Abstract
Colorectal cancer (CRC) is one of the most common cancer types around the world. The prognosis of patients with advanced diseases is still poor in spite of currently available therapeutic options. Regorafenib is an oral tyrosine kinase inhibitor (TKI) approved to treat refractory metastatic colorectal cancer (mCRC). We investigated Somatic mutations in several genes involved in immunological response and cancer progression in both long/short responder mCRC patients who underwent third-line therapy with regorafenib to identify predictive biomarkers of response using Ion Torrent PGM sequencing and bioinformatic tools. We found Somatic mutations in TGFBR1, TGFBR2, and TGFBR3 genes in primary tumor and metastases samples of long-responder patients. Furthermore, our bioinformatic results show that they were mainly enriched in immune response, cell junction, and cell adhesion in long responder patients, particularly in primary tumor and metastatic sites. These data suggest that the TGF-b pattern could be the leading actor of a prolonged response to this drug.
Keywords: colorectal cancer, TGF-b, regorafenib
1. Introduction
Colorectal cancer (CRC) is the third leading cause of cancer death globally, and its incidence is steadily rising in developing nations [1,2,3]. In recent years, many targeted therapeutic strategies have been proposed for metastatic colorectal cancer (mCRC) patients [4]. Regorafenib is an oral type II multi-kinase inhibitor that inhibits the activity of vascular endothelial growth factor receptor 1, 2, 3 (VEGFR-1, -2, -3), platelet-derived growth factor receptors, fibroblast growth factor receptors (FGFR), tyrosine kinase receptor with immunoglobulin-like and EGF-like domains 2 (TIE-2), and oncogenic receptor tyrosine kinases [5], showing an impact on angiogenesis and metastasis processes. Observational post-marketing studies have confirmed its efficacy in the face of a non-negligible toxicity profile [6,7]. It has been approved in the third or later lines of treatment for patients with chemorefractory mCRC, according to the results of two randomized phases III trials (CORRECT and CONCUR) [8,9]. Unlike anti-EGFR monoclonal antibodies for which well-defined molecular predictive factors are available [10], results for anti-angiogenic drugs are still inconclusive [11]. However, since its introduction into the clinical setting, single exceptional responders to this drug have been reported [12,13,14,15]. Thus far, identifying useful predictive factors to identify the best candidates for such a therapy represents a relevant clinical challenge.
A more remarkable progression-free survival (PFS) benefit for regorafenib has been observed in patients showing epithelial-mesenchymal transition (EMT) phenotype and higher TGF-β pathway activation [16]. TGF-β pathway promotes angiogenesis and EMT and inhibits the growth of epithelial and immune cells [17,18]. Loss of SMAD4, a tumor suppressor gene, disrupts R-SMAD-SMAD4 complexes in the canonical TGF-β signaling, leading to the deregulation of several SMAD4-related target genes, such as VEGF-A, VEGF-C, and β-catenin [19]. A strict and mutual regulation between vasculature normalization and immune activation has been described in the tumor microenvironment [20]. Consequently, in this study, we evaluate somatic mutations of several genes involved in immunological response and cancer progression, with the aim to identify, through a NGS platform, potential biomarkers of response to regorafenib in one very long and short responder mCRC patient.
2. Materials and Methods
2.1. Patients’ Characteristics
The clinical history of the 58-year-old mCRC who presented a prolonged progression-free survival (PFS) (16 months) to third-line regorafenib has been previously published [21].
The control case was a 54-year-old female mCRC patient who received the third-line treatment with regorafenib with a PFS of 4 months. This study was approved by the Local Ethical Committee (Prot. N.709/CE). Both patients provided informed consent.
2.2. Sample Processing
All surgical samples were formalin-fixed and paraffin-embedded (FFPE). Tumor sections were cut from each FFPE block: one section was stained by hematoxylin/eosin to confirm and locate the tumor, and consecutive sections were used for immunohistochemistry and gene expression analyses.
2.2.1. DNA and RNA Extraction
Three to six FFPE tissue sections (6 μm thick) with adequate tumor cellularity, selected by a pathologist (>50%), were macro dissected and subjected to the QIAamp DNA FFPE Tissue Kit (Qiagen, Venlo, The Netherlands) for DNA isolation and the RNeasy FFPE Kit (Qiagen) for RNA isolation, according to the manufacturer’s protocols. DNA was also isolated from blood samples using the QIAamp DNA Blood Midi Kit (Qiagen). DNA and RNA concentrations were measured using the Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.2.2. Ion Torrent PGM Sequencing
The sequencing which has been used in the current study has been reported in our previous study [22]. Briefly, two custom panels have been designed through the Ion Ampliseq designer tool, one including the coding region of 41 genes to detect Somatic mutations and one to study the gene expression of 95 genes. Both include genes involved in immune regulation and inflammation. Variant calling and filtering have been described in [22]. In particular, somatic variants were called when matching the following conditions: DP > 50, VD > 20, and QUAL > 30. Furthermore, the Cancer hotspot Panel V2 has been used to identify druggable alterations. Briefly, the call set has been generated merging results from the Somatic High-Stringency Variant Caller plugin of the Torrent Suite and the Vardict [23] algorithm. Germline variants were filtered out using a pool of healthy controls. Annovar [24] was used to annotate variants functionally. The Oncoprint plot has been designed with the ComplexHeatmap R package [25].
2.2.3. MSI Analysis
MSI Analysis was performed by using Real-Time PCR (Easy PGX Diatech) to detect the microsatellite region instability in tumor samples. The analysis of 8 mononucleotide markers (BAT-25, BAT-26, NR-21, NR-22, NR-24, NR-27, CAT-25, and MONO-27) was based on the denaturation profile and compared to a positive control with a stable profile and followed manufacture instructions (EasyPGX® ready MSI cat.no. RT033).
The EasyPGX® Analysis Software was used to analyze all the melting temperatures and to generate a melting profile from each sample and the positive control. The software automatically calculates the status of global instability (MSS, MSI-L, MSI-H) from the number of unstable markers.
A tumor with high instability (H-MSI) has ≥2 unstable markers, and a tumor with L-MSI (low instability) has 1 unstable marker. For all L-MSI tumors, it is needed to repeat the analysis comparing the melting profile of the tumor tissue to the melting profile of normal tissue to exclude possible germline instability.
2.3. Protein-Protein Interaction (PPI) Network and Pathway Enrichment Analysis
Metascape (Available online: https://metascape.org/gp/index.html#/main/step1 (accessed on 10 July 2020)), an online resource, has been used to depict biological networks, including the interaction between mutated genes and to perform functional enrichments.
3. Results
3.1. Mutational Pattern
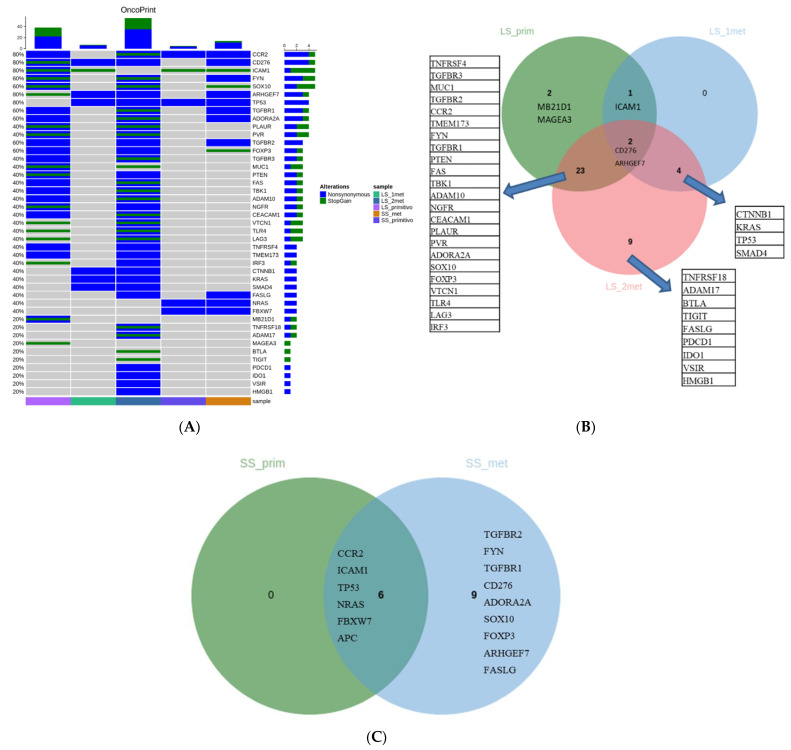
Through a custom targeted NGS panel including 41 genes, long-responder and short-responder samples were sequenced. In detail, long-responder samples included a primary tumor and two ovarian metachronous metastases. The control case included a primary tumor and one lung metastasis. The primary tumor and the first metastasis showed a distinct pattern of alterations (only CD276, ICAM1, and ARHGEF7 mutations are common) (Figure 1A). The second metastasis shared almost all mutations detected in the primary tumor: four alterations were detected in the first metastasis with 9 private mutations (Figure 1B). The mutational pattern of the short-responder in the primary and metastatic samples reflects a simple mechanism of clonal evolution (Figure 1C). Indeed, we observed that metastasis has the same alterations as the primary tumor samples with a further 9 private mutations. Microsatellite status has been checked in all samples, which were found to be stable (MSS).
Figure 1.
(A) Oncoprint including pathogenic alterations detected in the five analyzed samples. (B) Venn diagram of the alterations detected in the long survival samples (primary CRC and the two metastatic samples). (C) Venn diagram of the alterations detected in the short survival samples (primary CR Cand metastatic samples). LS_prim: primary tumor of the long survival case; LS_1met, LS_2met: metastatic samples of the long survival case; SS_prim: primary tumor of the short survival case; SS_met: metastatic sample of the short survival patient.
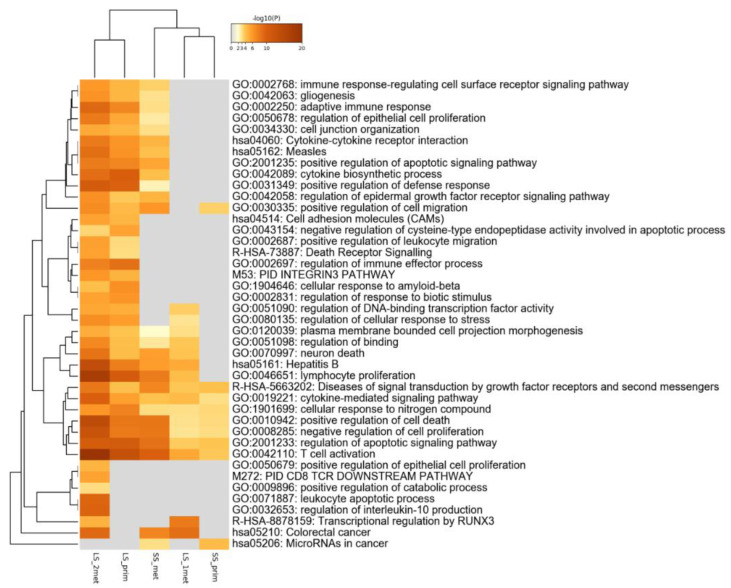
3.2. Functional Enrichment and PPI Network
The global pathway enrichment is displayed as a heatmap (Figure 2). Terms related to immune response, cell junction, and cell adhesion molecules are the most enriched in long-responder patients, particularly in the primary tumor and second metastasis samples.
Figure 2.
Global pathway enrichment of the detected alterations. LS_prim: primary tumor of the long survival case; LS_1met, LS_2met: metastatic samples of the long survival case; SS_prim: primary tumor of the short survival case; SS_met: metastatic sample of the short survival patient.
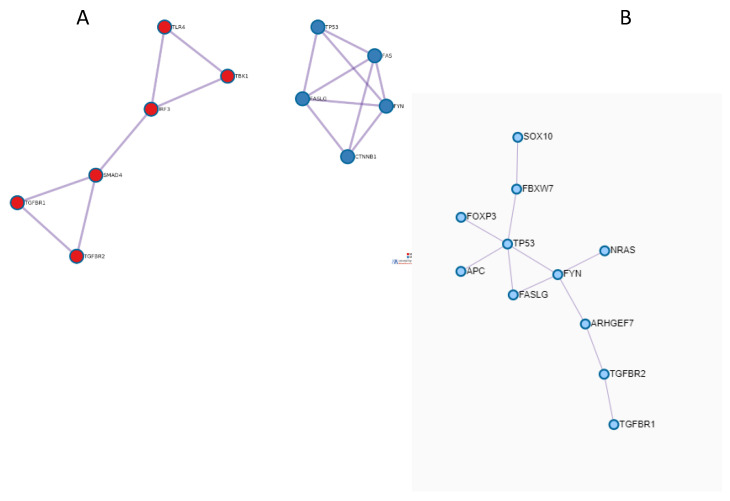
The PPI network, including proteins with relative genes which were found to be altered in the second metastatic samples, was built up. It can be observed that two sub-networks were identified, which were, in turn, functionally enriched (Figure 3A).
Figure 3.
Interaction networks of the alterations detected in (A) long survival samples and (B) in short survival samples. In Table 1, significantly enriched terms are shown according to the sub-network color scale. PPI network was built up also for the short responder case (Figure 3B) and functionally enriched (Table 1).
4. Discussion
Despite improvements in the management of mCRC, drug resistance remains a clinical challenge in the advanced stage. Regorafenib was the first approved multikinase inhibitor with survival benefits in unselected mCRC patients who had exhausted current standard therapies [8,9]. Regorafenib inhibits the activity of several protein kinases active in the regulation of angiogenesis, oncogenesis, and in the modulation of the tumor microenvironment. Although several clinical and biological parameters have been investigated, there are no useful predictive markers for regorafenib treatment [26,27]. In this study, we explored Somatic mutations of genes involved in immunological and inflammation response in one very long-responder and one short-responder mCRC patient to regorafenib. Recently, an immune profile that correlates with the outcome in mCRC patients treated with regorafenib has been reported, suggesting a cytokine signature able to discriminate patients who might derive a benefit from regorafenib treatment [28]. In particular, the plasma basal level of proteins TNF-α and TGF-β before treatment might be useful to identify mCRC patients that do not benefit from regorafenib and show the progression of the disease. According to these results, our data show most mutated genes involved in TGF-β signaling in long-responder mCRC patients to regorafenib therapy.
In particular, Somatic mutations in TGFBR1, TGFBR2, and TGFBR3 genes were found in the primary tumor and metastatic samples of our long-responder patient. TGF- β was identified as a major signaling pathway in CRC invasion and metastasis. Its activation generally promotes CRC invasion and metastasis through EMT, whereas it suppresses cancer immunity in the tumor microenvironment [29]. The relevance of TGF-β signaling in the acquisition of an invasive phenotype was also demonstrated by our PPI subnetwork obtained by protein products of altered genes found in the last metastasis of the long-responder mCRC patient. This subnetwork was enriched by terms related to “Signaling by TGF-beta Receptor Complex in Cancer” and “TGF-beta receptor signaling activates SMADs”. No similar terms were observed by the enrichment of the PPI network highlighted by protein products of altered genes found in metastases of the short-responder.
Interestingly, in a study, researchers showed a deleterious mutation in the SMAD4 gene in the long-responder metastatic patient [30]. Martinelli et al. [16] reported a greater PFS benefit for regorafenib therapy in patients with SMAD4 gene mutation characterized by the activation of TGFβ signaling and upregulation of an EMT pathway [31]. Authors observed mutation in SMAD4 in two long-responder patients, suggesting a key role of this gene in regorafenib response [16]. Down-regulation or mutation of SMAD4 underlies a more rapid protein degradation, leading to pancreatic cancer cell cycle arrest and apoptosis [32]. A key role of this member of TGFβ signaling has also been reported in CRC cells, in which deletion of SMAD4 decreased the number of TAMs in the tumor microenvironment, contributing to unfavorable prognoses [19].
Regorafenib appears to participate in the immune system with tumor interaction in different ways, including inhibition of tyrosine kinase receptor CSF1R, which is involved in macrophage proliferation [33]. Recently, a strict and mutual regulation between vasculature normalization and immune activation has been described in the tumor microenvironment [20]. In conclusion, we can hypothesize that the TGF-b pattern could be the leading actor of a longer response; however, all our results should be better explored in a larger cohort.
Table 1.
The significant genes enriched in the different pathways. In red are the enriched pathways for the network in Figure 3A; in blue is the sub-network in Figure 3B.
| Category | GO | Description | LogP |
|---|---|---|---|
| KEGG Pathway | hsa05161 | Hepatitis B | −10 |
| Reactome Gene Sets | R-HSA-3304349 | Loss of Function of SMAD2/3 in Cancer | −9.5 |
| Reactome Gene Sets | R-HSA-3304351 | Signaling by TGF-beta Receptor Complex in Cancer | −9.3 |
| GO Biological Processes | GO:0045351 | type I interferon biosynthetic process | −8.9 |
| Reactome Gene Sets | R-HSA-936964 | Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon | −8.2 |
| GO Biological Processes | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | −8 |
| GO Biological Processes | GO:0032727 | positive regulation of interferon-alpha production | −7.8 |
| Canonical Pathways | M185 | PID ALK1 PATHWAY | −7.7 |
| GO Biological Processes | GO:0003272 | endocardial cushion formation | −7.6 |
| GO Biological Processes | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | −7.5 |
| GO Biological Processes | GO:0032647 | regulation of interferon-alpha production | −7.5 |
| GO Biological Processes | GO:0032728 | positive regulation of interferon-beta production | −7.4 |
| GO Biological Processes | GO:0032607 | interferon-alpha production | −7.4 |
| Reactome Gene Sets | R-HSA-2173789 | TGF-beta receptor signaling activates SMADs | −7.4 |
| GO Biological Processes | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | −7.3 |
| GO Biological Processes | GO:0060317 | cardiac epithelial to mesenchymal transition | −7.2 |
| GO Biological Processes | GO:0003203 | endocardial cushion morphogenesis | −7.2 |
| GO Biological Processes | GO:2000826 | regulation of heart morphogenesis | −7 |
| GO Biological Processes | GO:0060412 | ventricular septum morphogenesis | −6.9 |
| GO Biological Processes | GO:0003197 | endocardial cushion development | −6.9 |
| GO Biological Processes | GO:1901216 | positive regulation of neuron death | −8.9 |
| GO Biological Processes | GO:2001233 | regulation of apoptotic signaling pathway | −8.9 |
| KEGG Pathway | hsa05162 | Measles | −8.4 |
| GO Biological Processes | GO:0097190 | apoptotic signaling pathway | −8 |
| Reactome Gene Sets | R-HSA-109581 | Apoptosis | −7.8 |
| Reactome Gene Sets | R-HSA-5357801 | Programmed Cell Death | −7.8 |
| KEGG Pathway | hsa05205 | Proteoglycans in cancer | −7.6 |
| GO Biological Processes | GO:0010942 | positive regulation of cell death | −7.6 |
| GO Biological Processes | GO:0043523 | regulation of neuron apoptotic process | −7.5 |
| GO Biological Processes | GO:2001234 | negative regulation of apoptotic signaling pathway | −7.4 |
| GO Biological Processes | GO:0051402 | neuron apoptotic process | −7.3 |
| GO Biological Processes | GO:0070266 | necroptotic process | −7.3 |
| GO Biological Processes | GO:0097300 | programmed necrotic cell death | −7.1 |
| GO Biological Processes | GO:0043525 | positive regulation of neuron apoptotic process | −6.9 |
| GO Biological Processes | GO:1901214 | regulation of neuron death | −6.9 |
| GO Biological Processes | GO:0070265 | necrotic cell death | −6.8 |
| GO Biological Processes | GO:0070997 | neuron death | −6.7 |
| KEGG Pathway | hsa01524 | Platinum drug resistance | −6.6 |
| KEGG Pathway | hsa05200 | Pathways in cancer | −6.5 |
| GO Biological Processes | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | −6.1 |
Author Contributions
S.D.S. and K.D., the first authors of the manuscript, designed the project, performed the experiment, analyzed the data, and wrote the initial version of the manuscript. R.F. and A.C. collected the samples and provided comments. B.P., G.M., R.L. and S.T. performed the surgical procedures. N.S. and A.A. interpreted the results, helped in data categorization, and critically reviewed the manuscript. O.B., the corresponding author of the manuscript, supervised the project and revised the main text of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was approved by the Ethics Committee of Cancer institute of Bari (Prot. n° 209/CE).
Informed Consent Statement
All participants signed written informed consent before the experiment.
Data Availability Statement
Not available.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rumpold H., Niedersüß-Beke D., Heiler C., Falch D., Wundsam H.V., Metz-Gercek S., Piringer G., Thaler J. Prediction of mortality in metastatic colorectal cancer in a real-life population: A multicenter explorative analysis. BMC Cancer. 2020;20:1–9. doi: 10.1186/s12885-020-07656-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derakhshani A., Hashemzadeh S., Asadzadeh Z., Shadbad M.A., Rasibonab F., Safarpour H., Jafarlou V., Solimando A.G., Racanelli V., Singh P.K., et al. Cytotoxic T-Lymphocyte Antigen-4 in Colorectal Cancer: Another Therapeutic Side of Capecitabine. Cancers. 2021;13:2141. doi: 10.3390/cancers13102414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asadzadeh Z., Mansoori B., Mohammadi A., Kazemi T., Mokhtarzadeh A., Shanehbandi D., Hemmat N., Derakhshani A., Brunetti O., Safaei S., et al. The combination effect of Prominin1 (CD133) suppression and Oxaliplatin treatment in colorectal cancer therapy. Biomed. Pharmacother. Biomed. Pharmacother. 2021;137:111364. doi: 10.1016/j.biopha.2021.111364. [DOI] [PubMed] [Google Scholar]
- 4.Xie Y.-H., Chen Y.-X., Fang J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020;5:1–30. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm S.M., Dumas J., Adnane L., Lynch M., Carter C.A., Schütz G., Thierauch K.H., Zopf D. Regorafenib (BAY 73-4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer. 2011;129:245–255. doi: 10.1002/ijc.25864. [DOI] [PubMed] [Google Scholar]
- 6.O’Connor J.M., Öhler L., Scheithauer W., Metges J.-P., Dourthe L.-M., de Groot J.W., Thaler J., Yeh K.-H., Lin J.-K., Falcone A. Real-world dosing of regorafenib in metastatic colorectal cancer (mCRC): Interim analysis from the prospective, observational CORRELATE study. Liver. 2017;260:52. doi: 10.1093/annonc/mdx263.024. [DOI] [Google Scholar]
- 7.Tougeron D., Desseigne F., Etienne P., Dourthe L., Mineur L., Paule B., Hollebecque A., Tresch E., Spaeth D., Michel P. REBECCA: A large cohort study of regorafenib (REG) in the real-life setting in patients (pts) previously treated for metastatic colorectal cancer (mCRC) Ann. Oncol. 2014;25:iv205. doi: 10.1093/annonc/mdu333.104. [DOI] [Google Scholar]
- 8.Grothey A., Van Cutsem E., Sobrero A., Siena S., Falcone A., Ychou M., Humblet Y., Bouché O., Mineur L., Barone C. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): An international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–312. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 9.Li J., Qin S., Xu R., Yau T.C., Ma B., Pan H., Xu J., Bai Y., Chi Y., Wang L. Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2015;16:619–629. doi: 10.1016/S1470-2045(15)70156-7. [DOI] [PubMed] [Google Scholar]
- 10.Silvestris N., Tommasi S., Santini D., Russo A., Simone G., Petriella D., Maiello E., Tonini G., Colucci G. KRAS mutations and sensitivity to anti-EGFR monoclonal antibodies in metastatic colorectal carcinoma: An open issue. Expert Opin. Biol. Ther. 2009;9:565–577. doi: 10.1517/14712590902870394. [DOI] [PubMed] [Google Scholar]
- 11.Hegde P.S., Wallin J.J., Mancao C. Seminars in Cancer Biology. Academic Press; Cambridge, MA, USA: 2018. Predictive markers of anti-VEGF and emerging role of angiogenesis inhibitors as immunotherapeutics; pp. 117–124. [DOI] [PubMed] [Google Scholar]
- 12.Rosati G., Del Gaudio N., Scarano E., Cifarelli R.A., Altucci L., Bilancia D. Unexpected and durable response with regorafenib in a metastatic colorectal cancer patient without KDR mutation: A case report. Medicine. 2018;97:e11178. doi: 10.1097/MD.0000000000011178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callebout E., Ribeiro S.M., Laurent S., De Man M., Ferdinande L., Claes K.B., Van der Meulen J., Geboes K.P. Long term response on Regorafenib in non-V600E BRAF mutated colon cancer: A case report. BMC Cancer. 2019;19:1–5. doi: 10.1186/s12885-019-5763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshino K., Manaka D., Kudo R., Kanai S., Mitsuoka E., Kanto S., Hamasu S., Konishi S., Nishitai R. Metastatic colorectal cancer responsive to regorafenib for 2 years: A case report. J. Med. Case Rep. 2017;11:1–5. doi: 10.1186/s13256-017-1366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberto M., Falcone R., Mazzuca F., Archibugi L., Castaldi N., Botticelli A., Osti M.F., Marchetti P. The role of stereotactic body radiation therapy in oligometastatic colorectal cancer: Clinical case report of a long-responder patient treated with regorafenib beyond progression. Medicine. 2017;96:e9023. doi: 10.1097/MD.0000000000009023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinelli E., Sforza V., Cardone C., Capasso A., Nappi A., Martini G., Napolitano S., Rachiglio A.M., Normanno N., Cappabianca S. Clinical outcome and molecular characterisation of chemorefractory metastatic colorectal cancer patients with long-term efficacy of regorafenib treatment. ESMO Open. 2017;2:e000177. doi: 10.1136/esmoopen-2017-000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang D., Sun W., Zhou Y., Li P., Chen F., Chen H., Xia D., Xu E., Lai M., Wu Y. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37:173–187. doi: 10.1007/s10555-017-9726-5. [DOI] [PubMed] [Google Scholar]
- 18.Derakhshani A., Silvestris N., Hemmat N., Asadzadeh Z., Abdoli Shadbad M., Nourbakhsh N.S., Mobasheri L., Vahedi P., Shahmirzaie M., Brunetti O. Targeting TGF-β-Mediated SMAD Signaling pathway via novel recombinant cytotoxin II: A potent protein from naja naja oxiana venom in Melanoma. Molecules. 2020;25:5148. doi: 10.3390/molecules25215148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D., Qiu X., Li J., Zheng S., Li L., Zhao H. TGF-β secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle. 2018;17:2766–2778. doi: 10.1080/15384101.2018.1556064. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Song Y., Fu Y., Xie Q., Zhu B., Wang J., Zhang B. Anti-angiogenic agents in combination with immune checkpoint inhibitors: A promising strategy for cancer treatment. Front. Immunol. 2020;11:1956. doi: 10.3389/fimmu.2020.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunetti O., Calabrese A., Palermo L., Solimando A.G., Argentiero A. Long-term survival of an advanced colorectal cancer patient treated with Regorafenib: Case report and literature review. Clin. Case Rep. 2019;7:2379–2383. doi: 10.1002/ccr3.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvestris N., Brunetti O., Pinto R., Petriella D., Argentiero A., Fucci L., Tommasi S., Danza K., De Summa S. Immunological mutational signature in adenosquamous cancer of pancreas: An exploratory study of potentially therapeutic targets. Expert Opin. Ther. Targets. 2018;22:453–461. doi: 10.1080/14728222.2018.1456530. [DOI] [PubMed] [Google Scholar]
- 23.Lai Z., Markovets A., Ahdesmaki M., Chapman B., Hofmann O., McEwen R., Johnson J., Dougherty B., Barrett J.C., Dry J.R. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinform. (Oxf. Engl. ) 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 26.Del Prete M., Giampieri R., Loupakis F., Prochilo T., Salvatore L., Faloppi L., Bianconi M., Bittoni A., Aprile G., Zaniboni A. Prognostic clinical factors in pretreated colorectal cancer patients receiving regorafenib: Implications for clinical management. Oncotarget. 2015;6:33982. doi: 10.18632/oncotarget.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabernero J., Lenz H.-J., Siena S., Sobrero A., Falcone A., Ychou M., Humblet Y., Bouché O., Mineur L., Barone C. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015;16:937–948. doi: 10.1016/S1470-2045(15)00138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci V., Granetto C., Falletta A., Paccagnella M., Abbona A., Fea E., Fabozzi T., Nigro C.L., Merlano M.C. Circulating cytokines and outcome in metastatic colorectal cancer patients treated with regorafenib. World J. Gastrointest. Oncol. 2020;12:301. doi: 10.4251/wjgo.v12.i3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itatani Y., Kawada K., Sakai Y. Transforming growth factor-β signaling pathway in colorectal cancer and its tumor microenvironment. Int. J. Mol. Sci. 2019;20:5822. doi: 10.3390/ijms20235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ullah I., Sun W., Tang L., Feng J. Roles of Smads family and alternative splicing variants of Smad4 in different cancers. J. Cancer. 2018;9:4018. doi: 10.7150/jca.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marisa L., de Reyniès A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.-C., Schiappa R., Guenot D., Ayadi M. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed S., Bradshaw A.-D., Gera S., Dewan M.Z., Xu R. The TGF-β/Smad4 signaling pathway in pancreatic carcinogenesis and its clinical significance. J. Clin. Med. 2017;6:5. doi: 10.3390/jcm6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grothey A., Prager G., Yoshino T. The mechanism of action of regorafenib in colorectal cancer: A guide for the community physician. Clin. Adv. Hematol. Oncol. USA. 2019;17:1–19. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not available.