Abstract
The use of the versatile cyanobacterial extracellular polymeric substances (EPS) for biotechnological/biomedical applications implies an extensive knowledge of their biosynthetic pathways to improve/control polymer production yields and characteristics. The multiple copies of EPS-related genes, scattered throughout cyanobacterial genomes, adds another layer of complexity, making these studies challenging and time-consuming. Usually, this issue would be tackled by generating deletion mutants, a process that in cyanobacteria is also hindered by the polyploidy. Thus, the use of the CRISPRi multiplex system constitutes an efficient approach to addressing this redundancy. Here, three putative Synechocystis sp. PCC 6803 kpsM homologues (slr0977, slr2107, and sll0574) were repressed using this methodology. The characterization of the 3-sgRNA mutant in terms of fitness/growth and total carbohydrates, released and capsular polysaccharides, and its comparison with previously generated single knockout mutants pointed towards Slr0977 being the key KpsM player in Synechocystis EPS production. This work validates CRISPRi as a powerful tool to unravel cyanobacterial complex EPS biosynthetic pathways expediting this type of studies.
Keywords: CRISPRi, cyanobacteria, Synechocystis, extracellular polymeric substances (EPS), KpsM
1. Introduction
Most of the cyanobacterial strains can produce extracellular polymeric substances (EPS), mainly composed of heteropolysaccharides, that can be released to the extracellular medium or remain associated to the cell surface as capsules, sheaths, or slime. [1,2]. These EPS are emerging as promising biomaterials for biotechnological and biomedical applications as they possess distinctive and advantageous characteristics compared to other natural and synthetic polymers [3,4,5]. In addition to the complexity/variety of polymers produced by cyanobacteria, their cultivation is inexpensive due to their photoautotrophic lifestyle, the growth rates are similar or higher than algae and plants, and often, the produced polymers can be easily functionalized and the producing strain engineered to obtain a product with the desired properties and/or enhanced performance [6]. However, this manipulation requires a comprehensive knowledge on the molecular mechanisms underlying EPS biosynthesis, assembly, and export. Such knowledge is therefore crucial to increase the production yields and to tailor polymer variants for a specific application. In addition, this knowledge can also aid efforts to redirect carbon fluxes from the high-energy-demanding EPS production towards the production of target/value-added compounds, when using cyanobacteria as green cell factories [7,8].
The last steps of the EPS biosynthetic pathways are relatively conserved throughout bacteria and often follow one of three major pathways—Wzy-, ABC transporter- or Synthase-dependent—ending with an assembled polymer outside the cell wall [9]. Previously, by performing a phylum-wide analysis, we showed that most cyanobacteria harbor genes encoding proteins related to these pathways but often not a complete set defining one pathway, and the EPS-related genes are scattered throughout the genomes or organized in small clusters, often in multiple copies [1,10,11]. Up until now, the study of the putative EPS-related genes/proteins has been performed, by us and others, mainly through the generation and characterization of knockout mutants using the model cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis). Previous works have confirmed the involvement of cyanobacterial homologues of key bacterial proteins belonging to both the ABC transporter- and Wzy-dependent pathways in EPS production. Regarding the Wzy-dependent pathway, knockout mutants of wza (sll1581), wzb (slr0328), and wzc (sll0923) exhibited fewer capsular polysaccharides (CPS), fewer released polysaccharides (RPS), or less of both, respectively [12,13]. Regarding the ABC transporter-dependent pathway, Fisher et al. reported knockout mutants of a putative transport permease (slr0977 (kpsM)) and its associated ATP-binding module (slr0982 (kpsT)), and a triple mutant (slr0977 (kpsM) and the putative pair sll0574 (kpsM)/sll0575 (kpsT)) produced EPS with different monosaccharidic composition compared to the EPS produced by the wild-type [14]. While Fisher et al. [14] did not report on the amounts of EPS produced by these mutants, a recent extensive characterization of a slr0977 (kpsM) mutant showed that the absence of Slr0977 resulted in a significant reduction of RPS (50%) and a smaller decrease of CPS (20%) [8]. In addition, a mutant lacking Slr2107 (another KpsM homologue) did not show significant differences in the total carbohydrates, RPS, and CPS compared to the wild-type [13]. Although one must bear in mind the growth conditions and the Synechocystis substrain used in those studies, disruption of slr0977 (kpsM) is thus responsible for one of the most significant reductions in the amount of RPS reported to date.
Nevertheless, it is important to highlight that kpsM has three putative homologues in Synechocystis: slr0977, slr2107, and sll0574 (Figure 1A), and it is necessary to clarify the role of the proteins, encoded by these genes, on EPS production. Traditionally, this would be tackled by generating a triple knockout mutant. However, the systematic knockout of multiple genes in Synechocystis is a time-consuming task due to its relatively slow growth rate compared to other bacteria, polyploidy [15], and the need to use different and increasing concentrations of antibiotics as selection markers. The use of a system such as CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR associated nuclease) allows faster and easier gene editing, cleavage, or inactivation [16]. In addition, CRISPRi (interference), which relies on the use of a nuclease-deficient Cas9 (dCas9) and a single-guide RNA (sgRNA), enables targeted gene regulation as the dCas9-sgRNA complex blocks the RNA polymerase binding or the elongation, resulting in gene repression. The main advantage of CRISPRi over traditional gene knockouts is the ability to repress multiple genes simultaneously, as elegantly demonstrated in 2016 by Yao et al. [17] that reported the repression of up to four genes, providing the CRISPRi multiplex proof-of-concept for cyanobacteria.
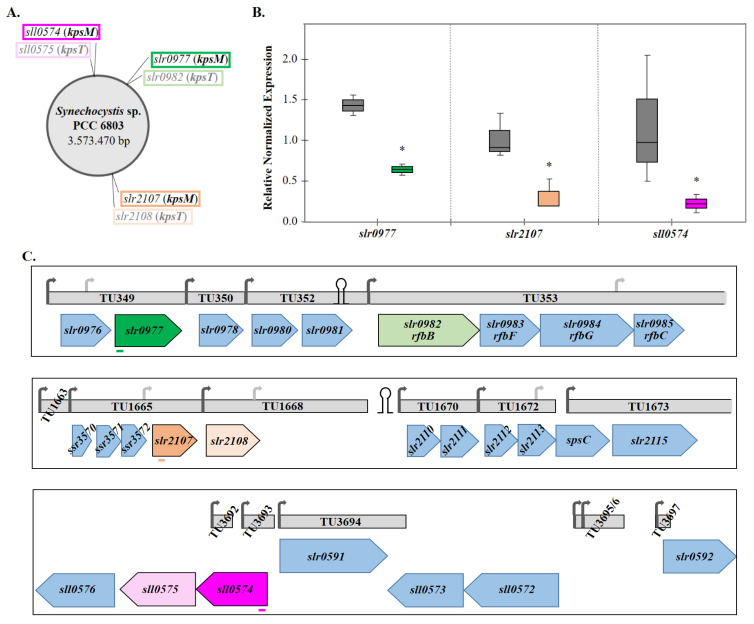
Figure 1.
EPS-related genes in Synechocystis sp. PCC 6803 encoding a putative transport permease of the ABC transporter-dependent pathway. (A) Location of the putative homologues of kpsM/kpsT in Synechocystis chromosome (kpsT is the second component of the two-protein complex, and is responsible for ATP-binding). (B) CRISPRi multiplex repression of three kpsM homologues (sll0574, slr0977, and slr2107), evaluated by RT-qPCR. The catalytically dead Cas9 (dCas9) and the 3 single guide-RNAs were constitutively expressed from promoters PpsbA2 and PL31, respectively. Expression of the target genes in the 3-sgRNA mutant (slr0977: green; slr2107: orange; sll0574: pink) relative to Synechocystis wild-type harboring dCas9 (grey). Data from at least two biological replicates and three technical replicates were normalized against three reference genes (rrn16S, rnpB, and petB), the whiskers represent the minimum and maximum non-outlier values in the data set. * p-value ≤ 0.05. (C) Schematic representation of the genomic context of the three target genes. sgRNA binding sites are depicted as colored lines below the target gene. Neighboring genes are annotated according to information available at the CyanoBase and KEGG databases. The transcriptional unit and transcription start sites (arrows; light grey indicate internal TSSs) are annotated according to Kopf et al. [25]. The predicted terminators (loops) were found using the FindTerm algorithm (Softberry).
In this work, to pursue the unravelling of cyanobacterial EPS assembly and export pathways, the CRISPRi system was employed as a tool for the multiplex repression of EPS-related genes in Synechocystis, namely for the three kpsM homologues (slr0977, slr2107 and sll0574). The generated mutant was characterized in terms of growth and carbohydrate production, and its phenotype compared to the conventional single knockout mutants generated by double homologous recombination.
2. Experimental Section
To construct the strain that will serve as a control and genetic background for the CRISPRi experiments (Syn dCas9), Synechocystis sp. PCC 6803 (Pasteur Culture Collection), substrain Kazusa [18,19] was transformed with the pMD19T vector harboring the sequence encoding the dCas9 from Streptococcus pyogenes under the control of the constitutive promoter PpsbA2. This construct was integrated into the psbA1 neutral site of the Synechocystis chromosome (Figure S1). For the simultaneous repression of the three putative kpsM homologues (slr0977, slr2107, and sll0574), an array of 3-sgRNAs was designed based on Larson et al. [20] and constructed as described [17]. Each 100 bp sgRNA unit comprised (i) its own promoter, (ii) the dCas9 binding handle and protospacer, and (iii) a terminator. The designed sgRNAs (Table 1) were evaluated for potential off-target binding sites using the CasOT software [21]. All the potential off-targets detected contained 6 or more mismatches compared to the sgRNA protospacer, including at least 1 in the 12 bp seed region (Table S1), so off-target binding was likely not significant. Therefore, these sgRNAs were expressed constitutively using the PL31 promoter (without a TetR repressor) and introduced into Syn dCas9 using a pLYK2-derived replicative plasmid (Table S2).
Table 1.
Sequences of the three sgRNAs used in this study.
| sgRNA Identifier/(Position) * | sgRNA Sequence Including PAM # |
|---|---|
| sll0574 (15) | GGGGACCAGTTCACCCTTGTCGG |
| slr0977 (16) | CCCCCAGAACTGATTATTGAAGCAGGAC |
| slr2107 (56) | CCCATGACTGGTTGCGATTGACGAT |
* sgRNA nucleotide binding position counting from the start codon. # underlined nucleotides indicate the protospacer adjacent motif (PAM).
The multiplex repression of the kpsM homologues was quantified by RT-qPCR, assessing the expression of slr0977, slr2107, and sll0574 in the 3-sgRNA kpsM mutant compared to the Syn dCas9. Sample collection (using three clones of the 3-sgRNA kpsM mutant), RNA extraction, cDNA synthesis, as well as the control measurements and PCRs were performed as previously described [22], except that three-fold standard dilutions of the cDNAs were made (1/3, 1/9, 1/27 and 1/81). The RT-qPCR reactions (10 µL) were setup as described by Pinto et al. [23] using the iTaq™ Universal SYBR® Green Supermix (Bio-Rad(Hercules, CA, USA)), 1 μL of template cDNA and the primers listed in Table S3. Validation of the reference genes (rrn16S, petB, and rnpB) and data analysis were performed using the Bio-Rad CFX Maestro™ 1.1 software. The single sll0574 knockout mutant was generated via double homologous recombination, by partially replacing the gene with a kanamycin (Km) resistance cassette, as described by Santos et al. [8]. The mutants were characterized in terms of growth (absorbance and chlorophyll a content) and carbohydrate production (total carbohydrates, CPS, and RPS) using the phenol-sulfuric acid method [24], as described previously by Santos et al. [8]. Data were statistically analyzed with GraphPad Prism v5 (GraphPad Software) using analysis of variance (ANOVA), followed by Tukey’s multiple-comparison test. For the qPCR data, analysis was performed using CFX Maestro Software (Bio-Rad) using analysis of variance (ANOVA).
3. Results & Discussion
Repression of Three kpsM Homologues in Synechocystis and Characterization of the 3-sgRNA Mutant
The kpsM homologues (slr0977, slr2107, and sll0574) were successfully repressed in the Synechocystis 3-sgRNA kpsM mutant compared to Syn dCas9 (control strain). The repression levels for the target genes were: ~60% for slr0977, ~70% for slr2107, and ~80% for sll0574 (Figure 1B). Since the repression of slr0977 was weaker compared to the other targets, the integrity of the sgRNAs in all clones was confirmed by sequencing. The repression level observed could be explained by slr0977 being the second gene in its operon (Figure 1C). The slr0977 predicted transcription start site (TSS) is 1175 bp upstream from the start codon [25] and 1191 bp upstream from the sgRNA binding site (Figure 1C). As mentioned by Yao et al. [17], the blocking of transcription elongation may not be as efficient if the gene of interest is part of an operon with a distant TSS. Nevertheless, the repression levels obtained in this study are within those previously reported for other targets in Synechocystis [17,26,27]. On the one hand, it is also possible to purposefully design the sgRNA further from the TSS, resulting in lower repression of the target gene(s) and enabling fine-tuning gene expression, as demonstrated by Shabestary et al. [26]. On the other hand, a partial repression will allow to sustain cell viability, even when essential genes are targeted, allowing the identification of new phenotypes.
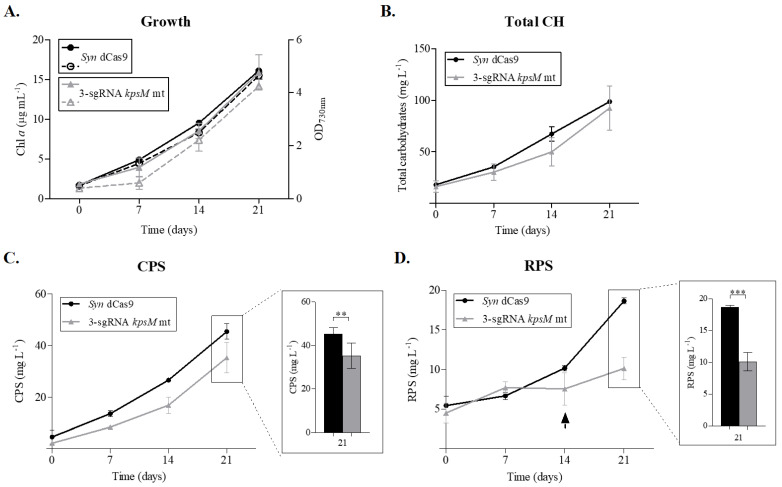
Subsequently, the 3-sgRNA kpsM mutant was characterized in terms of growth and carbohydrate production. Repression of the kpsM homologues did not significantly affect growth, compared to the Syn dCas9 (Figure 2A) and wild-type strains. However, similarly to the slr0977 knockout mutant, the 3-sgRNA kpsM mutant displayed a clumping phenotype at low cell densities [8]. Regarding total carbohydrates, the 3-sgRNA kpsM mutant produced approximately the same amount as the Syn dCas9 strain (Figure 2B). However, it had approximately 20% less CPS and 40% less RPS at 21 days of cultivation (Figure 2C,D).
Figure 2.
Growth curves and carbohydrate production by Synechocystis sp. PCC 6803 constitutively expressing the dead Cas 9 (Syn dCas9), and the 3 single guide-RNAs kpsM mutant targeting sll0574, slr0977 and slr2107 (3-sgRNA kpsM mt). Growth was monitored by measuring the optical density at 730 nm (full lines) and chlorophyll a (Chl a) (dashed lines) (A). Total carbohydrates (Total CH) (B), capsular polysaccharides (CPS) (C), and released polysaccharides (RPS) (D) were measured by the phenol-sulfuric acid method [24] and expressed as milligrams per liter of culture. The arrow indicates the point of divergence between the amount of RPS of Syn dCas9 and the 3-sgRNA kpsM mutant. Cells were grown in BG11 medium at 30 °C under a 12 h light (50 µE m−2 s−1)/12 h dark regimen, with orbital shaking at 150 rpm. Experiments were performed in triplicate, and statistical analysis is presented for the final time point (** p-value ≤ 0.01 *** p-value < 0.001).
This phenotype is very similar to the one observed for the slr0977 single knockout mutant [8], suggesting that the protein encoded by slr0977 could be the main KpsM homologue involved in RPS export, at least in the conditions tested. However, in the slr0977 single mutant, at 21 days, the amount of RPS is 50% less compared to the wild-type [8], while in the 3-sgRNA kpsM mutant, this difference only reaches 40%. In addition, while the amount of RPS for the slr0977 mutant is already reduced at the start of the experiment [8], in the 3-sgRNA kpsM mutant, this asymmetry is only noticeable after 14 days of cultivation (Figure 2D, arrow), which could be due to the weaker level of repression achieved for slr0977 (60%). This reduction on RPS production occurs without a significant change in the amount of total carbohydrates. In the single slr0977 mutant, this is associated with the intracellular accumulation of poly-hydroxybutyrate (PHB) [8], as it may happen in the 3-sgRNA kpsM mutant.
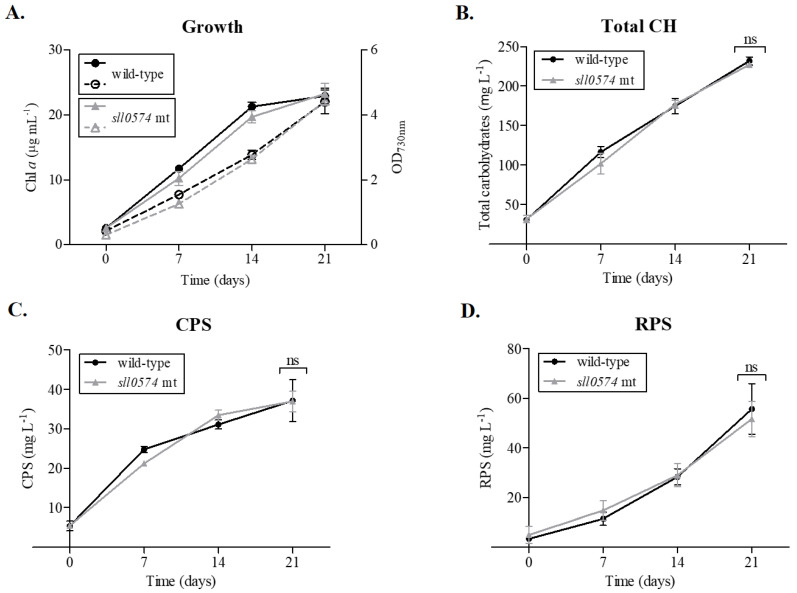
To our knowledge, no single mutant on the third kpsM homologue, sll0574, had been previously generated. Therefore, we generated a sll0574 knockout mutant by partially replacing the gene with a kanamycin (Km) resistance cassette via double homologous recombination (Table S2) and characterized it in terms of its growth and carbohydrate content. The sll0574 mutant did not show any significant differences in growth, total carbohydrates, RPS, or CPS compared to the wild-type (Figure 3), as it was previously reported for the slr2107 mutant [13]. CRISPRi mutants for each target gene were not generated, as the goal was to evaluate the effect of the simultaneous repression of the three kpsM homologues. Although each sgRNA could have off-target effects (detailed analysis in Table S1), Yao et al. have previously shown that the expression of dCas9 from various weak and moderate promoters with a non-targeting, “dummy” sgRNA does not significantly affect the growth of Synechocystis or its transcriptome, suggesting that off-target binding with phenotypical consequences is indeed infrequent [17,28]. A direct comparison between deletion and repression mutants is not straightforward; however, the phenotype shared by the 3-sgRNA kpsM and slr0977 mutants, together with the absence of an EPS-related phenotype for the slr2107 and sll0574 single knockout mutants, further supports our hypothesis that Slr0977 is the key KpsM homologue involved in RPS export, at least under the conditions tested. In agreement, a comparative analysis of the transcriptomes of Synechocystis under ten different conditions [25] showed that slr0977 was indeed the most expressed, while the slr2107 transcript levels increased under specific stress conditions (low temperature and nitrogen depletion). In Kopf et al., no data were reported for sll0574 (consistent with the low levels detected in our RT-qPCR experiment).
Figure 3.
Growth curves and carbohydrate production by Synechocystis sp. PCC 6803 wild-type and the kpsM sll0574 mutant (sll0574 mt). Growth was monitored by measuring the optical density at 730 nm (full lines) and chlorophyll a (Chl a) (dashed lines) (A). Total carbohydrates (Total CH) (B), capsular polysaccharides (CPS) (C), and released polysaccharides (RPS) (D) were measured by the phenol-sulphuric acid method [24] and expressed as milligrams per liter of culture. Cells were grown in BG11 medium at 30 °C under a 12 h light (50 µE m−2 s−1)/12 h dark regimen, with orbital shaking at 150 rpm. Experiments were performed in triplicate, and statistical analysis is presented for the final time point (ns: not significant; p-value > 0.05).
4. Conclusions
In summary, the use of CRISPRi in multiplex to repress the three putative kpsM homologues in Synechocystis established a novel approach to tackle the redundancy of EPS-related genes. The use of this methodology not only expands the possibilities to study other putative redundant components, but also the simultaneous evaluation of homologues that in other bacteria are associated with the different pathways. As we expect that the cyanobacterial EPS biosynthetic pathways will diverge from the well-characterized bacterial ones, the use of CRISPRi will enable a faster screening of the role of the different players, allowing to piece together these molecular mechanisms. Although the use of CRISPRi in cyanobacteria is not yet widespread, this platform is certainly a powerful tool and will become more relevant as it is more frequently used.
Acknowledgments
The authors acknowledge the technical and scientific support of the i3S Scientific Platform “Cell Culture and Genotyping” in the real-time qPCR experiments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/life11111198/s1: Figure S1: Synechocystis sp. PCC 6803 wild-type slr1181 (psbA1) genomic context and confirmation of the Syn dCas9 mutant generation. Table S1: list of potential off-target binding sites for the three sgRNAs used in this work; Table S2: list of organisms and plasmids used/generated in this work; Table S3: primer nucleotide sequences and annealing temperatures (Ta) used in RT-qPCR; Table S4: Primer nucleotide sequences used to verify the segregation of the Syn dCas9 strain.
Author Contributions
Conceptualization, M.S., C.C.P., E.P.H. and P.T.; methodology, M.S., C.C.P. and L.Y.; validation, M.S. and C.C.P.; formal analysis, M.S. and C.C.P.; investigation, M.S., L.Y. and C.C.P.; resources, E.P.H. and P.T.; writing—original draft preparation, M.S., C.C.P. and P.T.; writing—review and editing, M.S., C.C.P., E.P.H. and P.T.; visualization, M.S., C.C.P. and P.T.; supervision, E.P.H. and P.T.; project administration, P.T.; funding acquisition, P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by FEDER-Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020-Operacional Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through the Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior, in the framework of projects POCI-01-0145-FEDER-028779 (PTDC/BIA-MIC/28779/2017), and also supported by national funds through the FCT, I.P., under the projects UIDB/04293/2020 and UIDP/04293/2020. We also greatly acknowledge FCT for the PhD fellowship SFRH/BD/119920/2016 (M.S.) and the assistant researcher contract CEECIND/00259/2017 (C.C.P.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pereira S., Zille A., Micheletti E., Moradas-Ferreira P., De Philippis R., Tamagnini P. Complexity of cyanobacterial exopolysaccharides: Composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009;33:917–941. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- 2.Rossi F., De Philippis R. Exocellular polysaccharides in microalgae and cyanobacteria: Chemical features, role and enzymes and genes involved in their biosynthesis. In: Borowitzka M.A., Beardall J., Raven J.A., editors. The Physiology of Microalgae. 1st ed. Springer; Cham, Switzerland: 2016. pp. 565–590. [Google Scholar]
- 3.Flores C., Lima R.T., Adessi A., Sousa A., Pereira S.B., Granja P.L., De Philippis R., Soares P., Tamagnini P. Characterization and antitumor activity of the extracellular carbohydrate polymer from the cyanobacterium Synechocystis ΔsigF mutant. Int. J. Biol. Macromol. 2019;136:1219–1227. doi: 10.1016/j.ijbiomac.2019.06.152. [DOI] [PubMed] [Google Scholar]
- 4.Costa R., Costa L., Rodrigues I., Meireles C., Soares R., Tamagnini P., Mota R. Biocompatibility of the biopolymer Cyanoflan for applications in skin wound healing. Mar. Drugs. 2021;19:147. doi: 10.3390/md19030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierre G., Delattre C., Dubessay P., Jubeau S., Vialleix C., Cadoret J.-P., Probert I., Michaud P. What is in store for EPS microalgae in the next decade? Molecules. 2019;24:4296. doi: 10.3390/molecules24234296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira S.B., Sousa A., Santos M., Araújo M., Serôdio F., Granja P.L., Tamagnini P. Strategies to obtain designer polymers based on cyanobacterial extracellular polymeric substances (EPS) Int. J. Mol. Sci. 2019;20:5693. doi: 10.3390/ijms20225693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Woude A.D., Angermayr S.A., Puthan V.V., Osnato A., Hellingwerf K.J. Carbon sink removal: Increased photosynthetic production of lactic acid by Synechocystis sp. PCC 6803 in a glycogen storage mutant. J. Biotechnol. 2014;184:100–102. doi: 10.1016/j.jbiotec.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Santos M., Pereira S.B., Flores C., Príncipe C., Couto N., Karunakaran E., Cravo S.M., Oliveira P., Tamagnini P. Absence of KpsM (Slr0977) impairs the secretion of extracellular polymeric substances (EPS) and impacts carbon fluxes in Synechocystis sp. PCC 6803. mSphere. 2021;6:e00003-21. doi: 10.1128/mSphere.00003-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitfield C., Wear S.S., Sande C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu. Rev. Microbiol. 2020;74:521–543. doi: 10.1146/annurev-micro-011420-075607. [DOI] [PubMed] [Google Scholar]
- 10.Pereira S.B., Mota R., Vieira C.P., Vieira J., Tamagnini P. Phylum-wide analysis of genes/proteins related to the last steps of assembly and export of extracellular polymeric substances (EPS) in cyanobacteria. Sci. Rep. 2015;5:14835. doi: 10.1038/srep14835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda K., Okuda Y., Enomoto G., Watanabe S., Ikeuchi M. Biosynthesis of a sulfated exopolysaccharide, synechan, and bloom formation in the model cyanobacterium Synechocystis sp. strain PCC 6803. eLife. 2021;10:e66538. doi: 10.7554/eLife.66538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jittawuttipoka T., Planchon M., Spalla O., Benzerara K., Guyot F., Cassier-Chauvat C., Chauvat F. Multidisciplinary evidences that Synechocystis PCC 6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PLoS ONE. 2013;8:e55564. doi: 10.1371/journal.pone.0055564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira S.B., Santos M., Leite J.P., Flores C., Eisfeld C., Büttel Z., Mota R., Rossi F., De Philippis R., Gales L., et al. The role of the tyrosine kinase Wzc (Sll0923) and the phosphatase Wzb (Slr0328) in the production of extracellular polymeric substances (EPS) by Synechocystis PCC 6803. Microbiologyopen. 2019;8:e00753. doi: 10.1002/mbo3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher M.L., Allen R., Luo Y., Curtiss R., III Export of extracellular polysaccharides modulates adherence of the cyanobacterium Synechocystis. PLoS ONE. 2013;8:e74514. doi: 10.1371/journal.pone.0074514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerulla K., Ludt K., Soppa J. The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology. 2016;162:730–739. doi: 10.1099/mic.0.000264. [DOI] [PubMed] [Google Scholar]
- 16.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao L., Cengic I., Anfelt J., Hudson E.P. Multiple gene repression in cyanobacteria using CRISPRi. ACS Synth. Biol. 2016;5:207–212. doi: 10.1021/acssynbio.5b00264. [DOI] [PubMed] [Google Scholar]
- 18.Trautmann D., Voss B., Wilde A., Al-Babili S., Hess W.R. Microevolution in cyanobacteria: Re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 2012;19:435–448. doi: 10.1093/dnares/dss024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanesaki Y., Shiwa Y., Tajima N., Suzuki M., Watanabe S., Sato N., Ikeuchi M., Yoshikawa H. Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 2012;19:67–79. doi: 10.1093/dnares/dsr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao A., Cheng Z., Kong L., Zhu Z., Lin S., Gao G., Zhang B. CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014;30:1180–1182. doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- 22.Gonçalves C.F., Pacheco C.C., Tamagnini P., Oliveira P. Identification of inner membrane translocase components of TolC-mediated secretion in the cyanobacterium Synechocystis sp. PCC 6803. Environ. Microbiol. 2018;20:2354–2369. doi: 10.1111/1462-2920.14095. [DOI] [PubMed] [Google Scholar]
- 23.Pinto F., Pacheco C.C., Ferreira D., Moradas-Ferreira P., Tamagnini P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE. 2012;7:e34983. doi: 10.1371/journal.pone.0034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 25.Kopf M., Klähn S., Scholz I., Matthiessen J.K.F., Hess W.R., Voß B. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014;21:527–539. doi: 10.1093/dnares/dsu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabestary K., Anfelt J., Ljungqvist E., Jahn M., Yao L., Hudson E.P. Targeted repression of essential genes to arrest growth and increase carbon partitioning and biofuel titers in cyanobacteria. ACS Synth. Biol. 2018;7:1669–1675. doi: 10.1021/acssynbio.8b00056. [DOI] [PubMed] [Google Scholar]
- 27.Kirtania P., Hódi B., Mallick I., Vass I.Z., Fehér T., Vass I., Kós P.B. A single plasmid based CRISPR interference in Synechocystis 6803—A proof of concept. PLoS ONE. 2019;14:e0225375. doi: 10.1371/journal.pone.0225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao L., Shabestary K., Björk S.M., Asplund-Samuelsson J., Joensson H.N., Jahn M., Hudson E.P. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat. Commun. 2020;11:1666. doi: 10.1038/s41467-020-15491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.