Abstract
Depletion of ptk7 is associated with both congenital scoliosis (CS) and adolescent idiopathic scoliosis (AIS) in zebrafish models. However, only one human variant of PTK7 has been reported previously in a patient with AIS. In this study, we systemically investigated the variant landscape of PTK7 in 583 patients with CS and 302 patients with AIS from the Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study. We identified a total of four rare variants in CS and four variants in AIS, including one protein truncating variant (c.464_465delAC) in a patient with CS. We then explored the effects of these variants on protein expression and sub-cellular location. We confirmed that the c.464_465delAC variant causes loss-of-function (LoF) of PTK7. In addition, the c.353C>T and c.2290G>A variants identified in two patients with AIS led to reduced protein expression of PTK7 as compared to that of the wild type. In conclusion, LoF and hypomorphic variants are associated with CS and AIS, respectively.
Keywords: protein tyrosine kinase 7 (PTK7), congenital scoliosis, adolescent idiopathic scoliosis, whole exome sequencing
1. Introduction
Protein tyrosine kinase (PTK7), also known as colon carcinomakinase-4 (CCK-4), is an evolutionarily conserved atypical receptor tyrosine kinase. The PTK7 protein consists of seven extracellular immunoglobulin (Ig)-like domains, one transmembrane domain and one catalytically inactive kinase domain [1]. PTK7 plays an important role in vertebrate canonical and non-canonical Wnt (planar cell polarity, PCP), Semaphorin/Plexin and vascular endothelial growth factor (VEGF) signaling pathways [2,3,4,5]. These signaling pathways are important for embryonic developmental processes, including tissue specification, axial morphogenesis, formation of the cardiovascular, endocrine and immune systems, and regulation of neural crest migration and tumorigenesis [6,7,8,9,10].
Zebrafish models depleted of ptk7 presented various spinal curve phenotypes. Maternal zygotic ptk7 (MZptk7) and zygotic ptk7 (Zptk7) mutant zebrafish develop spinal curvatures that model congenital scoliosis (CS) and adolescent idiopathic scoliosis (AIS), respectively, due to differential timing of ptk7 loss-of-function [2,11]. Meanwhile, a novel sequence variant PTK7P545A has been reported in a patient with AIS, but without further in vitro investigation [11]. The association between human PTK7 variants and scoliotic phenotypes continue to be understudied.
In this study, we analyzed variants in PTK7 identified in a mixed cohort of patients with congenital scoliosis and adolescent idiopathic scoliosis, then performed in vitro experiments to determine the effects of these variants on protein expression and sub-cellular location.
2. Materials and Methods
2.1. Human Subjects
A total of 885 Han Chinese individuals who received a diagnosis of congenital scoliosis (CS, n = 583) and adolescent idiopathic scoliosis (AIS, n = 302) were recruited between 2009 and 2018 at Peking Union Medical College Hospital (PUMCH) for the Deciphering disorders Involving Scoliosis and COmorbidities (DISCO, http://discostudy.org/, accessed on 1 November 2021) project. Clinical manifestations, physical examination results, and detailed medical histories were obtained with the patients’ informed consent. Clinical diagnoses were confirmed by radiology imaging, including X-ray and computed tomography (CT). The criteria for the diagnosis of congenital scoliosis and adolescent idiopathic scoliosis were as follow: congenital scoliosis was caused by vertebral defects, and may be associated with rib anomalies, while idiopathic scoliosis was diagnosed by spinal curvature exceeding 10° on a plain antero-posterior X-ray image, with no other identifiable underlying disease. For a diagnosis of adolescent idiopathic scoliosis, patients were required to have an onset age of 10–18 years old. All radiographic evaluations were conducted by trained spine surgeons, while clinical reviews were performed by alternate observers blinded to the radiographic assessment. Patients with a prior molecular diagnosis such as a disease-causing genetic variant were excluded. Blood was obtained from all subjects and whole-exome sequencing (WES) was performed.
Written informed consent for both clinical data and the genetic exome sequencing was obtained from each participant prior to study participation. This study was approved by the Department of Scientific Research and Ethics Committee of PUMCH in China.
2.2. Bioinformatic Analysis and Variant Interpretation
Two DNA extraction and purification kits, Red Blood Cell (RBC) Lysis Buffer (R1010, Solarbio) and Circulating Nucleic Acid Kit (55114, Qiagen), were used in accordance with the manufacturers’ protocols. Approximately 4 mL of peripheral blood was transferred to an Eppendorf safe lock tube after sufficient centrifugation. 10 mL RBC lysis solution was added to each centrifuge tube for efficient lysis. 4.5 mL cell lysis solution and 250 µL proteinase K solution were added to each tube and placed at 56.5 °C constant temperature shaker digestion overnight. 1.5 mL protein precipitation solution was added to each tube and allowed to incubate for 10 min at −20 °C. After centrifugation, the supernatant was taken, and 7 mL precooled isopropanol was added into the supernatant until floccule was precipitated. Finally, 1 mL of 75% ethanol was used to wash the DNA pellet after inverting the tube several times, followed by centrifugation at 17,000× g for 10 min. The quality and quantity of the DNA was evaluated using a spectrophotometer (NanoPhotometer Pearl, Denville Scientific, Inc., Holliston, MA, USA) and fluorometer (Qubit® dsDNA High Sensitivity and dsDNA Broad Range assay, Life Technologies Corporation, Waltham, MA, USA). DNA samples were prepared in Illumina libraries and then underwent whole-exome capture with the SureSelect Human All Exon V6 + UTR r2 core design (91 Mb, Agilent, Santa Clara, CA, USA), followed by sequencing on the Illumina HiSeq 4000 platform in 150-bp paired-end reads mode (Illumina, San Diego, CA, USA). WES data processing was performed with the Peking Union Medical College Hospital Pipeline (PUMP) based on the reference genome GRCh37-v1.6 [12,13]. Combined Annotation Dependent Depletion (CADD PHRED-score) [14] and Polyphen-2 [15] were used to predict the pathogenicity of candidate variants. Genotype was filtered for read-depth (DP > 10×), genotype quality (GQ > 20), quality by depth (QD < 2), strand odds ratio (SOR > 9), and allele balance (AB > 0.25). The populational frequency of each QC-passed variant was obtained from the public population databases, including the 1000 Genomes Project, the Exome Sequencing Project [16], the Genome Aggregation Database (gnomAD) [17], and the in-house database of DISCO (Deciphering disorders Involving Scoliosis and COmorbidities, http://discostudy.org/, ≈8000 exomes/genomes, accessed on 1 November 2021) study. Rare variants (minor allele frequency < 0.001) were retained for further filtering. From these rare variants, we included the protein-altering or splice-region variants for subsequent analysis. Potential spicing variants were predicted using SpliceAI [18].
Candidate variants of PTK7 were selected based on the following criteria:
-
(1)
Predicted to alter the protein sequence;
-
(2)
Either absent or with a low frequency (<0.001) from the public database mentioned above.
-
(3)
The missense variants have a CADD score ≥15.
2.3. Site-Directed Mutagenesis
pcDNA3.1+ with C-terminal flag-tagged wild type (WT) and variant PTK7 cDNA (NM_152881.3) plasmids were acquired from Beijing Hitrobio Biotechnology (Beijing, China). The variant constructs were sequenced on both strands to verify nucleotide changes.
2.4. Cell Culture and Transfection Assay
HEK293T cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA), penicillin (50 U/mL), and streptomycin (50 μg/mL) in six-well plates. HEK293T cells were transfected with full-length WT or variant PTK7 constructs using Lipofectamine 3000 (Invitrogen).
2.5. Western Blot Assay
After a 48 h transfection, HEK293T cells were harvested in RIPA lysis buffer (Solarbio, Beijing, China) and whole-cell lysate was resolved on gels under reducing conditions, transferred to a Nitrocellulose Transfer (NC) membrane, blocked with non-fat milk for 30 min at room temperature, and probed with primary antibodies: mouse anti-DDK(FLAG) antibody (1:1000, ZSGB-BIO, TA-05) and mouse anti-β-actin antibody (1:5000, Proteintech, 66009-1-Ig) over-night at 4 °C, and then with a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:5000, ZSGB-BIO, ZB-2305). Immunoreactivity was visualized using Western Lighting chemiluminescence reagent (Beyotime, Shanghai, China). All Western blotting experiments were repeated three times.
2.6. Immuno-Fluorescence
Cells were grown on 35 mm glass bottom cell culture dishes for 48 h after transfection, washed three times, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and incubated with primary anti-DDDDK-tag mouse antibody (1:1000, MBL, M185-3L) at 4 °C overnight. Cells were incubated and stained with secondary antibody Alexa FlourTM 488 goat anti-mouse IgG (1:2000, Invitrogen, 1911843), then covered with DAPI (Solarbio, Beijing, China).
2.7. Statistical Analysis
The overall protein expression levels were normalized to WT (set as 1.0) and mean values of variant versus WT from all three experiments were compared using unpaired t-test. All charts are drawn and analyzed using GraphPad Prism 8 and p < 0.05 was considered significant for all analyses.
3. Results
A total of 885 genomes from patients with scoliosis were sequenced and eight PTK7 variants in nine patients were found. The mean age of the included nine patients with variants was 11.11 ± 5.51 years. In five CS patients, the mean Cobb angle of the coronal plane was 59.94° ± 25.85°. Among them, three patients displayed kyphosis with a mean angle of 50.53° ± 8.05°. The mean Cobb angle of structural curve in four AIS patients was 48.95° ± 4.77°. In the CS group (n = 583), four possibly deleterious variants were revealed in five patients, including one frameshift variant and three missense variants (c.464_465delAC, c.1394A>G, c.1879G>A, c. 1955G>T) (Table 1). One of the missense variants (c.1955G>T) was identified in two patients. In the AIS group (n = 302), four deleterious missense variants (c.49C>T, c.353C>T, c.2290G>A, c.2384G>A) were identified (Figure 1A, Table 2). No peripheral blood samples from the patients’ families were obtained, and no similar family history of spinal deformity was found after follow-up.
Table 1.
Summary of clinical features of the PTK7 variant carriers.
| Patient Number | Variant | Gender | Age of Onset | Diagnosis | Vertebral Malformations | Other skeletal Malformations or Deformities | Cardiac Abnormalities | Spinal Canal Deformities | Visceral Abnormalities |
|---|---|---|---|---|---|---|---|---|---|
| SCO1905P0038 | c.464_465delAC | M | 13 | CS | T12 butterfly vertebra, T9-T10 failure of segmentation | None | None | None | None |
| SCO2003P2127 | c.1394A>G | F | 21 | CS | None | Fusion of 4th and 5th ribs, 12th ribs absent | Postoperative of patent ductus arteriosus | None | None |
| SCO2003P0372 | c.1879G>A | F | 8 | CS | L2 Hemivertebra, L2-L3 fusion | None | None | None | None |
| SCO1908P0053 | c.1955G>T | F | 1 | CS | T11, T12 segmented wedge vertebrae | None | None | None | None |
| SCO2003P0541 | c.1955G>T | F | 7 | CS | T7-T11 failure of segmentation | Chest deformity | None | Diastematomyelia | None |
| SCO1907P0150 | c.49C>T | F | 11 | AIS | None | None | Ventricular septal defect | None | None |
| SCO2003P0632 | c.353C>T | F | 12 | AIS | None | None | None | None | None |
| SCO2003P2288 | c.2290G>A | F | 14 | AIS | None | None | None | None | None |
| SCO2003P2237 | c.2384G>A | F | 13 | AIS | None | None | None | None | None |
M, male; F, female; CS, congenital scoliosis; AIS, adolescent idiopathic scoliosis.
Figure 1.
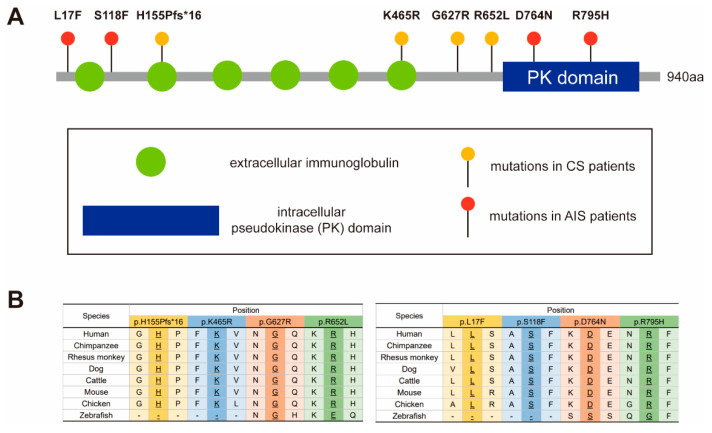
Mapping and conservation analysis of the identified variants. (A) Mapping of eight PTK7 variants revealed that p.L17F, p.S118F, p.H155Pfs*16 and p.K465R are located in the extracellular region of PTK7 protein. p.D764N and p.R795H are located in the intracellular pseudokinase domain. (B) The conservation of variants in human and other vertebrate species.
Table 2.
Sequence variants identified in PTK7.
| Patient Number | cDNA Change | Protein Change | Variant Type | Position | Gnomad_Exome_ALL | Gnomad_Exome_EAS | Gnomad_Genome_ALL | Gnomad_Genome_EAS | CADD | Ployphen-2 HDV Score | Evidence of Pathogenicity by ACMG | ACMG Classification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCO1905P0038 | c.464_465delAC | p.H155Pfs*16 | Frameshift | 43097560 | 0 | 0 | 0 | 0 | NA | NA | PVS1+PM2+PP3 | Pathogenic |
| SCO2003P2127 | c.1394A>G | p.K465R | Missense | 43109684 | 0 | 0 | 0.00003247 | 0.0006 | 26.2 | 0.997 | PP3 | Uncertain significance |
| SCO2003P0372 | c.1879G>A | p.G627R | Missense | 43112206 | 0.00004873 | 0.0006 | 0.00003232 | 0.0006 | 26.9 | 0.986 | PP3 | Uncertain significance |
| SCO1908P0053 SCO2003P0541 |
c.1955G>T | p.R652L | Missense | 43112282 | 0.00007718 | 0.0005 | 0 | 0 | 23.6 | 0.947 | PP3 | Uncertain significance |
| SCO1907P0150 | c.49C>T | p.L17F | Missense | 43044275 | 0 | 0 | 0 | 0 | 19.95 | 0.997 | PM2 | Uncertain significance |
| SCO2003P0632 | c.353C>T | p.S118F | Missense | 43096988 | 0 | 0 | 0 | 0 | 25.6 | 0.982 | PM2 | Uncertain significance |
| SCO2003P2288 | c.2290G>A | p.D764N | Missense | 43114395 | 0.00000814 | 0.000058 | 0 | 0 | 23.8 | 0.114 | PM1+PP3 | Uncertain significance |
| SCO2003P2237 | c.2384G>A | p.R795H | Missense | 43126607 | 0.00000406 | 0 | 0.00003231 | 0 | 30 | 0.986 | PM1+ +PP3 | Uncertain significance |
The RefSeq transcript sequence used for PTK7 is NM_152881.3. Abbreviations: NA, not available. ACMG, American College of Medical Genetics and Genomics. PVS, pathogenic very strong. PM, pathogenic moderate. PP, supporting pathogenicity.
3.1. Variant and Phenotypic Characteristics
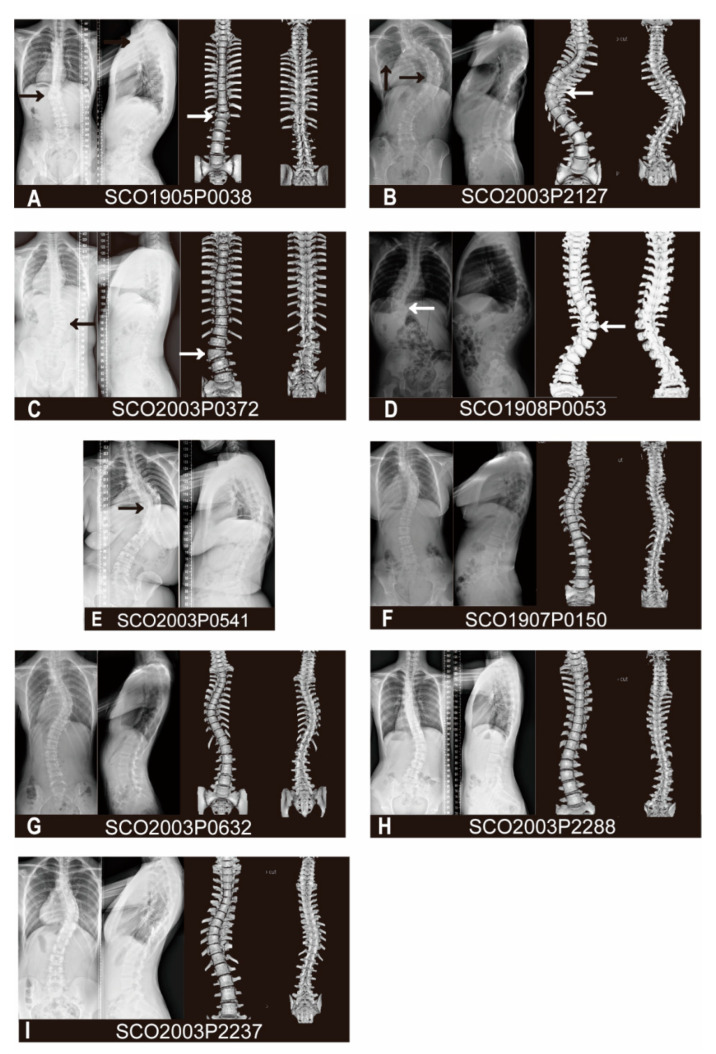
Patient SCO1905P0038 is a 13-year-old male with T12 butterfly vertebra and T9-T10 segmentation defect. The spinal plain radiograph shows not only a coronal curve to the left, but also has a severe thoracolumbar kyphosis in the sagittal plane with a 65° Cobb angle, both results of continuous deformity in the vertebral body (Figure 2A). The patient has a heterozygous deletion between nucleotide 464 and 465 (c.464_465delAC, p.H155Pfs*16). This frameshift variant was mapped to the extracellular immunoglobulin region of PTK7 protein (Figure 1A) and is predicted to cause the early termination of mRNA translation. It is a novel variant, previously undescribed in mutational databases and is highly conserved across different vertebral species except zebrafish (Figure 1B).
Figure 2.
Antero-posterior, lateral spinal X-ray and the spinal three-dimensional CT reconstruction of patient SCO1905P0038 (A), SCO2003P2127 (B), SCO2003P0372 (C), SCO1908P0053 (D), SCO1907P0150 (F), SCO2003P0632 (G), SCO2003P2288 (H), SCO2003P2237 (I). Antero-posterior, lateral spinal X-ray of patient SCO2003P0541 (E). Arrowheads point to the vertebral or rib deformities.
Patient SCO2003P2127 is a 21-year-old female with a diagnosis of congenital scoliosis. This patient presents with the failure of segmentation in the concave side of T9-T11, fusion of the 4th and 5th ribs, and absence of 12th ribs. As a result, the patient has a severe imbalance of the spine in the coronal plane, with a 92° Cobb angle of the main curve at T5-L1 and a compensatory curve of 45° at the lumbar level (Figure 2B). She also has a history of patent ductus arteriosus. The heterozygous missense variant c.1394A>G (p.K465R) of the PTK7 gene is mapped to the extracellular immunoglobulin region of the PTK7 protein (Figure 1A). This variant is previously unreported in mutational databases and is highly conserved (Figure 1B). It is predicted by CADD and PolyPhen-2 to be deleterious.
Patient SCO2003P0372 is an 8-year-old female with L2 hemivertebrae and L2-L3 segmentation defect. Her spine has a 38° right curve at the lumbar region with the hemivertebrae as its apex in the coronal plane, and a slight compensatory curve at the thoracic region (Figure 2C). She has a heterozygous missense variant c.1879G>A (p.G627R) of PTK7. This variant is located in the intracellular domain and adjacent to the transmembrane region of the PTK7 protein (Figure 1A). This variant is highly conserved (Figure 1B) and has been reported in the gnomAD database with low frequency. It is predicted by CADD and PolyPhen-2 to be deleterious.
Patient SCO1908P0053 is a 1-year-old female detected segmented wedge vertebrae in T11 and T12. This child displays severe imbalance in both the coronal and sagittal planes, in which the left curve reached 78° with the deformed vertebral body as the apex in the coronal plane, and severe kyphosis of 48° in the thoracolumbar region in the sagittal plane (Figure 2D). Due to her young age, long segment involvement, and the combination of thoracolumbar scoliosis and kyphosis, she received growth rod implantation and repeated growth rod extension processes over the past few years. Patient SCO2003P0541 is a 7-year-old female with segmentation failure from T7 to T11. Although the spinal deformity was discovered at the age of 7, the patient did not receive proper treatment until adulthood, leading to the continued progression of kyphoscoliosis. Similar to the patient SCO1908P0053, this patient has severe imbalance on both the coronal and sagittal planes in the thoracic spine, with a right curve of 87° and a thoracic kyphosis of 62°, which compared with the normal physiological thoracic kyphosis ranged from 10° to 40° (Figure 2E). She also suffered from chest deformity and diastematomyelia from MRI scans, which is consistant with the reported association between PTK7 variants and neural tube defects (NTDs) [19]. The patient showed no dyspnea, sensory or motor disorders in the lower limbs, nor did she show urinal or excretory dysfunction. These two individuals carry the same heterozygous missense variant c. 1955G>T (p.R652L). This variant is located in the intracellular domain of PTK7, close to the pseudokinase (PK) domain (Figure 1A). This variant is conserved across vertebral species besides zebrafish (Figure 1B), and has been reported in gnomAD database with low frequency.
Patients SCO1907P0150, SCO2003P0632, SCO2003P2288 and SCO2003P2237 all suffer from adolescent idiopathic scoliosis (AIS). Patient SCO1907P0150 is an 11-year-old female. The patient has two curves on the coronal plane, a 47° left curve in the upper thoracic segment (T2-T6) and a 51° right curve in the thoracic segment (T6-T12) (Figure 2F). She had a congenital ventricular septal defect that was surgically treated. WES analysis reveals a missense variant c. 49C>T (p.L17F) in PTK7, which has not been reported in the mutational databases. This variant is located at the beginning of the extracellular portion of the PTK7 protein (Figure 1A). It is predicted by CADD and PolyPhen-2 score to be deleterious.
Patient SCO2003P0632 displays a 50° right curve and a 42° left curve at the thoracic and thoracolumbar levels, respectively, without significant trunk deviation (Figure 2G). A missense variant c. 353C>T (p.S118F) is identified in patient SCO2003P0632, a 12-year-old girl, which is also absent from mutational databases. It is predicted to be located at the junction between the first and second Ig domains of the extracellular part of PTK7 protein (Figure 1A). This variant is predicted by CADD and Polyphen-2 scores to be deleterious.
Patient SCO2003P2288 is a 14-year-old female with a c. 2290G>A (p.D764N) variant in PTK7. X-rays showed that the main curve was a 46° right-sided lumbar scoliosis, with a long compensatory curve of 25° at T1-T11 to maintain basic balance of the trunk (Figure 2H). This missense variant has been reported in gnomAD database with low frequency and the altered amino acid is highly conserved in vertebrates except zebrafish (Figure 1B). The variant is mapped to the PK domain of the intracellular portion of the PTK7 protein (Figure 1A). The pathogenicity assessment has contradictory results using CADD and Polyphen-2 scores.
Patient SCO2003P2237 is a 13-year-old girl. On the coronal plane, the patient presents a single thoracic curve from T7 to T12 with a Cobb Angle of 52°. Due to the severe vertebral rotation, a relatively obvious razor-back deformity is seen on the sagittal plane (Figure 2I). WES analysis reveals that she has a missense variant c.2384G>A (p.R795H) in PTK7. The variant amino acid is highly conserved in vertebrates (Figure 1B) and mapped to the PK domain of the intracellular section of the PTK7 protein (Figure 1A). Reported in gnomAD database with low frequency, it is predicted by both CADD and Polyphen-2 scores to be deleterious.
3.2. Western Blot and Immunocytochemistry Analyses
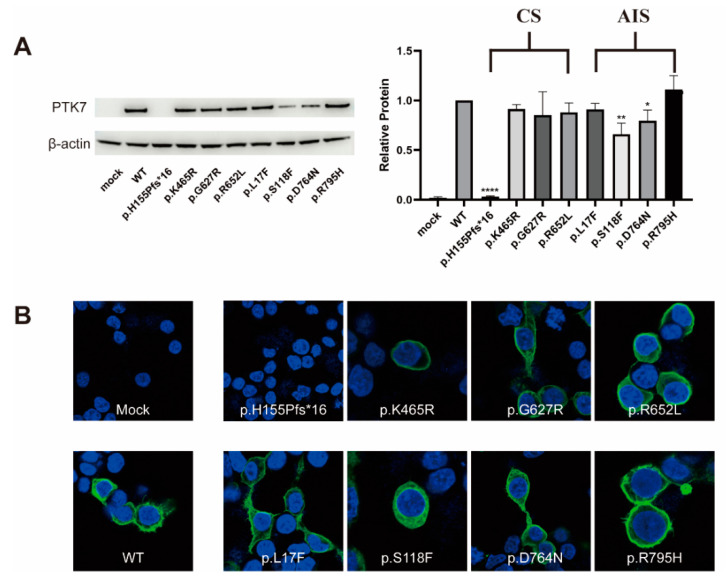
To identify the influence of the identified variants on PTK7 protein function, we evaluated overall protein expression and sub-cellular location of the variants of PTK7 compared to the WT. Western blotting analysis identified the immunoreactive-specific band for flag-tagged WT PTK7 at 150kDa and β-actin at 42kDa. The overall expression level was quantified by estimating bands from PNGase-treated samples and normalizing to WT. As anticipated, the overall expression of the frameshift variant (c.464_465delAC, p.H155Pfs*16) was significantly decreased compared to that of WT (p < 0.0001) (Figure 3A), indicating the loss-of-function effect of this variant. The expression level of two missense variants (p.S118F and p.D764N) were partially reduced (p = 0.0061 and p = 0.0293, respectively) (Figure 3A). Interestingly, these two variants were both identified in patients with AIS but not CS. There were no significant differences in the overall protein expression of the other missense variants (p.K465R, p.G627R, p.R562L, p.L17F, p.R795H) compared with the WT.
Figure 3.
Western blot and ICC analyses. (A) HEK293T cells transiently transfected with WT or variant PTK7 constructs revealed diminished PTK7 protein expression levels of p.H155Pfs*16, p.S118F and p.D764N. * p value < 0.05, ** p value < 0.01, **** p value < 0.0001. (B) ICC assays showed a faint signal of the frameshift variant PTK7 protein. No difference in PTK7 protein sub-cellular location was detected.
We also performed immunocytochemistry (ICC) assays and found there was a faintest signal from the frameshift variant of the PTK7 protein (Figure 3B), which supported the results of Western blotting assays. However, the sub-cellular location of all missense variant PTK7 protein did not change compared to WT (Figure 3B).
4. Discussion
Here, we performed WES on the genomes of 885 scoliosis patients, including 583 CS patients and 302 AIS patients, and identified seven missense variants and one frameshift variant in PTK7. Loss-of-function of PTK7 resulted in skeletal phenotypes in both zebrafish and mice. In zebrafish models, embryos with the PTK7hsc9 variant (a mutant allele harboring a 10bp deletion that results in a frameshift and the incorporation of multiple premature termination codons) had defects in the convergence of both neuroectoderm and axial mesoderm tissues, and abnormal three-dimensional spinal curvature with growth and development [2]. In 2016, Grimes et al. reported that mutated ptk7 zebrafish exhibited spinal curvature as well as hydrocephalus, ependymal cell (EC) ciliary dysfunction and abnormal rate and pattern of the cerebrospinal fluid (CSF) flow [20]. The chuzhoi mutant mice embryo showed several congenital abnormalities including neural tube, heart and lung defects caused by the disruption of PTK7 protein expression [21]. In human, LoF mutations in PTK7 and the double-heterozygous mutation of PTK7 and VANGL2 were associated with spina bifida and increased the genetic risk of NTDs [19]. However, only one case of scoliosis with a PTK7 mutation has been previously reported [11], and our identification of these eight variants expands the variant and phenotypic spectrum of PTK7.
The PTK7 protein consists of an extracellular continuous immunoglobulin domain, a transmembrane domain and an intracellular PK domain [1]. PTK7 interacts with several molecules in both canonical and non-canonical Wnt signaling as a co-receptor. These molecules include Wnt ligands, Wnt receptors as well as intracellular components such as Dvls and β-catenin [22]. According previous studies, the extracellular domains and intracellular domains of PTK7 play distinct roles. By constructing and studying PTK7 with deletion of different domains, Hayes et al. found that the plasma membrane-tethered Ptk7 extracellular domain (Ptk7ΔICD) was sufficient to promote normal PCP as well as the inhibition of canonical Wnt signaling [2], while the intracellular domains may play a specific role in oriented cell division, radial intercalation and cilia orientation [23,24,25,26]. The intracellular PK domain of PTK7 may act as a scaffold promoting the binding of intracellular proteins and other receptors. In Xenopus, the ptk7 intracellular domain was stimulated by Wnt5A and induced its translocation into the nucleus, which promoted cell and tissue movements [27]. Additionally, the C-terminal PTK7 species up-regulated cadherin-11 expressed in the mesoderm-derived tissues, and which regulates osteogenesis. It is possible that PTK7 and Cadherin-11 might interact in embryogenesis and regulate similar developmental processes [28,29]. In our study, the frameshift variant and three missense variants (p.L17F, p.S118F, and p.K465R) were located in the extracellular domains of the PTK7 protein, while the other four missense variants (p.G627R, p.R652L, p.D764N, and p.R795H) were mapped to the intracellular portion, with p.D764N and p.R795H variants being located in the pseudokinase domain. In our expression assay, one variant in the extracellular domain and one variant in the intracellular domain were shown to alter the expression of PTK7, suggesting that both domains are critical for the integrity of PTK7 protein.
Interestingly, the frameshift variant that caused almost no expression of PTK7 was found in the CS patient, while the two missense variants that resulted in the significant decreases in protein levels were both found in patients with AIS. The patient carrying the truncating variant was diagnosed to have a mixed subtype of CS, with failure of formation (T12 butterfly vertebra) and segmental disorder (T9-T10) and no other developmental malformations. The two patients with the missense variants (p.S118F and p.D764N) did not have congenital developmental deformities of the vertebral body or other systems, presenting simple thoracolumbar scoliosis. However, due to the complex etiology of AIS, the role of PTK7 in AIS still warrants further validation and investigation.
Taken together, we hypothesize that the pathogenesis of vertebral deformities and the onset time of spinal curvature may be due to the effect of PTK7 variants on the protein expression. In other words, congenital and adolescent idiopathic scoliosis may have a common genetic basis. Although some missense variants in our study did not show abnormalities in protein expression or sub-cellular location, it is possible that the altered amino acids can affect the structure of the protein and subsequently affect downstream signaling pathways. Therefore, it is necessary to further explore the possible downstream pathways of PTK7, such as the canonical and non-canonical Wnt pathways.
5. Conclusions
In conclusion, we identified eight PTK7 variants in our mixed scoliosis cohort, including one frameshift variant (c.464_465delAC) and seven missense variants (c.49C>T, c.353C>T, c.1394A>G, c.1879G>A, c. 1955G>T, c.2290G>A, c.2384G>A). The frameshift variant resulted in a depleted expression of PTK7 protein, and two of missense variants caused reduced expression of PTK7. Our study extended the variant and phenotype spectrums of PTK7 and suggested a common genetic basis of CS and AIS.
Acknowledgments
We thank the patients and their families who participated in this research. We also thank the Nanjing Geneseq Technology Inc. for the technical help in sequencing, the Ekitech Technology Inc. for the technical support in database and data management. The members of the DISCO study group are provided in http://discostudy.org/.
Author Contributions
Conceptualization, Z.S., N.W. and S.Z.; methodology, Z.S., S.Z. and H.Z.; software, S.Z. and Y.N.; validation, Z.S., S.W., S.Z. and X.L.; formal analysis, Z.S. and Y.Y.; investigation, Z.S.; resources, S.W., Y.Y. and Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) Study Group; data curation, Z.S., S.Z. and H.Z.; writing—original draft preparation, Z.S., Y.Y.; writing—review and editing, S.W., S.Z., X.L. and N.W.; visualization, Z.S.; supervision, N.W., Z.W., G.Q. and T.J.Z.; project administration, N.W., Z.W., G.Q. and T.J.Z.; funding acquisition, N.W., Z.W., G.Q. and T.J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the National Natural Science Foundation of China (81902178 to S.W., 81972037 and 82172382 to J.Z., 81822030 and 82072391 to N.W., 81930068 and 81772299 to Z.W., 81772301 and 81972132 to G.Q.), Beijing Natural Science Foundation (No. L192015 to J.Z., JQ20032 to N.W., 7191007 to Z.W.), Capital’s Funds for Health Improvement and Research (2020-4-40114 to N.W.), Tsinghua University-Peking Union Medical College Hospital Initiative Scientific Research Program, Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2019PT320025), and the PUMC Youth Fund & the Fundamental Research Funds for the Central Universities (No. 3332019021 to Shengru W.).
Institutional Review Board Statement
Approval for the study was obtained from the ethics committee at Peking Union Medical College Hospital (JS-098).
Informed Consent Statement
Written informed consent was provided by each participant.
Data Availability Statement
Data are available upon reasonable request. The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mossie K., Jallal B., Alves F., Sures I., Plowman G.D., Ullrich A. Colon carcinoma kinase-4 defines a new subclass of the receptor tyrosine kinase family. Oncogene. 1995;11:2179–2184. [PubMed] [Google Scholar]
- 2.Hayes M., Naito M., Daulat A., Angers S., Ciruna B. Ptk7 promotes non-canonical Wnt/PCP-mediated morphogenesis and inhibits Wnt/beta-catenin-dependent cell fate decisions during vertebrate development. Development. 2013;140:1807–1818. doi: 10.1242/dev.090183. [DOI] [PubMed] [Google Scholar]
- 3.Lee H.K., Chauhan S.K., Kay E., Dana R. Flt-1 regulates vascular endothelial cell migration via a protein tyrosine kinase-7-dependent pathway. Blood. 2011;117:5762–5771. doi: 10.1182/blood-2010-09-306928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X., Borchers A.G., Jolicoeur C., Rayburn H., Baker J.C., Tessier-Lavigne M. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430:93–98. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
- 5.Wagner G., Peradziryi H., Wehner P., Borchers A. PlexinA1 interacts with PTK7 and is required for neural crest migration. Biochem. Biophys. Res. Commun. 2010;402:402–407. doi: 10.1016/j.bbrc.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 6.Capparuccia L., Tamagnone L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment--two sides of a coin. J. Cell Sci. 2009;122:1723–1736. doi: 10.1242/jcs.030197. [DOI] [PubMed] [Google Scholar]
- 7.Gay C.M., Zygmunt T., Torres-Vazquez J. Diverse functions for the semaphorin receptor PlexinD1 in development and disease. Dev. Biol. 2011;349:1–19. doi: 10.1016/j.ydbio.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neufeld G., Kessler O. The semaphorins: Versatile regulators of tumour progression and tumour angiogenesis. Nat. Rev. Cancer. 2008;8:632–645. doi: 10.1038/nrc2404. [DOI] [PubMed] [Google Scholar]
- 9.Moon R.T., Brown J.D., Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/S0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 10.Wodarz A., Nusse R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 11.Hayes M., Gao X., Yu L.X., Paria N., Henkelman R.M., Wise C.A., Ciruna B. ptk7 mutant zebrafish models of congenital and idiopathic scoliosis implicate dysregulated Wnt signalling in disease. Nat. Commun. 2014;5:4777. doi: 10.1038/ncomms5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S., Zhang Y., Chen W., Li W., Wang S., Wang L., Zhao Y., Lin M., Ye Y., Lin J., et al. Diagnostic yield and clinical impact of exome sequencing in early-onset scoliosis (EOS) J. Med. Genet. 2021;58:41–47. doi: 10.1136/jmedgenet-2019-106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen N., Zhao S., Jolly A., Wang L., Pan H., Yuan J., Chen S., Koch A., Ma C., Tian W., et al. Perturbations of genes essential for Mullerian duct and Wolffian duct development in Mayer-Rokitansky-Kuster-Hauser syndrome. Am. J. Hum. Genet. 2021;108:337–345. doi: 10.1016/j.ajhg.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 19.Lei Y., Kim S.E., Chen Z., Cao X., Zhu H., Yang W., Shaw G.M., Zheng Y., Zhang T., Wang H.Y., et al. Variants identified in PTK7 associated with neural tube defects. Mol. Genet. Genom. Med. 2019;7:e00584. doi: 10.1002/mgg3.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimes D.T., Boswell C.W., Morante N.F., Henkelman R.M., Burdine R.D., Ciruna B. Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science. 2016;352:1341–1344. doi: 10.1126/science.aaf6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paudyal A., Damrau C., Patterson V.L., Ermakov A., Formstone C., Lalanne Z., Wells S., Lu X., Norris D.P., Dean C.H., et al. The novel mouse mutant, chuzhoi, has disruption of Ptk7 protein and exhibits defects in neural tube, heart and lung development and abnormal planar cell polarity in the ear. BMC Dev. Biol. 2010;10:87. doi: 10.1186/1471-213X-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger H., Wodarz A., Borchers A. PTK7 Faces the Wnt in Development and Disease. Front. Cell Dev. Biol. 2017;5:31. doi: 10.3389/fcell.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Y., Mo C., Fraser S.E. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430:689–693. doi: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- 24.Roszko I., Sawada A., Solnica-Krezel L. Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Semin. Cell Dev. Biol. 2009;20:986–997. doi: 10.1016/j.semcdb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Happe H., de Heer E., Peters D.J. Polycystic kidney disease: The complexity of planar cell polarity and signaling during tissue regeneration and cyst formation. Biochim. Biophys. Acta. 2011;1812:1249–1255. doi: 10.1016/j.bbadis.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Jessen J.R., Topczewski J., Bingham S., Sepich D.S., Marlow F., Chandrasekhar A., Solnica-Krezel L. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golubkov V.S., Strongin A.Y. Downstream signaling and genome-wide regulatory effects of PTK7 pseudokinase and its proteolytic fragments in cancer cells. Cell Commun. Signal. 2014;12:15. doi: 10.1186/1478-811X-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehler A., Schlupf J., Schneider M., Kraft B., Winter C., Kashef J. Loss of Xenopus cadherin-11 leads to increased Wnt/beta-catenin signaling and up-regulation of target genes c-myc and cyclin D1 in neural crest. Dev. Biol. 2013;383:132–145. doi: 10.1016/j.ydbio.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Martinez S., Scerbo P., Giordano M., Daulat A.M., Lhoumeau A.C., Thome V., Kodjabachian L., Borg J.P. The PTK7 and ROR2 Protein Receptors Interact in the Vertebrate WNT/Planar Cell Polarity (PCP) Pathway. J. Biol. Chem. 2015;290:30562–30572. doi: 10.1074/jbc.M115.697615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request. The datasets analyzed during the current study are available from the corresponding author on reasonable request.