Abstract
The rapid identification of bacteria in blood cultures and other clinical specimens is important for patient management and antimicrobial therapy. We describe a rapid (<4 h) detection and identification system that uses universal PCR primers to amplify a variable region of bacterial 23S ribosomal DNA, followed by reverse hybridization of the products to a panel of oligonucleotides. This procedure was successful in discriminating a range of bacteria in pure cultures. When this procedure was applied directly to 158 unselected positive blood culture broths on the day when growth was detected, 125 (79.7%) were correctly identified, including 4 with mixed cultures. Nine (7.2%) yielded bacteria for which no oligonucleotide targets were present in the oligonucleotide panel, and 16 culture-positive broths (10.3%) produced no PCR product. In seven of the remaining eight broths, streptococci were identified but not subsequently grown, and one isolate of Staphylococcus aureus was misidentified as a coagulase-negative staphylococcus. The accuracy, range, and discriminatory power of the assay can be continually extended by adding further oligonucleotides to the panel without significantly increasing complexity or cost.
The isolation of bacteria from blood cultures (bacteremia) is usually indicative of a serious invasive infection requiring urgent antimicrobial therapy. Different organisms have different antimicrobial susceptibilities, and successful treatment is dependent on the prompt administration of the correct drug (6, 14, 23, 25, 26, 33). Blood culture broths usually become positive 8 to 24 h after inoculation. At this time, some indication of bacterial identity can be obtained by Gram staining, but definitive identification and antibiotic susceptibilities are usually not available until 24 to 48 h later. This delay has two consequences: first, the patient may suffer if ineffective therapy is given for antibiotic-resistant organisms, and second, antibiotic resistance may be encouraged if unnecessary antibiotics are given for sensitive organisms (3, 21). Although in our hospital nine bacterial groups (coagulase-negative staphylococci [CoNS], Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Enterococcus spp., Klebsiella spp., Enterobacter spp., Proteus spp., and Streptococcus pneumoniae) account for more than half of all clinically significant blood culture isolates, as many as 50 species may be involved, and there are usually few clinical clues as to the specific causative organism. Rapid species detection and identification would facilitate earlier effective therapy.
Rapid diagnosis can be achieved by the direct detection of characteristic bacterial genes in clinical specimens, and many primer sets have been developed to detect species-specific genes in simple PCRs (10, 18, 31). These systems are usually designed to confirm the diagnosis of specific clinical syndromes and include the identification of Burkholderia pseudomallei in melioidosis (9, 34), S. pneumoniae in pneumococcal pneumonia and meningitis (7, 22), Coxiella burnettii in Q fever (46), Listeria monocytogenes in listeriosis (5), Rhodococcus equi in rhodococcosis in horses (42), Mycobacterium tuberculosis in tuberculosis (12, 36), Salmonella enterica serovar Typhi in typhoid fever (38, 44), Mycoplasma pneumoniae in mycoplasmal pneumonia (30), Neisseria meningitidis in meningococcal meningitis (31), and Borrelia burgdorferi in Lyme disease (15). Similar test systems have been designed for yeasts (29). In most cases, the PCR has been performed on positive blood culture bottle samples, but some have been successful with direct blood samples, serum, buffy coat specimens, or negative blood culture bottle samples; these results suggest that DNA-based methods may be more sensitive than conventional bacteriology methods.
However, the use of different primers for different species is impractical for the routine analysis of blood cultures that may contain one or more of many possible pathogens. Either a complex PCR with a mixture of large numbers of primers is needed, or a large series of individual PCRs must be run in parallel or sequentially. Multiplex PCR may be effective for a limited number of organisms, but as more primers are added, the sensitivity decreases and the chance that two unrelated primers will produce spurious products increases. Multiple individual PCRs increase the expense and complexity of the assay and, if they are run sequentially, the processing time increases for less common or unexpected pathogens.
These problems can be avoided by using a single pair of universal primers designed to amplify conserved stretches of DNA from any bacterium present, followed by sequence analysis of the PCR product to determine the species. Previous investigators have usually chosen the 16S ribosomal DNA (rDNA) or the 16S-23S rDNA spacer region as a target for universal primers (35). The 16S rDNA is highly conserved, and sequences from it are now used in bacterial taxonomy (45). In contrast, the 16S-23S rDNA spacer region is highly variable within many species, frequently containing tRNA genes, and this variation has been used for typing clinical isolates (2, 18, 41). There have been a few reports of the use of 16S rDNA variation for the detection and identification of bacteria causing bacteremia or meningitis (8, 16) or for the differentiation of bacteremia from other causes of the sepsis syndrome (24). Detection of variation within fungal rDNA spacer regions by hybridization has been shown to be effective for the identification of yeast species in clinical specimens (43).
Recently, sequence data for the large subunit (23S rDNA) have become available for a few bacterial species. Analysis of these sequences (4, 17, 20, 27, 28, 37) suggests that this region shows more variation between species of medical importance than 16S rDNA; therefore, universal primers designed to amplify this region might be more useful for clinical diagnosis. These sequences can be analyzed by hybridizing the labeled PCR product to an array of oligonucleotides immobilized on a solid support (membrane or glass slides) or synthesized in situ on silicon wafers (32). As both the target and the probe are present at much higher concentrations than is typical for Southern blots, these hybridization reactions can be carried out in very short periods of time (less than 1 h). This method is often referred to as reverse hybridization because the probes are immobilized and the target is in solution.
Thus, organisms of bacteremia could be identified directly from blood culture bottles by amplification of bacterial 23S rDNA, followed by reverse hybridization to an oligonucleotide array designed to differentiate the sequence variation of the species. With this method, all specimens can be processed in the same way, and an unlimited number of bacterial species or sequence variants can be incorporated without affecting the complexity or speed of the assay. However, the method is dependent on the successful amplification of bacterial DNA directly from all positive blood cultures and the sensitivity and specificity of the oligonucleotide array.
In this study, we designed primers to amplify 23S rDNA from a wide range of bacterial genera and tested their ability to amplify DNA directly from positive blood culture bottles. Using published data and our own sequencing results, we constructed an oligonucleotide array to interrogate PCR amplicons from a collection of blood culture isolates. By sequencing amplicons that failed to hybridize or gave incorrect identifications with early versions of the assay, we continuously extended the array to improve discrimination. We then applied the test to positive blood culture broths. In this report, we describe primer, amplicon, and oligonucleotide sequences, the methodology for DNA extraction, amplification, and hybridization, and preliminary results obtained with both pure bacterial cultures and clinical blood culture specimens.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study were mainly from collections of blood culture and other clinical isolates at our laboratory. S. aureus NCTC657, Staphylococcus epidermidis NCTC11047, and E. coli NCTC8879 from the National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom, were also included. The strains were chosen to include a wide range of species and many of the common organisms causing bacteremia (11); the species selected are listed in Table 1. Organisms were identified by conventional methods and the appropriate API test system (bioMerieux SA, Lyon, France); streptococci were identified to the species level using the BBL crystal system (Becton Dickinson and Co., Paramus, N.J.)
TABLE 1.
Results of 23S PCR amplification and hybridization for pure cultures of 95 medically significant organismsa
| Phenotypic identification | No. of strains that were PCR positive/no. tested | Hybridization profile (oligonucleotide/no. of strains hybridizing) |
|---|---|---|
| Staphylococcus epidermidis | 5/5 | 7b/5, 8a/4, 8b/1 |
| Staphylococcus warneri | 2/2 | 7b/2, 6c/2 |
| Staphylococcus saprophyticus | 1/1 | 7b/1, 7c/1 |
| Staphylococcus xylosus | 1/1 | 7b/1 |
| Staphylococcus cohnii | 1/1 | 7b1, 8c/1 |
| Staphylococcus simulans | 1/1 | 7b/1 |
| Staphylococcus hominis | 1/1 | 7b/1 |
| Staphylococcus haemolyticus | 2/2 | 7b/2, 8c/1 |
| Staphylococcus aureus | 5/5 | 7a5, 7b/5 |
| Proteus mirabilis | 2/2 | 1a/2, 1b/2 |
| Proteus vulgaris | 1/1 | 1a/1, 1b/1 |
| Klebsiella pneumoniae | 3/3 | 2b/3, 3a/3, 3b/3 |
| Klebsiella oxytoca | 3/3 | 2a/3, 3a/3, 3b/3 |
| Enterobacter cloacae | 3/3 | 2b/3, 3a/3, 3b/3 |
| Enterobacter aerogenes | 1/1 | 2b/1, 3a/1, 3b/1 |
| Citrobacter freundii | 1/1 | 2b/1, 3a/1, 3b/1 |
| Escherichia coli | 5/5 | 1c/5, 2b/5, 2c/5, 3a/5, 3b/5 |
| Salmonella typhimurium | 1/1 | 1c/1, 2b/1, 2c/1, 3a/1, 3b/1 |
| Serratia marcesens | 1/1 | 2a/1, 2b/1, 3a/1, 3b/1 |
| Listeria monocytogenes | 1/1 | 7b/1, 9c/1 |
| Enterococcus faecium | 8/8 | 6b/8 |
| Enterococcus faecalis | 10/10 | 5b/10 |
| Aeromonas hydrophila | 1/1 | 5c/1 |
| Haemophilus influenzae | 1/1 | 4c/1 |
| Pseudomonas aeruginosa | 8/8 | 4b/8 |
| Stenotrophomonas maltophilia | 2/2 | 9b/2 |
| Burkholderia cepacia | 4/4 | 9a/4 |
| Viridans group Streptococcus | 11/11 | 4a/1, 5a/11, 6a/11, 7b/11, 10a/3, 10c/1 |
| Streptococcus anginosus | 2/2 | 5a/2, 7b/2, 10b/2 |
| Streptococcus pneumoniae | 1/1 | 4a/1, 5a/1, 6a/1, 7b/1, 10a/1 |
| Streptococcus pyogenes | 2/2 | 5a/2, 6a/2, 7b/2 |
| Corynebacterium spp. | 2/2 | No hybridization |
| Candida albicans | 0/2 | No hybridization |
A patent for the use of the oligonucleotides in this assay has been applied for.
Blood cultures.
Blood cultures are performed in our laboratory by using the Vital automated system (bioMerieux). In this method, up to 10 ml of blood is placed in anaerobic and aerobic Vital blood culture bottles. The bottles are then incubated in the Vital machine and continuously monitored for evidence of bacterial growth. When possible, growth is identified, the bottle is removed from the incubator, and a sample is taken for Gram staining and subculturing to agar plates. During this study, an additional sample of 100 μl for DNA extraction was taken from 158 unselected positive blood culture bottles as described below. The DNA assay was performed without knowledge of the patient details or the initial Gram stain result.
Conventional microbiological identification of clinical isolates.
Organisms were identified by conventional methods and, except for the CoNS, by the appropriate API test system.
Extraction of bacterial DNA from pure bacterial cultures.
Stored organisms were subcultured onto Columbia blood agar plates (Oxoid). A single colony of overnight growth at 37°C was suspended in 100 μl of distilled water containing 1 μl of a 1-mg/ml solution of lysostaphin (Sigma Chemical Co.) and incubated at 37°C for 10 min. The tubes were then transferred to a thermal cycler (Perkin-Elmer 2400 GeneAmp PCR system) and heated to 95°C for 10 min. Finally, they were spun at 10,000 × g for 2 min in a microcentrifuge, and 1 μl of the supernatant was used in the 23S PCR described below.
Extraction of bacterial DNA directly from Vital blood culture bottles.
DNA was extracted from all positive blood culture bottles in a class II safety cabinet using the following protocol. Two to four drops of broth was transferred into 0.5 ml of sterile distilled water at the time of aspiration for Gram staining and subculturing. The tubes were spun at 10,000 × g in a microcentrifuge for 4 min, and the supernatant was discarded. The pellet was resuspended in 100 μl of distilled water containing 1 μl of a 1-mg/ml solution of lysostaphin and incubated at 37°C for 20 min in a dry block (Scotlab). The temperature was then raised to 95°C, and the tubes were incubated for a further 15 min. Finally, the tubes were spun at 10,000 × g for 2 min in a microcentrifuge, and 1 μl of the supernatant was used in the 23S PCR described below.
Design of primers to amplify 23S bacterial rDNA.
The primers chosen were based on, but did not exactly match, conserved regions (region 6 and region 10) previously reported within the bacterial 23S rDNA (17). Based on the E. coli 23S rDNA (17), primer 6 starts at nucleotide 130 rather than nucleotide 132 at the 3′ end to avoid a wobble at the 3′ end and was extended 5′ to nucleotide 108 to increase the annealing temperature. Primer 10 starts at nucleotide 457 rather than nucleotide 456 at the 3′ end, again to avoid a wobble at the 3′ end, and was extended 3 nucleotides 5′ to provide a higher annealing temperature. The sequences of the primers used were as follows: forward primer 6, 5′-GCGATTTCYGAAYGGGGRAACCC; and reverse primer 10, 5′-digoxigenin-TTCGCCTTTCCCTCACGGTACT (where Y is C or T and R is A or G).
Primers were commercially synthesized (Amersham Pharmacia Biotech, Amersham, United Kingdom). A PCR master mix containing DnaZyme buffer (Flowgen), 1 μM primer 6, 2 μM primer 10, and 150 μM each deoxynucleoside triphosphate was made up in 5-ml quantities. Forty-microliter aliquots of the master mix were dispensed into 100-μl PCR tubes. When the DNA extracts were available, 1 μl of the appropriate extract and 1 U of DnaZyme DNA polymerase (Flowgen) were added to each tube. The PCR mixtures were then subjected to 5 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s, followed by 25 cycles of 95°C for 15 s and 65°C for 30 s. The initial five cycles with an annealing temperature of 55°C were included to allow a small amount of mispriming in these cycles and thus to initiate the amplification of DNA from bacteria with 23S DNA which did not exactly match the sequences of the primers. The subsequent 25 cycles were carried out as a rapid two-step PCR with a high annealing temperature, as the amplicon generated in the first 5 cycles was able to act as a template in the PCR for all strains. The presence of a PCR product was confirmed by agarose electrophoresis and visualization with ethidium bromide.
Sequence determination for primary pathogens and identification of potential reverse hybridization targets.
The sequence variability of the 23S rDNA target region was initially determined by interrogation of public databases (1). When species information was not available, we sequenced PCR products from selected isolates in our organism collection. This information was supplemented by sequence data from products that failed to hybridize with the early oligonucleotide arrays or gave erroneous identifications. The 23S PCR products were sequenced by the cycle sequencing method (Amersham Pharmacia) using fluorescein-labeled primers on an automated sequencer (Amersham Pharmacia Alf system). Accurate sequences were reproducibly obtained in both the forward and the reverse directions between conserved regions seven and eight (17), and all the oligonucleotides chosen were targeted at sequences within this area. Alignments were performed between the sequences obtained and those available from public databases. Using this information, 30 oligonucleotides with similar melting temperatures were designed (Table 2), and their ability to discriminate stored isolates was assessed. Oligonucleotides that failed to show detectable hybridization to target DNA or gave only weak hybridization signals (6c, 7c, 8a, and 8c) were resynthesized with an additional five 3′ thymine residues in order to increase binding to the nylon membrane and thus the hybridization intensity (39).
TABLE 2.
Oligonucleotides used in this studya
| Oligonucleotide | Species from which the sequence was derived | GenBank accession no. | Sequence 5′ to 3′ |
|---|---|---|---|
| 1a | Proteus mirabilis | AF146762b | AATAGCAGTGTCAGGAGAACGGTCT |
| 1b | Proteus mirabilis | AF146762b | ATAGCCCCGTATCTGAAGATGCT |
| 1c | Escherichia coli | M25458 | CCAGAGCCTGAATCAGTGTGT |
| 2a | Klebsiella oxytoca | AF146763b | TCCCGTACACTAAAACGCACAGG |
| 2b | Klebsiella pneumoniae | x87284 | TCCCGTACACCAAAATGCACAGG |
| 2c | Escherichia coli | L10328 | CCAGAGCCTGAATCAGTATGTG |
| 3a | Enterobacter cloacae | AF146764b | TCCCGTACACGAAAATGCACAGG |
| 3b | Escherichia coli | v00331 | CCCGTACACAAAAATGCACA |
| 3c | Salmonella enterica | U77923 | AGAGCCTGAATCAGCATGTGT |
| 4a | Streptococcus pneumoniae | m60763 | AGAAGAATGATTTGGGAAGATC |
| 4b | Pseudomonas aeruginosa | y00432 | GCTTCATTGATTTTAGCGGAAC |
| 4c | Haemophilus influenzae | U32742 | GTGAGGAGAATGTGTTGGGAAG |
| 5a | Streptococcus pneumoniae | M60763 | GGTTGTAGGACTGCAATGTGGACTC |
| 5b | Enterococcus faecalis | AF146765b | GGTAGTCTGTTAGTATAGTTGAAG |
| 5c | Aeromonas hydrophila | X87281 | TGGAACGGTCCTGGAAAGGC |
| 6a | Streptococcus oralis | X68427 | GCAGGAGGGCAAACCGAAGAGTT |
| 6b | Enterococcus faecium | AF146766b | GGTAGTTCTTTCAGATAGTCGG |
| 6c | Staphylococcus warneri | AF146768b | ACGGAGTTACAAAAGTATATATTAGTTTTT |
| 7a | Staphylococcus aureus | X68425 | ACGGAGTTACAAAGGACGACATTA |
| 7b | Staphylococcus aureus | x68425 | GGTTGTAGGACACTCTATACGGAGTT |
| 7c | Staphylococcus saprophyticus | AF146769b | ACGGAGTTACAAAAGAACAGACTAGTTTTT |
| 8a | Staphylococcus epidermidis | AF146770b | ACGGAGTTACAAAAGAACATGTTAGTTTTT |
| 8b | Staphylococcus epidermidis | AF146771b | ACGGAGTTACAAAAGAATTTGTTAGTTTTT |
| 8c | Staphylococcus haemolyticus | AF146772b | ACGGAGTTACAAAGGAATATATTAGTTTTT |
| 9a | Burkholderia cepacia | X16368 | CGTATTGTTAGCCGAACGCTCT |
| 9b | Stenotrophomonas maltophilia | AF146773b | AGCCCTGTATCTGAAAGGGCCA |
| 9c | Listeria spp. | X64533 | ACGGAGTTACAAAAGAAAGTTATAA |
| 10a | Streptococcus oralis | X68427 | AGAAGAATGATTTGGGAAGATC |
| 10b | Streptococcus anginosus | AF146774b | AGAAGAAGACCTTGGGAAAGG |
| 10c | Streptococcus thermophilus | X68429 | AGAAGAACTACCTGGGAAGGT |
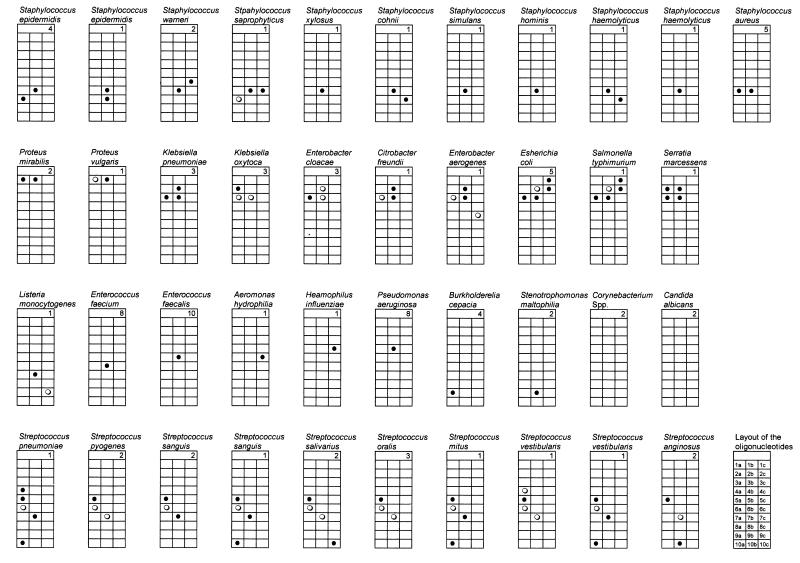
Oligonucleotides were arranged on the arrays as indicated in Fig. 1.
Determined in this study.
Production of the hybridization membranes.
The 30 oligonucleotides were bound to nylon strips as described below. A 3-mm grid was printed on a nylon membrane (MAGNA Micron Separations Inc.) with a bubble jet printer to allow the spots to be more accurately positioned. Strips were made in batches of 20. Oligonucleotides were purchased from Amersham Pharmacia, and 50 pg of each in 0.3 μl of water was spotted onto a specific position on the nylon membrane. Once all the oligonucleotides had been applied, the strips were dried and exposed to shortwave UV in an Amplirad light box (Genetic Research Instruments, Essex, United Kingdom) for 30 s. The length of exposure was found to have a marked effect on the intensity of the resulting spots: with our UV illuminator, 30 s was found to give the optimal spot intensity. After the oligonucleotides had been cross-linked to the membrane, any unbound oligonucleotides were removed by two washes in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) for 2 min at 37°C. The strips were dried and stored at room temperature, ready for use.
Hybridization protocol.
The digoxigenin-labeled 23S rDNA amplicons were hybridized to the oligonucleotide arrays using the following protocol. Each strip was numbered and placed in a separate 2.5-ml screw-top microcentrifuge tube containing 0.5 ml of 5× SSC, 0.1% N-laurylsarcosine, 0.02% SDS, and 1% blocking reagent (Boehringer GmbH, Mannheim, Germany). The digoxigenin-labeled PCR products were heated to 95°C in a thermal cycler, and the appropriate PCR product was added directly to each tube. Hybridization was continued for 45 min at 50°C with gentle agitation. The strips were then removed from the tubes and washed four times in 25 ml of 0.25× SSC–0.1% SDS for each batch of 20 strips at 37°C for 2 min. Any hybridization was detected using a colorimetric detection system according to the manufacturer's instructions (Boehringer). Color development was clearly visible between 15 min and 1 h after the start of the reaction.
Interpretation of the hybridization results.
All samples obtained from blood culture bottles were processed on the day on which they were identified, and the resulting strips were visually compared with the results previously obtained from pure cultures. A report predicting the bacterial species present was then produced. These predictions were then compared with subsequent identification of organisms by routine laboratory testing.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined in this study are AF146762, AF146763, AF146764, AF146765, AF146766, AF146768, AF146769, AF146770, AF146771, AF146772, AF146773, and AF146774 (Table 2).
RESULTS
Assessment of the primers.
The effectiveness of the primers was first assessed with DNA extracts from 93 stored bacterial isolates representing 36 bacterial species, together with 2 isolates of Candida albicans (Table 1). The organisms were chosen to include examples of the common organisms causing bacteremia, several species of CoNS and viridans group streptococci, and several nonfermenting gram-negative species. All the bacterial isolates tested produced PCR products. A band of approximately 400 bp was produced with gram-positive bacteria, and a band of 350 bp was produced with gram-negative bacilli. The isolates of C. albicans did not produce PCR products. No bands were seen in the DNA-negative amplification controls.
Sequencing of the products and initial choice of oligonucleotides.
The initial choice of oligonucleotides was based on information in public databases. This information was expanded by sequencing products from organisms in our collection of pure cultures. From these results, 30 oligonucleotides were constructed (Table 2). The amplicons obtained from DNA extracts of pure cultures were then hybridized to arrays of these oligonucleotides. The results are shown in Table 1 and summarized schematically in Fig. 1. The results were in close agreement with those predicted from the DNA sequences. Interpretation of the hybridization reactions was clear-cut, with the exception of some Enterobacteriaceae and some streptococci. All the S. aureus isolates were correctly identified. The CoNS isolates were clearly distinguished from S. aureus species, but the species could not be determined with the current array. No oligonucleotides targeted at the coryneform group were present in the array, and the two coryneform isolates tested produced no detectable hybridization.
FIG. 1.
Summary of hybridization detected for the 23S rDNA PCR products of 95 pure cultures. The positions of the oligonucleotides are indicated in the lower right-hand strip. Their sequences are indicated in Table 2. The number of strains tested is indicated in the top portion of each strip. Filled circles represent strong hybridization, empty circles represent weak hybridization, and empty cells represent no hybridization detected.
Hybridization from enrichment broths.
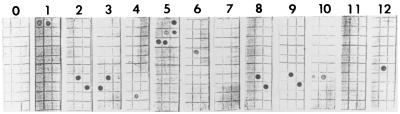
Samples from 158 blood culture broths identified as positive by the Vital system were subjected to PCR on the day on which they became positive. The results are shown in Table 3, and examples of the strips obtained are shown in Fig. 2. Six bottles (3.8%) produced no growth and no hybridization and, for the purposes of this study, were regarded as having been correctly designated by the PCR system. A further 119 culture-positive bottles (75.3%) produced correct identifications. These included four (2.5%) in which mixed cultures were correctly identified (one contained P. aeruginosa plus Enterococcus faecalis, one contained P. aeruginosa plus Stenotrophomonas maltophilia, and two contained S. aureus plus E. faecalis). Sixteen (10.3%) bottles produced no PCR product and no hybridization but subsequently grew bacteria that would have been expected to hybridize to the strips (nine CoNS, three E. faecalis, one Streptococcus sp., and one Enterobacter cloacae). This result was due to a failure of the PCR. Eight bottles (5.1%) produced incorrect identifications when compared with conventional tests. One was reported as a CoNS but yielded S. aureus, and seven were incorrectly identified by PCR as containing streptococci (hybridization to oligonucleotide 5a). The nine remaining bottles (5.6%) yielded a range of organisms for which oligonucleotides had not been constructed. Negative controls containing no DNA extract were included in parallel with each day's tests, and all gave negative results. Oligonucleotides targeted at Burkholderia cepacia, Aeromonas hydrophila, and Enterococcus faecium, species that were not recovered during this study, did not hybridize to any of the amplicons.
TABLE 3.
Comparison of identifications based on the hybridization assay and conventional bacteriology methods for 158 unselected blood culture bottles flagged as positive by the Vital system
| Hybridization report | No. (%) | Subsequent growth | No. |
|---|---|---|---|
| Correct identification | 119 (75.3) | ||
| CoNS | 62 | CoNS | 62 |
| CoNS | 2 | CoNS + Corynebacterium spp. | 2 |
| Staphylococcus aureus | 18 | Methicillin-resistant Staphylococcus aureus | 8 |
| S. aureus | 10 | ||
| S. aureus + Enterococcus faecalis | 2 | S. aureus + Enterococcus faecalis | 2 |
| E. faecalis | 4 | E. faecalis | 4 |
| Pseudomonas aeruginosa | 5 | Pseudomonas aeruginosa | 5 |
| P. aeruginosa + E. faecals | 1 | P. aeruginosa + E. faecalis | 1 |
| P. aeruginosa + Stenotrophomonas maltophilia | 1 | P. aeruginosa + Stenotrophomonas maltophilia | 1 |
| S. maltophilia | 1 | S. maltophilia | 1 |
| Streptococcus spp. | 5 | Streptococcus spp. | 5 |
| Proteus spp. | 3 | Proteus mirabilis | 2 |
| Proteus vulgaris | 1 | ||
| Escherichia coli or Salmonella spp. | 12 | Salmonella spp. | 6 |
| Escherichia coli | 6 | ||
| Klebsiella pneumoniae or Enterobacter spp. | 1 | Enterobacter cloacae | 1 |
| Haemophilus influenzae | 1 | Haemophilus influenzae | 1 |
| Listeria spp. | 1 | Listeria monocytogenes | 1 |
| No hybridization | 31 (19.6) | ||
| Expected result | 15 (9.5) | No growth | 6 |
| Corynebacterium spp. | 6a | ||
| Moraxella spp. | 1a | ||
| Propionibacterium spp. | 1a | ||
| Pseudomonas pickettii | 1a | ||
| PCR failure | 16 (10.3) | ||
| CoNS | 9 | ||
| E. faecalis | 3 | ||
| E. cloacae | 1 | ||
| E. coli | 1 | ||
| Streptococcus spp. | 2 | ||
| Incorrect identification | 8 (5.1) | ||
| Streptococcus spp. | 7 | Ralstonia pickettii | 1a |
| No growth | 3 | ||
| CoNS | 2 | ||
| Corynebacterium spp. + CoNS | 1 | ||
| CoNS | 1 | S. aureus | 1 |
No oligonucleotides targeted at this species were present in the array, so no hybridization was expected.
FIG. 2.
Results of the hybridization assay for 23S PCR amplifications from 12 blood culture bottles. Strip 0 was a DNA-negative PCR control. Strips 1 to 12 were PCR amplifications from bottles which subsequently grew bacteria identified as Proteus mirabilis (strip 1), CoNS (strips 2, 3, 7, 8, and 9), S. maltophilia (strip 4), E. coli (strip 5), P. aeruginosa (strip 6), S. aureus (strip 10), Corynebacterium spp. (strip 11), and E. faecium (strip 12). No hybridization is visible on strip 7 due to failure of the PCR. The oligonucleotides are listed in Table 2 and were arranged as shown in Fig. 1.
Thus, the positive predictive value of this hybridization assay was 100% for all organism groups except streptococci (positive predictive value, 50%) and CoNS (positive predictive value, 96%). The misidentification of the S. aureus isolate was based on a weak hybridization reaction; when the assay was repeated with a culture of this organism, the typical hybridization profile for S. aureus (hybridization to oligonucleotides 7a and 7b) was seen.
DISCUSSION
We have investigated a novel PCR method for the identification of organisms causing bacteremia. We used universal primers to amplify a conserved region of bacterial 23S rDNA, followed by characterization of the PCR products by reverse hybridization to an oligonucleotide array. The use of a single protocol for all bacterial species is essential for testing blood cultures, where the bacterial diagnosis is usually uncertain. We first designed primers capable of producing PCR products from common organisms causing bacteremia and then developed a protocol for direct application to blood culture broths. The use of relatively long, moderately degenerate universal primers with a high annealing temperature allowed the 23S rDNA to be targeted while avoiding nonspecific amplification. High annealing temperatures also allowed the annealing and extension stages of the later PCR cycles to be combined, resulting in shorter cycle times, approximately 50 min with a Perkin-Elmer 2400 thermal cycler.
The primers were based on previously described conserved regions of the bacterial 23S rDNA (region 6 and region 10) (17). Amplification products were obtained from all 91 stored bacterial isolates tested (approximately 350 bp from gram-negative species and 400 bp from gram-positive species) but, as expected, not from 2 strains of C. albicans.
We sequenced the PCR products of species for which there were no published 23S rDNA gene data and constructed a series of complementary oligonucleotides for species identification. These were successful in identifying all 91 stored bacterial isolates to at least the genus level in a reverse hybridization assay. The two organisms not identified were coryneforms and produced PCR products that did not hybridize to the array used.
With an ideal array, each oligonucleotide should hybridize only to one bacterial species and to all members of that species. In practice, two or more oligonucleotides are often required. Multiple target oligonucleotides would also facilitate the identification of clinical isolates of species that show minor sequence variations within the target region (19). For example, Listeria spp. were found to hybridize to oligonucleotide 7b, so it was necessary to add an additional oligonucleotide to the array to allow these species to be discriminated from CoNS isolates. An important feature of this identification system is that the panel of oligonucleotides can be continually extended to include sequences for additional species or variant isolates as they are characterized. A single S. aureus isolate was incorrectly identified as a CoNS in the blood culture study; this identification was based on weak hybridization to oligonucleotide 7b alone. When the assay was repeated with a pure culture of this organism, hybridization to oligonucleotides 7a and 7b was clearly visible. Since the membranes were manually prepared, it is possible that this oligonucleotide was inadvertently not included on the membrane. If the identification of all species of CoNS had been based on hybridization to at least two oligonucleotides, this identification based on a single spot would have been immediately recognized as doubtful.
The array that was developed could clearly discriminate the important organisms causing bacteremia. P. aeruginosa, S. maltophilia, B. cepacia, Proteus spp., E. faecium, Haemophilus influenzae, and E. faecalis were all unambiguously identified, and S. aureus was distinguished from CoNS.
In this study, we limited ourselves to the common species causing bacteremia plus some less frequent species that were isolated from the blood culture bottles that we examined. The range of organisms that can be identified can be expanded by increasing the number of oligonucleotide targets in the array. The hybridization results will become more robust as more targets are added.
The system was successful in identifying mixtures of organisms in polymicrobial bacteremias. This result represents an important advantage of using a single parallel identification procedure. In hierarchical detection systems, investigations are often terminated once one species is identified, and such studies often eliminate samples which appear to be mixed by Gram staining (8).
We have not yet investigated sufficient numbers and species of streptococci or CoNS to accurately interpret hybridization profiles seen with these organisms. As more isolates are studied, interpretation of variations in hybridization will be possible and their taxonomic significance can be determined.
In this study, the Enterobacteriaceae (E. coli and Proteus, Klebsiella, Citrobacter, Enterobacter, Serratia, and Salmonella spp.) were clearly identified as a group. The Proteus spp. tested were readily identified to the genus level, and some discrimination between other Enterobacteriaceae was demonstrated with the present oligonucleotide targets. Klebsiella pneumoniae was differentiated from Klebsiella oxytoca but produced a hybridization profile that could not be distinguished from those of the Enterobacter sp. or Citrobacter freundii isolates studied with the present array. E. coli and Salmonella isolates produced indistinguishable hybridization profiles with the array. The Serratia marcescens isolate tested produced a unique hybridization profile. The Enterobacteriaceae have similar and in many cases multiple 23S rDNA sequences which may not be identical, and many more strains need to be studied before we can design additional oligonucleotides to identify species for this group of organisms. With the present limited number of oligonucleotides, the hybridization results were visually analyzed. For some organisms, there appeared to be consistent differences in the degree of hybridization to different targets. In order to confirm this variation and for the analysis of a larger array, computer analysis of quantitative hybridization fingerprints may be required.
Seven of the eight bottles that produced incorrect identifications were identified by hybridization as containing streptococci. This did not appear to be the result of sequence similarity with other pathogens, since in three of these bottles no organisms were recovered. We have tested noninoculated broth from a series of bottles, all of which gave negative results (results not shown). Unfortunately, this finding does not exclude the possibility that only a proportion of the bottles are contaminated or that the contamination is at such a low level that detectable amplification does not invariably occur. False-positive PCR identifications of S. pneumoniae have previously been reported from blood specimens. Other workers (7, 40) have used PCR methods specific for the pneumococcal pneumolysin gene and reported between 6 and 17% apparently false-positive results for healthy controls. Our false-positive rate of streptococcal hybridization was 4.4%. Although we cannot rule out nonspecific hybridization or contamination and the presence of bacterial DNA in commercial enrichment bottles has been reported (13), it is possible that streptococcal DNA is present in blood samples from colonized patients.
The failure of the PCR in 16 (10.3%) of the amplifications carried out from enrichment broth was almost certainly due to the presence of inhibitors in our DNA preparations from the blood or enrichment broth, since products were reproducibly obtained from purified isolates cultured on standard media. Effective methods for the removal of these substances have been developed but often require significant manipulation of the sample, increasing the processing time and the chance of contamination (13). The extraction procedure used here was simple, rapid, and effective for more than 80% of samples, but modification of the extraction protocol, the composition of the PCR mixture, or the composition of enrichment media may be required to ensure successful amplification for all samples.
The inclusion of a PCR control template and oligonucleotide target would identify all potentially false-negative PCR results due to enzymatic inhibition. These results could then be discarded. With the rigorous use of controls and the physical separation of the DNA extraction, amplification, and detection steps, this technique has the potential to be incorporated into the routine microbiology laboratory in the near future. The availability of species identifications from the majority of positive blood cultures within 4 h would improve patient management and potentially reduce the inappropriate use of antibiotics. Conventional phenotypic identification and susceptibility testing would still be required, allowing isolates for which the assay failed to be identified and providing accurate information on antibiotic susceptibility.
In conclusion, we have shown that a simple, rapid, DNA-based method can identify a wide range of clinically significant bacterial species in blood cultures. This method can be applied directly to positive blood culture broths and can identify mixtures of organisms. Results are available within 4 h, and with improvements in technology, this time may be substantially reduced. These tests will not replace conventional bacteriology methods, which are still required for susceptibility tests, but will provide rapid clinical information relevant to patients. This approach is more practical for clinical laboratories than species-specific PCR, since all samples can be processed identically and the method lends itself to automation. Studies are currently under way to extend the range of species that can be identified and to apply the system directly to blood specimens and other clinical specimens. The accuracy, range, and discriminatory power of the assay can be continually extended by adding further oligonucleotides to the panel without significantly increasing complexity or cost.
ACKNOWLEDGMENTS
We thank G. Rao for the isolates from Lewisham Hospital, A. King for the determination of the species of the CoNS isolates, and T. Bathgate for the determination of the species of the streptococci. We also thank the clinical laboratory staff, who have shown patience and enthusiasm throughout this project, and Sophie Masliah and Victor de Benito, whose work was greatly appreciated.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacot C M, Reeves R H. Novel tRNA gene organization in the 16S-23S intergenic spacer of the Streptococcus pneumoniae rRNA gene cluster. J Bacteriol. 1991;173:4234–4236. doi: 10.1128/jb.173.13.4234-4236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron M G, Ouellette M. Preventing antibiotic resistance using rapid DNA-based diagnostic tests. Infect Control Hosp Epidemiol. 1998;19:560–564. doi: 10.1086/647873. [DOI] [PubMed] [Google Scholar]
- 4.Burgin A B, Parodos K, Lane D J, Pace N R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 5.Cocolin L, Manzano M, Cantoni C, Comi G. A PCR-microplate capture hybridization method to detect Listeria monocytogenes in blood. Mol Cell Probes. 1997;11:453–455. doi: 10.1006/mcpr.1997.0133. [DOI] [PubMed] [Google Scholar]
- 6.Cunney R J, McNamara E B, Alansari N, Loo B, Smyth E G. The impact of blood culture reporting and clinical liaison on the empiric treatment of bacteraemia. J Clin Pathol. 1997;50:1010–1012. doi: 10.1136/jcp.50.12.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagan R, Shriker O, Hazan I, Leibovitz E, Greenberg D, Schlaeffer F, Levy R. Prospective study to determine clinical relevance of detection of pneumococcal DNA in sera of children by PCR. J Clin Microbiol. 1998;36:669–673. doi: 10.1128/jcm.36.3.669-673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis T E, Fuller D D. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dharakul T, Songsivilai S, Viriyachitra S, Luangwedchakarn V, Tassaneetritap B, Chaowagul W. Detection of Burkholderia pseudomallei DNA in patients with septicemic melioidosis. J Clin Microbiol. 1996;34:609–614. doi: 10.1128/jcm.34.3.609-614.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engleberg N C, Eisenstein B I. Detection of microbial nucleic acids for diagnostic purposes. Annu Rev Med. 1992;43:147–155. doi: 10.1146/annurev.me.43.020192.001051. [DOI] [PubMed] [Google Scholar]
- 11.Eykyn S, Gransden W R, Phillips I. The causative organisms of septicaemia and their epidemiology. J Antimicrob Chemother. 1990;25(Suppl. C):41–58. doi: 10.1093/jac/25.suppl_c.41. [DOI] [PubMed] [Google Scholar]
- 12.Folgueira L, Delgado R, Palenque E, Aguado J M, Noriega A R. Rapid diagnosis of Mycobacterium tuberculosis bacteremia by PCR. J Clin Microbiol. 1996;34:512–515. doi: 10.1128/jcm.34.3.512-515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredricks D N, Relman D A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French G L, Cheng A F B, Ling A M L, Mo P, Donnan S. Hong Kong strains of methicillin-resistant and methicillin-sensitive Staphylococcus aureus have similar virulence. J Hosp Infect. 1990;15:117–125. doi: 10.1016/0195-6701(90)90120-d. [DOI] [PubMed] [Google Scholar]
- 15.Goodman J L, Bradley J F, Ross A E, Goellner P, Lagus A, Vitale B, Berger B W, Luger S, Johnson R C. Bloodstream invasion in early Lyme disease: results from a prospective, controlled, blinded study using the polymerase chain reaction. Am J Med. 1995;99:6–12. doi: 10.1016/s0002-9343(99)80097-7. [DOI] [PubMed] [Google Scholar]
- 16.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 18.Hall L M, Duke B, Urwin G. An approach to the identification of the pathogens of bacterial meningitis by the polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1995;14:1090–1094. doi: 10.1007/BF01590946. [DOI] [PubMed] [Google Scholar]
- 19.Harvey S, Hill C W, Squires C L. Loss of the spacer loop sequence from the rrnB operon in the Escherichia coli K-12 subline that bears the relA1 mutation. J Bacteriol. 1998;170:1235–1238. doi: 10.1128/jb.170.3.1235-1238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopfl P, Ludwig W, Schleifer K H, Larsen N. The 23S ribosomal RNA higher-order structure of Pseudomonas cepacia and other prokaryotes. Eur J Biochem. 1989;185:355–364. doi: 10.1111/j.1432-1033.1989.tb15123.x. [DOI] [PubMed] [Google Scholar]
- 21.Hospital Infection Control Practices Advisory Committee. Recommendations for preventing the spread of vancomycin resistance. Am J Infect Control. 1995;23:87–94. doi: 10.1016/0196-6553(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 22.Isaacman D J, Zhang Y, Reynolds E A, Ehrlich D G. Accuracy of a polymerase chain reaction-based assay for detection of pneumococcal bacteremia in children. Pediatrics. 1998;101:813–816. doi: 10.1542/peds.101.5.813. [DOI] [PubMed] [Google Scholar]
- 23.Jamulitrat S, Meknavin U, Thongpiyapoon S. Factors affecting mortality outcome and risk of developing nosocomial bloodstream infection. Infect Control Hosp Epidemiol. 1994;15:163–170. doi: 10.1086/646884. [DOI] [PubMed] [Google Scholar]
- 24.Kane T D, Alexander J W, Johannigman J A. The detection of microbial DNA in the blood: a sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1998;227:1–9. doi: 10.1097/00000658-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebovici L S, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik S D. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med. 1998;244:379–386. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 26.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1998;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig W, Kirchhof G, Klugbauer N, et al. Complete 23S ribosomal RNA sequences of Gram-positive bacteria with a low DNA G+C content. Syst Appl Microbiol. 1992;15:487–501. [Google Scholar]
- 28.Ludwig W, Rossello-Mora R, Aznar R, Klugbauer S, Spring S, Reetz K, Beimfohr C, Brockmann E, Kirchhof G, Dorn S, Bachleitner M, Klugbauer N, Springer N, Lane D, Nietupsky R, Weizenegger M, Schleifer K H. Comparative sequence analysis of 23S rRNA from proteobacteria. Syst Appl Microbiol. 1995;18:164–188. [Google Scholar]
- 29.Morace G, Sanguinetti M, Posteraro B, Lo Cascio G, Fadda G. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J Clin Microbiol. 1997;35:667–672. doi: 10.1128/jcm.35.3.667-672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narita M, Matsuzono Y, Itakura O, Togashi T, Kikuta H. Survey of mycoplasmal bacteremia detected in children by polymerase chain reaction. Clin Infect Dis. 1996;23:522–525. doi: 10.1093/clinids/23.3.522. [DOI] [PubMed] [Google Scholar]
- 31.Newcombe J, Cartwright K, Palmer W H, McFadden J. PCR of peripheral blood for diagnosis of meningococcal disease. J Clin Microbiol. 1996;134:1637–1640. doi: 10.1128/jcm.34.7.1637-1640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pease A C, Solas D, Sullivan E J, Cronin M T, Holmes C P, Fodor S P. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen G, Schonheyder H C, Sorensen H T. Antibiotic therapy and outcome of monomicrobial gram-negative bacteremia: a 3-year population-based study. Scand J Infect Dis. 1997;29:601–606. doi: 10.3109/00365549709035903. [DOI] [PubMed] [Google Scholar]
- 34.Rattanathongkom A, Sermswan R W, Wongratanacheewin S. Detection of Burkholderia pseudomallei in blood samples using polymerase chain reaction. Mol Cell Probes. 1997;11:25–31. doi: 10.1006/mcpr.1996.0072. [DOI] [PubMed] [Google Scholar]
- 35.Relman D A. Universal bacterial 16S rDNA amplification and sequencing. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 489–495. [Google Scholar]
- 36.Richter C, Kox L F, Van Leeuwen J V, Mtoni I, Kolk A H. PCR detection of mycobacteremia in Tanzanian patients with extrapulmonary tuberculosis. Eur J Clin Microbiol Infect Dis. 1996;15:813–817. doi: 10.1007/BF01701525. [DOI] [PubMed] [Google Scholar]
- 37.Roller C, Ludwig W, Schleifer K H. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J Gen Microbiol. 1992;138:1167–1175. doi: 10.1099/00221287-138-6-1167. [DOI] [PubMed] [Google Scholar]
- 38.Rubin F A, McWhirter P D, Punjabi N H, Lane E, Sudarmono P, Pulungsih S P, Lesmana M, Kumala S, Kopecko D J, Hoffman S L. Use of a DNA probe to detect Salmonella typhi in the blood of patients with typhoid fever. J Clin Microbiol. 1989;27:1112–1114. doi: 10.1128/jcm.27.5.1112-1114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saiki R K, Walsh P S, Levenson C H, Erlich H A. Genetic analysis of amplified DNA with immobilized sequence specific oligonucleotide probes. Proc Natl Acad Sci USA. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salo P, Ortqvist A, Leinonen M. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J Infect Dis. 1995;171:479–482. doi: 10.1093/infdis/171.2.479. [DOI] [PubMed] [Google Scholar]
- 41.Saruta K, Matsunaga T, Kono M, Hoshina S, Ikawa S, Sakai O, Machida K. Rapid identification and typing of Staphylococcus aureus by nested PCR amplified ribosomal DNA spacer region. FEMS Microbiol Lett. 1997;146:271–278. doi: 10.1111/j.1574-6968.1997.tb10204.x. [DOI] [PubMed] [Google Scholar]
- 42.Sellon D C, Walker K, Suyemoto M, Altier C. Nucleic acid amplification for rapid detection of Rhodococcus equi in equine blood and tracheal wash fluids. Am J Vet Res. 1997;58:1232–1237. [PubMed] [Google Scholar]
- 43.Shin J H, Nolte F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song J H, Cho H, Park M Y, Na D S, Moon H B, Pai C H. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J Clin Microbiol. 1993;31:1439–1443. doi: 10.1128/jcm.31.6.1439-1443.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang G Q, Nguyen S V, To H, Ogawa M, Hotta A, Yamaguchi T, Kim H J, Fukushi H, Hirai K. Clinical evaluation of a new PCR assay for detection of Coxiella burnetii in human serum samples. J Clin Microbiol. 1998;36:77–80. doi: 10.1128/jcm.36.1.77-80.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]