Abstract
Herpes simplex virus (HSV) causes several clinical manifestations in both normal and immunocompromised hosts; this agent is the most frequently detected virus in diagnostic laboratories. Recovery of the virus in cell culture is considered the “gold standard” for detection of this virus from sources other than cerebrospinal fluid. LightCycler is a newly developed, commercially available system designed to rapidly perform PCR, with real-time detection of PCR products by a fluorescence resonance energy transfer assay. We compared the detection of HSV for 200 specimens (number of genital specimens, 160; number of dermal specimens, 38; number of ocular specimens, 2) by shell vial cell cultures (MRC-5) and by LightCycler PCR. Of a total of 88 (44%) HSV strains detected, 69 (78%) were detected by both shell vial cell cultures and LightCycler PCR (DNA polymerase target). A total of 19 (22%) specimens were detected exclusively by LightCycler PCR. No specimens were positive by the shell vial assay only. All 19 discrepant samples had HSV DNA detected by an independent PCR directed to the thymidine kinase gene of the virus. The melting curve analysis feature of the LightCycler instrument identified identical genotype results for HSV type 1 (HSV-1) and HSV-2 from 84 of 88 (96%) positive samples. Specimens can be extracted, target HSV DNA can be amplified, and HSV PCR products can be identified by genotype within 2 h after receipt of specimen into the laboratory. The increased level of accurate identification (all 88 positive samples) compared with that of shell vial cell culture (69 of 88 samples identified as positive) and the agreement of LightCycler PCR results with all shell vial positive results indicate the potential for routine implementation of this technology for laboratory diagnosis of HSV infections.
Herpes simplex virus (HSV) causes a variety of clinical syndromes; anatomical sites infected include the skin, lips, oral cavity, eyes, genital tract, and central nervous system. Generalized or disseminated HSV infection may occur in patients immunologically compromised by neoplasia, organ transplantation, inherited immunodeficiency disease, or AIDS and through neonatal infection acquired by transmission of the virus through an infected birth canal. Most disseminated disease is fatal (23).
HSV is the virus most commonly detected in most diagnostic laboratories (18). In our practice, over a 25-year period HSV has accounted for over 40% of the viruses that we detect in cell cultures. The virus replicates optimally in human diploid fibroblast cell cultures, producing detection rates that typically exceed 30%, especially with specimens from genital and dermal sources; this diagnostic method is the “gold standard” for detection of HSV, except in cerebrospinal fluid (CSF) specimens obtained from patients with central nervous system disease (1, 3, 13). For example, of 425 viral isolates recovered from CSF at the Mayo Clinic over a 12-year period (1984 to 1986), only 9 (2%) were identified as HSV positive. Alternatively, using PCR at our institution from 1993 through 1997, we detected HSV DNA in 409 CSF specimens from 6,607 patient samples (6.2%) (20).
Modifications of cell culture detection of HSV, shell vial assay, and genetically engineered host cells reduce diagnostic time to 24 to 48 h postinoculation but require supplemental use of conventional tube cell cultures to ultimately achieve maximum diagnostic sensitivity (6, 14). Similarly, attempts at direct detection of HSV from clinical specimens by enzyme-linked immunosorbent assay (ELISA) and latex agglutination, nucleic acid probe, and fluorescent antibody methods generally fail when low titers of HSV are present in specimens that are inoculated into cell cultures (4, 5, 7, 9, 10–12, 19, 22, 24).
Several recent studies have indicated the potential for increased detection of HSV infections by PCR compared to antigen detection or cell culture methods; however, routine implementation of nucleic acid amplification techniques in the clinical laboratory for specimens from dermal, genital, and other sites has not been practical because of concerns of amplicon carryover contamination and technically cumbersome PCR product detection methods (2, 15–17).
We compared the detection of HSV from genital, dermal, and ocular sources by automated PCR with the LightCycler instrument (Roche Molecular Biochemicals, Indianapolis, Ind.) with that by shell vial and cell culture methods. The increased sensitivity of the LightCycler PCR compared to cell culture methods and a configuration for containment and detection of the amplified product by the instrument indicate the feasibility for implementation of this technology for routine diagnosis of HSV infection in the clinical laboratory.
MATERIALS AND METHODS
Specimens and shell vial assay.
Genital (n = 160), dermal (n = 38), and ocular (n = 2) swab specimens from patients suspected of having HSV infections were extracted into 2-ml volumes of serum-free medium, and the specimen extract volumes were divided into two equal aliquots. Each of two shell vial MRC-5 cell cultures received 0.2 ml of inoculum from one aliquot. The vials were centrifuged, incubated overnight at 36°C, and stained by the indirect immunofluorescence test as previously described (8). Nucleic acids were extracted from the second aliquot and processed for amplification of HSV DNA by PCR.
Nucleic acid extraction.
Nucleic acids were extracted from a 0.2-ml volume of serum-free extract of genital, dermal, or ocular swab specimens by the IsoQuick procedure (Orca Research, Inc., Bothell, Wash.), according to the manufacturer's instructions. The sample and an equal volume of lysis buffer were placed in a 1.5-ml microcentrifuge tube. A 700-μl volume of extraction matrix and a 400-μl volume of extraction buffer were added, and the tube was centrifuged for 5 min at 13,000 rpm (Eppendorf model 5417C; Fisher, Eden Prairie, Minn.). The top aqueous layer was placed in a fresh tube and sodium acetate (1/10), and 2 μl each of glycogen and isopropyl alcohol were added. The tube was then centrifuged for 10 min at 13,000 rpm (Eppendorf model 5417C; Fisher). The alcohol was poured off, and 2 volumes of 70% ethanol were added; the tube was then centrifuged for 5 min at 13,000 rpm (Eppendorf model 5417C; Fisher). The ethanol was aspirated from the tube, and the pellet was resuspended in 60 μl of RNase-free water.
LightCycler PCR.
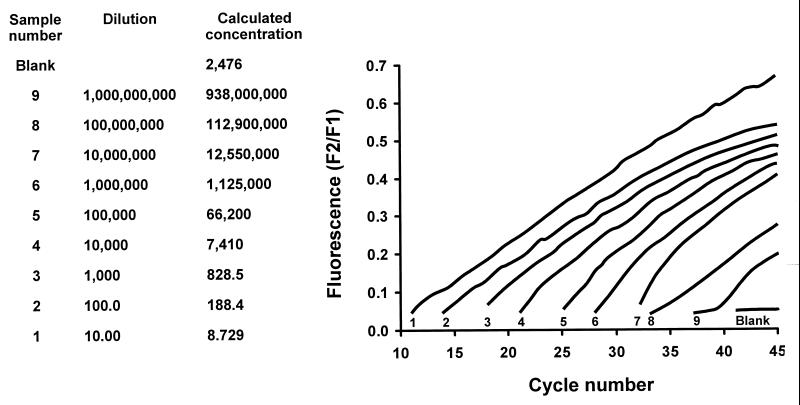
The LightCycler instrument (Roche Molecular Biochemicals) amplifies and monitors by fluorescence assay the development of target nucleic acid after each cycle (denaturation, annealing, and extension). This instrument provides rapid (30- to 40-min) automation of PCR by precise air-controlled temperature cycling and capillary cuvettes; the continuous monitoring of amplicon development after the annealing step is based on the fluorescence resonance energy transfer (FRET) principle. Primers directed to target HSV DNA in the polymerase gene generate a product of 215 bp (Table 1). For FRET product detection, a hybridization probe with a donor fluorophore, fluorescein, on the 3′ end is excited by an external light source and emits light that is absorbed by a second hybridization probe with an acceptor fluorophore, LC-Red 640, at the 5′ end. The acceptor fluorophore then emits light of a different wavelength that can be measured with a signal that is proportional to the amount of specific PCR product. We were able to detect as few as 20 genomic copies of HSV with the LightCycler assay. Ten 10-fold dilutions of a plasmid containing a portion of the HSV DNA polymerase gene were used to determine the sensitivity (20 genomic copies/PCR) of the LightCycler assay. A total of 28 specimens, including the controls, can be processed in a single run.
TABLE 1.
Characteristics and nucleotide base sequences of primers and probes used to detect target DNA of HSV
| Gene target | GenBank accession no. (HSV type) | Product size (bp) | Sequencesa |
|---|---|---|---|
| DNA polymerase | M16721 | 215 | GCT CGA GTG CGA AAA AAC GTT C (primer) and CGG GGC GCT CGG CTA AC (primer) |
| 56 | GTA CAT CGG CGT CAT CTG CGG GGG CAA G-FLUOR (probe) and LC-Red 640-T GCT CAT CAA GGG CGT GGA TCT GGT GC-Phos (probe)b | ||
| TK | X03764 (HSV-1) and X01712 (HSV-2) | 335 | GAC MAG CGC CCA GAT AAC AA (primer), MCA GCA TRG CCA GGT CAA GC (primer), AGG CGG TCG ATG TGT CTG TC (HSV-1 capture probe), and AGG CGG TCG GCG TGT TCG GC (HSV-2 capture probe) |
Sequences shown are in the 5′ to 3′ direction. The sequence type is indicated in parentheses. Hybridization probes. FLUOR, fluorescein; Phos, phosphate.
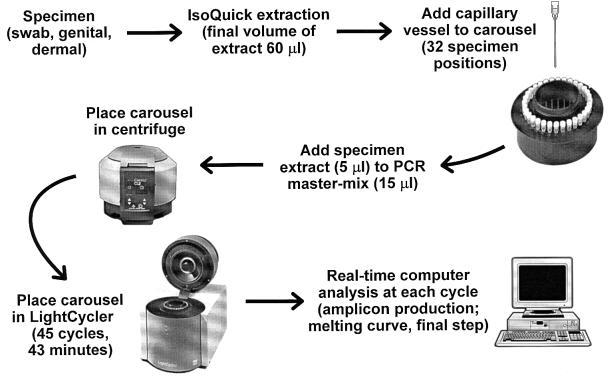
For the assay, a 5-μl aliquot of extracted nucleic acid was added to 15 μl of PCR mixture in each reaction capillary (Fig. 1). A no-target control received 15 μl of reaction mixture with 5 μl of water. A master mix was optimized for the LightCycler and contained the following: a 0.2 mM concentration of each of the deoxyribonucleoside triphosphates (50 mM KCl, 10 mM Tris-Cl [pH 8.3]), 3 mM MgCl2, 0.7 μM concentrations of the primers for DNA polymerase gene, 0.025% bovine serum albumin, 2% dimethyl sulfoxide, 0.2 μM fluorescein probe, 0.2 μM LC-Red 640 probe, and 0.03 U of platinum Taq (Perkin-Elmer Corp., Branchburg, N.J.) per ml. The PCR reagents and specimen extracts were centrifuged in the capillary to facilitate mixing. All capillaries were then sealed and amplified using the following protocol: 95°C for 2 min for one cycle, followed by denaturation at 95°C, 10 s of annealing at 62°C, and 12 s of primer extension at 72°C for 45 cycles.
FIG. 1.
Scheme for processing specimens for the diagnosis of HSV infections by LightCycler PCR.
Melting curve for HSV genotype analysis.
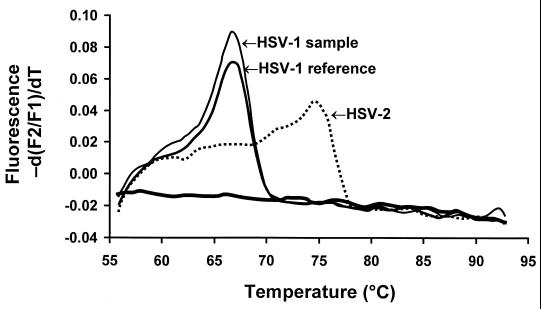
LightCycler hybridization probes were designed for HSV-2, and sequence differences between HSV-2 and HSV-1 were detected by melting curve analysis. Melting curve analysis was performed following PCR amplification. Starting at 54°C, the temperature in the thermal chamber was slowly raised to 95°C, and the fluorescence was measured at frequent intervals. Sequence differences between the PCR product and hybridization probes resulted in shifts in the melting temperatures (66.7°C for HSV-1 and 74.7°C for HSV-2) which were detected (see Fig. 4). Analysis of the PCR amplification and probe melting curves was accomplished through the use of LightCycler software.
FIG. 4.
Melting curve analysis of HSV-1 and HSV-2 genotypes determined by LightCycler PCR using FRET assay.
PCR discrepant analysis.
Specimens yielding shell vial-negative, LightCycler PCR-positive results were resolved as true-positive samples for HSV DNA by using primers directed to a thymidine kinase (TK) gene target that generated a 335-bp product (Table 1). This assay has been validated as sensitive and specific for detection of HSV DNA from CSF samples (13).
PCR amplification.
The PCR mixtures for amplification of the 335-bp TK gene target contained the following: 200 μM (each) adenine, cytosine, and guanosine; 100 μM thymidine, 90 μM uracil, and 10 μM digoxigenin 11-uracil deoxyribonucleoside triphosphate; and 10× buffer (500 mM KCl, 100 mM Tris-Cl [pH 8.3], 15 mM MgCl, 2.5 mg of bovine serum albumin per ml), 50 pmol (each) of the appropriate primers (TK gene), 10 μl of a 50% glycerol solution, 1 U of uracil N-glycosylase per μl and 1.25 U of AmpliTaq polymerase (PE Applied Biosystems, Foster City, Calif.). Each reaction tube received 45 μl of the reaction mixture plus 5 μl of target. A no-target control received 50 μl of the reaction mixture only. Reaction tubes were amplified in a DNA thermal cycler (model 9600; PE Applied Biosystems), using the following protocol: 50°C for 5 min, 94°C for 3 min for one cycle, followed by 15 s of denaturation at 94°C, 30-s of annealing or primer extension at 60°C for 50 cycles, followed by 10 min at 72°C for one cycle (13).
Identification of PCR product (335 bp) and genotype determination.
Detection of the PCR-amplified products was performed using a commercially available PCR ELISA microtiter detection format assay (PCR ELISA [DIG-DETECTION]; Roche Molecular Biochemicals). A portion of the denatured amplicon was mixed with a hybridization solution containing a 5′ biotin-labeled DNA capture probe specific for each of the two types of HSV. The probe hybridized to the corresponding target DNA sequence if present, and the resulting biotinylated DNA complex was captured on the streptavidin-coated microtiter plate wells. HSV-1- or -2-specific DNA complexes were detected by anti-digoxigenin-peroxidase conjugate, which recognized digoxigenin 11-dUTP substitutions incorporated into the amplicon during PCR. The peroxidase substrate, 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS), was added and color was allowed to develop. A positive result was indicated by an A405/A490 ratio of >0.1, as calculated after correction for the extinction coefficient of the ABTS blank (21).
RESULTS
HSV was detected in 88 (44%) of 200 specimens. A total of 69 (43%) of 160 genital specimens and 18 (47%) of 38 dermal specimens were positive for HSV DNA. Only two ocular specimens were tested, yielding one positive result. A total of 69 specimens were positive for detection of HSV by both shell vial assay and LightCycler PCR. Nineteen additional specimens were identified as HSV positive by the LightCycler assay (total number of positive specimens, 88). There were no specimens for which the shell vial assay result was positive and LightCycler result was negative (specificity, 100%). Of the 19 discrepant results (negative by shell vial assay but positive by LightCycler assay), all were confirmed as positive for HSV DNA by an independent PCR protocol that generated a 335-bp product from the TK gene of the virus; genotype-specific amplicons were subsequently identified by an ELISA.
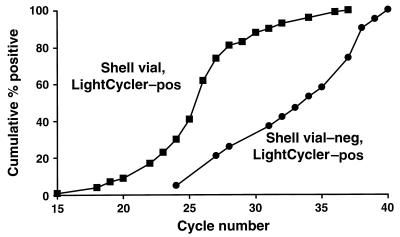
Specimens positive by both the shell vial and LightCycler assays (n = 69) were detected by PCR at an average of 26 cycles (range, 18 to 37 cycles). Discrepant specimens (n = 19) were positive after an average of 33 cycles by LightCycler assay (range, 24 to 40 cycles). The cumulative rate of detection of the 69 specimens with concordant results reached 100% after 37 PCR cycles, but 81% by cycle 28, whereas the 19 specimens with discrepant results required 40 PCR cycles to achieve positive results for all samples, and only 26% of these samples were detected by cycle 28 (Fig. 2). Therefore, as expected, specimens positive by both shell vial assay and LightCycler PCR apparently have higher copy numbers of HSV DNA than those specimens detected exclusively by the LightCycler assay. These results are confirmed experimentally in that the lowest dilution of a suspension of HSV target DNA yields PCR product in an earlier cycle and in direct proportion to 10-fold less-concentrated dilutions of the viral genome (Fig. 3).
FIG. 2.
Detection of HSV DNA from clinical specimens by LightCycler PCR.
FIG. 3.
Detection of serially diluted suspensions of HSV DNA by LightCycler PCR using FRET assay. The sequential numbers indicated at the base of each signal designation refer to the corresponding sample number and dilution (two lefthand columns).
Probes designed to detect nucleotide polymorphisms in two base pairs of the 215-bp product of LightCycler PCR correctly identified the genotype (HSV-1 or HSV-2) by melting curve analysis in 66 of 69 specimens, whereas monoclonal antibody differentiation of the two serotypes by the shell vial assay was less accurate (Table 1; Fig. 4). Of the 19 specimens with discrepant results analyzed by PCR directed to the TK gene of HSV, 14 were HSV-2 and 5 were HSV-1. The LightCycler assay gave concordant genotype results for 18 of 19 (95%) specimens. The melting curves for the four specimens with discrepant results (HSV-1 or HSV-2) overlapped and did not produce distinctive patterns that provided easy visual differentiation of the two genotypes.
DISCUSSION
Because of the frequency of infection and the risk of disseminated disease in the immunologically compromised host and the importance of sexual and perinatal transmission of HSV, rapid laboratory detection of this virus has been a pervasive goal in diagnostic virology. Generally, the sensitivity for detection of HSV with rapid assays, such as immunostaining of early antigens by the shell vial assay and assays of enzymatic activity expressed in HSV-infected genetically engineered cells, and an extensive number of home-brew and commercially available ELISAs have been, at best, equivalent in performance to the standard, but slower, tube cell culture isolation method (4, 5, 7, 9, 10–12, 19, 22, 24).
Although nucleic acid amplification techniques, particularly PCR, have demonstrated superior sensitivity to all other diagnostic methods for the detection of HSV infections, routine implementation in clinical laboratories has been impeded by problems of amplicon carryover contamination and the time-consuming gel electrophoresis and Southern blot techniques that have only recently yielded to more rapid PCR product detection methods (21). Importantly, with the LightCycler assay, detection of amplified nucleic acid products is accomplished in a closed system; that is, the capillary reaction vessels are never opened after the cycling process has started. Thus, there is no opportunity for carryover contamination to occur postamplification. Certainly, as in all PCR assays, there are steps in the extraction and processing procedures that may be susceptible to cross contamination of target nucleic acid between specimens. To address this potential problem, specimen (nucleic acid) extraction and target loading areas were physically separated; master mix preparation was performed in a room in which specimens and target nucleic acid had never been present. Disposable gloves, gowns, and barrier pipette tips were used at all times. Thymidine was replaced with 3× uracil in the master mix together with uracil glycosylase.
The LightCycler technology is a significant breakthrough in PCR cycling and amplicon detection compared to the open and contamination-susceptible system of thermocycling with subsequent transfer of the amplified product for gel electrophoresis and Southern blot analysis. In contrast, the LightCycler is a closed system in which the formation of amplicons is measured in real time without transfer to some other product detection system. The capillary reaction vessels are made of plastic and glass and can break during insertion or removal from the sample carousel. To address this concern, capillary vessels were inserted into the carousel, and then master mix and sample were added to the capillaries with the subsequent centrifugation step. After thermocycling and product analysis, the capillary vessels were removed from the carousel in a biosafety cabinet (P-2) and placed into a solution of bleach for amplicon inactivation.
We feel that these important characteristics of the LightCycler assay parallel those of the LCx (Ligase Chain Reaction) system, which is a closed system which was incorporated into our P-2 safety laboratory over 3 years ago for the diagnosis of Chlamydia trachomatis and Neisseria gonorrhoeae. We have not experienced obvious specimen or amplicon contamination in this system.
The LightCycler system is a newly developed, commercially available system designed to decrease the time needed to achieve PCR results by monitoring amplification of target amplicons in real time by a FRET assay. The LightCycler assay achieves these goals in the following manner: cycling temperatures are achieved by alternating heated air with air of ambient temperature, producing cycling times significantly faster than those achieved by conventional block or water bath cyclers. The reaction vessels are plastic and glass capillaries, which ensure rapid equilibration between the air and the reaction components because of the high surface area-to-volume ratio of the capillaries. The combination of the use of air for rapid thermal cycling and the high surface area-to-volume ratio of the capillaries reduces the time required for a single PCR cycle to fewer than 30 s. An entire run (32 specimens [including controls]/run) of 45 cycles can be completed in 30 to 40 min. Amplified products are monitored every cycle. Importantly, sequence differences in target amplicons can be detected by melting curve analysis, performed by the instrument after PCR amplification is completed and the PCR product is detected.
Specificity of PCR detection of amplified HSV DNA products is highly relevant for clinical microbiology practice, especially for the laboratory diagnosis of the infections involved in sexually transmitted diseases. Specimens positive by both shell vial assay and LightCycler PCR (69 of 88 specimens [78%]) in our study likely reflect high-titer specimens which were detectable by both systems; these concordant results were considered specific based on past evaluations and our current experience in which the shell vial assay results were never positive when the LightCycler results were negative (100% specific) (8). Importantly, LightCycler PCR was experimentally demonstrated to be specific for target HSV DNA and did not amplify DNA from varicella-zoster virus, cytomegalovirus, Epstein-Barr virus, or human herpesviruses 6, 7, and 8. Conversely, we detected HSV DNA in 19 specimens that were not found to be positive by the shell vial assay. These samples probably contain low levels of HSV detectable only by the sensitive PCR. We considered these true-positive results based on confirmation by another independent PCR generating a 335-bp product, amplified from a TK gene target, with demonstrated specificity for detection of HSV in CSF specimens (20). In addition, of the 19 samples with discrepant results (negative by shell vial assay but positive by LightCycler assay), 7 came from patients with dermal lesions that were clinically apparent and from which the specimens were collected.
Because of the manual methods and general technical demands of PCR in past years, our laboratory has focused on the development of these assays for viral infections for which laboratory methods for detection were limited and ineffective. The best example has been PCR detection of HSV DNA in CSF specimens from patients with central nervous system disease (13, 20). In contrast, application of LightCycler PCR in the routine clinical laboratory provides a system that has a 22% (69 of 88 positive by shell vial assay compared with 88 of 88 positive by LightCycler assay) increased sensitivity and 100% specificity compared with shell vial cell culture, together with rapid processing (∼2 h) (nucleic acid extraction plus PCR analysis) and genotype identification (HSV-1 versus HSV-2), in a closed and contained reaction vessel incorporated into an automated instrument in which carryover contamination is virtually eliminated. Our future goal is to implement automated PCR into our diagnostic laboratory for routine detection of HSV from clinical specimens.
REFERENCES
- 1.Arvin A M, Prober C G. Herpes simplex viruses. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 878–887. [Google Scholar]
- 2.Beards G, Graham C, Pillay D. Investigation of vesicular rashes for HSV and VZV by PCR. J Med Virol. 1998;54:155–157. doi: 10.1002/(sici)1096-9071(199803)54:3<155::aid-jmv1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Callihan D R, Menegus M A. Rapid detection of herpes simplex virus in clinical specimens with human embryonic lung fibroblast and primary rabbit kidney cell cultures. J Clin Microbiol. 1984;19:563–565. doi: 10.1128/jcm.19.4.563-565.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dascal A, Chan-Thim J, Morahan M, Portnoy J, Mendelson J. Diagnosis of herpes simplex virus infection in a clinical setting by a direct antigen detection enzyme immunoassay kit. J Clin Microbiol. 1989;27:700–704. doi: 10.1128/jcm.27.4.700-704.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espy M J, Smith T F. Detection of herpes simplex virus in conventional tube cell cultures and in shell vials with a DNA probe kit and monoclonal antibodies. J Clin Microbiol. 1988;26:22–24. doi: 10.1128/jcm.26.1.22-24.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espy M J, Wold A D, Jespersen D J, Jones M F, Smith T F. Comparison of shell vials and conventional tubes seeded with rhabdomyosarcoma and MRC-5 cells for the rapid detection of herpes simplex virus. J Clin Microbiol. 1991;29:2751–2753. doi: 10.1128/jcm.29.12.2701-2703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleaves C A, Rice D H, Lee C F. Evaluation of an enzyme immunoassay for the detection of herpes simplex virus (HSV) antigen from clinical specimens in viral transport media. J Virol Methods. 1990;28:133–139. doi: 10.1016/0166-0934(90)90027-d. [DOI] [PubMed] [Google Scholar]
- 8.Gleaves C A, Wilson D J, Wold A D, Smith T F. Detection and serotyping of herpes simplex virus in MRC-5 cells by use of centrifugation and monoclonal antibodies 16 h postinoculation. J Clin Microbiol. 1985;21:29–32. doi: 10.1128/jcm.21.1.29-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston S L, Siegel C S. Comparison of enzyme immunoassay, shell vial culture, and conventional cell culture for the rapid detection of herpes simplex virus. Diagn Microbiol Infect Dis. 1990;13:241–244. doi: 10.1016/0732-8893(90)90066-5. [DOI] [PubMed] [Google Scholar]
- 10.Kok T, Michau L, Schepetuik S. Rapid detection, culture-amplification and typing of herpes simplex viruses by enzyme immunoassay in clinical specimens. Clin Diagn Virol. 1998;10:67–74. doi: 10.1016/s0928-0197(98)00003-8. [DOI] [PubMed] [Google Scholar]
- 11.Kudesia G, Van Hegan A, Wake S, Van Hegan R J, Kinghorn G R. Comparison of cell culture with an amplified enzyme immunoassay for diagnosing genital herpes simplex infection. J Clin Pathol. 1991;44:778–780. doi: 10.1136/jcp.44.9.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafferty W E, Krofft S, Remington M, Giddings R, Winter C, Cent A, Corey L. Diagnosis of herpes simplex virus by direct immunofluorescence and viral isolation from samples of external genital lesions in a high-prevalence population. J Clin Microbiol. 1987;25:323–326. doi: 10.1128/jcm.25.2.323-326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell P S, Espy M J, Smith T F, Toal D R, Rys P N, Berbari E F, Osmon D R, Persing D H. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J Clin Microbiol. 1997;35:2873–2877. doi: 10.1128/jcm.35.11.2873-2877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivo P D. Transgenic cell lines for detection of animal viruses. Clin Microbiol Rev. 1996;9:321–334. doi: 10.1128/cmr.9.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risbud A, Chan-Tack K, Gadkari D, Gangakhedkar R R, Shepherd M E, Bollinger R, Mehendale S, Gaydos C, Divekar A, Rompalo A, Quinn T C. The etiology of genital ulcer disease by multiplex polymerase chain reaction and relationship to HIV infection among patients attending sexually transmitted disease clinics in Pune, India. Sex Transm Dis. 1999;26:55–62. doi: 10.1097/00007435-199901000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Safrin S, Shaw H, Bolan G, Cuan J, Chiang C S. Comparison of virus culture and the polymerase chain reaction for diagnosis of mucocutaneous herpes simplex virus infection. Sex Transm Dis. 1997;24:176–180. doi: 10.1097/00007435-199703000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Slomka M J, Emery L, Munday P E, Moulsdale M, Brown D W. A comparison of PCR with virus isolation and direct antigen detection for diagnosis and typing of genital herpes. J Med Virol. 1998;55:177–183. [PubMed] [Google Scholar]
- 18.Smith T F, Wold A D, Espy M J, Marshall W F. New developments in the diagnosis of viral diseases. Infect Dis Clin N Am. 1993;7:183–201. [PubMed] [Google Scholar]
- 19.Storch G A, Reed C A, Dalu Z A. Evaluation of a latex agglutination test for herpes simplex virus. J Clin Microbiol. 1988;26:787–788. doi: 10.1128/jcm.26.4.787-788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Y-W, Mitchell P S, Espy M J, Smith T F, Persing D H. Molecular diagnosis of herpes simplex virus infections in the central nervous system. J Clin Microbiol. 1999;37:2127–2136. doi: 10.1128/jcm.37.7.2127-2136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y-W, Rys P N, Rutledge B J, Mitchell P S, Smith T F, Persing D H. Comparative evaluation of colorimetric microtiter plate systems for detection of herpes simplex virus in cerebrospinal fluid. J Clin Microbiol. 1998;36:2714–2717. doi: 10.1128/jcm.36.9.2714-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verano L, Michalski F J. Comparison of a direct antigen enzyme immunoassay, Herpchek, with cell culture for detection of herpes simplex virus from clinical specimens. J Clin Microbiol. 1995;33:1378–1379. doi: 10.1128/jcm.33.5.1378-1379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2297–2342. [Google Scholar]
- 24.Zimmerman S J, Moses E, Sofat N, Bartholomew W R, Amsterdam D. Evaluation of a visual, rapid, membrane enzyme immunoassay for the detection of herpes simplex virus antigen. J Clin Microbiol. 1991;29:842–845. doi: 10.1128/jcm.29.4.842-845.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]