sp. nov., an anamorphic yeast species found in the guts of wood-boring insect larvae. Int. J. Syst. Evol. Microbiol. 2013;63:1174–1178. doi: 10.1099/ijs.0.048637-0. [DOI] [PubMed] [Google Scholar]
- 53.Wlckerham L.J., Kurtzman C.P. Two new saturn-spored species of Pichia. Mycologia. 1971;63:1013–1018. doi: 10.1080/00275514.1971.12019197. [DOI] [PubMed] [Google Scholar]
- 54.Yarrow D. Pichia onychis sp. n. Antonie Van Leeuwenhoek. J. Microbiol. Serol. 1965;31:465–467. doi: 10.1007/BF02045923. [DOI] [PubMed] [Google Scholar]
- 55.Cioch-Skoneczny M., Satora P., Skoneczny S., Skotniczny M. Biodiversity of yeasts isolated during spontaneous fermentation of cool climate grape musts. Arch. Microbiol. 2021;203:153–162. doi: 10.1007/s00203-020-02014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abu-Mejdad N.M.J.A., Al-Badran A.I., Al-Saadoon A.H. New record of ascomycetous yeasts strains from soil in Basrah, Iraq. Drug Invent. Today. 2019;11:3073–3080. [Google Scholar]
- 57.Bah A., Ferjani R., Fhoula I., Gharbi Y., Najjari A., Boudabous A., Ouzari H.I. Microbial community dynamic in tomato fruit during spontaneous fermentation and biotechnological characterization of indigenous lactic acid bacteria. Ann. Microbiol. 2019;69:41–49. doi: 10.1007/s13213-018-1385-4. [DOI] [Google Scholar]
- 58.Ooi T.S., Sepiah M., Khairul Bariah S. Diversity of yeast species identified during spontaneous shallow box fermentation of cocoa beans in Malaysia. Int. J. Eng. Innov. Technol. 2016;3:379–385. [Google Scholar]
- 59.Maciel N.O., Johann S., Brandão L.R., Kucharíková S., Morais C.G., Oliveira A.P., Freitas G.J., Borelli B.M., Pellizzari F.M., Santos D.A., et al. Occurrence, antifungal susceptibility, and virulence factors of opportunistic yeasts isolated from Brazilian beaches. Mem. Inst. Oswaldo Cruz. 2019;114:114. doi: 10.1590/0074-02760180566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nasr S., Nguyen H.D., Soudi M.R., Fazeli S.A.S., Sipiczki M. Wickerhamomyces orientalis fa, sp. nov.: An ascomycetous yeast species belonging to the Wickerhamomyces clade. Int. J. Syst. Evol. Microbiol. 2016;66:2534–2539. doi: 10.1099/ijsem.0.001096. [DOI] [PubMed] [Google Scholar]
- 61.de García V., Brizzio S., Libkind D., Rosa C.A., van Broock M. Wickerhamomyces patagonicus sp. nov., an ascomycetous yeast species from Patagonia, Argentina. Int. J. Syst. Evol. Microbiol. 2010;60:1693–1696. doi: 10.1099/ijs.0.015974-0. [DOI] [PubMed] [Google Scholar]
- 62.van Walt J.P., Tscheuschner I.T. Three new yeasts. Antonie Van Leeuwenhoek. 1957;23:184–190. doi: 10.1007/BF02545868. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen D.S., Teniola O.D., Ban-Koffi L., Owusu M., Andersson T.S., Holzapfel W.H. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int. J. Food Microbiol. 2007;114:168–186. doi: 10.1016/j.ijfoodmicro.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Abdel-Sater M.A., Moubasher A.H., Zeinab S.M. Diversity of yeasts and filamentous fungi in five fresh fruit juices in Egypt. Curr. Res. Environ. 2017;7:356–386. doi: 10.5943/cream/7/4/12. [DOI] [Google Scholar]
- 65.Shimizu Y., Konno Y., Tomita Y. Wickerhamomyces psychrolipolyticus fa, sp. nov., a novel yeast species producing two kinds of lipases with activity at different temperatures. Int. J. Syst. Evol. 2020;70:1158–1165. doi: 10.1099/ijsem.0.003894. [DOI] [PubMed] [Google Scholar]
- 66.Rosa C.A., Morais P.B., Lachance M.A., Santos R.O., Melo W.G., Viana R.H., Bragança M.A., Pimenta R.S. Wickerhamomyces queroliae sp. nov. and Candida jalapaonensis sp. nov., two yeast species isolated from Cerrado ecosystem in North Brazil. Int. J. Syst. Evol. Microbiol. 2009;59:1232–1236. doi: 10.1099/ijs.0.006411-0. [DOI] [PubMed] [Google Scholar]
- 67.Soneda M., Uchida S. A survey on the yeasts. Bull. Nat. Science Museum. 1971;14:451–453. [Google Scholar]
- 68.Junyapate K., Jindamorakot S., Limtong S. Yamadazyma ubonensis fa, sp. nov., a novel xylitol-producing yeast species isolated in Thailand. Antonie Van Leeuwenhoek. 2014;105:471–480. doi: 10.1007/s10482-013-0098-8. [DOI] [PubMed] [Google Scholar]
- 69.Ninomiya S., Mikata K., Kajimura H., Kawasaki H. Two novel ascomycetous yeast species, Wickerhamomyces scolytoplatypi sp. nov. and Cyberlindnera xylebori sp. nov., isolated from ambrosia beetle galleries. Int. J. Syst. Evol. Microbiol. 2013;63:2706–2711. doi: 10.1099/ijs.0.050195-0. [DOI] [PubMed] [Google Scholar]
- 70.Kaewwichian R., Kawasaki H., Limtong S. Wickerhamomyces siamensis sp. nov., a novel yeast species isolated from the phylloplane in Thailand. Int. J. Syst. Evol. Microbiol. 2013;63:1568–1573. doi: 10.1099/ijs.0.050013-0. [DOI] [PubMed] [Google Scholar]
- 71.Bien S., Damm U. Prunus trees in Germany—A hideout of unknown fungi? Mycol. Prog. 2020;19:667–690. doi: 10.1007/s11557-020-01586-4. [DOI] [Google Scholar]
- 72.Hyun S.H., Min J.H., Lee H.B., Kim H.K., Lee J.S. Characteristics of two unrecorded yeasts from wildflowers in Ulleungdo, Korea. Kor. J. Mycol. 2014;42:170–173. doi: 10.4489/KJM.2014.42.2.170. [DOI] [Google Scholar]
- 73.Masiulionis V.E., Pagnocca F.C. Wickerhamomyces spegazzinii sp. nov., an ascomycetous yeast isolated from the fungus garden of Acromyrmex lundii nest (Hymenoptera: Formicidae) Int. J. Syst. Evol. Microbiol. 2016;66:2141–2145. doi: 10.1099/ijsem.0.001001. [DOI] [PubMed] [Google Scholar]
- 74.Ramírez C., Boidin J. Saccharomyces chambardi, nouvelle espèce de levure isolée de liqueur tannante. De La Société Linnéenne De Lyon. 1954;23:151–152. doi: 10.3406/linly.1954.7652. [DOI] [Google Scholar]
- 75.Kurtzman C.P. Synonomy of the yeast genera Hansenula and Pichia demonstrated through comparisons of deoxyribonucleic acid relatedness. Antonie Van Leeuwenhoek. 1984;50:209–217. doi: 10.1007/BF02342132. [DOI] [PubMed] [Google Scholar]
- 76.Moubasher A.H., Abdel-Sater M.A., Soliman Z. Yeasts and filamentous fungi inhabiting guts of three insect species in Assiut, Egypt. Mycosphere. 2017;8:1297–1316. doi: 10.5943/mycosphere/8/9/4. [DOI] [Google Scholar]
- 77.Scott D.B., Van der Walt J.P. Hansenula sydowiorum sp. n. Antonie Van Leeuwenhoek. 1970;36:45–48. doi: 10.1007/BF02069006. [DOI] [PubMed] [Google Scholar]
- 78.Boonmak C., Jindamorakot S., Kawasaki H., Yongmanitchai W., Suwanarit P., Nakase T., Limtong S. Candida siamensis sp. nov., an anamorphic yeast species in the Saturnispora clade isolated in Thailand. FEMS Yeast Res. 2009;9:668–672. doi: 10.1111/j.1567-1364.2009.00490.x. [DOI] [PubMed] [Google Scholar]
- 79.Samagaci L., Ouattara H., Niamké S., Lemaire M. Pichia kudrazevii and Candida nitrativorans are the most well-adapted and relevant yeast species fermenting cocoa in Agneby-Tiassa, a local Ivorian cocoa producing region. Int. Food Res. J. 2016;89:773–780. doi: 10.1016/j.foodres.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 80.Echeverrigaray S., Scariot F.J., Foresti L., Schwarz L.V., Rocha R.K.M., da Silva G.P., Moreira J.P., Delamare A.P.L. Yeast biodiversity in honey produced by stingless bees raised in the highlands of southern Brazil. Int. J. Food Microbiol. 2021;347:109200. doi: 10.1016/j.ijfoodmicro.2021.109200. [DOI] [PubMed] [Google Scholar]
- 81.Francesca N., Carvalho C., Almeida P.M., Sannino C., Settanni L., Sampaio J.P., Moschetti G. Wickerhamomyces sylviae f.a., sp. nov., an ascomycetous yeast species isolated from migratory birds. Int. J. Syst. Evol. Microbiol. 2013;63:4824–4830. doi: 10.1099/ijs.0.056382-0. [DOI] [PubMed] [Google Scholar]
- 82.Nakase T., Jindamorakot S., Am-In S., Ninomiya S., Kawasaki H. Wickerhamomyces tratensis sp. nov. and Candida namnaoensis sp. nov., two novel ascomycetous yeast species in the Wickerhamomyces clade found in Thailand. J. Gen. Appl. Microbiol. 2012;58:145–152. doi: 10.2323/jgam.58.145. [DOI] [PubMed] [Google Scholar]
- 83.Ravasio D., Carlin S., Boekhout T., Groenewald M., Vrhovsek U., Walther A., Wendland J. Adding flavor to beverages with non-conventional yeasts. Fermentation. 2018;4:15. doi: 10.3390/fermentation4010015. [DOI] [Google Scholar]
- 84.Yan S., Xiangsong C., Xiang X. Improvement of the aroma of lily rice wine by using aroma-producing yeast strain Wickerhamomyces anomalus HN006. AMB Express. 2019;9:89. doi: 10.1186/s13568-019-0811-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li W., Shi C., Guang J., Ge F., Yan S. Development of Chinese chestnut whiskey: Yeast strains isolation, fermentation system optimization, and scale-up fermentation. AMB Express. 2021;11:17. doi: 10.1186/s13568-020-01175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Czarnecka M., Żarowska B., Połomska X., Restuccia C., Cirvilleri G. Role of biocontrol yeasts Debaryomyces hansenii and Wickerhamomyces anomalus in plants’ defence mechanisms against Monilinia fructicola in apple fruits. Food Microbiol. 2019;83:1–8. doi: 10.1016/j.fm.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 87.Limtong S., Into P., Attarat P. Biocontrol of rice seedling rot disease caused by Curvularia lunata and Helminthosporium oryzae by epiphytic yeasts from plant leaves. Microorganisms. 2020;8:647. doi: 10.3390/microorganisms8050647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lima J.R., Gondim D.M.F., Oliveira J.T.A., Oliveira F.S.A., Gonçalves L.R.B., Viana F.M.P. Use of killer yeast in the management of postharvest papaya anthracnose. Postharvest Biol. Technol. 2013;83:58–64. doi: 10.1016/j.postharvbio.2013.03.014. [DOI] [Google Scholar]
- 89.Fernandez-San Millan A., Farran I., Larraya L., Ancin M., Arregui L.M., Veramendi J. Plant growth-promoting traits of yeasts isolated from Spanish vineyards: Benefits for seedling development. Microbiol. Res. 2020;237:126480. doi: 10.1016/j.micres.2020.126480. [DOI] [PubMed] [Google Scholar]
- 90.Walker G.M. Pichia anomala: Cell physiology and biotechnology relative to other yeasts. Antonie Van Leeuwenhoek. 2011;99:25–34. doi: 10.1007/s10482-010-9491-8. [DOI] [PubMed] [Google Scholar]
- 91.Passoth V., Olstorpe M., Schnürer J. Past, present and future research directions with Pichia anomala. Antonie Van Leeuwenhoek. 2011;99:121–125. doi: 10.1007/s10482-010-9508-3. [DOI] [PubMed] [Google Scholar]
- 92.Golubev W.I. Antifungal activity of Wickerhamomyces silvicola. Microbiology. 2015;84:610–615. doi: 10.1134/S0026261715050100. [DOI] [PubMed] [Google Scholar]
- 93.Golubev W.I. Taxonomic specificity of the sensitivity to the Wickerhamomyces bovis fungistatic mycocin. Microbiology. 2016;85:444–448. doi: 10.1134/S0026261716040081. [DOI] [PubMed] [Google Scholar]
- 94.Silva C.F., Schwan R.F., Dias Ë.S., Wheals A.E. Microbial diversity during maturation and natural processing of coffee cherries of Coffea arabica in Brazil. Int. J. Food Microbiol. 2000;60:251–260. doi: 10.1016/S0168-1605(00)00315-9. [DOI] [PubMed] [Google Scholar]
- 95.Zhou N., Schifferdecker A.J., Gamero A., Compagno C., Boekhout T., Piškur J., Knecht W. Kazachstania gamospora and Wickerhamomyces subpelliculosus: Two alternative baker’s yeasts in the modern bakery. Int. J. Food Microbiol. 2017;250:45–58. doi: 10.1016/j.ijfoodmicro.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 96.Adelabu B., Kareem S., Oluwafemi F., Adeogun A. Consolidated bioprocessing of ethanol from corn straw by Saccharomyces diastaticus and Wikerhamomyces chambardii. Food Appl. Biosci. J. 2018;6:1–17. [Google Scholar]
- 97.Adelabu B., Kareem S.O., Adeogun A.I., Wakil S.M. Optimization of cellulase enzyme from sorghum straw by yeasts isolated from plant feeding-termite Zonocerus variegatus. Food Appl. Biosci. J. 2019;7:81–101. [Google Scholar]
- 98.Dejwatthanakomol C., Anuntagool J., Morikawa M., Thaniyavarn J. Production of biosurfactant by Wickerhamomyces anomalus PY189 and its application in lemongrass oil encapsulation. Sci. Asia. 2016;42:252–258. doi: 10.2306/scienceasia1513-1874.2016.42.252. [DOI] [Google Scholar]
- 99.Fernandes N.D.A.T., de Souza A.C., Simoes L.A., Dos Reis G.M.F., Souza K.T., Schwan R.F., Dias D.R. Eco-friendly biosurfactant from Wickerhamomyces anomalus CCMA 0358 as larvicidal and antimicrobial. Microbiol. Res. 2020;241:126571. doi: 10.1016/j.micres.2020.126571. [DOI] [PubMed] [Google Scholar]
- 100.Samadlouie H.R., Nurmohamadi S., Moradpoor F., Gharanjik S. Effect of low-cost substrate on the fatty acid profiles of Mortierella alpina CBS 754.68 and Wickerhamomyces siamensis SAKSG. Biotechnol. Biotechnol. Equip. 2018;32:1228–1235. doi: 10.1080/13102818.2018.1471360. [DOI] [Google Scholar]
- 101.Palladino F., Rodrigues R.C., Cadete R.M., Barros K.O., Rosa C.A. Novel potential yeast strains for the biotechnological production of xylitol from sugarcane bagasse. Biofuels Bioprod. Biorefining. 2021;15:690–702. doi: 10.1002/bbb.2196. [DOI] [Google Scholar]
- 102.Seymour F.R., Slodki M.E., Plattner R.D., Stodola R.M. Methylation and acetolysis of extracellular D-mannans from yeast. Carbohydr. Res. 1976;48:225–237. doi: 10.1016/S0008-6215(00)83218-8. [DOI] [PubMed] [Google Scholar]
- 103.Dufour J.P., Verstrepen K., Derdelinckx G. Brewing yeasts. In: Boekhout T., Robert V., editors. Yeasts in Food. Elsevier; Amsterdam, The Netherlands: 2003. pp. 347–388. [Google Scholar]
- 104.Timke M., Wang-Lieu N.Q., Altendorf K., Lipski A. Fatty acid analysis and spoilage potential of biofilms from two breweries. J. Appl. Microbiol. 2005;99:1108–1122. doi: 10.1111/j.1365-2672.2005.02714.x. [DOI] [PubMed] [Google Scholar]
- 105.Bonjean B., Guillaume L.D. Yeasts in bread and baking products. In: Boekhout T., Robert V., editors. Yeasts in Food. Elsevier; Amsterdam, The Netherlands: 2003. pp. 289–307. [Google Scholar]
- 106.Lanciotti R., Sinigaglia M., Gardini F., Guerzoni M.E. Hansenula anomala as spoilage agent of cream-filled cakes. Microbiol. Res. 1998;153:145–148. doi: 10.1016/S0944-5013(98)80032-3. [DOI] [PubMed] [Google Scholar]
- 107.Paula C.R., Krebs V.L., Auler M.E., Ruiz L.S., Matsumoto F.E., Silva E.H., Diniz E.M., Vaz F.A. Nosocomial infection in newborns by Pichia anomala in a Brazilian intensive care unit. Med. Mycol. 2006;44:479–484. doi: 10.1080/13693780600561809. [DOI] [PubMed] [Google Scholar]
- 108.Linton C.J., Borman A.M., Cheung G., Holmes A.D., Szekely A., Palmer M.D., Bridge P.D., Campbell C.K., Johnson E.M. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom Mycology Reference Laboratory. J. Clin. Microbiol. 2007;45:1152–1158. doi: 10.1128/JCM.02061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oliveira V.K.P., Ruiz L.d.S., Oliveira N.A.J., Moreira D., Hahn R.C., Melo A.S.d.A., Nishikaku A.S., Paula C.R. Fungemia caused by Candida species in a children’s public hospital in the city of São Paulo, Brazil: Study in the period 2007–2010. Rev. Do Inst. De Med. Trop. De São Paulo. 2014;56:301–305. doi: 10.1590/S0036-46652014000400006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kamoshita M., Matsumoto Y., Nishimura K., Katono Y., Murata M., Ozawa Y., Shimmura S., Tsubota K. Wickerhamomyces anomalus fungal keratitis responds to topical treatment with antifungal micafungin. J. Infect. Chemother. 2015;21:141–143. doi: 10.1016/j.jiac.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 111.Dutra V.R., Silva L.F., Oliveira A.N.M., Beirigo E.F., Arthur V.M., da Silva R.B., Ferreira T.B., Andrade-Silva L., Silva M.V., Fonseca F.M., et al. Fatal case of fungemia by Wickerhamomyces anomalus in a pediatric patient diagnosed in a teaching hospital from Brazil. J. Fungi. 2020;6:147. doi: 10.3390/jof6030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Hoog G.S. Risk assessment of fungi reported from humans and animals. Mycoses. 1996;39:407–417. doi: 10.1111/j.1439-0507.1996.tb00089.x. [DOI] [PubMed] [Google Scholar]
- 113.Hyde K.D., Norphanphoun C., Chen J., Dissanayake A.J., Doilom M., Hongsanan S., Jayawardena R.S., Jeewon R., Perera R.H., Thongbai B., et al. Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 2018;93:215–239. doi: 10.1007/s13225-018-0415-7. [DOI] [Google Scholar]
- 114.Kumla J., Suwannarach N., Wannathes W. Hymenagaricus saisamornae sp. nov. (Agaricales, Basidiomycota) from northern Thailand. Chiang Mai J. Sci. 2021;48:827–836. [Google Scholar]
- 115.Nakase T., Suzuki M. Bullera intermedia sp. nov. and Sporobolomyces oryzicola sp. nov. isolated from dead leaves of Oryza sativa. J. Gen. Appl. Microbiol. 1986;32:149–155. doi: 10.2323/jgam.32.149. [DOI] [Google Scholar]
- 116.Hagler A.N., Ahearn D.G. Rapid diazonium blue B test to detect basidiomycetous yeasts. Int. J. Syst. Evol. 1981;31:204–208. doi: 10.1099/00207713-31-2-204. [DOI] [Google Scholar]
- 117.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 118.Kurtzman C.P. DNA relatedness among saturn-spored yeasts assigned to the genera Williopsis and Pichia. Antonie Van Leeuwenhoek. 1991;60:13–19. doi: 10.1007/BF00580436. [DOI] [PubMed] [Google Scholar]
- 119.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miller M.A., Pfeiffer W., Schwartz T. Creating the cipres science gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing EnvironmentsWorkshop (GCE); New Orleans, LA, USA. 14 November 2010; Manhattan, NY, USA: IEEE; pp. 1–8. [Google Scholar]
- 121.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 123.Rambaut A. FigTree Tree Figure Drawing Tool Version 131, Institute of Evolutionary 623 Biology, University of Edinburgh. [(accessed on 20 August 2021)]. Available online: http://treebioedacuk/software/figtree/
- 124.Vu D., Groenewald M., Szöke S., Cardinali G., Eberhardt U., Stielow B., de Vries M., Verkleij G.J.M., Crous P.W., Boekhout T., et al. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016;85:91–105. doi: 10.1016/j.simyco.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rivera F.N., Gómez Z., González E., López N., Rodríguez C.H.H., Zúñiga G. Yeasts associated with bark beetles of the genus Dendroctonus erichson (Coleoptera: Curculionidae: Scolytinae): Molecular identification and biochemical characterization; Proceedings of the Third Workshop on Genetics of Bark Beetles and Associated Microorganisms; Asheville, NC, USA. 20 May 2006; pp. 45–48. [Google Scholar]
- 126.Sitepu I., Enriquez L., Nguyen V., Fry R., Simmons B., Singer S., Simmons C., Boundy-Mills K.L. Ionic liquid tolerance of yeasts in family Dipodascaceae and genus Wickerhamomyces. Appl. Biochem. Biotechnol. 2020;191:1580–1593. doi: 10.1007/s12010-020-03293-y. [DOI] [PubMed] [Google Scholar]
- 127.Schoch C.L., Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., Meyer W., Nilsson R.H., Hughes K., Miller A.N., et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for fungi. Database. 2014;2014:bau061. doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kurtzman C.P., Robnett C.J. Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 129.Wang Y., Ren Y.C., Zhang Z.T., Ke T., Hui F.L. Spathaspora allomyrinae sp. nov., a D-xylose-fermenting yeast species isolated from a scarabeid beetle Allomyrina dichotoma. Int. J. Syst. Evol. 2016;66:2008–2012. doi: 10.1099/ijsem.0.000979. [DOI] [PubMed] [Google Scholar]
- 130.Barnett J.A. A history of research on yeasts 2: Louis Pasteur and his contemporaries, 1850–1880. Yeast. 2000;16:755–771. doi: 10.1002/1097-0061(20000615)16:8<755::AID-YEA587>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 131.Cavicchioli R., Ripple W.J., Timmis K.N., Azam F., Bakken L.R., Baylis M., Behrenfeld M.J., Boetius A., Boyd P.W., Classen A.T., et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019;17:569–586. doi: 10.1038/s41579-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jain P.K., Purkayastha S.D., De Mandal S., Passari A.K., Govindarajan R.K. Effect of climate change on microbial diversity and its functional attributes. In: De Mandal S., Bhatt P., editors. Recent Advancements in Microbial Diversity. Academic Press; San Diego, CA, USA: 2020. pp. 315–331. [Google Scholar]
- 133.van der Putten W.H. Climate change, aboveground-belowground interactions, and species’ range shifts. Annu. Rev. Ecol. Evol. Syst. 2012;43:365–383. doi: 10.1146/annurev-ecolsys-110411-160423. [DOI] [Google Scholar]
- 134.Wookey P.A., Aerts R., Bardgett R.D., Baptist F., BraThen K.A., Cornelissen J.H.C., Shaver G.R. Ecosystem feedbacks and cascade processes: Understanding their role in the responses of Arctic and alpine ecosystems to environmental change. Glob. Change Biol. 2009;15:1153–1172. doi: 10.1111/j.1365-2486.2008.01801.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DNA sequence data obtained from this study have been deposited in GenBank under accession numbers; D1/D2 domains (MT639220, MT623569, MT613722, MT613875 and MT623569) and ITS (OK135750, OK135752, OK135753, OK143510 and OK143511).