Abstract
For many pathogenic microbes that utilize mainly asexual modes of reproduction, it is unknown whether epidemics are due to either the emergence of pathogenic clones or environmentally determined increases in the population size of the organism. Descriptions of the genetic structures of epidemic populations, in conjunction with analyses of key environmental variables, are able to distinguish between these competing hypotheses. A major epidemic of coccidioidomycosis (etiologic agent, Coccidioides immitis) occurred between 1991 and 1994 in central California, representing an 11-fold increase above the mean number of cases reported from 1955 to 1990. Molecular analyses showed extensive genetic diversity, a lack of linkage disequilibria, and little phylogenetic structure, demonstrating that a newly pathogenic strain was not responsible for the observed epidemic. Epidemiological analyses showed that morbidity caused by C. immitis was best explained by the interaction between two variables, the lengths of droughts preceding epidemics and the amounts of rainfall. This shows that the principal factors governing this epidemic of C. immitis are environmental and not genetic. An important implication of this result is that the periodicity of cyclical environmental factors regulates the population size of C. immitis and is instrumental in determining the size of epidemics. This knowledge provides an important tool for predicting outbreaks of this pathogen, as well as a general framework that may be applied to determine the causes of epidemics of other fungal diseases.
The microbes of interest in this report, pathogenic fungi, cause severe disease epidemics in both animals and plants, illustrated by recently reported mass mortalities in sea fan coral and amphibian populations (3, 19). However, it is usually not known why fungal epidemics occur. Epidemics of many species of microbes have their origin in the differential success of highly fit individuals (e.g., Staphylococcus aureus [14], Mycobacterium tuberculosis [15], Vibrio cholerae [22], and Phytophthora sp. [5]). Within phytopathogenic fungi, strong selection by host resistance alleles (20) causes differential success between fungal clones and this selection has been shown to result in epidemics as highly fit clones emerge (7). Due to the low rates of sexual recombination in microbe populations, such epidemics can be identified by the characteristically high levels of nonrandom association between alleles (genetic linkage disequilibrium [26]). However, epidemics have other causes and may arise due to the occurrence of favorable environmental conditions releasing a pathogen from environmental constraints. This is typified by the epidemicity of malaria in Central America resulting from a cyclicity of environmental variables (4). Under these conditions, the genetic structures of epidemic populations are not expected to be composed of highly fit clones that have arisen due to selection (26). To demonstrate the principal cause underlying any epidemic of infectious disease, it is necessary to distinguish between these environmental and genetic hypotheses. This task may be done by comparing the population genetic structures of epidemic pathogen isolates against predictive statistical models that incorporate known or potentially important environmental variables. We demonstrate this approach by using combined molecular and epidemiological analyses to describe the factors that have resulted in an unexplained epidemic of the pathogenic fungus Coccidioides immitis (12).
C. immitis is endemic to semiarid soils of the New World. The fungus is dimorphic, with the parasitic phase causing a systemic infection in humans and other vertebrate hosts once arthrospores have been inhaled (32). Molecular analysis has shown that C. immitis consists of two genetically isolated cryptic species, the California and non-California species, that have been reproductively isolated from one another for an estimated 11 million years (23, 24). These species occur with largely nonoverlapping geographical distributions (23, 24), and although no sexual state has ever been described for C. immitis, the non-California species has been shown to be recombining in nature (8). Clinical isolates of C. immitis from both geographical areas have been shown to exhibit a wide range of degrees of virulence for mice, demonstrating the existence of virulence-determining factors (18).
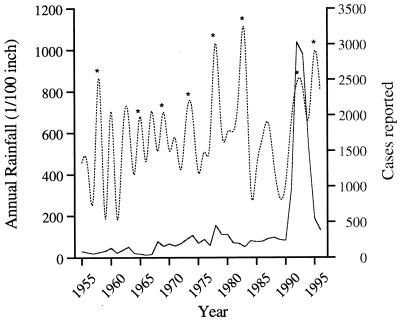
Between 1991 and 1994, California experienced an epidemic of coccidioidomycosis when the numbers of reported cases increased from a 1950 to 1990 average of ≈400 cases per year to more than 4,500 cases per year (2) (Fig. 1). More than 80% of these cases occurred in one location, Bakersfield, Calif. While previous epidemics of C. immitis had been observed in the previous 40 years, these were attributable to dust storms and earthquakes causing arthroconidia to become wind-borne (17, 30, 33). However, the 1991-1994 epidemic was unusual in that it was an order of magnitude larger than those previously seen, was localized in distribution, and persisted for 3 years (Fig. 1), compared to the previous maximum of 16 weeks for an outbreak in 1978 (17). Epidemiological analyses of the 1991-1994 epidemic found no contributing factors among environmental or sociological variables (annual rainfall, particulate matter, wind speed, occupation) and only the expected biases of the risk factors ethnicity, sex, and diabetes (2). On the other hand, recent molecular data had shown no genetic variation in 25 isolates collected during the epidemic using 12 loci that were polymorphic in a Tucson population of C. immitis (10). This lack of variation was consistent with the hypothesis that the 1991-1994 epidemic was caused by the evolution and expansion of a single virulent C. immitis clone. However, as argued by Burt et al. (10), an alternative explanation for the lack of polymorphism seen in Bakersfield at these loci is that genetic drift, occurring over the 12 million years since speciation between California and non-California C. immitis, fixed alleles that were originally polymorphic in the ancestral population (36). Thus, markers that are polymorphic in non-California C. immitis are not polymorphic in California C. immitis and vice versa, giving rise to the observed clonality in the epidemic population. Because the markers found to be polymorphic in the non-California species are monomorphic in the California species, they cannot be used to address the questions of clonality and recombination.
FIG. 1.
Annual number of cases of coccidioidomycosis reported from Bakersfield (solid line) and annual rainfall (dotted line). El Niño years are marked with an asterisk. These data were supplied by the Division of Communicable Disease Control, California Department of Health Services.
Therefore, to further investigate the causes behind this epidemic, we found additional genetic markers that were polymorphic in the California species of C. immitis and used them to reanalyze the original 25 Bakersfield isolates and 12 more clinical isolates collected from Bakersfield during the epidemic. Also included were two representative outgroup non-California isolates from Arizona and Guatemala. Further, we collected an expanded data set of environmental variables and used retrospective statistical analyses to ascertain if these factors are correlated with the epidemicity of C. immitis.
MATERIALS AND METHODS
C. immitis isolates.
C. immitis isolates were cultured from patients suffering from respiratory and disseminated coccidioidomycosis during the 1993-1994 phase of the epidemic by the Kern County Public Health Laboratory. Isolates were cultured in liquid medium, and genomic DNA was extracted as described previously (9).
Isolation of polymorphic genetic loci.
Two classes of genetic markers were used. Firstly, slowly evolving single-nucleotide polymorphisms (SNPs; mutation rate, ≈10−9) were isolated to provide data on the genetic diversity and reproductive mode of C. immitis isolated from the epidemic. Secondly, rapidly evolving multiallelic short tandem repeats (STRs; mutation rate, ≈10−2 to 10−5) were used to increase the statistical power for differentiation between isolates that were identical at all SNP loci.
To search for SNPs, seven tester strains were randomly chosen from the 37 Bakersfield C. immitis isolates and genomic DNA was amplified by PCR using arbitrary primers and low-stringency conditions. Amplified bands were used as template DNA in a new PCR, except that 0.1 μl of [α-35S]thio-dATP (12.5 mCi ml−1; 1 mCi = 37 MBq) was added and the amplicons were electrophoresed on an MDE gel (AT Biochem, Malvern, Pa.) to reveal single-strand conformational polymorphisms (SSCPs) (8). Polymorphic amplicons were sequenced to determine the genetic basis of the variation, and specific primers were designed to amplify the locus. We also searched for polymorphisms in C. immitis sequences from the GenBank database for (i) CTS1, (ii) pyrG (OR), (iii) ITS, (iv) BL, and (v) SP and unpublished sequences of HSP60 (HSP) and BGL2 provided by G. Cole (Medical College of Ohio, Toledo). Restriction endonuclease assays were designed to score each polymorphic site. If no suitable restriction site existed, then SSCP was used to score the polymorphism. The primer sequences used have been published previously (16).
All isolates were genotyped at two loci containing polymorphic STRs, locus 621r, containing an (AC)n (n = 6 to 18) microsatellite, and locus B34, containing a (TAA ACA AAC)n (n = 1 to 6) minisatellite (16). The sequence for locus 621r was provided by D. Carter, amplified with TAMRA-labeled primers, and genotyped using an automated sequencer and GENESCAN software (Applied Biosystems). Locus B34 was found as described above and genotyped by analysis of SSCPs.
Genetic analyses.
If the epidemic was due to the spread of an epidemic clone of C. immitis, then all loci must share a recent common evolutionary history due to their inheritance as a single linkage group. This clonal mode of reproduction is expected to result in (i) low levels of genetic diversity and (ii) strong nonrandom associations between alleles at different loci, based on the assumption that extensive genetic recombination has not occurred between the epidemic clone and nonepidemic individuals of C. immitis. Due to the short time scale of the epidemic, this assumption is probably warranted. In all subsequent statistical analyses, isolates from outside California (1036 and 3272) were excluded due to their outgroup status.
(i) Population genetic analyses.
The probability of sampling a particular genotype more than once in this data set may be calculated using the binomial expression
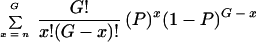 |
where G is the number of genotyped C. immitis isolates, n is the number of isolates with the same genotype as that in question (and equals 1 for this data set), and P is the probability of observing the original genotype (P = Πfr, where fr is the frequency of each allele found at a locus). This method assumes that (i) different genotypes arise by recombination and not mutation, (ii) mating is random, and (iii) loci are at linkage equilibrium. Demonstration of low linkage disequilibria in the data set validates assumptions ii and iii, and low sequence diversity between loci suggests that mutation rates in C. immitis are not unusually high. The disequilibrium coefficients for alleles A and B at two loci were calculated as DAB = pAB − pApB, where pAB is the gametic frequency of AB and pA and pB are the respective allele frequencies. The disequilibrium coefficients for 78 pairs of loci were calculated, and Fisher's exact test was used to assess the significance of the association (37). The index of association (IA) measures the degree of association among all loci based on the variation of the genetic distance between individuals (6, 26). The IA was calculated, and its significance was assessed by comparison with values calculated from 1,000 artificially recombined data sets (8).
(ii) Phylogenetic analyses.
Alleles at each locus were treated as phylogenetic characters with two character states. Maximum parsimony was used to find the shortest tree that fitted the data using the program PAUP 4.062a and the strength of branches was examined by bootstrapping the data 1,000 times. In order to test whether the observed data set contained more phylogenetic signal than a population undergoing complete recombination, 500 artificially recombined data sets were created and the lengths of their most parsimonious trees were compared to those found for the observed data set (1). As alleles are subject to reversals at STR loci (29), 621 and B34 were only used to measure genotypic diversity and were not used in the genetic analysis unless specified.
Epidemiological analyses.
C. immitis morbidity data for 1955 to 1995 in Kern County were supplied by the California Department of Health Services, Sacramento. To account for changes in population size over time and to highlight variation between years, the data were transformed by calculating the relative change in C. immitis morbidity (RCM) as the numbers of cases in year n divided by the numbers of cases in year n − 1.
Generalized linear models were constructed with RCM as the dependent variable and the following as independent variables: (i) mean annual rainfall, (ii) the occurrence of type I El Niño southern oscillation (ENSO) events, (iii) the annual Palmer drought severity index (PDSI), (iv) the length of moderate to extreme droughts preceding any particular year, (v) mean annual temperature, (vi) mean annual 10-μm particulate matter concentration (PM10), (vii) mean annual total suspended particle concentration (TSP), and (viii) yearly 1970 to 1996 Kern County population size. Type I ENSO events (ii) are known to be the principal determinants of above-normal rainfall in California and are defined as an equatorial Pacific sea surface anomaly of +2.0°C (Fig. 1) (J. Null, http://www.nws.mbay.net/cal_enso.html). Droughts (iii) are defined as having a PDSI of −1 to −2.9 for 9 months or more following National Climatic Data Center guidelines (28). Rainfall and temperature data were obtained from the Bakersfield Meadows meteorological station, and drought severity indicators were for the Central Valley region (California Division 5). These data were collated from the National Climatic Data Center databases (28). PM10 (vi) and TSP (vii) data were for the San Joaquin valley air basin for the years 1986 to 1996 and 1965 to 1991, respectively (11).
The fit of each linear regression model was assessed by inspection of the regression r values with their associated residuals and tested with the F statistic. Noncontributing variables were rejected in a stepwise fashion, and interactions between contributing variables were tested to find the most parsimonious model.
RESULTS
Genetic analyses.
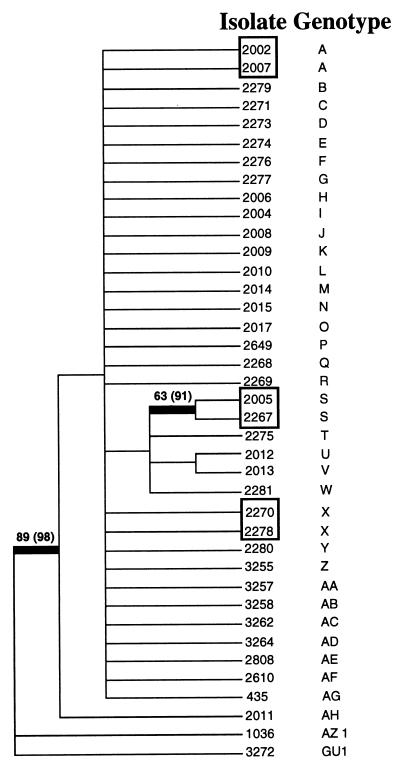
All loci were biallelic and contained a single allele within an isolate, confirming the haploid state of C. immitis. The alleles grouped all isolates from Bakersfield into a single clade, with the exception of isolates 1036 and 3272, which formed a separate clade with high bootstrap support (Fig. 2). Where isolates from this study had been typed using other genetic markers in previous studies (10), they corresponded exactly to the assignment of C. immitis to one of two species, California or non-California (23). In this study, all isolates from Bakersfield were of the California species and isolates 1036 and 3272 were of the non-California species, as expected from previous data (10, 23).
FIG. 2.
Strict consensus of 46,072 most-parsimonious phylogenetic trees. Heavy bars signify branches with strong bootstrap support. Values in parentheses are for analyses with the STR loci included. Multilocus genotype designations are shown, and C. immitis isolates with identical genotypes are boxed.
Considerable allelic diversity was observed at both loci containing STRs, with eight alleles identified for 621r and five alleles for B34 (Table 1). Multilocus genotypic diversity was high, with a total of 34 unique multilocus genotypes observed in the sample of 37 California isolates. Three pairs of isolates had identical genotypes at all loci (Fig. 2). Of these, two pairs were expected to be observed by chance due to their sharing of alleles that had a high frequency in the data set (binomial probability, P > 0.01) but one pair (isolates 2005 and 2267, genotype S) was unlikely to be observed in this data set (binomial probability, P < 0.001) and could be considered genetically identical clones. While the extensive genetic diversity seen in the data set suggests that these isolates are not descended from a single clonal ancestor, it is necessary to examine the structure of the variation in order to determine the degree of mixis within the parental population of C. immitis. Nonrandom associations were rare, with only 4 of 78 pairs of loci (STR loci excluded) showing significant linkage disequilibria (P < 0.05), and no statistical associations were observed after controlling for type I error with a Bonferroni correction (31). The multilocus IA for all loci was not significantly greater than that expected for a fully recombined population (P = 0.28, STR loci excluded).
TABLE 1.
Multilocus genotypes of C. immitis isolatesa
| Locus | Occurrence of polymorphism in isolated:
|
Site (base)b | Enzymec | ||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2002B | 2004B | 2005B | 2006B | 2007B | 2008B | 2009B | 2010B | 2011B | 2012B | 2013B | 2014B | 2015B | 2017B | 2649B | 2267B | 2268B | 2269B | 2270B | 2271B | 2273B | 2274B | 2275B | 2276B | 2277B | 2278B | 2279B | 2280B | 2281B | 3255B | 3257B | 3258B | 3262B | 3264B | 2808B | 2610B | 435B | 1036 | 3272 | |||
| SNP | |||||||||||||||||||||||||||||||||||||||||
| VL | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 70 (t/c) | HinfI |
| CTS1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1441 (g/c) | MnlI |
| OR | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 69 (a/g) | MaeIII |
| ITS | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 461 (t/c) | HaeIII |
| VK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 290 (g/c) | MnlI |
| BL | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 215 (–/a) | HinfI |
| CAG | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 377 (t/g) | HaeIII |
| CNS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 64 (c/t) | MspI |
| HSP | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 252 (t/c) | EcoRV |
| BOR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 135 (t/c) | TaqI |
| BGL2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 253 (t/c) | SSCP |
| RAN1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 192 (t/c) | NruI |
| 621 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | — | 1 | 1 | 1 | 1 | 1 | 1 | 255 (g/a) | Sau96I |
| SP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 425 (g/c) | MnlI |
| STR | |||||||||||||||||||||||||||||||||||||||||
| 621r | 3 | 1 | 8 | 4 | 3 | 4 | 2 | 3 | 4 | 5 | 5 | 8 | 4 | 7 | 5 | 8 | 4 | 4 | 2 | 4 | 7 | 3 | 6 | 4 | 2 | 2 | 3 | 5 | 7 | 7 | 3 | 7 | 7 | 2 | 3 | 7 | 4 | 0 | 0 | 121 | SV |
| B34 | 2 | 2 | 6 | 2 | 2 | 3 | 5 | 3 | 3 | 1 | 3 | 3 | 5 | 5 | 3 | 6 | 2 | 3 | 3 | 2 | 3 | 3 | 2 | 3 | 2 | 3 | 6 | 3 | 3 | 2 | 3 | 2 | 3 | 3 | 3 | 2 | 1 | 1 | 1 | 100 | SSCP |
All isolates came from Bakersfield, except 1036 and 3272, which were from patients who gained their infections in Arizona and Guatemala, respectively. These are representatives of the non-California species of C. immitis. SNP loci are loci containing single-nucleotide polymorphisms; STR loci are loci containing short tandem repeats.
Position of the polymorphic nucleotide in GenBank sequence. Allele 0/allele 1 nucleotides are shown. For loci 621r and B34, the numbers of repeats found in the STR sequence are shown.
Enzymes used to score the alleles are shown. BGL2, B34, and 621r were genotyped by SSCP or size variation (SV).
Reported as presence (1) or absence (0) for SNP loci or as number of repeats for STR loci. —, no data were obtained.
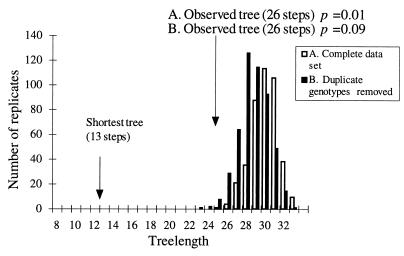
A second approach used to assess the population structure was phylogenetic analysis. If the epidemic population is clonal in origin, then the data will fit a short, well-resolved tree with minimal homoplasy. On the other hand, if the epidemic isolates are derived from a genetically diverse recombining population, then multiple homoplasies will result and the tree(s) will be poorly resolved. No single tree could be fitted to the SNP data; instead, 46,072 trees of 32 steps were identified, all of which had low consistency indices (0.44). The strict consensus of these trees, shown in Fig. 2, is poorly resolved, with few internal branches and most isolates falling into a polycotomy of 31 isolates. Two C. immitis isolates of the non-California cryptic species (1036 and 3272) were included as outgroup taxa and fell outside the California clade with strong bootstrap support, as expected. Randomization of alleles within California between isolates but within each locus mimics mixis by creating artificially recombined data sets. Comparison of the observed tree length with those found for 500 recombined data sets shows that the observed tree is 16 steps longer than the shortest possible tree length of 13 (expected under a model of complete clonal evolution of C. immitis) but significantly shorter than those of the recombined data sets (Fig. 3). This is largely due to the effect of the three duplicated genotypes A, S, and X. Collapsing these duplicates into single genotypes (clone correcting [26]) eliminates the effect of resampling of individuals of these genotypes and renders the observed tree length statistically indistinguishable from that of the simulated randomly mating populations.
FIG. 3.
Distribution of tree lengths for 500 artificially recombined data sets using all data (A) and clone-corrected data with the identical genotypes A, S, and X represented only once (B).
Epidemiological analyses.
Observed seasonal rainfall and occurrence of type I ENSO events are shown in Fig. 1. No periodicity is obvious for the 1955-1996 morbidity data, suggesting that the ENSO influence on central Californian rainfall is not a primary factor in determining whether epidemics of C. immitis occur. This lack of ENSO influence is borne out by analysis of variance of the El Niño years that occurred between 1955 and 1993, during which period no association was seen between the eight ENSO events and the mean RCM (F4,30 = 0.792, P = 0.540). Multivariate regression excluded from consideration the variables mean annual rainfall, mean annual temperature, ENSO, PDSI, PM10, TSP, and population size. However, a significant correlation was observed between the length of drought and RCM (r = 0.439, P = 0.004). Post hoc analysis identified an outlying data point with high influence (Studentized residual = 0.64). This data point has been inflated by the transformation used here and corresponds to an unusually low incidence of coccidioidomycosis in 1967 rather than an epidemic in 1968 (Fig. 1). Exclusion of this data point and reanalysis normalized the residuals and increased the fit of length of drought and RCM, as well as introducing mean annual rainfall as a significant variable (multiple r = 0.673; rainfall, P < 0.01; drought, P < 0.001).
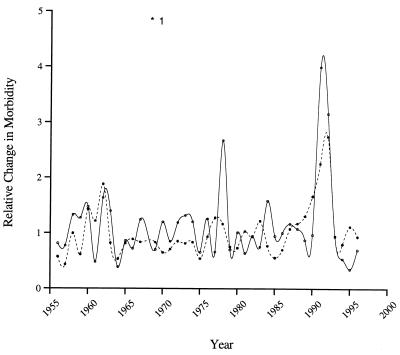
The regression equation y = 0.295x1 + 0.001x2 − 0.16 was used to predict relative change in coccidioidomycosis in the past time series as a function of the variables length of drought (x1) and rainfall (x2). The modeled change in RCM is shown in Fig. 4 together with that observed. The regression explains 45% of the variation in the yearly series and successfully predicts the 1991-1994 epidemic. The model predictions differ markedly from observations only during 1966 to 1975, where the RCM is underestimated, and 1978. The period 1966 to 1975 is unusual in that there were a number of wet years in rapid succession and it is possible that factors others than those included in this model predominated during this time period. The unexplained peak in infections in 1978 is due to a singular event, a windstorm that blew through Kern County on 20 December 1977, exposing a large number of people and resulting in a spike of infections that were recorded for the subsequent year (30).
FIG. 4.
Relative change in the number of cases of coccidioidomycosis in Bakersfield from 1956 to 1996 (solid line). The dotted line shows the modeled change using the regression equation y = 0.295x1 + 0.001x2 − 0.16 with the variables length of drought (x1) and mean annual rainfall (x2). The asterisk beside the number one signifies the outlier data point.
DISCUSSION
Here, we have shown that a lengthy and persistent epidemic of coccidioidomycosis is attributable to environmental, and not genetic, causes. We can rule out the amplification of a virulent clone of C. immitis as a cause of the epidemic because isolates of C. immitis collected during the epidemic are genetically diverse and have undergone extensive interlocus recombination. While it may be argued that the genetic diversity in this population may have accrued due to our isolation of markers containing hypervariable SNPs, we can show that this is not the case. Sequencing of the SNPs demonstrated that the polymorphic sites were rare (>1% of nucleotides were polymorphic in Bakersfield genomes), and sequencing of multiple isolates showed that no more than two nucleotides were ever found at a single polymorphic site. Moreover, genotyping of these loci in non-California C. immitis showed that all SNP loci had drifted to fixation (unpublished data), a situation that would not be expected to have occurred if these loci were hypervariable. Therefore, it appears that the 1991-1994 epidemic was not caused by the emergence of a single clonal lineage of C. immitis. Rather, the epidemic was due to infection by multiple progeny from a parental fungal population that was close to panmixia. It is apparent that (as among its sister taxon, non-California C. immitis) recombination is occurring in nature between individuals of the California type of C. immitis. No sexual reproduction (the production of ascospores and meiosis) has ever been directly observed in C. immitis, and it is reasonable to assume that when sex does occur, it is rare (8). For unlinked loci, the medium time to genetic equilibrium in a randomly mating population with a recombination rate (c) of 0.01 is 69 generations and that for c = 0.001 is about 693 generations (13). Given that the generation frequency of C. immitis is, at the most, 1 to 2 per year (J. W. Taylor, personal communication), we would not expect genetic equilibrium to be established within the time scale of this epidemic. This rules out the evolution of a virulent C. immitis clone as a cause of the 1991-1994 epidemic. However, there is also direct evidence for clonal propagation, and subsequent infection, by C. immitis within this data set. Two patients were infected by C. immitis isolates with identical multilocus genotypes, 2005 and 2267. The patients who contributed these isolates lived and worked in the same town, but the C. immitis isolates were collected by separate clinicians. This is strong evidence that these people had been infected by the local dispersion of asexually produced C. immitis spores from a single fungal individual and is the first data demonstrating this occurrence.
Statistical analyses discarded several variables as factors contributing to the epidemic, including the long-term population increase in Bakersfield, mean annual temperature, and TSP. It has long been recognized that coccidioidomycosis is a seasonal disease, with rates of new cases peaking in October to November (21, 34), the dustiest months of the year. It has also been observed that the number of cases is sometimes greatest after a heavy winter rainfall (34). Our analysis found the first statistical evidence to link the amount of rainfall with the number of cases of coccidioidomycosis. Unexpectedly, the occurrence of ENSO was not found to be a significant factor in determining the number of cases of coccidioidomycosis in this region. Rather, the number of cases was related to the length of the drought preceding rainfall, showing that the timing of rainfall, and not simply its amount, is important in determining infection rates. Most significantly, the drought preceding the 1991-1994 epidemic was the most sustained seen since 1956, implicating it in the development of this uncharacteristically large epidemic. How the drought length may affect the growth of C. immitis is a matter of conjecture. It is known that C. immitis is a poor competitor on nutrient media and is easily overgrown by common soil fungi but is resistant to dessication and high temperatures (35). We can hypothesize that this particular extended drought suppressed fungal competitors relative to C. immitis to the extent that constraints on the growth of C. immitis were released when the 1992 ENSO increased rainfall in California. However, this is a purely speculative scenario and further studies of the natural soil ecology of C. immitis are necessary to further this argument. We conclude that a fortuitous conjunction of climatic variables appears to have allowed the epidemic to occur. Further analyses across the geographic range of C. immitis will demonstrate the generality of this finding and establish whether the predictive model developed here is of use as an early warning system for epidemics of this particular fungal pathogen.
The emergence of aggressive fungal pathogens and the consequent epidemics in their host organisms are currently receiving much attention. It has been demonstrated that a chytrid, Batrachochytrium dendrobatidis (25), is responsible for a pandemic in amphibian populations and that Aspergillus sydowii is causing mass mortality of sea fan corals in the Caribbean (19). However, in both cases, it is not known whether environmental change impacting host fitness or increases in virulence of the pathogen are to blame (3, 27). That a recent Phytophthora epidemic on British alder (Alnus sp.) was due to the emergence of an aggressive hybrid (5) shows the potential for damaging genetic change in fungal species. However, our data show equally how environmental factors have to be accounted for. Here, we have shown the practical utility of using well-characterized loci and population genetic analysis in conjunction with multivariate analyses to answer biologically relevant epidemiological questions. These approaches will aid in dissecting the factors behind epidemics of fungal populations, as well as in elucidating fundamental features of microbial natural history.
ACKNOWLEDGMENTS
We thank R. Talbot for supplying the isolates; G. Cole and D. Carter for the information on unpublished sequences; D. Geiser, D. Greene, and S. Kroken for comments and assistance; and J. J. Bull and F. J. Ayala for comments.
This work was supported by grants to J. W. Taylor from the NIH and the Novartis Agricultural Discovery Institute, Inc.
REFERENCES
- 1.Archie J W. A randomization test for phylogenetic information in systematic data. Syst Zool. 1989;38:239–252. [Google Scholar]
- 2.Arsura E, Caldwell J, Johnson R, Einstein H, Welch G, Talbot R, Affentranger H. Coccidioidomycosis epidemic of 1991: epidemiologic features. In: Einstein H E, Catanzaro A, editors. Coccidioidomycosis. Washington, D.C.: National Foundation for Infectious Diseases; 1996. pp. 98–107. [Google Scholar]
- 3.Berger L, Speare R, Daszak P, Green D. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouma M J, Dye C. Cycles of malaria associated with El Nino in Venezuela. JAMA. 1997;278:1772–1774. [PubMed] [Google Scholar]
- 5.Brasier C M, Cooke D E L, Duncan J M. Origin of a new Phytophthora pathogen through interspecific hybridization. Proc Natl Acad Sci USA. 1999;96:5878–5883. doi: 10.1073/pnas.96.10.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown A H D, Feldman M W, Nevo E. Multilocus structure of natural populations of Hordeum spontaneum. Genetics. 1980;96:523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown J K M, Simpson C G. Genetic analysis of DNA fingerprints and virulences in Erysiphe graminis f. sp. hordei. Curr Genet. 1994;26:172–178. doi: 10.1007/BF00313807. [DOI] [PubMed] [Google Scholar]
- 8.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. A safe method of extracting DNA from Coccidioides immitis. Fungal Genet Newsl. 1995;42:23. [Google Scholar]
- 10.Burt A, Dechairo B M, Koenig G L, Carter D A, White T J, Taylor J W. Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Mol Ecol. 1997;6:781–786. doi: 10.1046/j.1365-294x.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 11.California Air Resources Board. California Air Resources Board, Air Quality Data Review Section. 1998. Sacramento. [Google Scholar]
- 12.Centers for Disease Control. Update: coccidioidomycosis—California, 1991–1993. Morbid Mortal Weekly Rep. 1994;43:421–423. [PubMed] [Google Scholar]
- 13.Crow J F, Kimura M. An introduction to population genetics. New York, N.Y: Harper & Row, Publishers; 1970. [Google Scholar]
- 14.De Sousa A, Sanches S, Ferro M L, Vaz M J, De Lancastre H. The intercontinental spread of a drug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edlin B R, Tokars J I, Grieco M H, Crawford J T, Williams J, Holmberg S D. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N Engl J Med. 1992;326:1514–1521. doi: 10.1056/NEJM199206043262302. [DOI] [PubMed] [Google Scholar]
- 16.Fisher M C, Koenig G L, White T J, Taylor J W. Primers for genotyping single nucleotide polymorphisms and microsatellites in the pathogenic fungus Coccidioides immitis. Mol Ecol. 1999;8:1082–1084. doi: 10.1046/j.1365-294x.1999.00655_5.x. [DOI] [PubMed] [Google Scholar]
- 17.Flynn N M, Hoeprich P D, Kawachi M M. An unusual outbreak of wind-borne coccidioidomycosis. N Engl J Med. 1979;301:358–361. doi: 10.1056/NEJM197908163010705. [DOI] [PubMed] [Google Scholar]
- 18.Friedman L, Smith S E, Roessleb W G, Berman R J. The virulence and infectivity of twenty-seven strains of Coccidioides immitis. Am J Hyg. 1956;64:198–210. doi: 10.1093/oxfordjournals.aje.a119834. [DOI] [PubMed] [Google Scholar]
- 19.Geiser D M, Taylor J W, Ritchie K B, Smith G W. Cause of sea fan death in the West Indies. Nature. 1998;394:137–138. [Google Scholar]
- 20.Gentzbittel L, Mouzeyar S, Badaoui S, Mesties E, Vear F, Tourvieille De Labrouhe D, Nicolas P. Cloning of molecular markers for disease resistance in sunflower, Helanthus annuus L. Theor Appl Genet. 1998;96:519–525. doi: 10.1007/s001220050769. [DOI] [PubMed] [Google Scholar]
- 21.Jinadu B A. Valley Fever Task Force report on the control of Coccidioides immitis. 1995. [Google Scholar]
- 22.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1998;95:8414. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1997;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longcore J E, Pessier A P, Nichols D K. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia. 1999;91:219–227. [Google Scholar]
- 26.Maynard-Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morell V. Are pathogens felling frogs? Science. 1999;5415:728–731. doi: 10.1126/science.284.5415.728. [DOI] [PubMed] [Google Scholar]
- 28.National Climatic Data Center. Vol. I. Asheville, N.C: National Climatic Data Center; 1998. [Google Scholar]
- 29.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pappagianis D, Einstein H. Tempest from Tehachapi takes toll or Coccidioides conveyed aloft and afar. West J Med. 1978;129:527–530. [PMC free article] [PubMed] [Google Scholar]
- 31.Rice W R. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 32.Saubolle M A. Life cycle and epidemiology of Coccidioides immitis. In: Einstein H E, Catanzaro A, editors. Coccidioidomycosis. Washington, D.C.: National Foundation for Infectious Diseases; 1996. pp. 1–9. [Google Scholar]
- 33.Schneider E, Hajjeh R A, Spiegel R A, Jibson R W, Harp E L, Marshall G A, Gunn R A, McNeil M M, Pinner R W, Baron R C, Burger R C, Hutwagner L C, Crump C, Kaufman L, Reef S E, Feldman G M, Pappagianis D, Werner S B. A coccidioidomycosis outbreak following the Northridge, California, earthquake. JAMA. 1997;277:904–908. [PubMed] [Google Scholar]
- 34.Smith C E, Beard R R, Rosenberger H G, Whiting E G. Effect of season and dust control on coccidioidomycosis. JAMA. 1946;132:833–838. doi: 10.1001/jama.1946.02870490011003. [DOI] [PubMed] [Google Scholar]
- 35.Swatek F E, Omieczynski D T. Coccidioidomycosis. Tucson: The University of Arizona Press; 1967. [Google Scholar]
- 36.Taylor J W, Jacobson D J, Fisher M C. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 37.Weir B S. Genetic data analysis II. Sunderland, Mass: Sinauer Associates, Inc., Publishers; 1996. [Google Scholar]