Abstract
We report on the first case of fatal septicemia caused by Bordetella hinzii. The causative organism exhibited a biochemical profile identical to that of Bordetella avium with three commercial identification systems (API 20E, API 20 NE, and Vitek GNI+ card). However, its cellular fatty acid profile was not typical for either B. avium or previously reported strains of B. hinzii. Presumptive identification of the patient's isolate was accomplished by traditional biochemical testing, and definitive identification was achieved by 16S rRNA gene sequence analysis. Phenotypic features useful in distinguishing B. hinzii from B. avium were production of alkali from malonate and resistance to several antimicrobial agents.
The genus Bordetella comprises seven species including three recent additions over the past 5 years. Bordetella pertussis and Bordetella parapertussis are strict human pathogens that cause the respiratory tract infection called whooping cough. Bordetella bronchiseptica, a commensal organism of the respiratory tract in many animals, also causes respiratory infections. It is less pathogenic in humans, among whom it has been associated with respiratory and systemic infections mostly in immunocompromised patients (21). Bordetella avium has consistently been associated with rhinotracheitis in poultry but has never been reported to cause infection in humans (9). Bordetella holmesii and Bordetella trematum exclusively infect humans, causing either septicemia and endocarditis or ear and wound infections, respectively (17, 20). Lastly, Bordetella hinzii colonizes the respiratory tracts of poultry (18). Five strains of B. hinzii have so far been isolated from human sources (3, 7, 17, 18), and three of these strains have been documented to cause infection (3, 7). We report on the first human infection with B. hinzii that resulted in fatal septicemia.
CASE REPORT
A 69-year-old black man presented with a 3-week history of painless jaundice accompanied by decreased appetite, dark urine, and loose stools. He denied fever, nausea, or vomiting. He had attended a cookout at a farm 2 weeks before admission. He had had a 15-lb weight loss in the previous month. His past surgical and medical histories were negative, and he was not taking any medicines. He denied intravenous drug use and had no other risk factors for human immunodeficiency virus infection. He was not on any medications. On admission, he was afebrile and icteric. Neck, lung, and cardiovascular examinations were normal. The abdomen was soft and nontender. Bowel sounds were present. There were no palpable masses. The liver span was 10 cm, and there was no splenomegaly or lymphadenopathy. Rectal examination revealed no masses and a normal-sized prostate. Stools were negative for occult blood. Hepatitis B surface antigen, hepatitis B core antibody, and hepatitis C virus-specific antibody testing by enzyme-linked immunosorbent assay performed 2 weeks prior to admission were negative. Laboratory results were as follows: white blood cell count, 5.3 × 103/mm3 with 52% segmented neutrophils, 40% lymphocytes, and 8% monocytes; red blood cell count, 3.2 × 106/mm3; hemoglobin concentration, 12.5 g/dl; hematocrit, 33%; mean corpuscular volume, 102 fl; platelet count, 199 × 103/mm3; glucose concentration, 85 mg/dl; creatinine concentration, 0.3 mg/dl; urea nitrogen concentration, 11 mg/dl; total protein concentration, 6.3 g/dl; alkaline phosphatase concentration, 525 mU/ml (reference range, 43 to 130); serum glutamic oxalacetic transaminase level, 109 mU/ml (reference range, 8 to 40); serum glutamic pyruvic transaminase level, 144 mU/ml (reference range, 8 to 45); total bilirubin concentration, 28 mg/dl (reference range, 0.2 to 1.2); direct bilirubin concentration, 17.8 mg/ml (reference range, 0.0 to 2.0); amylase level, 269 U/liter (reference range, 44 to 128), lipase level, 1,520 U/liter (reference range, 4 to 240 U/liter). An ultrasound of the abdomen done 1 week before admission showed intrahepatic and extrahepatic ductal dilatation and gallbladder distension with the presence of sludge but no stones or masses. A computed tomographic scan of the abdomen done on the day of admission revealed dilatation of the intrahepatic biliary system and gallbladder distension, but no pancreatic abnormality or lymphadenopathy was noted. Chest X rays showed no infiltrates. The patient was started empirically on antibiotics, initially ampicillin-sulbactam but was soon switched to cefotetan. On day 4, he underwent endoscopic retrograde cholangiopancreatography which revealed a stricture at the ampulla of Vater and showed a 5-cm stricture of the common biliary duct. Multiple attempts to place a stent failed, and cytology brushings of the stricture showed inflammation but no evidence of malignancy. On hospital day 6, the patient spiked a temperature of 102°F. The white blood cell count was 15.2 × 103/mm3, with 96% segmented neutrophils, and blood samples were taken for culture. On day 8, the antibiotics were changed to ampicillin, gentamicin, and metronidazole. On day 9, the patient had a cardiopulmonary arrest and was resuscitated, and antibiotics were again changed to ticarcillin-sulbactam and ciprofloxacin. The patient died from sepsis on day 9. Postmortem examination and testing for human immunodeficiency virus infection were not authorized. Two sets of blood samples for culture each taken on day 6 and day 9 became positive within 24 h and grew only gram-negative rods in four of four aerobic bottles (BACTEC 9240 with PLUS AEROBIC/F and PLUS ANAEROBIC/F bottles; Becton Dickinson, Sparks, Md.).
MATERIALS AND METHODS
Bacterial strains.
The strains studied included the current isolate (strain BC-306), B. hinzii type strain LMG 13501, which was kindly provided by Peter Vandamme (Laboratory of Microbiology, University of Ghent, Ghent, Belgium), a B. hinzii strain obtained from cultures of blood from a human immunodeficiency virus-infected patient, strain BC-305 (LMG 13505) (3, 18), and B. avium type strain ATCC 35086.
Biochemical evaluation.
Gram staining, flagellar staining (Remel, Lenexa, Kans.), colonial morphology, and biochemical testing were evaluated from 24-h cultures on Trypticase soy agar base supplemented with 5% sheep blood incubated at 35°C in 5% CO2 unless specified otherwise. Slide catalase activity was tested from a 24-h growth on MacConkey agar. Tube catalase activity was determined from a 24-h growth on Trypticase-soy-yeast extract agar. Oxidase activity was tested with the Difco (Detroit, Mich.) oxidase slide. The API 20E and API 20NE strips (bioMérieux-Vitek, Hazelwood, Mo.) were used according to the recommendations of the manufacturer and were read at 24 and 48 h. The VITEK AMS GNI+ card (gram-negative identification card, software version R06.01; bioMérieux, Hazelwood, Mo.) was read automatically by the instrument within 24 h. The conventional biochemical tests used for the identification of gram-negative rods and the genus Bordetella (18, 19) included the triple sugar iron agar, oxidative-fermentative carbohydrate utilization (glucose, maltose, xylose, 10% lactose), urea, Voges-Proskauer, acetamide, and malonate tests. All standard biochemical media purchased from Remel were heavily inoculated, kept for 7 days at 35°C in 5% CO2, and examined daily.
Fatty acid methyl ester analysis.
Bacteria grown on Trypticase soy agar with 5% sheep blood at 35°C in 5% CO2 were harvested after 24 and 48 h. Whole-cell fatty acid (CFA) methyl esters were prepared, separated by gas-liquid chromatography, and identified with the Microbial Identification System (Sherlock; software version 1.06; MIDI, Inc., Newark, Del.) as described previously (10). The organism profiles were searched against the clinical library of the MIDI system (version 4.0), and similarity indices for each match were computed by the instrument. Similarity indices below 0.5 represent poor matches (below 3 standard deviations from the mean profile in the database). Similarity indices that exceed 0.5 are generally considered reliable for species identification.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed by the disk diffusion method, the E-test (AB BIODISK, Solna, Sweden), and the broth microdilution method (gram-negative panel; Dade MicroScan, Sacramento, Calif.). Most antibiotics were tested by at least two methods (see Table 2 in the Results section). Results were interpreted according to the National Committee for Clinical Laboratory Standards (NCCLS) (13) breakpoints for nonfermenting gram-negative rods or breakpoints for members of the family Enterobacteriaceae for antibiotics without breakpoints for nonfermenting rods (ampicillin, amoxicillin-clavulanic acid, cefazolin, cephalothin, cefotetan, and cefuroxime).
TABLE 2.
Antimicrobial susceptibility testing of B. hinzii and B. avium by three methods
| Antibiotica | Method | Susceptibilityb
|
|||
|---|---|---|---|---|---|
| B. hinzii BC-306 | B. hinzii LMG13501T | B. hinzii BC-305 | B. avium ATCC 35086T | ||
| Ampicillin* | Disk | R (9) | R (13) | R (11) | S (30) |
| Ampicillin* | E-test | I (16) | I (16) | I (16) | S (0.5) |
| Ampicillin* | Micro brothc | R (32) | R (32) | R (32) | S (<1) |
| Ampicillin-sulbactam | Disk | R (11) | I (14) | I (14) | S (33) |
| Ampicillin-sulbactam | E-test | I (16/8) | I (16/8) | I (16/8) | S (0.5) |
| Ampicillin-sulbactam | Micro broth | I (16/8) | R (32/16) | R (32/16) | S (<2/1) |
| Amoxicillin-clavulanic acid* | Disk | I (17) | S (21) | S (20) | S (34) |
| Amoxicillin-clavulanic acid* | E-test | R (32) | R (16) | R (16) | S (0.5) |
| Ticarcillin | Disk | I (17) | S (21) | S (20) | S (36) |
| Ticarcillin | Micro broth | I (32) | I (32) | I (64) | S (<4) |
| Ticarcillin-clavulanic acid | Disk | S (21) | S (23) | S (23) | S (36) |
| Ticarcillin-clavulanic acid | E-test | I (32) | I (32) | I (64) | S (0.75) |
| Ticarcillin-clavulanic acid | Micro broth | S (16) | S (16) | S (16) | S (<4) |
| Piperacillin | Disk | S (35) | S (37) | S (36) | S (37) |
| Piperacillin | Micro broth | S (<4) | S (<4) | S (<4) | S (<4) |
| Piperacillin-tazobactam | Disk | S (35) | S (37) | S (36) | S (38) |
| Piperacillin-tazobactam | E-test | S (1) | S (1) | S (1) | S (1) |
| Piperacillin-tazobactam | Micro broth | S (<4/4) | S (<4/4) | S (<4/4) | S (<4/4) |
| Cefazolin* | Disk | R (10) | R (10) | R (13) | I (17) |
| Cephalotin* | Disk | S (22) | S (24) | S (25) | S (31) |
| Cefotetan* | Disk | R (6) | R (6) | R (6) | S (20) |
| Cefuroxime* | Disk | R (6) | R (6) | R (6) | I (16) |
| Ceftriaxone | Disk | I (14) | R (13) | I (15) | S (28) |
| Ceftriaxone | E-test | R (64) | R (64) | R (64) | S (4) |
| Ceftriaxone | Micro broth | I (32) | I (32) | I (32) | S (2) |
| Cefotaxime | Micro broth | I (32) | I (32) | I (32) | S (2) |
| Ceftazidime | Disk | S (26) | S (26) | S (27) | S (31) |
| Ceftazidime | E-test | S (4) | S (4) | S (4) | S (2) |
| Ceftazidime | Micro broth | S (2) | S (2) | S (2) | S (<1) |
| Cefepime | Disk | S (25) | S (27) | S (27) | S (30) |
| Cefepime | E-test | S (8) | S (8) | S (8) | S (4) |
| Cefepime | Micro broth | S (4) | S (4) | S (4) | S (2) |
| Imipenem | Disk | S (30) | S (30) | S (30) | S (26) |
| Imipenem | E-test | S (2) | S (2) | S (2) | S (2) |
| Imipenem | Micro broth | S (1) | S (1) | S (1) | S (1) |
| Aztreonam | Disk | R (6) | R (6) | R (6) | I (17) |
| Aztreonam | Micro broth | R (32) | R (>32) | R (32) | I (16) |
| Chloramphenicol | Disk | R (10) | I (13) | R (10) | S (2) |
| Tetracycline | Disk | S (19) | S (25) | S (25) | S (32) |
| Trimethoprim-sulfamethoxazole | Disk | S (33) | S (34) | S (35) | S (35) |
| Trimethoprim-sulfamethoxazoled | E-test | S (0.047) | S (0.047) | S (0.023) | S (0.047) |
| Ciprofloxacin | Disk | I (17) | I (19) | I (18) | S (23) |
| Ciprofloxacin | E-test | R (4) | R (4) | R (4) | I (2) |
| Ciprofloxacin | Micro broth | I (2) | I (2) | I (2) | S (1) |
| Levofloxacin | Disk | S (20) | S (23) | S (22) | S (26) |
| Levofloxacin | E-test | S (2) | S (2) | S (2) | S (0.5) |
| Gentamicin | Disk | S (22) | S (19) | S (20) | S (24) |
| Gentamicin | E-test | S (2) | S (4) | S (4) | S (2) |
| Gentamicin | Micro broth | S (4) | S (4) | S (4) | S (2) |
| Amikacin | Disk | S (24) | S (23) | S (22) | S (22) |
| Amikacin | Micro broth | S (8) | S (8) | S (8) | S (<4) |
| Tobramycin | Disk | S (18) | S (15) | S (15) | S (22) |
| Tobramycin | Micro broth | R (8) | R (16) | R (16) | S (2) |
| Netilmycin | Disk | S (25) | S (23) | S (22) | S (26) |
Results with antibiotics marked by asterisk were interpreted according to NCCLS breakpoints for Enterobacteriaceae; all others were interpreted according to NCCLS breakpoints for nonfermenting gram-negative rods; S, susceptible; I, intermediate; R, resistant.
Numbers in parentheses for the disk diffusion method indicate the diameters of inhibition zones (in millimeters); numbers in parentheses for the E-test and broth microdilution method indicate the MICs (in micrograms per milliliter).
Micro broth, broth microdilution method.
For trimethoprim-sulfamethoxazole, the result is expressed as the MIC of trimethoprim.
16S rRNA gene sequencing and phylogenetic analysis.
Genomic DNA was extracted from bacterial colonies by alkaline lysis. The rRNA gene was amplified by PCR with universal primers 8FPL and DG74 (positions 8 to 27 and 1522 to 1540, respectively; Escherichia coli numbering) (2, 6, 8) as described previously (8) on a Perkin-Elmer model 9700 thermocycler (Perkin-Elmer Applied Biosystems, Foster City, Calif.). After denaturation at 95°C for 10 min, DNA samples were subjected to 30 cycles of amplification with denaturation at 95°C for 30 s, annealing at 68°C for 30 s, and extension at 72°C for 45 s, followed by a final elongation step at 72°C for 10 min. The PCR product was column purified (Microcon 100; Amicon, Beverly, Mass.), and cycle sequencing was subsequently performed (6, 14) with the Big Dye terminator kit (Perkin-Elmer Applied Biosystems) as recommended by the manufacturer on a model 9700 thermocycler with the preprogrammed Big Dye cycle sequencing parameters on the instrument. Sequences assembled and edited with the Sequencher software (version 3.1) were used to generate 1,522-bp consensus sequences for the four strains corresponding to positions 8 to 1540 in the 16S rRNA gene of E. coli. The consensus sequences were searched against the GenBank database by using the BLAST tool (1) and were submitted to the Ribosomal Database Project for similarity ranking (11). The 16S rRNA gene sequences from the bordetellae, related members of the family Alcaligenaceae, and other species phenotypically related to the bordetellae were retrieved from GenBank and were aligned by use of CLUSTAL_X software (16). A phylogenetic tree was constructed by using PHYLIP, version 3.573 (J. Q. Felsenstein, PHYLIP—phylogeny inference package, version 3.573, Department of Genetics, University of Washington, Seattle, WA. [http://evolution.genetics.washington.edu/phylip.html]), with Pseudomonas aeruginosa as the outgroup in the dendrogram. A total of 1,341 bases were used for phylogenetic analysis with four treeing algorithms: neighbor-joining, Fitch-Margoliash, maximum parsimony, and maximum likelihood, assuming a molecular clock model. With each treeing algorithm, the branching pattern was also tested with 1,000 bootstraps by the SEQBOOT program in the PHYLIP package (4).
Nucleotide sequence accession numbers.
The sequences for the B. hinzii and B. avium type strains have been deposited in GenBank under the following accession numbers: AF177667 (B. hinzii LMG 13501) and AF177666 (B. avium ATCC 35086).
RESULTS
The patient isolate (strain BC-306) was compared with B. hinzii LMG 13501, B. hinzii BC-305, and B. avium ATCC 35086. Each consisted of pleomorphic gram-negative coccobacilli and slender rods that were motile with peritrichous flagellae. Colonies were visible after 24 h of aerobic incubation at 35 or 42°C on Trypticase soy agar with 5% sheep blood and on MacConkey agar with crystal violet either with or without 5% CO2. B. hinzii colonies were approximately 2 mm in diameter, nonhemolytic, round, glistening, and raised gray-white with entire edges. Colonies of B. avium ATCC 35086 were also nonhemolytic and a darker gray and measured approximately 1.2 mm in diameter. The B. hinzii strains were weakly slide catalase positive, and catalase activity was better demonstrated by the tube catalase test. Compared to the other three strains, slide catalase activity was more vigorous with the B. avium type strain. Oxidase activity was positive for all strains. On a triple sugar iron agar slant, they produced an alkaline slant and alkaline butt reaction with no gas or H2S. When tested with the API 20E and API 20NE strips and VITEK AMS GNI+ card, the isolates yielded identical profiles. Results with the three commercial systems were as follows: (i) numerical code 0000067 with API 20NE, identifying all isolates as B. avium (percent identification, 95.6%; good identification); these organisms were negative for caprate and assimilated adipate, malate, citrate, and phenylacetate; (ii) numerical code 020000441 with API 20E, corresponding to Bordetella species (percent identification, 38.7%) in which the organisms were positive only for citrate hydrolysis; and (iii) numerical code 40200000040 on the VITEK GNI+ card with no identification (84% nonfermenting gram-negative rods; 7% B. bronchiseptica); the strains were only citrate positive.
Results for CFA analysis are shown in Table 1. The closest “match” to the current isolate, BC-306, was B. avium, with a low similarity index (0.249). We suspected that isolate BC-306 might be B. hinzii, originally reported by our group to cause human infection (3), whereas human infections with B. avium have never been reported. B. hinzii is not included in the database of the MIDI system. CFA profiles previously distinguished B. hinzii from B. avium after 24 h of growth (3, 7), but the profile for isolate BC-306 was atypical (Table 1). Notably, the proportions of C17:0 cyc and its precursor, C16:1ω7c, were markedly different compared to those for the type strain and previously reported isolates of B. hinzii. The CFA profiles of B. hinzii and B. avium after 48 h of culture incubation were indistinguishable.
TABLE 1.
Proportions of short-chain fatty acids of B. hinzii and B. avium
| Isolate | Culture period (h) | % Short-chain fatty acids
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C12:0 | 2OH-C12:0 | C14:0 | 2OH-C14:0 | 3OH C14:0 | C16:1W7c | C16:0 | C17:0 cyc | C17:0 | 2OH-C16:0 | 3OH-C16:0 | C18:1W7c | C18:0 | C19:0 iso | C19:0 cyclo w8c | Organism match | SIa | ||
| B. hinzii BC-306b | 24 | 0.48 | 2.34 | 0.54 | 3.51 | 8.00 | 7.31 | 39.01 | 26.67 | 0.63 | 1.50 | 0.52 | 1.29 | 8.33 | 0.37 | B. avium | 0.249 | |
| B. hinzii LMG13501T | 24 | 0.47 | 2.28 | 0.65 | 3.35 | 7.75 | 18.50 | 35.66 | 18.51 | 0.74 | 0.85 | 0.37 | 2.79 | 7.66 | Alcaligenes xylos-oxidans | 0.294 | ||
| B. hinzii BC-305c | 24 | 0.53 | 2.03 | 0.65 | 3.14 | 6.57 | 20.58 | 36.95 | 19.13 | 0.73 | 0.57 | 0.48 | 2.42 | 5.95 | 0.39 | 0.48 | B. avium | 0.385 |
| B. avium ATCC 35086Td | 24 | 0.63 | 2.54 | 1.03 | 3.17 | 7.57 | 5.22 | 38.42 | 30.11 | 0.54 | 0.89 | 1.59 | 6.71 | 0.8 | 0.77 | B. avium | 0.521 | |
| B. hinziie | 24 | NAf | NA | NA | NA | NA | 17–24 | 38–40 | 16–22 | NA | NA | NA | NA | NA | NA | NA | NA | |
| B. hinzii BC-306 | 48 | 0.43 | 2.16 | 0.59 | 3.17 | 7.12 | 4.50 | 35.74 | 31.65 | 0.59 | 2.29 | 0.42 | 0.86 | 8.42 | 0.5 | 1.31 | B. avium | 0.45 |
| B. hinzii LMG13501T | 48 | 0.4 | 2.34 | 0.61 | 3.38 | 7.82 | 3.80 | 34.53 | 33.24 | 0.84 | 1.48 | 0.48 | 1.65 | 7.60 | 1.26 | B. avium | 0.59 | |
| B. hinzii BC-305c | 48 | 0.41 | 2.25 | 0.60 | 3.28 | 7.40 | 4.16 | 35.08 | 32.64 | 0.87 | 1.22 | 0.43 | 1.07 | 7.87 | 0.58 | 1.43 | B. avium | 0.533 |
| B. avium ATCC 35086T | 48 | 0.53 | 2.51 | 1.56 | 3.07 | 7.11 | 2.10 | 36.45 | 35.22 | 0.92 | 1.45 | 0.73 | 6.23 | 2.10 | B. avium | 0.579 | ||
SI, similarity index.
BC-306 is the strain under investigation.
Results for B. hinzii BC-305 from a 24-h culture were reported previously (3).
Results for the type strain of B. avium are similar to those reported previously for six strains of B. avium (3).
B. hinzii strains from a cystic fibrosis patient cultured under different conditions (7).
NA, not available.
By standard biochemical testing, all four strains were asaccharolytic and did not metabolize maltose, xylose, glucose, or 10% lactose either with or without exposure to air; they were urea and Voges-Proskauer negative. Strain BC-306 and the two B. hinzii strains were positive for acetamide and malonate after 5 days. The B. avium type strain was positive for acetamide but negative for malonate. Importantly, the acetamide and malonate tests on the VITEK GNI+ card were read by the instrument as negative within 24 h for all strains. On the basis of the biochemical profile, the bacterium BC-306 was presumptively identified as B. hinzii. Production of alkali from malonate by B. hinzii distinguishes it from B. avium, which is negative (18).
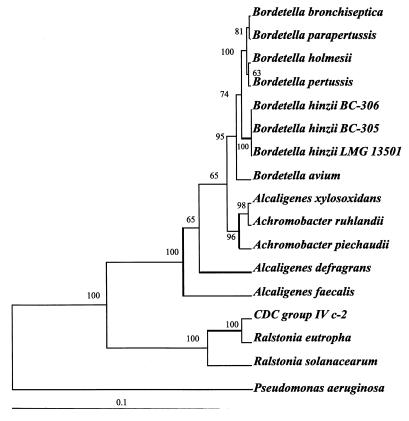
To reconcile the biochemical data with an atypical CFA profile, we definitively identified isolate BC-306 as B. hinzii by comparing its 16S rRNA gene sequence with the sequences of the other two strains and the B. avium type strain. The sequences obtained from all three strains of B. hinzii were 100% identical. The closest match to B. hinzii in both GenBank and RDP searches was B. bronchiseptica. The phylogenetic relationships of isolate BC-306 to the other bordetellae and phenotypically related species are shown in Fig. 1. With all treeing algorithms, the bordetellae were monophyletic. The branching order among B. pertussis, B. parapertussis, B. holmesii, and B. bronchiseptica differed among the various algorithms; however, in all trees, these species formed a single cluster. B. hinzii occupied an intermediate position between that cluster and B. avium (bootstrap values of 100%). With all algorithms, B. hinzii was most closely related to B. bronchiseptica and B. parapertussis. Pairwise sequence comparisons among the 16S rRNA sequences of the bordetellae showed that the B. hinzii sequence differed by 8 bp (0.5%) from B. parapertussis, 9 bp (0.6%) from B. bronchiseptica, 11 bp (0.7%) from B. avium, 12 bp (0.8%) from B. pertussis, and 19 bp (1.2%) from B. holmesii. Sixty-six percent of the variability in the 16S rRNA gene sequence among the bordetellae segregated in the first 500 bp. The full-length sequence that we obtained from the B. avium type strain diverged from the previously published partial sequence at six positions (20).
FIG. 1.
Maximum likelihood consensus dendrogram based on 1,341 consecutive positions of 16S rRNA of bordetellae and related species. The lower bar indicates the genetic distance. The sequence accession numbers retrieved from GenBank for phylogenetic tree construction are as follows: B. bronchiseptica, U04948; B. parapertussis, U04949; B. pertussis, AF142326; B. holmesii, U04820; Achromobacter ruhlandii, AB010840; Achromobacter piechaudii, AB010841; Alcaligenes xylosoxidans subsp. xylosoxidans, M22509; Alcaligenes defragrans, AJ005450; Alcaligenes faecalis, M22508; Ralstonia eutropha, AF027407; Ralstonia solanacearum, X67036; CDC Group IV C-2, AF067657; and P. aeruginosa (used as the outgroup), Z76651.
Results of antimicrobial susceptibility testing are shown in Table 2. MICs were very similar for the three B. hinzii strains. Overall, testing by the three methods consistently gave concordant results, with discrepancies observed with four antibiotics. B. hinzii strains were susceptible to ticarcillin and tobramycin by the disk diffusion method but resistant by the broth microdilution method, susceptible to amoxicillin-clavulanic acid by the disk diffusion method but resistant by the E-test, and susceptible to ticarcillin-clavulanic acid by the disk diffusion method and the broth microdilution method but intermediate by the E-test. For some antibiotics, the MICs determined by the E-test were within 1 log2 dilution of the MICs determined by the broth microdilution method. B. hinzii strains were all sensitive to piperacillin, piperacillin-tazobactam, cephalothin, ceftazidime, cefepime, imipenem, tetracycline, trimethoprim-sulfamethoxazole, levofloxacin, gentamicin, amikacin, and netilmicin. They were either intermediate or resistant to ampicillin, ampicillin-sulbactam, cefazolin, cefotetan, cefuroxime, ceftriaxone, cefotaxime, aztreonam, chloramphenicol, and ciprofloxacin. These data and biochemical characterization provide phenotypic coherency (15) among the B. hinzii strains. This supports our conclusion, based upon the 100% identity of the 16S rRNA gene (5), that the unknown organism is B. hinzii.
DISCUSSION
B. hinzii was proposed as a new species in 1995 by Vandamme et al. (18). Most strains included in their study were derived from the respiratory tract of turkey poults. The pathogenicity of B. hinzii in the poultry population has never been demonstrated, yet the organism is capable of colonizing turkeys (18). Excluding the strain described in this report, five strains of B. hinzii have been isolated from human sources. The first reported strain was the cause of febrile bacteremia associated with an indwelling device in a patient with human immunodeficiency virus syndrome and no history of animal contact (3). The second strain was identified from the sputum of a patient in France, but further details were not available (18). The third and fourth strains were obtained from cultures of consecutive sputum samples from a cystic fibrosis patient over a 3-year period (7). In most of these sputum sample cultures, B. hinzii was associated with other respiratory pathogens, but in some cultures, B. hinzii was the sole organism isolated. The fifth strain was obtained from an endotracheal aspirate of a farmer but apparently did not cause disease (17). Although our patient did have cholestasis, our patient is unusual in that B. hinzii was solely responsible for his bacteremia and eventually his demise. His persistent septicemia could be attributed to the resistance of this organism (Table 2) to most antimicrobial agents used for empirical therapy. The patient had attended a cookout at a farm 2 weeks prior to his illness; however, no further detail regarding contact with poultry or birds was available. It is possible that B. hinzii may have colonized the respiratory tract of our patient at that time without causing a respiratory infection and subsequently spread into the bloodstream. A gastrointestinal portal of entry is also possible, resulting in intestinal colonization, ascending cholangitis, and bacteremia. This case also indicates that B. hinzii is capable of causing lethal infection. Previous case reports demonstrate some level of virulence. B. hinzii may be underrecognized as a pathogen because the organism is biochemically inert and is difficult to identify by the routine phenotypic methods used in most microbiology laboratories. Biochemical methods rely upon inoculum size and duration of incubation. The proportions of some CFAs in B. hinzii vary depending on whether bacterial cells are harvested after 24 or 48 h of incubation (3, 7, 18). The CFA profile of B. hinzii when it was harvested after 24 h of incubation discriminated between B. hinzii and the phenotypically related species B. avium in previous studies (3, 7). However, as noted previously by Vandamme et al. (18), we found that the CFA profiles of B. hinzii and B. avium are indistinguishable after 48 h of culture (Table 1). Our isolate showed an atypical CFA content, indicating that CFA analysis is not consistently reliable for identification of B. hinzii. In contrast, we have obtained a unique 16S rRNA sequence which was identical among all three strains examined. Our data from three epidemiologically unrelated isolates suggest that there is little interstrain polymorphism in the 16S rRNA sequence of B. hinzii, and therefore, 16S rRNA sequencing is useful for identification and differentiation of B. hinzii from related species. Moreover, we found that 66% of the interspecies variability in the 16S rRNA sequence among the bordetellae lies in the first 500 bp. This observation suggests that partial sequencing of 16S rRNA gene would be sufficient for identification of B. hinzii.
By 16S rRNA sequence analysis B. hinzii clusters with the bordetellae and is most closely related to B. bronchiseptica and B. parapertussis. Our phylogenetic data thus confirm the taxonomic position of B. hinzii and corroborate the findings of others on the basis of 23S rRNA-DNA hybridization and whole-cell protein analysis (7, 18).
The antimicrobial susceptibility profile of B. hinzii parallels that observed by Funke et al. (7) for their isolates from a cystic fibrosis patient and underscores the fact that it is a multidrug-resistant organism. Moreover, the MICs of different antimicrobial agents resemble those reported for most strains of B. bronchiseptica (21) and are in contrast to the susceptibility profiles of B. avium, which is a more sensitive bacterium (12). Possibly, B. hinzii and B. bronchiseptica share similar antimicrobial resistance mechanisms. Together with the 16S rRNA sequence analysis data, these two species appear to be more genetically related to one another than either is to B. avium.
In summary, our case and previous reports support the notion that B. hinzii is a potential pathogen in humans. It should be suspected as a pathogen in patients when cultures of clinical specimens grow an organism that phenotypically resembles B. avium and that is malonate positive and resistant to several antimicrobial agents. 16S rRNA sequence analysis allows accurate identification of this organism.
ACKNOWLEDGMENT
We thank Marie B. Coyle for reviewing the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cookson B T, Vandamme P, Carlson L C, Larson A M, Sheffield J V, Kersters K, Spach D H. Bacteremia caused by a novel Bordetella species, “B. hinzii.”. J Clin Microbiol. 1994;32:2569–2571. doi: 10.1128/jcm.32.10.2569-2571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 5.Fox G E, Wisotzkey J D, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 6.Fredricks D N, Relman D A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke G, Hess T, von Graevenitz A, Vandamme P. Characteristics of Bordetella hinzii strains isolated from a cystic fibrosis patient over a 3-year period. J Clin Microbiol. 1996;34:966–969. doi: 10.1128/jcm.34.4.966-969.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersters K, Hinz K-H, Hertle A, Segers P, Lievens A, Siegmann O, De Ley J. Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Int J Syst Bacteriol. 1984;34:56–70. [Google Scholar]
- 10.Leonard R B, Nowowiejski D J, Warren J J, Finn D J, Coyle M B. Molecular evidence of person-to-person transmission of a pigmented strain of Corynebacterium striatum in intensive care units. J Clin Microbiol. 1994;32:164–169. doi: 10.1128/jcm.32.1.164-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortensen J E, Brumbach A, Shryock T R. Antimicrobial susceptibility of Bordetella avium and Bordetella bronchiseptica isolates. Antimicrob Agents Chemother. 1989;33:771–772. doi: 10.1128/aac.33.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 9th informational supplement. M100-S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 14.Relman D A. Universal bacterial 16S rRNA amplification and sequencing. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: Principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 489–495. [Google Scholar]
- 15.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 16.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandamme P, Heyndrickx M, Vancanneyt M, Hoste B, De Vos P, Falsen E, Kersters K, Hinz K H. Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int J Syst Bacteriol. 1996;46:849–858. doi: 10.1099/00207713-46-4-849. [DOI] [PubMed] [Google Scholar]
- 18.Vandamme P, Hommez J, Vancanneyt M, Monsieurs M, Hoste B, Cookson B, Wirsing von Konig C H, Kersters K, Blackall P J. Bordetella hinzii sp. nov., isolated from poultry and humans. Int J Syst Bacteriol. 1995;45:37–45. doi: 10.1099/00207713-45-1-37. [DOI] [PubMed] [Google Scholar]
- 19.Weyant R S, Moss C W, Weaver R E, Hollis D G, Jorda J G, Cook E C, Daneshvar M I. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria. 2nd ed. Baltimore, Md: The Williams & Wilkins Co.; 1996. [Google Scholar]
- 20.Weyant R S, Hollis D G, Weaver R E, Amin M F, Steigerwalt A G, O'Connor S P, Whitney A M, Daneshvar M I, Moss C W, Brenner D J. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol. 1995;33:1–7. doi: 10.1128/jcm.33.1.1-7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woolfrey B F, Moody J A. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev. 1991;4:243–255. doi: 10.1128/cmr.4.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]