Abstract
A genome-wide association study (GWAS) was performed to elucidate genetic architecture of growth traits in Braunvieh cattle. Methods: The study included 300 genotyped animals by the GeneSeek® Genomic Profiler Bovine LDv.4 panel; after quality control, 22,734 SNP and 276 animals were maintained in the analysis. The examined phenotypic data considered birth (BW), weaning (WW), and yearling weights. The association analysis was performed using the principal components method via the egscore function of the GenABEL version 1.8-0 package in the R environment. The marker rs133262280 located in BTA 22 was associated with BW, and two SNPs were associated with WW, rs43668789 (BTA 11) and rs136155567 (BTA 27). New QTL associated with these liveweight traits and four positional and functional candidate genes potentially involved in variations of the analyzed traits were identified. The most important genes in these genomic regions were MCM2 (minichromosome maintenance complex component 2), TPRA1 (transmembrane protein adipocyte associated 1), GALM (galactose mutarotase), and NRG1 (neuregulin 1), related to embryonic cleavage, bone and tissue growth, cell adhesion, and organic development. This study is the first to present a GWAS conducted in Braunvieh cattle in Mexico providing evidence for genetic architecture of assessed growth traits. Further specific analysis of found associated genes and regions will clarify its contribution to the genetic basis of growth-related traits.
Keywords: association, candidate gene, growth, quantitative trait loci, single nucleotide polymorphism
1. Introduction
The identification of causal genetic variability is one of the main goals in the genetic improvement of cattle. Commonly, liveweight traits are used as the primary selection criterion in cow-calf production systems in Mexico [1]. Usually, farms use these traits as efficiency and meat potential production indicators, and they are used for genetic evaluations in most of the registered breeds [2].
Braunvieh is a worldwide cattle breed used in the beef industry that has been used in both specialized beef and dual-purpose production herds [3,4]. Due to its initial dual-purpose origin, most of the available information about the Braunvieh deals with dairy production traits. However, during the last 15 years, Braunvieh cattle have been selected and genetically improved for beef production traits [4,5]. In Mexico, Braunvieh is one of the breeds most utilized for the beef production industry either as purebred or in crosses with Bos indicus cattle [3,6]. However, despite its extensive use, there is scarce information on the breed’s productive performance, and the available information is mainly related to growth traits coming from genetic evaluations or isolated studies [5,6,7]. The selection, management, and genetic improvement programs of the Braunvieh cattle could be enhanced using high-throughput genotyping technologies.
The use of microarrays of thousands of SNP markers in genome-wide association studies (GWAS) has allowed discovering the genetic basis of complex traits and diseases by detecting genotype–phenotype associations in a group of individuals [8]. GWAS approaches have confirmed many QTL for growth traits in beef and crossbred cattle [1,9,10], some of which have been used as the basis for the search for specific causal variation [11] and a better understanding of the genetic architecture of these complex traits. Many of these QTL, genomic regions and genes, affecting production traits in beef cattle have been reported [1,12,13,14], but most of the association studies focus on specialized beef breeds and only a few studies have been implemented in breeds such as Braunvieh [15,16]. The present study is aimed at performing a GWAS to identify QTLs and candidate genes related to liveweight traits in a Braunvieh cattle population.
2. Materials and Methods
2.1. Population and Phenotypic Data
Hair follicle samples from 236 females and 64 males registered in the Mexican Braunvieh Cattle Association database were collected. The cattle were born between 2000 and 2015. This population came from herds located in the east, west, and central highlands of Mexico. Herds from west and east were raised under extensive production systems, while central highlands herds were under intensive regimens. The sampled population’s genetic background included Austrian, Swiss, Canadian, American, and Mexican animals. Phenotypic data were provided by the breeding association and included records of birth weight (BW, kg), weaning weight (WW, kg), yearling weight (YW, kg). Weaning and yearling weights were adjusted to perform the GWAS analysis.
2.2. Genotyping and Quality Control
The animals were genotyped using 30,125 SNP markers from the GeneSeek® Genomic Profiler Bovine LDv.4 panel (Neogen Corp., Lincoln, NE, USA). Before association analysis, the genotypic data quality was verified using the SNPQC program [17]. The genotypes were considered successful if they presented a GenCall value greater than 0.50, and all SNPs with lower values were discarded (n = 1623). Those SNPs that were monomorphic (n = 3604), presented call rates of less than 90% (n = 1290) or minor allele frequencies < 0.01 (n = 1325), or deviated from the Hardy–Weinberg equilibrium according to Fisher’s exact test and exhibited p-values >1 × 10−15 (n = 0) were also eliminated. Additionally, SNPs with unknown coordinates in the assembly of the bovine genome UMD v3.1 [18] (n = 1484) and SNPs that were not located on autosomal chromosomes (n = 1820) were discarded.
Samples were also eliminated if they exhibited call rates of less than 80% (n = 0) or levels of heterozygosity (HE) above 3 SD (n = 1), considering that the mean and SD of the observed HE were 0.32 and 0.019, respectively. A Pearson correlation was computed for detecting potentially duplicate samples, considering a maximum of r = 0.98, according to their genotype information obtaining an average of r = 0.817 and minimum and maximum values of 0.66 and 0.90, respectively. A total of 22,734 SNPs and 276 samples passed the quality control procedure and were retained for further analysis. Quality control and subsequent analyses were performed in the R environment.
2.3. Population Structure and Association Analysis
Population structure was analyzed, calculating first a genomic relationships matrix using the information on genotypes [19], in addition to performing a singular value decomposition and a principal components (PC) analysis.
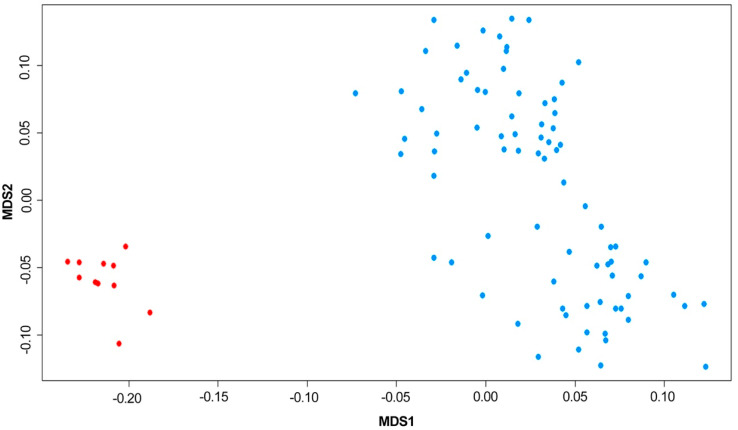
The PC analysis indicated that the first two PCs explained 28.6% of the variance in the data. A multidimensional scaling analysis confirmed this structure (Figure 1). Therefore, the genome-wide association analysis was performed using the PC method proposed by Price et al. [20]. For this analysis, the egscore function from the GenABEL package [21] was employed. This function accounts for population stratification and uses the genomic kinship matrix to derive axes of genetic variation, and then both phenotypes and genotypes are adjusted onto these axes.
Figure 1.
Muldimensional scaling analysis showing population genomic structure in the studied Braunvieh population.
A linear model for each trait was fitted, including the first two PC as covariates. For the analysis of BW, the model also included the contemporary group (CG) and the linear and quadratic effects of cow age at the birth and weaning of her calf. The CG included herd, sex, year, and calving season. The statistical model used to adjust the other traits only included the CG and the PCs as covariates; cow age was excluded because it was not significant in the previous analysis. Finally, the association between corrected genotypes and phenotypes was assessed via correlation. p-values were obtained by calculating the square of the correlation multiplied by (N-K-1), where N was the number of genotyped individuals, and K was the number of PCs.
Minimum allele frequencies, allele substitution effect (β), and percentage of phenotypic variance explained by the SNP were estimated. SNP with p-values < 5 × 10−5 were considered significantly associated with studied traits. The proportion of phenotypic variance explained by the SNPs was estimated by dividing the x2 value for a df by the number of individuals used to analyze each SNP marker, followed by multiplication by 100. All described analyses and estimations were performed using the GenABEL package [21].
2.4. Analysis of Genomic Regions with Significant SNPs
The closest genes to significant markers and those located within a 250 kb window on both SNP location sides were identified. The list of genes was obtained using the snp2gene.LD function from the Postgwas package [22]. Distance between SNPs and genes was calculated as the difference between the marker position and the beginning or end of the gene, according to coordinates from bovine genome assembly UMD v3.1. Gene functions were investigated in the UniProt database [23].
Annotations from humans or mice were used when there was no information on the genes in cattle. Genes were considered functional and positional candidates if they were biologically related to the trait under study, supported by experimental evidence in the literature. Finally, we determined whether significant SNPs mapped against QTLs previously associated with growth-related traits such as BW, carcass, and reproduction traits, deposited in the cattle AnimalQTLdb [24]. For this purpose, SNP positions according to the Btau4.6 genome sequence were used because many of the previously reported QTLs had no well-defined positions in the bovine genome assembly UMD v3.1.
3. Results
A total of 30,125 SNP markers from the GeneSeek® Genomic Profiler Bovine LD v4 microarray panel (Neogen Corp. Lincoln, NE, USA) were used for association with live weight traits of Braunvieh cattle. On average, 1004 SNP markers were evaluated in each BTA. Bos taurus chromosomes 1 and 27 exhibited the highest (1602) and lowest (512) SNP numbers. The average distance between adjacent SNP was 87,641 bp, the minimum distance (0 bp) between adjacent SNP were found on BTA 1, 6, 7, 12, 17, 18, 22, 25, 26, 28, and 29, while the maximum distance (1,962,000 bp) was found on BTA 6. Table 1 show the descriptive statistics for each trait.
Table 1.
Descriptive statistics for studied live weight traits (kg) of Braunvieh cattle.
| Trait 1 | N | n(QC) 2 | Mean | SD | Minimum | Maximum |
|---|---|---|---|---|---|---|
| BW | 300 | 266 | 38.007 | 4.067 | 22 | 50 |
| WW | 300 | 263 | 212.399 | 27.426 | 128 | 308 |
| YW | 300 | 244 | 313.165 | 45.473 | 176 | 440 |
1 BW: birth weight; WW: weaning weight; YW = yearling weight; SD: Standard deviation; 2 n(QC) = n after quality control.
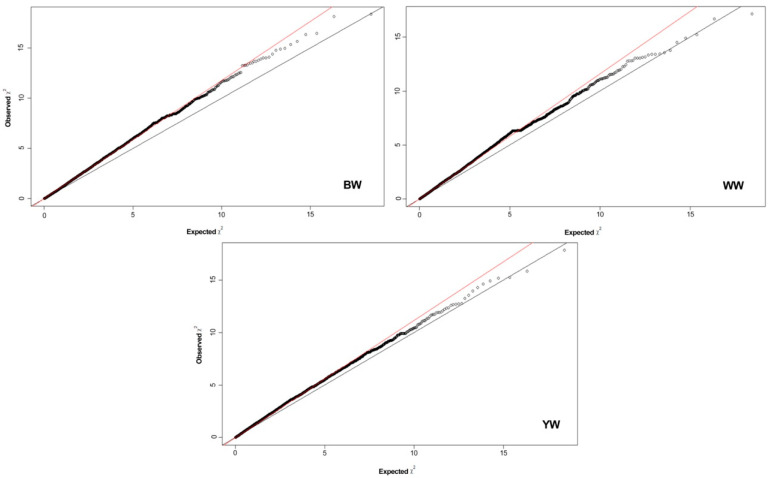
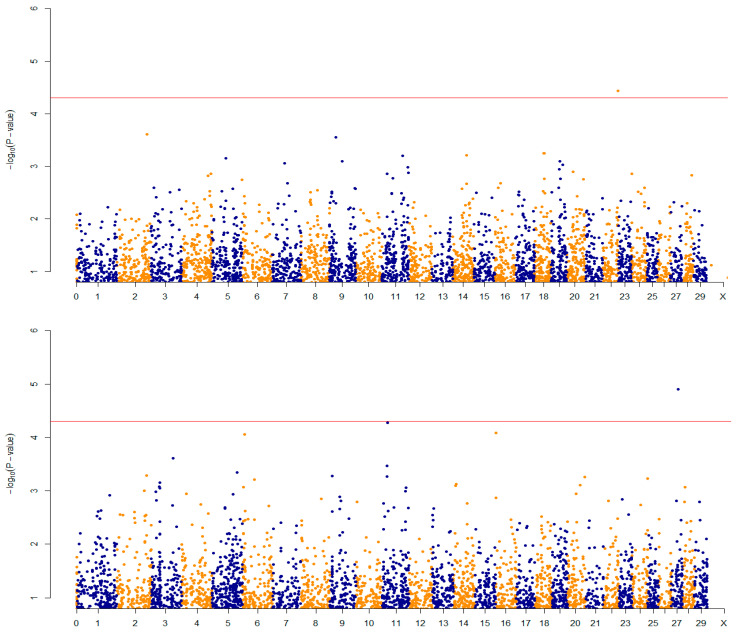
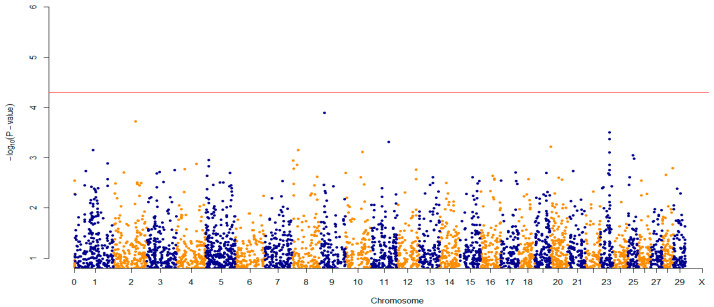
Figure 2 shows quantile–quantile plots for each GWAS analysis performed. According to the significance threshold considered (p < 5 × 10−5), 3 SNP were associated with the studied growth traits. Two SNP was associated with BW and one SNP was associated to WW. The rs133262280 located in BTA22 was associated with BW, showing an allelic substitution effect of 0.320 ± 0.02 kg. The rs43668789 and rs136155567, located in BTA11 and 27, respectively, were associated with WW. These markers showed allelic substitution effects of −9.590 ± 0.25 and 1.110 ± 0.72 kg, respectively (Table 2, Figure 3).
Figure 2.
Quantile–quantile (QQ) plots for the genome-wide association study of birth (BW), weaning (WW) and yearling (YW) weight traits in Braunvieh cattle. The straight line in the QQ plots indicates the distribution of SNP markers under the null hypothesis, and the skew at the edge indicates that these markers are more strongly associated with the traits than would be expected by chance. BW = birth weight; WW = weaning weight; YW = yearling weight.
Table 2.
Parameters and statistics of SNP associated with liveweight traits of Braunvieh cattle.
| Trait | SNP ID 2 | BTA | UMD 3 bp | Btau4.6,4 bp | Allele | MAF 5 | Β 6 | SE | Var% 7 | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 BW | rs133262280 | 22 | 60,759,211 | 127,745,473 | C/T | 0.18 | 0.320 | 0.02 | 0.1 | 2.74 × 10−5 |
| WW | rs43668789 | 11 | 21,312,462 | 22,502,811 | C/T | 0.17 | −9.590 | 0.25 | 2.98 | 5.28e − 5 |
| rs136155567 | 27 | 27,056,807 | 29,944,194 | A/G | 0.20 | 1.110 | 0.72 | 1.1 | 1.27e − 5 |
Figure 3.
Manhattan plots of the p-values for the genome-wide association study of birth, weaning and yearling weights of Braunvieh cattle. The horizontal line indicates the significance threshold for significant associations (p < 5 × 10−5). Blue and orange differentiate chromosomes.
Table 3, presents QTLs identified near the associated SNPs for BW and WW. Figure 3, shows the Manhattan plots in which the −log10 transformations of the p-values are plotted for each GWAS.
Table 3.
Previously reported QTL1 found near the SNP associated with growth traits of Braunvieh cattle.
| Trait_SNP ID 2_BTA_Mb | QTL | QTL ID | QTL in Btau4.6, 3 bp | QTL Reference |
|---|---|---|---|---|
| BW_rs133262280_22_60.7 | — | — | — | — |
| WW_rs43668789_11_21.3 | SOUND | 3591 | 18,215,471–23,417,727 | Buitenhuis et al. [26] |
| RFI | 5281 | 8,076,786–33,430,175 | Sherman et al. [27] | |
| RANGLE | 3447 | 16,291,959–80,096,141 | Boichard et al. [28] | |
| WWTMM | 10894 | 16,291,959–80,096,141 | McClure et al. [29] | |
| WW_rs136155567_27_27.0 | BQ | 3598 | 24,473,016–31,018,770 | Buitenhuis et al. [26] |
| SOUND | 3594 | 24,473,016–31,018,770 | Buitenhuis et al. [26] | |
| ADFI | 21028 | 27,034,490–29,073,970 | Rolf et al., [14] | |
| ADG | 20979 | 27,034,490–29,073,970 | Rolf et al., [14] | |
| RFI | 21095 | 27,034,490–29,073,970 | Rolf et al. [14] | |
| CALEASE | 11259 | 21,801,052–31,012,980 | McClure et al. [29] |
ADG = average daily gain; ADFI = average daily feed intake; BQ = bone quality; CALEASE = calving ease; RFI = residual feed intake; RANG = rump angle; SOUND = structural soundness; WWTMM = weaning weight–maternal milk; 2 ID = identification; 3 Elsik et al. [25].
Table 4 and Table 5, show complete descriptions of genes close to the SNP associated with BW and WW of Braunvieh cattle, including the identifier number and exact location identified in this study.
Table 4.
Genes close to the SNP rs133262280_22 associated with birth weight of Braunvieh cattle.
| SNP_BTA | Gene in ±250 kb 1 | Gene ID 2 | Distance, 3 kb | Description |
|---|---|---|---|---|
| rs133262280 | PODXL2 | 532521 | U 202.2 | Podocalyxin-like 2 |
| MCM2 | 510120 | U 177.6 | Minichromosome maintenance complex component 2 | |
| TPRA1 | 617772 | U 160.1 | Transmembrane protein adipocyte-associated 1 | |
| LOC10105309 | 109905309 | U 57.8 | Uncharacterized LOC101905309 | |
| PLXNA1 | 531240 | D 192.2 | Plexin A1 | |
| CHCHD6 | 615934 | D 200.9 | Coiled-coil helix coiled-coil helix domain-containing 6 |
1 rs136155567: gene in ±600 kb; 2 ID = identification; 3 D = downstream; U = upstream.
Table 5.
Genes close to the SNPs associated to weaning weight of Braunvieh cattle.
| SNP_BTA | Gene in ±250 kb 1 | Gene ID 2 | Distance, 3 kb | Description |
|---|---|---|---|---|
| rs43668789 | GALM | 616676 | U 217.4 | Galactose mutarotase |
| SRSF7 | 507066 | U 201.6 | Serine and arginine rich splicing factor 7 | |
| GEMIN6 | 525263 | U 160.6 | Gem nuclear organelle associated protein 6 | |
| LOC107132913 | 107132913 | U 156.0 | Uncharacterized LOC107132913 | |
| DHX57 | 540993 | U 86.1 | Dexh-box helicase 57 | |
| MORN2 | 616607 | U 77.8 | MORN repeat containing 2 | |
| ARHGEF33 | 100335703 | Cover | Rho guanine nucleotide exchange factor 33 | |
| SOS1 | 537682 | D 17.0 | SOS Ras/Rac guanine nucleotide exchange factor 1 | |
| MIR2284Z-2 | 102465308 | D 62.5 | Microrna 2284z-2 | |
| LOC104973309 | 104973309 | D 121.0 | Ubiquitin-40S ribosomal protein S27a pseudogene | |
| CDKL4 | 517478 | D 207.4 | Cyclin dependent kinase like 4 | |
| LOC782845 | 782845 | D 241.7 | 60S ribosomal protein L23a pseudogen | |
| rs136155567_27 | LOC104976093 | 104976093 | D 470.9 | Uncharacterized LOC104976093 |
| NRG1 | 281361 | D 567.1 | Neuregulin 1 |
1 rs136155567: gene in ±600 kb; 2 ID = identification; 3 D = downstream; U = upstream.
4. Discussion
The inclusion of the population’s genetic structure and fixed effect into the analysis model allowed the better fitting of the GWAS model for all traits, as showed by quantile–quantile plots (Figure 2). This genetic structure was expected because tested herds presented different selection criteria, and perhaps, ancestors from the imported genetic material (i.e., semen, sires). Stratification results could include extensive use of sires or semen that breeders usually choose in their genetic improvement programs. Some studies [30,31] have used subdivisions to estimate QTLs using genome-wide association studies (GWAS). Smitz et al. [32] concluded that the stratification in the studied populations needs to be considered in genetic improvement programs to conserve those populations “genetic health”. Jemaa et al. [33] indicated that some QTLs found in GWAS could not be present in all the studied animals due to the population’s stratification.
Birth weight in Braunvieh cattle represents an important trait to consider in the genetic improvement programs due to its association with calving difficulty in young heifers, especially when the Braunvieh is used as a sire for smaller-size breeds [34]. In the present study, the rs133262280 was identified as the only marker associated with BW, located at 60.7 Mb of BTA 22. This SNP showed an allelic substitution effect of 0.320 kg, explaining 0.1% of the phenotypic variance of BW. Genes located closer to this SNP included CHCHD6 (coiled-coil helix coiled-coil helix domain-containing 6), MCM2 (mini-chromosome maintenance complex component 2), PLXNA1 (plexin A1), PODXL2 (podocalyxin like 2), TPRA1 (transmembrane protein adipocyte associated 1), and uncharacterized LOC10105309 (Table 4). The most important genes identified in this region were MCM2 and TPRA1. The MCM2 gene is located at 177.6 kb and TPRA1 at 160.1 kb; both genes are upstream of the rs133262280 SNP. MCM2 acts as a component of the MCM2-7 complex (MCM complex) which is the putative replicative helicase essential for “once per cell cycle” DNA replication initiation and elongation in eukaryotic cells [31]. Additionally, it plays a role in cell division and apoptosis [35]. Gao et al. [36] reported MCM2 protein expression in the cochlea of rats and guinea pigs slightly increase the apoptosis rate of the cells without any changes in proliferation or cell cycle. Recently, Khan et al. [37] found by a transcriptomic analysis that supplementation with folic acid in perinatal Holstein cows significantly increases the expression of the MCM2 gene.
The other associated gene with biological importance was the TPRA1 gene belonging to the G protein-coupled receptor (GPCR) family. Functions related to this gene include regulating early embryonic cleavage and enhancing the hedgehog signaling pathway [38,39]. Several studies have highlighted its importance in pre- and perinatal tissue development in mice. Aki et al. [38] determined that the TPRA1 gene influenced the hedgehog signaling pathway, which plays an essential role in vertebrate embryonic tissue patterning of many developing organs, showing differences of around 50% in the signaling levels comparing homozygotes and heterozygotes animals.
This evidence suggests that MCM2 and TPRA1 could participate in the early stages of cattle development and, therefore, influence BW. There were no quantitative trait loci previously located in this region, which could be a specific QTL of the studied population.
The present study identified two regions (Table 3, WW_rs43668789_11_21.3 and WW_rs136155567_27_27.0) previously reported by McClure et al. [29] as associated with weaning weight and calving ease in Angus cattle. Furthermore, Boichard et al. [28] and Buitenhuis et al. [26] reported associations between the identified regions in this study and conformation traits, explaining between 5.9 and 8.9 % of the structural soundness in ten European dairy cattle breeds. On the other hand, Sherman et al. [27] and Rolf et al. [14] reported associations with allele substitution effects between -0.319 to 2.199 kg for feeding traits like average daily gain and residual feed intake in Angus, Charolais, and Canadian beef hybrid cattle.
From the associated WW rs43668789 associated SNP, genes located closer or covering this SNP included ARHGEF33 (Rho guanine nucleotide exchange factor 33), CDKL4 (cyclin-dependent kinase-like 4), DHX57 (DExH-box helicase 57), GALM (galactose mutarotase), GEMIN6 (gem nuclear organelle associated protein 6), LOC104973309 (ubiquitin-40S ribosomal protein S27a pseudogene), LOC107132913 (uncharacterized LOC107132913), LOC782845 (60S ribosomal protein L23a pseudogene), MAP4K3 (mitogen-activated protein kinase 3), MIR2284Z-2 (microRNA 2284z-2), MORN2 (MORN repeat containing 2), SOS1 (SOS Ras/Rac guanine nucleotide exchange factor 1), and SRSF7 (serine and arginine-rich splicing factor 7) (Table 5). Interestingly, the position of rs43668789 in intronic ARHGEF33 gene need further attention. Although unreported effects of intronic regulation in this gene were found, some evidence indicates that transcriptional regulations by intronic SNPs is possible [40]. Moreover, intronic variation might carry deeper functional effects since they are related to different types of noncoding RNAs (ncRNAs) in genomes including miRNAs, siRNAs, piwi-interacting RNAs (piRNAs), long noncoding RNAs (lncRNAs), and small nucleolar RNAs (snoRNAs), and they are known to be located in the intron regions within genes [41].
The most important gene identified in this region was GALM. This gene is located 217.4 kb upstream of the rs43668789 and belongs to the proteins that convert the α-aldose to β-anomer. GALM is involved in the pathway hexose metabolism, which is part of carbohydrate metabolism [42]. McClure et al. [29] reported a positive association of GALM with the weaning weight in Angus cattle. Shin et al. [43] mentioned that the association between GALM and the weaning weight in Holstein and Hanwoo cattle lies in quantity and the quality of the calves’ milk consumption. Quantitative trait loci located in this region have been previously associated with weaning weight in Angus [29], conformation in dairy cattle breed [26,28], and residual feed intake in Canadian beef synthetic cattle [27].
The second marker associated with WW was rs136155567, located at 27.0 Mb of BTA 27, and its allele substitution effect was 1.110 kg which explains 1.1% of the phenotypic variance. Genes located closer to this SNP (±600 kb) included LOC104976093 (uncharacterized LOC104976093) and NRG1 (neuregulin 1) (Table 5). NRG1 was the most important gene identified. This gene is located at 567.1 kb downstream of the rs136155567. It is considered the direct ligand for ERBB3 and ERBB4 tyrosine kinase receptors. The multiple isoforms perform diverse functions, such as inducing growth and differentiation of epithelial, glial, neuronal, and skeletal muscle cells, and influence motor and sensory neuron development [44,45]. In cattle, NRG1 has been highly associated with organ development [46]. Zhao [47] mentioned that this gene could influence the weaning weight as an emerging regulator of prolactin secretion.
In general, the phenotypic variance explained by the SNPs identified in this study was marginal (1.39% on average). In growth trait studies, it is expected that most SNP markers will explain only a tiny proportion of the observed phenotypic variance due to the polygenic control over such traits and because individual genes only slightly influence a phenotype. However, consideration of SNPs’ sets that are significantly associated with each trait may allow a greater proportion of phenotypic variance to be explained. For example, the two SNPs associated with WW could explain 4.08% of the variance in that trait. It is important to note the lack of significant association for YW. Although not a highly strict threshold was chosen, the amount of data reduction from BW and WW to YW might be related to these outcomes. However, the present outcomes increase knowledge of the genetic architecture of growth traits important in beef cattle production.
5. Conclusions
In conclusion, in the present study, three SNPs were associated with the assessed growth traits of Braunvieh cattle. Two SNPs were located in intergenic regions, and one was located in an intronic region of the ARHGEF33 gene. Additionally, evidence shows that some of the genes closer to the three identified SNPs markers are functionally related to growth through embryonic cleavage, bone and tissue growth, cell adhesion, and organ development. There were four candidate genes with potential associations with assessed live weight traits in Braunvieh cattle, including MCM2, TPRA1, GALM, and NRG1. Subsequent studies examining these genomic regions could lead to the identification of polymorphisms with potential uses in the marker-assisted selection, providing a deeper understanding of the genetic basis and genetic architecture of growth traits in cattle. This study represents the first study to describe a GWAS conducted in Braunvieh cattle in Mexico. Further analysis using the present information would allow to conduct assessments on the ontogeny and specific search of causative mutations for live weight traits. Furthermore, examining particular and general genic effects would indicate the possibility of including genomic information into current genetic evaluations.
Acknowledgments
Authors want to thank the Comisión de Operación y Fomento de Actividades Académicas del IPN for the support in funding the open access publication.
Author Contributions
Conceptualization, J.L.Z.-B., R.R.-V. and G.M.P.-B.; Formal analysis, J.L.Z.-B. and F.J.J.-M.; Resources, R.N.-D., R.R.-V. and G.M.P.-B.; Supervision, R.R.-V., G.M.P.-B.; Writing—original draft preparation, J.L.Z.-B. wrote the first draft of the manuscript; Review and editing, G.M.P.-B., J.B.H.-O., R.R.-V. and R.N.-D.; Funding acquisition, R.N.-D., R.R.-V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by research grant DGIP-166701012 by the Universidad Autónoma Chapingo.
Institutional Review Board Statement
Not applicable. Approval from the ethical committee for animal care and use was unnecessary because the samples used in this study consisted of hair follicles, and all analyses were performed using pre-existing databases.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data could be made available upon reasonable request to the authors.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jahuey-Martinez F., Parra-Bracamonte G.M., Sifuentes-Rincón A.M., Martínez-González J.C., Gondro C., García-Pérez C.A., López-Bustamante L.A. Genomewide association analysis of growth traits in Charolais beef cattle. J. Anim. Sci. 2016;94:4570–4582. doi: 10.2527/jas.2016-0359. [DOI] [PubMed] [Google Scholar]
- 2.Parra-Bracamonte G.M., Sifuentes-Rincón A.M., Martínez-Gonzalez J.C., Magaña-Monforte J.G., Jahuey-Martínez F.J. Biotecnologías para el desarrollo de los sistemas pecuarios, aspectos aplicados a la ganadería bovina para carne. In: Núñez D.R., Ramírez V.R., Fernández R.S., Araujo F.O., García W.M., Díaz M.T.E., editors. La ganadería en América Latina y el Caribe, Alternativas Para la Producción Competitiva, Sustentable e Incluyente de Alimentos de Origen Animal. 1st ed. Volume I. Biblioteca Básica de Agricultura; Mexico City, Mexico: 2015. pp. 391–416. [Google Scholar]
- 3.Orantes-Zebadúa M.A., Platas-Rosado D., Córdova-Avalos V., Santos-Lara M.C., Córdova-Avalos A. Characterization of dual purpose livestock in a Region of Chiapas, Mexico. Ecosistemas Recur. Agropecu. 2014;1:49–58. [Google Scholar]
- 4.Phillips W., Holloway J., Warrington B., Venuto B. Stocker and Feedlot Performance of Beef Heifers Sired by Braunvieh and Wagyu Bulls from Angus-, Brahman-, Senepol-, and Tuli-sired Dams1. Prof. Anim. Sci. 2009;25:809–814. doi: 10.15232/S1080-7446(15)30793-2. [DOI] [Google Scholar]
- 5.Chin-Colli R.C., Estrada-León R., Magaña-Monforte J., Segura-Correa J., Núñez-Domínguez R. Genetic parameters for growth and reproductive traits of Brown Swiss cattle from Mexico. Ecosistemas Recur. Agropecu. 2016;3:11–20. [Google Scholar]
- 6.AMCGSR Asociación Mexicana de Criadores de Ganado Suizo de Registro. 2017. [(accessed on 23 August 2016)]. Evaluaciones Genéticas. Available online: https//amcgsr.com.mx/evaluaciones-geneticas/
- 7.Silva C., Aké R., Valle R. Edad y crecimiento a la pubertad en toros Suizo Pardo en condiciones tropicales. [(accessed on 13 September 2021)];Cuban J. Agric. Sci. 2002 36:205–210. Available online: http://www.redalyc.org/articulo.oa?id=193018103002. [Google Scholar]
- 8.Tian D., Wang P., Tang B., Teng X., Li C., Liu X., Zou D., Song S., Zhang Z. GWAS Atlas: A curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020;48:D927–D932. doi: 10.1093/nar/gkz828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu D., Miller S., Sargolzaei M., Kelly M., Voort G.V., Caldwell T., Wang Z., Plastow G., Moore S. Genome-wide association analyses for growth and feed efficiency traits in beef cattle1. J. Anim. Sci. 2013;91:3612–3633. doi: 10.2527/jas.2012-5716. [DOI] [PubMed] [Google Scholar]
- 10.Martínez R., Bejarano D., Gómez Y., Dasoneville R., Jiménez A., Even G., Sölkner J., Mészáros G. Genome-wide association study for birth, weaning and yearling weight in Colombian Brahman cattle. Genet. Mol. Biol. 2017;40:453–459. doi: 10.1590/1678-4685-gmb-2016-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takasuga A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2015;87:159–167. doi: 10.1111/asj.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Londoño-Gil M., Flórez J.C.R., Lopez-Herrera A., Gonzalez-Herrera L.G. Genome-wide association study for growth traits in blanco orejinero (bon) cattle from colombia. Livest. Sci. 2021;243:104366. doi: 10.1016/j.livsci.2020.104366. [DOI] [Google Scholar]
- 13.Purfield D.C., Bradley D.G., Evans R.D., Kearney F.J., Berry D.P. Genome-wide association study for calving performance using high-density genotypes in dairy and beef cattle. Genet. Sel. Evol. 2015;47:47. doi: 10.1186/s12711-015-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolf M.M., Taylor J.F., Schnabel R.D., McKay S.D., McClure M.C., Northcutt S.L., Kerley M.S., Weaber R.L. Genome-wide association analysis for feed efficiency in Angus cattle. Anim. Genet. 2012;43:367–374. doi: 10.1111/j.1365-2052.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J., Jorjani H., Carlborg Ö. A genome-wide association study using international breeding-evaluation data identifies major loci affecting production traits and stature in the Brown Swiss cattle breed. BMC Genet. 2012;13:82. doi: 10.1186/1471-2156-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxa J., Neuditschko M., Russ I., Förster M., Medugorac I. Genome-wide association mapping of milk production traits in Braunvieh cattle. J. Dairy Sci. 2012;95:5357–5364. doi: 10.3168/jds.2011-4673. [DOI] [PubMed] [Google Scholar]
- 17.Gondro C., Porto-Neto L.R., Lee S.H. snpqc- an R pipeline for quality control of Illumina SNP genotyping array data. Anim. Genet. 2014;45:758–761. doi: 10.1111/age.12198. [DOI] [PubMed] [Google Scholar]
- 18.Zimin A.V., Delcher A.L., Florea L., Kelley D.R., Schatz M.C., Puiu D., Hanrahan F., Pertea G., Van Tassell C.P., Sonstegard T.S., et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanRaden P. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 20.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Aulchenko Y.S., Ripke S., Isaacs A., Van Duijn C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 22.Hiersche M., Rühle F., Stoll M. Postgwas: Advanced GWAS Interpretation in R. PLoS ONE. 2013;8:e71775. doi: 10.1371/journal.pone.0071775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The UniProt Consortium UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z.-L., Park C.A., Wu X.-L., Reecy J.M. Animal QTLdb: An improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2012;41:D871–D879. doi: 10.1093/nar/gks1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsik C.G., Unni D., Diesh C.M., Tayal A., Emery M.L., Nguyen H., Hagen D.E. Bovine Genome Database: New tools for gleaning function from the Bos taurusgenome. Nucleic Acids Res. 2015;44:D834–D839. doi: 10.1093/nar/gkv1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buitenhuis A., Lund M., Thomasen J., Thomsen B., Nielsen V.H., Bendixen C., Guldbrandtsen B. Detection of Quantitative Trait Loci Affecting Lameness and Leg Conformation Traits in Danish Holstein Cattle. J. Dairy Sci. 2007;90:472–481. doi: 10.3168/jds.S0022-0302(07)72649-8. [DOI] [PubMed] [Google Scholar]
- 27.Sherman E.L., Nkrumah J.D., Li C., Bartusiak R., Murdoch B., Moore S.S. Fine mapping quantitative trait loci for feed intake and feed efficiency in beef cattle1. J. Anim. Sci. 2009;87:37–45. doi: 10.2527/jas.2008-0876. [DOI] [PubMed] [Google Scholar]
- 28.Boichard D., Grohs C., Bourgeois F., Cerqueira F., Faugeras R., Neau A., Rupp R., Amigues Y., Boscher M.Y., Levéziel H. Detection of genes inuencing economic traits in three French dairy cattle breeds. Gen. Sel. Evol. 2003;35:77–101. doi: 10.1186/1297-9686-35-1-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClure M.C., Morsci N.S., Schnabel R.D., Kim J.W., Yao P., Rolf M.M., McKay S.D., Gregg S.J., Chapple R.H., Northcutt S.L., et al. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim. Genet. 2010;41:597–607. doi: 10.1111/j.1365-2052.2010.02063.x. [DOI] [PubMed] [Google Scholar]
- 30.Erbe M., Hayes B., Matukumalli L., Goswami S., Bowman P., Reich C., Mason B., Goddard M. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 2012;95:4114–4129. doi: 10.3168/jds.2011-5019. [DOI] [PubMed] [Google Scholar]
- 31.Plieschke L., Edel C., Pimentel E.C., Emmerling R., Bennewitz J., Götz K.-U. A simple method to separate base population and segregation effects in genomic relationship matrices. Genet. Sel. Evol. 2015;47:53. doi: 10.1186/s12711-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smitz N., Cornélis D., Chardonnet P., Caron A., De Garine-Wichatitsky M., Jori F., Mouton A., Latinne A., Pigneur L.-M., Melletti M., et al. Genetic structure of fragmented southern populations of African Cape buffalo (Syncerus caffer caffer) BMC Evol. Biol. 2014;14:203. doi: 10.1186/s12862-014-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben Jemaa S., Boussaha M., Ben Mehdi M., Lee J.H., Lee S.H. Genome-wide insights into population structure and genetic history of tunisian local cattle using the illumina bovinesnp50 beadchip. BMC Genom. 2015;16:677. doi: 10.1186/s12864-015-1638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagger C., Hofer A. Genetic analyses of calving traits in the swiss black and white, braunvieh and simmental breeds by REML and MAPP procedures. Livest. Prod. Sci. 1990;24:93–107. doi: 10.1016/0301-6226(90)90070-M. [DOI] [Google Scholar]
- 35.Todorov I., Pepperkok R., Philipova R., Kearsey S., Ansorge W., Werner D. A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J. Cell Sci. 1994;107:253–265. doi: 10.1242/jcs.107.1.253. [DOI] [PubMed] [Google Scholar]
- 36.Gao J., Wang Q., Dong C., Chen S., Qi Y., Liu Y. Whole Exome Sequencing Identified MCM2 as a Novel Causative Gene for Autosomal Dominant Nonsyndromic Deafness in a Chinese Family. PLoS ONE. 2015;10:e013352. doi: 10.1371/journal.pone.0133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan M.Z., Liu L., Zhang Z., Khan A., Wang D., Mi S., Usman T., Liu G., Guo G., Li X., et al. Folic acid supplementation regulates milk production variables, metabolic associated genes and pathways in perinatal Holsteins. J. Anim. Physiol. Anim. Nutr. 2019;104:483–492. doi: 10.1111/jpn.13313. [DOI] [PubMed] [Google Scholar]
- 38.Aki T., Funakoshi T., Nishida-Kitayama J., Mizukami Y. TPRA40/GPR175 regulates early mouse embryogenesis through functional membrane transport by Sjögren’s syndrome-associated protein NA14. J. Cell. Physiol. 2008;217:194–206. doi: 10.1002/jcp.21492. [DOI] [PubMed] [Google Scholar]
- 39.Singh J., Wen X., Scales S.J. The Orphan G Protein-coupled Receptor Gpr175 (Tpra40) Enhances Hedgehog Signaling by Modulating cAMP Levels. J. Biol. Chem. 2015;290:29663–29675. doi: 10.1074/jbc.M115.665810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi J.-W., Park C.-S., Hwang M., Nam H.-Y., Chang H.S., Park S.G., Han B.-G., Kimm K., Kim H.L., Oh B., et al. A common intronic variant of CXCR3 is functionally associated with gene expression levels and the polymorphic immune cell responses to stimuli. J. Allergy Clin. Immunol. 2008;122:1119–1126.e7. doi: 10.1016/j.jaci.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 41.Jo B.-S., Choi S.S. Introns: The Functional Benefits of Introns in Genomes. Genom. Informatics. 2015;13:112–118. doi: 10.5808/GI.2015.13.4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoden J.B., Timson D., Reece R.J., Holden H.M. Molecular Structure of Human Galactose Mutarotase. J. Biol. Chem. 2004;279:23431–23437. doi: 10.1074/jbc.M402347200. [DOI] [PubMed] [Google Scholar]
- 43.Shin D.-H., Lee H.-J., Cho S., Kim H.J., Hwang J.Y., Lee C.-K., Jeong J., Yoon D., Kim H. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genom. 2014;15:240. doi: 10.1186/1471-2164-15-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ieguchi K., Fujita M., Ma Z., Davari P., Taniguchi Y., Sekiguchi K., Wang B., Takada Y.K., Takada Y. Direct Binding of the EGF-like Domain of Neuregulin-1 to Integrins (αvβ3 and α6β4) Is Involved in Neuregulin-1/ErbB Signaling. J. Biol. Chem. 2010;285:31388–31398. doi: 10.1074/jbc.M110.113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plowman G.D., Green J.M., Culouscou J.-M., Carlton G.W., Rothwell V.M., Buckley S. Heregulin induces tyrosine phosphorylation of HER4/p180erbB4. Nat. Cell Biol. 1993;366:473–475. doi: 10.1038/366473a0. [DOI] [PubMed] [Google Scholar]
- 46.Sweeney C., Fambrough D., Huard C., Diamonti A.J., Lander E.S., Cantley L., Iii K.L.C. Growth Factor-specific Signaling Pathway Stimulation and Gene Expression Mediated by ErbB Receptors. J. Biol. Chem. 2001;276:22685–22698. doi: 10.1074/jbc.M100602200. [DOI] [PubMed] [Google Scholar]
- 47.Zhao W. Neuregulin-1 (Nrg1): An Emerging Regulator of Prolactin (PRL) Secretion. Prolactin. 2013;23:83–96. doi: 10.5772/54716. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data could be made available upon reasonable request to the authors.