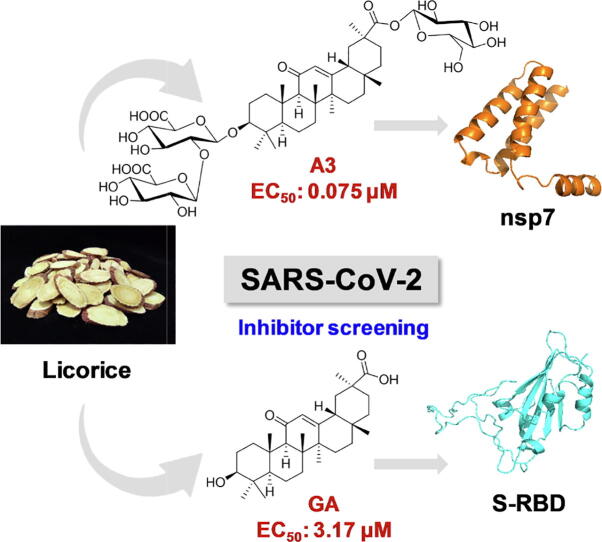
Graphical abstract
This work screened 125 compounds from the Chinese herbal medicine licorice. Licorice-saponin A3 and glycyrrhetinic acid potently inhibit SARS-CoV-2 infection, targeting the nsp7 protein and the spike protein receptor binding domain, respectively, with EC50 of 0.075 and 3.17 µM.
Keywords: COVID-19, SARS-CoV-2, Licorice, Licorice-saponin A3, Glycyrrhetinic acid
Highlights
-
•
Systemic approach to discover SARS-CoV-2 inhibitors from the herbal medicine licorice.
-
•
Licorice-saponin A3 and glycyrrhetinic acid inhibit SARS-CoV-2 by targeting nsp7 and the S-RBD, respectively.
-
•
Drug potential of these compounds was confirmed by cell models and PK studies.
-
•
The “multi-component, multi-target” mode of action was demonstrated for herbal medicines.
-
•
Licorice triterpenoids could be promising candidates for anti-SARS-CoV-2 drug development.
Abstract
Introduction
The COVID-19 global epidemic caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2) is a great public health emergency. Discovering antiviral drug candidates is urgent for the prevention and treatment of COVID-19.
Objectives
This work aims to discover natural SARS-CoV-2 inhibitors from the traditional Chinese herbal medicine licorice.
Methods
We screened 125 small molecules from Glycyrrhiza uralensis Fisch. (licorice, Gan-Cao) by virtual ligand screening targeting the receptor-binding domain (RBD) of SARS-CoV-2 spike protein. Potential hit compounds were further evaluated by ELISA, SPR, luciferase assay, antiviral assay and pharmacokinetic study.
Results
The triterpenoids licorice-saponin A3 (A3) and glycyrrhetinic acid (GA) could potently inhibit SARS-CoV-2 infection, with EC50 of 75 nM and 3.17 µM, respectively. Moreover, we reveal that A3 mainly targets the nsp7 protein, and GA binds to the spike protein RBD of SARS-CoV-2.
Conclusion
In this work, we found GA and A3 from licorice potently inhibit SARS-CoV-2 infection by affecting entry and replication of the virus. Our findings indicate that these triterpenoids may contribute to the clinical efficacy of licorice for COVID-19 and could be promising candidates for antiviral drug development.
Introduction
Coronaviruses (CoVs) represent the largest RNA viruses identified thus far, and are a group of enveloped viruses with a positive single-stranded RNA genome [1], [2], [3]. The COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus (SARS-CoV-2) has been a great threat to global public health. Specifically, SARS-CoV-2 Lambda and Delta variants exhibit higher infectivity and immune resistance [4], [5] and Delta variant possessing highly transmissible contains mutations that confer partial immune escape [5]. To develop clinically available SARS-CoV-2 inhibitors, a number of repurposed chemical or biological drugs have been evaluated [6], [7]. Unfortunately, recent clinical trials organized by the World Health Organization indicated that several promising drugs including remdesivir, hydroxychloroquine, and lopinavir had little effect on hospitalized patients with COVID-19 [8]. Therefore, the development of antiviral drugs is still a great demand.
In China, traditional Chinese medicine (TCM) treatment has been playing an important role in the effective control of COVID-19 [9], [10], [11], [12]. Glycyrrhiza uralensis Fisch. (licorice, Gan-Cao) is the most frequently used Chinese herbal medicines (top 1 among 179 herbs) for the treatment of COVID-19, and appears in 84% of 166 clinical herbal formulas [13]. Its major compound glycyrrhizic acid have been reported to possess antiviral activities against H1N1 influenza and SARS-CoV viruses [14], [15], [16]. However, little is known on the inhibitory activities of licorice compounds on SARS-CoV-2.
The entry step of the SARS-CoV-2 viral particles — encompassing attachment to the host cell membrane and fusion — are mediated by the spike protein [17], [18], [19]. The RBD is a key functional component within the S protein that is responsible for binding of SARS-CoV-2 with ACE2 [20]. The complex crystal structure of the SARS-CoV-2 spike protein receptor-binding domain (RBD) bound to its receptor ACE2 (angiotensin-converting enzyme 2) has been resolved [20], [21], [22]. Meanwhile, it has been reported that certain peptides such as dalbavacin [23] and human intestinal defensin 5 [24] could directly bind to ACE2 with high affinity (KD 147 nM and 76.2 nM, respectively) to block its interaction with the SARS-CoV-2 spike protein. However, very few small molecules have been reported to target the RBD, thus far.
Natural and biosynthetic SARS-CoV-2 inhibitors against 3CLpro and PLpro have been reported in recent years [25]. This work aims to discover small-molecule inhibitors targeting SARS-CoV-2 spike protein RBD from licorice (Fig. 1A). Here we report two licorice triterpenoids, namely licorice-saponin A3 (A3) and glycyrrhetinic acid (GA), which could potently inhibit SARS-CoV-2 with EC50 values of 75 nM and 3.17 μM, respectively. This finding reveals that triterpenoids from licorice could be promising drug candidates for COVID-19.
Fig. 1.
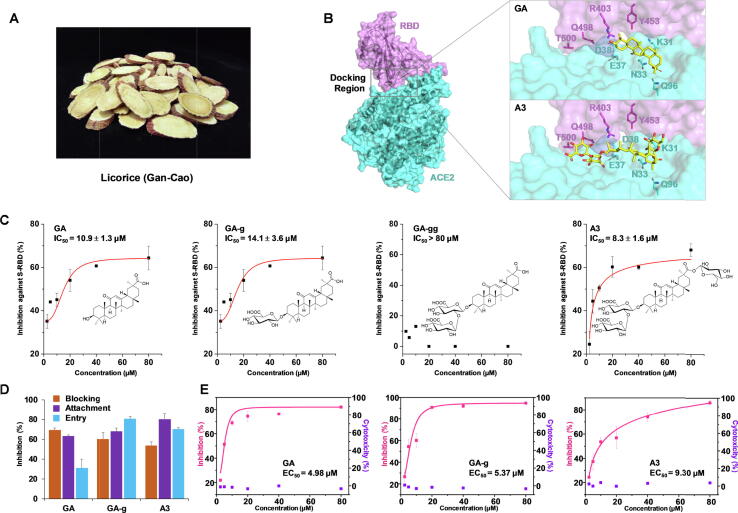
Screening of licorice compounds by molecular docking, ELISA, and pseudovirus infection assay. (A) Picture of the Chinese herbal medicine licorice (Gan-Cao). (B) The active pocket of SARS-CoV-2 spike protein RBD and ACE2, showing the binding of glycyrrhetinic acid (GA) and licorice-saponin A3 (A3) with surrounding amino acid residues. (C) Inhibitory activities of GA, GA-g, GA-gg, and A3 against SARS-CoV-2 spike protein by ELISA, n ≥ 3. Spike protein (0.5 µg/mL) was coated onto 96-well microplates. After incubation with test drugs (1 h), ACE2 (0.5 µg/mL) was added and incubated with the spike protein for 1 h. Structures of the test compounds are shown. (D) Inhibitory activities of GA, GA-g, and A3 at 10 µM at different steps of pseudovirus infection in Vero E6 cells, n ≥ 3. Luminescence was captured to display the viral infection and the inhibition efficiency. (E) Inhibition of pseudovirus infection of GA, GA-g, and A3 in the blocking assay. Cytotoxicities of the drugs against the Vero E6 cells were tested by CCK-8 assay. For each group, n ≥ 3.
Materials and methods
Virtual screening
Density functional theory (DFT) approach was used to optimize small molecules [26], [27], [28], [29]. All the structures were simulated by B3LYP/6-311G(d) method in the gas phase. All calculations for these small molecules were performed by utilizing the Gaussian09 suite of codes [30] and GaussView. Structures of the RBD (PDB ID: 6M0J) and nsp7 (PDB ID: 7JIT) were applied for protein-ligand calculations. Molecular Docking was performed by AutoDock Vina and AutoDocktools v1.5.6 softwares [31].
Cell strains, virus, and reagents
Vero E6 cells (ATCC) and Caco-2 cells were cultured in high glucose DMEM (GIBCO) supplemented with 10% FBS (GIBCO), penicillin (100 IU/mL), and streptomycin (100 μg/mL) in a 5% CO2 incubator at 37 °C, and were passaged every 2-3 days. Luciferase Assay System (E1500) was purchased from Promega (USA), and CCK8 kit (CK04) was purchased from Dojindo (Japan) for cytotoxicity test. SARS-CoV-2 strain (nCoV-2019BetaCoV/Wuhan/WIV04/2019) was propagated in Vero E6 or Caco-2 cells and titrated by standard plaque assay following the standard procedure. All experiments related to authentic SARS-CoV-2 were conducted at the Zhengdian Biosafety Level 3 (BSL3) facility of Wuhan Institute of Virology. S-trimer protein, ACE2 and 3-chymotrypsin-like protease (3CLpro) were from Novoprotein Technology Co., Ltd. RBD, RBD mutant (Y453F), RBD (L452Q, F490S), RBD (L452R), RBD (T478K) and nsp12 proteins were from Sino Biological Inc. All the drugs used in this study were from the compound library of the authors’ laboratory.
Protein expression and purification of nsp7 and nsp8
The cDNA encoding residues 11843-12091 (nsp7) and 12092-12685 (nsp8) of ORF1ab inserted into pET-28b vector was synthesized. Then it was expressed in E. coli strain BL21 (DE3) as soluble proteins at 37 °C for 10 h, until OD600 reached 0.5–0.7. Then, isopropyl-β-D-thiogalactopyranoside was added (final concentration was 0.1 mM). Express the protein at 18 °C for 18–20 h. Then harvest the cells at 7,000 rpm, 3 min, 4 °C. Lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole) was used for resuspension, and sonication in an ice bath was applied to disrupt cells. Finally, the cell debris was removed by centrifugation at 9,000 rpm for 30 min at 4 °C. The supernatant was collected and Ni-NTA affinity chromatography (Transgen, China) was used for protein purification. His-tag of the protein was digested and cut by thrombin. Then Ni-NTA was used to purify the protein for the second time. After ultrafiltration centrifugation, solvent of the protein solution was changed to the molecular sieve buffer (20 mM Tris-HCl, 100 mM NaCl, pH 7.5). A Superdex column (SuperdexTM 200 Increase 10/300 GL) was used for further purification, eluted with a buffer (20 mM Tris-HCl, 100 mM NaCl, pH 7.5) at a flow rate of 0.5 mL/min.
ELISA
Briefly, the spike protein (0.5 μg/mL) was diluted with coating solution (Na2CO3, 1.59 mg/mL; NaHCO3, 2.93 mg/mL; pH 9.6) and seeded on 96-well microplates at 4 °C overnight. After washing the microplates using the wash buffer (PBS buffer with 0.05% Tween-20 (v/v)) three times, 2% bovine serum albumin in the wash solution was used to block the plates for 40 min at 37 °C. The small molecules at a final concentration of 80, 40, 20, 10, 5, 2.5 μM were then added (one hour before ACE2) after washing the microplates three times. ACE2 (0.5 μg/mL) was added and incubated for another one hour. Then enzyme-linked antibody SA-HRP (1:15000) (Sangon Biotech Co., Ltd, Shanghai) was added after ACE2 was washed. Tetramethyl benzidine (TMB) was used as the chromogenic agent. Finally, 1 M HCl was used to stop the reaction and the enzyme activity was determined at OD450 nm.
Cell viability assay
Cell viability was measured by Cell Counting Kit-8 (CCK-8, Dojindo, Japan) assay. Firstly, Vero E6 cells were planted in 96-well plates. The density was 1 × 104 cells/well (in DMEM + 10 %FBS). Vero E6 cells were incubated under conditions of 5% CO2 at 37 °C for 24 h. Then, testing molecules (maximum concentration at 100 μM, less than1% DMSO) were diluted in DMEM culture medium and added to each well for another 24 h. The organic solvent in DMEM was less than 0.1%. 24 h after drug addition, the contents of the wells were substituted with fresh medium containing 10% CCK-8 solution and incubated at 37 °C for 3 h. The final optical density at OD450 was determined using Synergy Microplate reader.
Pseudovirus infection assay
SARS-CoV-2 pseudovirus preparation and infection assay were conducted according to published literature [32]. Vero E6 cells were planted in 96-well plates with 20,000 cells/well and then cells were infected with viral inocula of 650 TCID50/well. After 24 h, the cells were lysed, then the mixture was prepared by 20 µL of samples and 100 µL of Luciferase Assay Reagent (Promega, USA). Luminescence was measured with a microplate luminometer (PerkinElmer, USA).
Pseudovirus blocking, attachment, and entry assays
For the “blocking” assay, Vero E6 cells were planted at 96-well plates with 20,000 cells/well. 4 mg/mL breastmilk A17 and drugs at the following concentration 80, 40, 20, 10, 5, 2.5 μM were added into the wells. After 24 h, free drugs were washed away by PBS. The cells infected with viral inocula of 650 TCID50/well were inoculated with SARS-CoV-2 pseudovirus and incubated for other 24 h. Luminescence was measured to exhibit viral infection efficiency. For the “attachment” assay, the drugs were mixed with the pseudovirus at 4 °C for 1 h. The complex of drugs-pseudovirus was then added to the cells and incubate at 4 °C for 2 h for viral attachment to cells. PBS was used to wash away the free viruses and cells were then incubated at 37 °C for 24 h. For the “entry” assay, the drugs were mixed with the pseudovirus at 4 °C for 1 h. And Vero E6 cells were infected to the mixture at 37 °C for 1 h for viral entry into cells. PBS was used to wash away the free viruses and cells were then incubated at 37 °C for 24 h. All assays of blocking, attachment and entry assay were evaluated by a microplate luminometer (PerkinElmer, USA).
Pseudovirus-based neutralization assay
SARS-CoV-2-EGFP pseudovirus was provided by Prof. Ningshao Xia (Xiamen University, China) [33]. Vero E6 cells were planted in 96-well plates at 20,000 cells/well. The drugs at different concentrations and skimmed breastmilk A17 (4 mg/mL, the positive control) were respectively grouped with diluted VSV-SARS-CoV-2-EGFP virus (MOI = 0.5 PFU/ml) and cultivated at 37 °C for 1 h. The mixture was put into to seeded Vero E6 cells. After 36 h of incubation, fluorescence images were captured by an Echo Laboratories fluorescence microscope (Revolve, FL, USA).
Antiviral assay
Vero E6 or Caco-2 cells were planted in 48-well plates. The density was 1 × 104 cells/well. Cells were exposed to different concentrations of drugs for 8 h, and then inoculated with SARS-CoV-2 (nCoV-2019BetaCoV/Wuhan/WIV04/2019) at an MOI = 0.01. At 24 h post-infection, RNA from the cell supernatant was extracted by QIAamp viral RNA mini kit (52906, Qiagen) in accordance to the manufacturer’s instructions. qRT-PCR applying RT-PCR Kit (E3006, Invitrogen) was performed to quantify the viral RNA with primers ORF1ab-F (5′-CCCTGTGGGTTTTACACTTAA-3′) and ORF1ab-R (5′-ACGATTGTGCATCAGCTGA-3′) combined with the probe 5′-FAM246 CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′.
Immunofluorescence assay (IFA)
Vero E6 cells were planted on glass slides. The density was 1 × 105 cells/well. Then Vero E6 cells was infected with SARS-CoV-2 at an MOI = 0.01 in the presence of various concentrations of GA (0, 3, and 10 µM) or A3 (0, 0.11, and 0.33 µM). After 24 h post-infection, cells were exposed with 4% paraformaldehyde for 15 min at room temperature. Cells were incubated with 1:1000 diluted primary antibody against the spike protein of SARS-CoV-2 for 1 h at room temperature. After a thorough wash for 15 min to remove unbound antibodies, cells were mixed with 1:500 diluted FITC-conjugated goat anti-human IgG antibodies (Invitrogen, Thermo Fisher Scientific) for 1 h. The slides were washed for another 15 min, followed by nuclei staining with 4,6-diamidino-2-phenylindole (Sigma), then imaged by a confocal fluorescence microscope (UltraVIEW VoX; PerkinElmer, USA).
Surface plasmon resonance (SPR) assay
ACE2, RBD, the spike protein, nsp7, nsp8, and nsp12 were respectively immobilized to the sensor chip CM5 according to primary amine coupling reaction. Biacore T200 was used and running buffer was PBS containing 5% DMSO with 0.05% P20. The proteins were diluted with sodium acetate solution at 10 mM, pH 4.5 for ACE2 and nsp12, and pH 5.5 for RBD, spike protein, nsp7 and nsp8. The final concentration was 25 µg/mL and the final immobilized levels for ACE2, RBD, and spike protein were typically ∼ 10000, 10000, 13000, 8700, 10,000 and 6000 RU (response units), respectively. For binding tests, analytes were used at corresponding concentrations as running buffer at a flow rate of 30 µL/min with a contact time of 60 s and a dissociation time of 60 s. Data were analyzed by the Biacore evaluation software. The affinity constant KD values were calculated by the kinetics analysis or the steady state affinity method.
3CLpro enzymatic activity assay
The proteolytic activity of SARS-CoV-2 3CLpro was measured by fluorogenic substrate Dabcyl-KTSAVLQSGFRKME-Edans. The final reaction mixture contained 12.5 µg/mL purified enzyme, 50 µg/mL TCM extract or 8 µM compound sample, and 3 mM substrate. 20 mM TRIS-HCl buffer (pH 7.0) was applied in reaction system. Finally, reactions were detected with continuous monitoring of fluorescence at 25 °C for 10 min. The increase of the emission fluorescence at a wavelength of 535 nm upon excitation at 340 nm on FlexStation 3 Multi-Mode Microplate Reader was applied to determine the enzyme activity. The remaining enzymatic activity was calculated based on the following logistic derivative equation:
A = enzyme activity, df = dilution factor, ΔOD = absorption change/min, Vs = sample volume, C = concentration of 3CLpro.
Pharmacokinetic study
Male Sprague-Dawley rats (8–10 weeks, 180–220 g) were provided by the Experimental Animal Center of Peking University Health Science Center (Beijing, China). Lianhua Qingwen Capsules (1200 mg/kg), licorice extract (200 mg/kg), glycyrrhetinic acid (GA, 40 mg/kg) and glycyrrhizic acid (GA-gg, 40 mg/kg) were given by intragastric administration, and licorice-saponin A3 (A3, 20 mg/kg) was given by intravenous injection.
Blood samples were collected from rat orbit into heparinized tubes at 1, 4, 8, 10, 12, 24, and 48 h, respectively, and centrifuged at 15,000 rpm (4 °C) for 30 min to obtain the plasma. Then 100 μL of plasma sample, 200 μL of methanol and 100 μL IS solution (internal standard, 5 µg/mL oleanolic acid in methanol) were mixed. The mixture was vortexed and then centrifuged at 15,000 rpm for 30 min. The supernatant was separated, dried, and then dissolved in 100 μL of 50% methanol. The sample was centrifuged (15,000 rpm for 30 min), and then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). For pharmacokinetic analysis of the plasma samples, the regression equations for GA-gg, GA-g, GA, and A3 were y = 1.2 × 10-3x + 0.0175 (r = 0.9990), y = 6.27 × 10-3x + 0.0095 (r = 0.9979), y = 3.27 × 10-3x + 0.0319 (r = 0.9992), and y = 3.035 × 10-3x + 0.039 (r = 0.9997), respectively. In these equations, y represents the peak area, x the concentration (ng/mL), and r the correlation coefficient. Samples were separated on a Waters Xbridge C18 column (3.5 μm, 2.1 × 100 mm) (Waters, USA). The column temperature was 40 °C. The mobile phase consisted of water containing 0.1% formic acid (v/v, A) and acetonitrile containing 0.1% formic acid (v/v, B). The gradient elution program was as follows: 0–4 min, 15–100%; 4–5 min, 100% B; 5–6 min, 100–15% B; 6–10 min, 15% B. The flow rate was 0.3 mL/min. The injection volume was 2 μL. Mass spectrometry analysis was performed on an API 4000 QTRAP mass spectrometer equipped with a TurboIonSpray source (Foster City, USA). GA-gg, GA-g, GA, A3, and oleanolic acid (IS) were determined by multiple reaction monitoring (MRM) in the negative ion mode, with MRM transitions of m/z 921.3 → 821.4, m/z 645.5 → 469.4, m/z 469.4 → 425.3, m/z 983.5 → 821.3, and m/z 455.4 → 407.1, respectively. The instrument was controlled by Analyst 1.4.1 software.
The lung tissue samples were collected at 1, 2, 8, 10, 24 and 48 h, respectively. An aliquot of 100 mg sample in 200 μL deionized water was ground with grinding bead for 2 min, then centrifuged at 8,000 rpm (4 °C) for 15 min and the supernatant was obtained. Then 100 μL sample, 300 μL of pre-cooled methanol (-80 °C) and 100 μL IS solution (internal standard, 5 µg/mL oleanolic acid in methanol) were mixed. The mixture was vortexed and kept at −20 °C overnight, and centrifuged at 15,000 rpm (4 °C) for 15 min. The supernatant was separated, dried, and dissolved in 100 μL of 50% methanol. The sample was centrifuged (15,000 rpm for 30 min), and then analyzed by LC/MS/MS. The regression equation for GA was y = 0.000917x-0.00175 (r = 0.9990). Samples were separated on a Waters Acquity UPLC BEH C18 (1.7 μm × 50 mm) column at 25 °C. The mobile phase consisted of water containing 0.1% formic acid (v/v, A) and acetonitrile (v/v, B). The gradient elution program was as follows: 0–1 min, 70–90% B; 1–1.5 min, 90–100% B; 1.5–3 min, 100% B; 3–3.1 min, 100–70% B; 3.1–5 min, 70% B. The flow rate was 0.3 mL/min. The other parameters were the same as described above.
Ethics statement
All experiments involving animals were conducted according to the ethical policies and procedures approved by the ethics committee of the Faculty of Pharmacy, Cairo University, Egypt (Approval no. MI-2364).
Results
Virtual screening of 125 licorice compounds
Firstly, we screened 125 licorice compounds by molecular docking using AutoDock VINA (Supplementary Table S1). Structures of the natural products were optimized by density functional theory (DFT), and the RBD structure extracted from the RBD/ACE2 complex (PDB ID: 6M0J) was used for initial screening [20]. The location of residues R403, Q498, and T500 was determined as the binding site. Noticeably, majority of the compounds with high binding affinities were triterpenoid saponins (the glycosides of triterpenoids), together with the free triterpenoid glycyrrhetinic acid (GA). Then, we set the interactive area of RBD and ACE2 as the binding site to further screen 21 triterpenoids with high RBD affinities (<−7.0 kcal/mol or −29.2 kJ/mol) using the RBD/ACE2 complex structure (Fig. 1B, Supplementary Fig. S1; Table S2). Interestingly, GA, 3-O-β-D-glucuronosyl-glycyrrhetinic acid (GA-g), and licorice-saponin A3 (A3) showed remarkably higher affinities than glycyrrhizic acid (GA-gg), which had been reported as a SARS-CoV-2 inhibitor [34].
Inhibition of licorice triterpenoids against SARS-CoV-2 spike protein
Next, we determined inhibitory activities of the licorice compounds against SARS-CoV-2 spike protein by ELISA. At 10 µM, GA, GA-g, and A3 exhibited inhibition rates of 51.9%, 50.2% and 45.1%, respectively, which were remarkably higher than the other licorice compounds including GA-gg (Supplementary Fig. S2A). We also tested six non-licorice triterpenoids with similar structures to GA (Supplementary Fig. S2B). Majority of the triterpenoids showed noticeable inhibitory activities, with ursolic acid and betulinic acid as the most potent ones. The IC50 values for GA, GA-g, A3, betulinic acid, and ursolic acid were 10.9, 14.1, 8.3, 15.1, and 9.0 µM, respectively (Fig. 1C; Supplementary Fig. S2C).
Inhibition of licorice triterpenoids against SARS-CoV-2 pseudovirus
Then we tested the inhibitory activities of the above five hit compounds against the infection of SARS-CoV-2 pseudovirus on Vero E6 cells. Skimmed breastmilk A17 was used as the positive control [32]. Three assays, including “blocking”, “attachment”, and “entry”, were designed to investigate the inhibition mode. GA, GA-g, and A3 showed significant inhibitory activities in all the three procedures at 10 µM (Fig. 1D). In the “blocking” assay, GA, GA-g, and A3 exhibited remarkable anti-viral infection activities, with EC50 values of 4.98, 5.37, and 9.30 µM, respectively (Fig. 1E). Although ursolic acid and betulinic acid also showed noticeable effects, they exhibited remarkable cytotoxicity and were not further evaluated (Supplementary Fig. S3A-B). Consistent with the ELISA experiments, GA-gg showed weak activities in the pseudovirus system (Supplementary Fig. S3C-D). We also constructed a neutralization assay to visualize the anti-viral infections caused by GA, GA-g, and A3 in Vero E6 cells (Supplementary Fig. S4). GA and A3 inhibited SARS-CoV-2 pseudovirus with an inhibition rate of > 90% at 80 µM, which was similar to A17 at 4 mg/mL. Given the relatively weak activity of GA-g, we focused on GA and A3 for follow-up experiments.
Y453 is a key residue for the binding of GA and A3 with spike RBD
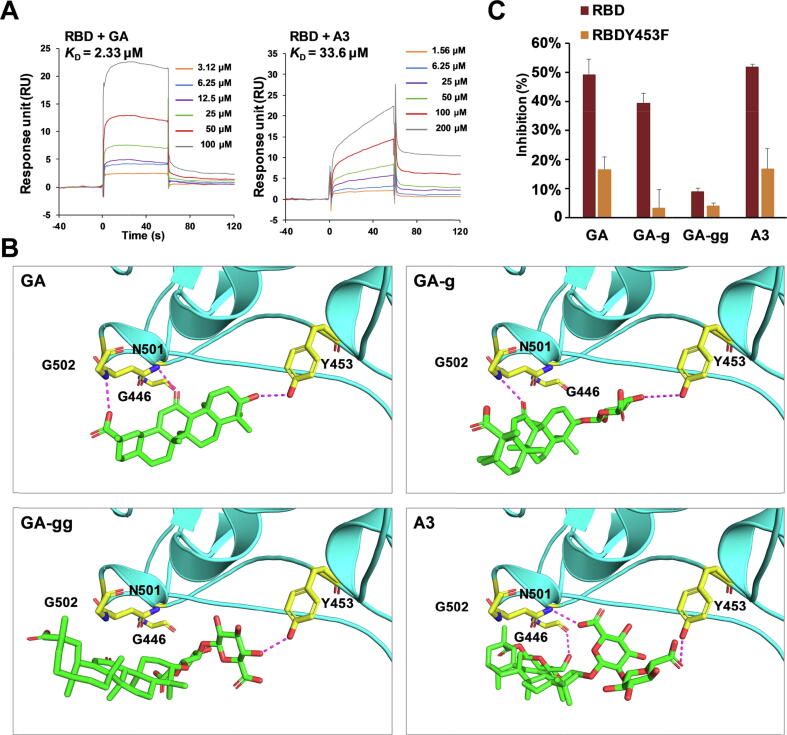
SPR (surface plasmon resonance) analysis indicated that GA and A3 could tightly bind to the RBD of SARS-CoV-2 spike protein, but GA-g and GA-gg only showed very weak bindings (Supplementary Fig. S5). The KD values of GA and A3 with RBD were 2.33 and 33.6 μM, respectively (Fig. 2A). Molecular docking analysis indicated that GA, GA-g, GA-gg, and A3 formed three, two, one, and three hydrogen bonds with surrounding amino acids, respectively (Fig. 2B). The 3-OH, 11-C O, and 30-COOH of GA formed hydrogen bonds with Y453, N501, and G502, respectively. Due to the existence of 3-O-glucuronyl residue, two hydrogen bonds could be formed for GA-g, i.e., 4′-OH/Y453, and 11-C O/G502. For GA-gg, the 3-OH is substituted with two glucuronyl groups, and 11-C O and 30-COOH are extruded from the binding pocket. Thus, only one hydrogen bond was formed, i.e., 4′′-OH and Y453. Interestingly, when the 30-COOH is attached with a glucosyl group, the structure of A3 exactly matches the binding pocket. The 6′′-COOH of 2′-O-glucuronyl, 6′-COOH of 3-O-glucuronyl, and 6′′′-OH of 30-glucosyl residue interact with Y453, N501, and G446, respectively. These hydrogen bond interactions determined the binding affinities, and were consistent with the structure-activity relationships of SPR analysis. We noticed that Y453 could bind to all the four triterpenoids. Therefore, we determined the inhibitory activities of the drugs at 10 μM by ELISA against the spike RBD and its Y453F mutant, respectively. All four triterpenoids showed remarkably decreased inhibitions, indicating that Y453 is a key residue for the binding of triterpenoids with spike RBD (Fig. 2C). Given the crucial role of L452, we also determined the inhibitory activities of GA and A3 against RBD L452Q/F490S, L452R and T478K mutants related to Lambda (C.37) and Delta (B.1.617.2) variants, respectively. Fortunately, both GA and A3 possessed inhibitory activities against these mutants although the inhibitions decreased by around 10% compared to RBD (Supplementary Fig. S6).
Fig. 2.
Structure-activity relationship for the binding of licorice triterpenoids with the spike RBD. (A) SPR analysis of GA and A3 binding to the RBD, respectively. The affinity constant KD values of GA and A3 with RBD were calculated by kinetics analysis. (B) Molecular docking of GA, GA-g, GA-gg, and A3 with RBD (PDB ID: 6M0J), showing the hydrogen bonds (magenta dashes). (C) Inhibitory activities of GA, GA-g, GA-gg and A3 (10 µM) against RBD and the Y453F mutant determined by ELISA, n ≥ 3. RBD and RBD-Y453F (0.5 µg/mL), ACE2 (0.5 µg/mL) were used.
Inhibition of GA and A3 against SARS-CoV-2 virus
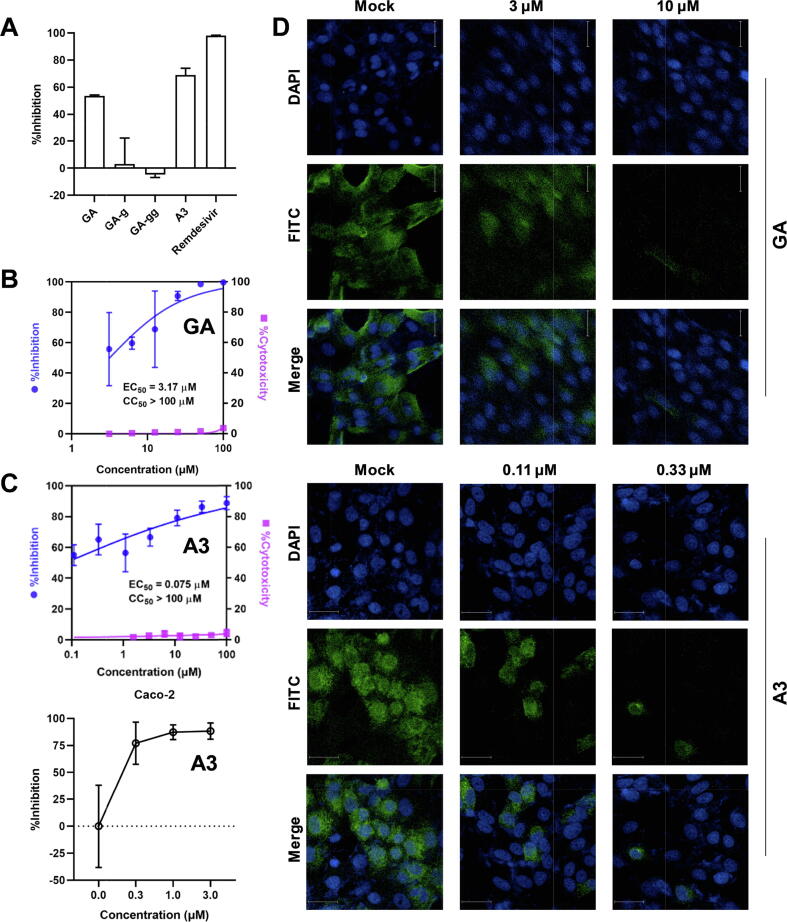
Subsequently, we tested inhibitory activities of the triterpenoids against SARS-CoV-2 virus in Vero E6 cells. Remdesivir was used as the positive control. Compared to GA-g and GA-gg, both GA and A3 showed significant inhibitory activities at 3 µM (Fig. 3A). The viral RNA level in cells decreased in a dose-dependent manner upon the treatment of GA with EC50 of 3.17 µM (Fig. 3B). Unexpectedly, A3 showed very potent inhibitory activity with EC50 of 75 nM (Fig. 3B). The inhibition could still be observed at 48 h post-infection (Supplementary Fig. S7). We further tested the antiviral activities of A3 in the human colon adenocarcinoma Caco-2 cells. A3 showed an inhibition rate of >75% at 0.3 μM (Fig. 3C). An indirect immunofluorescence assay (IFA) was conducted to verify the antiviral effects. Consistently, GA and A3 remarkably inhibited the SARS-CoV-2 virus in a dose-dependent manner (Fig. 3D), when compared with the drug-free mock cells.
Fig. 3.
Glycyrrhetinic acid (GA) and licorice-saponin A3 inhibit the infection of SARS-CoV-2 in vitro. (A) Antiviral activities of GA, GA-g, GA-gg, and A3 against SARS-CoV-2 in Vero E6 cells at 3 μM, n ≥ 3. Remdesivir (3 μM) was the positive control. (B) and (C) Dose-dependent inhibition of GA and A3 on SARS-COV-2 infection, n ≥ 3. Vero E6 cells and Caco-2 cells incubated with SARS-CoV-2 at an MOI = 0.01 were treated with increasing concentrations of GA and A3 for 24 h, respectively. qRT-PCR was applied to quantify the copy number of viral ORF1ab RNA in the culture medium. (D) IFA analysis of the inhibition of GA and A3 on SARS-CoV-2 replication. Infected cells were fixed at 24 h post-infection after treatment with 3 µM and 10 µM of GA and 0.11 µM and 0.33 µM of A3, respectively, and were subjected to IFA using the primary antibody against viral spike protein.
Nsp7 may be the target for A3
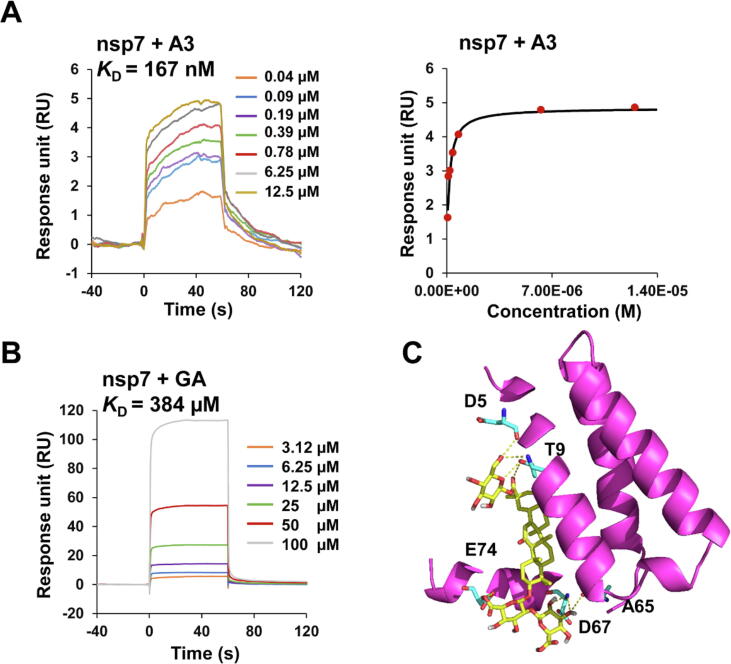
Given its nanomolar level inhibitory activity against SARS-CoV-2, A3 should have other targets than the spike protein [35]. Thus, we tested the activities of A3 against 3CLpro, the other important target for SARS-CoV-2 infection [36]. A3 only showed an inhibition of 14.37% at 10 µM, indicating 3CLpro was not a target for A3. RNA-dependent RNA polymerase (RdRp) is also a critical target for SARS-CoV-2. The nonstructural proteins nsp12, nsp7, and nsp8 are essential components of RdRp. Nsp12 is the catalytic subunit, and nsp7 and nsp8 are cofactors [37]. While we failed to establish the RdRp assay, we determined the binding of A3 and GA with nsp12, nsp7, and nsp8, respectively. The SPR results showed that A3 could bind with nsp7 much more tightly than with the other two proteins, with KD values of 167 nM, 15.5 μM, and 67.5 μM for nsp7, nsp8, and nsp12, respectively (Fig. 4A; Supplementary Fig. S8). In contrast, GA only showed very weak binding affinities, with KD values of 384, 91.2, and 416 μM, respectively (Fig. 4B; Supplementary Fig. S8). Molecular docking of A3 with nsp7 extracted from the nsp7-nsp8 complex crystal structure (PDB ID:7JIT) demonstrated seven hydrogen bonds, and the binding energy was −8.7 kcal/mol (Fig. 4C). These data indicated that nsp7 may be the target for A3.
Fig. 4.
Binding of A3 with nsp7. (A) and (B) SPR analysis of A3 and GA binding to nsp7, respectively. The affinity constant K D values of A3 and GA with nsp7 were calculated by the steady state affinity method and kinetics analysis, respectively. (C) Molecular docking of A3 with nsp7 (PDB ID:7JIT). Seven hydrogen bonds (yellow dashes) were formed by A3 and residues D5, T9, A65, D67 and E74.
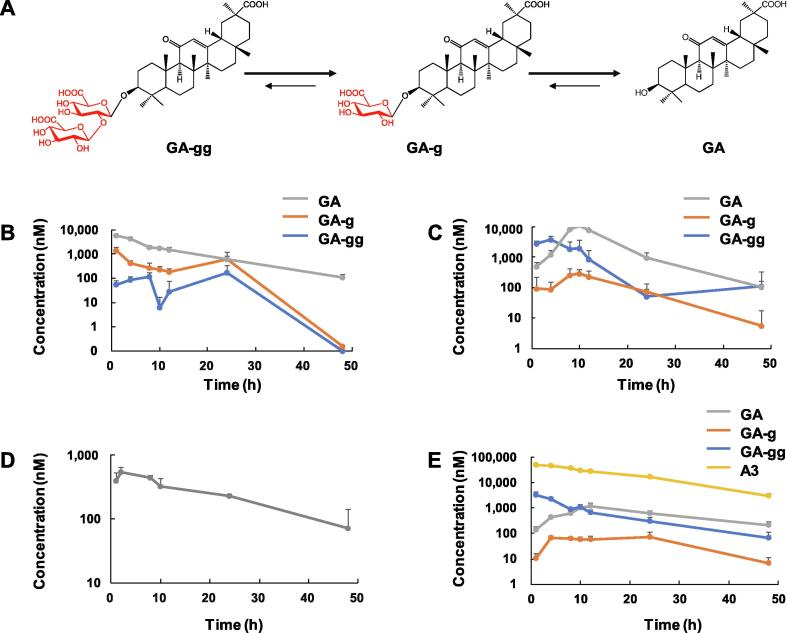
Pharmacokinetics of GA and A3 in rats
The rats maximum plasma concentration was 5.68, 10.23, 7.70, and 1.28 µM after oral administration (i.g.) of GA (40 mg/kg), GA-gg (40 mg/kg), licorice extract (200 mg/kg), and Lianhuaqingwen Capsules (1,200 mg/kg, a 13-component TCM patent drug containing licorice popularly used for clinical treatment of COVID-19), respectively (Fig. 5A-C; Supplementary Fig. S9). These results indicated both GA and GA-gg could be absorbed into circulation, and that they could be inter-converted. Although the concentration of GA in licorice or Lianhuaqingwen is very low, the abundant GA-gg could be readily metabolized into GA, which is active for SARS-CoV-2. More importantly, the elimination of GA in rats was slow, and the concentration remained at high levels between 8 and 24 h. GA could also be detected in the lung tissue, which is a key target organ for COVID-19 (Fig. 5D).
Fig. 5.
Metabolism and pharmacokinetics of licorice triterpenoids after oral administration in rats. (A) The metabolic pathway of GA and GA-gg. (B) and (C) Time-plasma concentration curves of GA-gg, GA-g, and GA in rats after oral administration of GA (B, 40 mg/kg, i.g., GA, AUCtotal = 0.026 h*g/L, t1/2 = 9.91 h), and GA-gg (C, 40 mg/kg, i.g., GA, Tmax = 8 h, AUCtotal = 0.046 h*g/L, t1/2 = 8.21 h). (D) Time-lung tissue concentration curves of GA in rats after oral administration of GA (40 mg/kg, i.g., Tmax = 2 h, AUCtotal = 0.006 h*g/L, t1/2 = 18.17 h). For each group, n = 4. (E) Time-plasma concentration curves of GA-gg, GA-g, GA, and A3 in rats after intravenous injection of A3 (20 mg/kg, i.v., A3, AUCtotal = 0.99 h*g/L, t1/2 = 10.31 h).
Given that A3 may be metabolized into GA-gg by gastrointestinal microbiota, it was given to rats by intravenous injection (i.v., 20 mg/kg). A3 showed a first-order elimination and the elimination rate was slow, with the plasma concentration decreased from 48.9 µM at 1 h to 27.3 µM at 24 h (Fig. 5E). Only a small portion of A3 was metabolized into GA-gg, GA-g, and GA.
Discussion
Although traditional Chinese medicines have been widely used in China for the prevention and treatment of COVID-19, very few antiviral compounds have been discovered and investigated in-depth. Meanwhile, licorice is the most frequently used herb in TCM formulas against COVID-19 [13], and its bioactive compounds remain unknown.
Our study screened 125 compounds from licorice and showed that licorice-saponin A3 and glycyrrhetinic acid are potent inhibitory compounds against SARS-CoV-2 virus. Moreover, this work provides important experimental evidences for presenting the clinical efficacy of licorice and related TCM products.
To our knowledge, A3 (EC50 75 nM) possesses promising anti-SARS-CoV-2 activity, compared to 11a (0.53 µM), remdesivir (0.77 µM), calpeptin (72 nM), and MI-30 (0.54 µM in Vero E6 cells, and 1.1 nM in HPAEpiC cells) [6], [36], [38], [39]. Natural triterpenoids have been reported to be antiviral agents against a wide spectrum of viruses including HIV-1 and influenza A [35]. While the abundant licorice compound GA-gg shows weak antiviral activities [40] and may reduce ACE2 expression in the lung [16], it could be readily metabolized into the active form GA. This may be one mechanism for the clinical therapeutic effects of licorice and its compound formulas like Lianhuaqingwen Capsules. Given the significant antiviral activities and appropriate pharmacokinetic behaviors, both A3 and GA could be promising drug candidates for the treatment of SARS-CoV-2. On the other hand, GA-gg is a clinical drug, and its effects against COVID-19 as a pro-drug of GA deserve to be further evaluated in both animal and clinical studies.
Considering that GA, GA-g and A3 could all inhibit S-RBD, and that GA-gg could be metabolized into the S-RBD inhibitor GA, S-RBD could be a critical target for licorice to inhibit SARS-CoV-2. Since S-RBD is responsible for entry of the virus into the host cells, licorice may be effective for both treatment and prevention of COVID-19. Our pharmacokinetic data showed the highest plasma concentration of GA at 7.7 μM when 200 mg/kg licorice extract was administered. Compared to the EC50 (3.17 μM) of GA, it further supported the potential of licorice to prevent and treat COVID-19 after oral administration.
Interestingly, although the S-RBD inhibitory effects of A3 and GA are similar (IC50 8.3 vs 10.9 µM), their antiviral activities are remarkably different (EC50 0.075 vs 3.17 µM). One possible explanation is that A3 targets other proteins in SARS-CoV-2 besides S-RBD. Our results suggest that A3 could strongly bind to nsp7, an essential component of the popular SARS-CoV-2 target RdRp. Thus, licorice may inhibit SARS-CoV-2 infection by affecting both entry and replication of the virus. The results also demonstrate the “multi-component, multi-target” feature of herbal medicines.
Conclusion
In this work, we find that A3 and GA could potently inhibit SARS-CoV-2 by targeting nsp7 and the spike protein RBD, respectively. Although the potential of GA had been reported by molecular docking calculations and protein inhibition assay, the anti-SARS-CoV-2 activities of GA and A3 were reported in this work for the first time. It is interesting that A3 and GA, which have the same triterpenoid skeleton, have different targets. This “multi-components, multi-targets” mode of action is a unique feature for herbal medicines and contributes to the overall clinical effects of licorice in the treatment of COVID-19.
Compliance with Ethics requirements
The animal facilities and protocols were approved by the Animal Care and Use Committee of Peking University Health Science Center (SYXK 2016-0041). All animal care and experimental procedures in this work were in accordance with Guide for the Care and Use of Laboratory Animals (National Institutes of Health).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81891010/81891011, 81725023, 82173950, 82003614, 81802002, 81873579, 31770192, and 32070187), the National Key Research and Development Program of China (No. 2017YFC1700405), the Science & Technology Department of Xinjiang Uygur Autonomous Region (2018AB012), and the Youth Innovation Promotion Association CAS. The authors thank Dr. Jing Wang and Dr. Qian Wang at State Key Laboratory of Natural and Biomimetic Drugs of Peking University and Dr. Lijun Zhong at Peking University Medical and Health Analysis Center for technical help. We thank Yue Chai, Huifei Su, Lulu Xu, and Dilaram Nijat of the Ye group and Qingyu Yao and Pingyao Luo of Prof. Tianyan Zhou’s group for technical assistance and discussions, the Wrocƚaw Centre for Networking and Super-computing for providing computer time, and Tao Du, Jin Xiong, Lun Wang and all the running team from Zhengdian Biosafety Level 3 Laboratory of Wuhan Institute of Virology for their critical support.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.11.012.
Contributor Information
Kuanhui Xiang, Email: kxiang@bjmu.edu.cn.
Hang Yang, Email: yangh@wh.iov.cn.
Min Ye, Email: yemin@bjmu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Cavanagh D. In: Coronaviruses with special emphasis on first insights concerning SARS. Schmidt A., Weber O., Wolff M.H., editors. Birkhauser Basel; Basel: 2005. Coronaviridae: a review of coronaviruses and toroviruses; pp. 1–54. [Google Scholar]
- 2.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura I, Kosugi1 Y, Wu J. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. bioRxiv preprint 2021. 10.1101/2021.07.28.454085. [DOI] [PMC free article] [PubMed]
- 5.Riemersma KK, Grogan, BE, Kita-Yarbro A, Shedding of Infectious SARS-CoV-2 Despite Vaccination. medRxiv preprint 2021. 10.1101/2021.07.31.21261387. [DOI] [PMC free article] [PubMed]
- 6.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young B., Tan T.T., Leo Y.S. The place for remdesivir in COVID-19 treatment. Lancet Infect Dis. 2021;21:20–21. doi: 10.1016/S1473-3099(20)30911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan H., Peto R., Karim Q.A., et al. Repurposed antiviral drugs for COVID-19–interim WHO SOLIDARITY trial results. New Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xian Y., Zhang J., Bian Z., et al. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm Sin B. 2020;10:1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C., Pan X., Xu X., et al. Salvianolic acid C potently inhibits SARS-CoV-2 infection by blocking the formation of six-helix bundle core of spike protein. Sig Trans Targ Therap. 2020;5:220. doi: 10.1038/s41392-020-00325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Runfeng L.i., Yunlong H., Jicheng H., Weiqi P., Qinhai M.a., Yongxia S., et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Wu Y., Chen C., et al. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm Sin B. 2021;11:222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo L., Jiang J., Wang C., et al. Analysis on herbal medicines utilized for treatment of COVID-19. Acta Pharm Sin B. 2020;10:1192–1204. doi: 10.1016/j.apsb.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji S., Li Z., Song W., et al. Bioactive constituents of Glycyrrhiza uralensis (Licorice): discovery of the effective components of a traditional herbal medicine. J Nat Prod. 2016;79:281–292. doi: 10.1021/acs.jnatprod.5b00877. [DOI] [PubMed] [Google Scholar]
- 15.Wu C., Liu Y., Yang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harald M. Symptomatic protective action of glycyrrhizin (Licorice) in COVID-19 Infection? Front Immunol. 2020 doi: 10.3389/fimmu.2020.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Mol Cell Biol. 2021 doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 21.Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370(6513):203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurini E., Marson D., Aulic S., Fermeglia M., Pricl S. Computational alanine scanning and structural analysis of the SAR-S-CoV-2 spike protein/Angiotensin-Converting Enzyme 2 complex. ACS Nano. 2020;14(9):11821–11830. doi: 10.1021/acsnano.0c04674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G., Yang M.L., Duan Z.L., et al. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and its effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C., Wang S., Li D., Wei D.-Q., Zhao J., Wang J. Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology. 2020;159(3):1145–1147.e4. doi: 10.1053/j.gastro.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan F., Gao F. An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2. Comput Struct Biotechnol J. 2021;19:4868–4883. doi: 10.1016/j.csbj.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98(7):5648–5652. [Google Scholar]
- 27.Lee C., Yang W., Parr R.G., et al. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1998;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 28.Miehlich B., Savin A., Stoll H., et al. Results obtained with the correlation-energy density functionals of Becke and Lee, Yang and Parr. Chem Phys Lett. 1989;157:200–206. [Google Scholar]
- 29.McLean A.D., Chandler G.S. Contracted Gaussian-basis sets for molecular calculations. J Chem Phys. 1980;72:5639–5648. [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, et al. Gaussian 09, Revision E.01, Inc., Wallingford CT; 2013.
- 31.Morris G.M., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan H., Hong B., Luo Y., et al. The effect of whey protein on viral infection and replication of SARS-CoV-2 and pangolin coronavirus in vitro. Sig Trans Targ Therap. 2020;5:275. doi: 10.1038/s41392-020-00408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong H.-L., Wu Y.-T., Cao J.-L., Yang R., Liu Y.-X., Ma J., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg Microbes Infect. 2020;9(1):2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S., Zhu Y., Xu J., Yao G., Zhang P., Wang M., et al. Glycyrrhizic acid exerts inhibitory activity against the spike protein of SARS-CoV-2. Phytomedicine. 2021;85:153364. doi: 10.1016/j.phymed.2020.153364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Sun J., Xiao S., et al. Triterpenoid-mediated inhibition of virus-host interaction: is now the time for discovering viral entry/release inhibitors from nature? J Med Chem. 2020;63:15371–15388. doi: 10.1021/acs.jmedchem.0c01348. [DOI] [PubMed] [Google Scholar]
- 36.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Günther S., Reinke P.Y.A., Fernández-García Y., Lieske J., Lane T.J., Ginn H.M., et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372(6542):642–646. doi: 10.1126/science:abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiao J., Li Y.-S., Zeng R., Liu F.-L., Luo R.-H., Huang C., et al. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371(6536):1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.