Abstract
A mild and versatile method based on Cu-catalyzed [2+3] cycloaddition (Huisgen-Meldal-Sharpless reaction) was developed to tether 3,3’-((4-(prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-ones) with 4-azido-2-quinolones in good yields. This methodology allowed attaching three quinolone molecules via a triazole linker with the proposed mechanism. The products are interesting precursors for their anti-proliferative activity. Compound 8g was the most active one, achieving IC50 = 1.2 ± 0.2 µM and 1.4 ± 0.2 µM against MCF-7 and Panc-1 cell lines, respectively. Moreover, cell cycle analysis of cells MCF-7 treated with 8g showed cell cycle arrest at the G2/M phase (supported by Caspase-3,8,9, Cytochrome C, BAX, and Bcl-2 studies). Additionally, significant pro-apoptotic activity is indicated by annexin V-FITC staining.
Keywords: click, azido, quinolones, triazole, anti-proliferative, apoptosis
1. Introduction
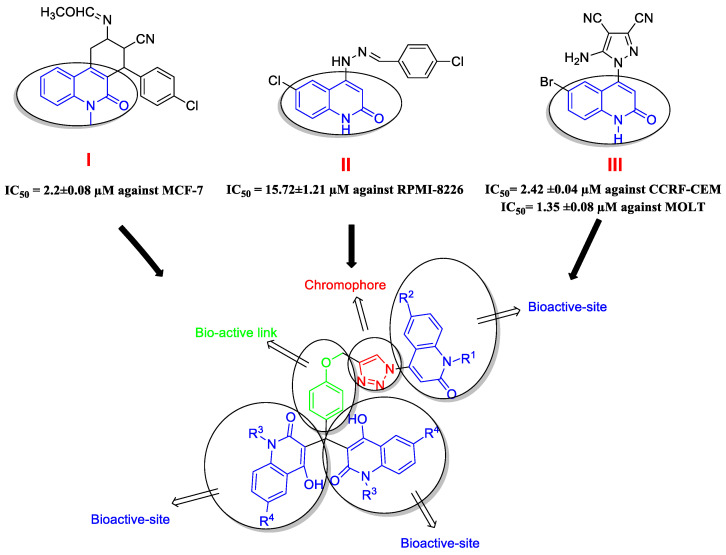
One of the most common heterocycles in drug development is the quinolone (oxo-quinoline) ring system [1,2]. Quinolone derivatives have been widely used in medicinal chemistry due to their unique structure, which exhibits a variety of pharmacological activities [3], especially antibacterial [4] and anti-cancer [5], with a promising role in the improvement of anti-cancer drug resistance [6]. Many quinolone derivatives exhibit excellent results in various operations, including growth inhibition via cell cycle arrest, apoptosis, and angiogenesis inhibition (Compounds I, II, and III, Figure 1) [7,8,9].
Figure 1.
Bioactive sites and chromophore group in the designed compounds 8a–l.
Triazole derivatives have a wide range of biological activities [10]. Furthermore, some drugs currently in clinical use are based on triazoles’ backbone, especially 1,2,3 triazole moiety [11,12]. Combining the quinolone scaffold with the triazole moiety will result in new leads with complementary activities and/or multiple pharmacological goals, and/or one component will counterbalance the side effects caused by the other [13,14]. Click chemistry is a simple method for assembling new molecular entities. The wide scope of CuAAC is firmly demonstrated by its use in different areas of life and material sciences fields, including drug discovery and bioconjugation [15,16].
Based on the preceding discussion and our efforts to produce quinoline derivatives as anti-cancer agents [17,18], as well as our work in the synthesis of heterocyclic molecules with potential biological activity [19], we used click chemistry to design and synthesize two bioactive sites in one molecule (Figure 1). The first is made up of three molecules of 4-hydroxy-2-quinolones, while the second is made up of a triazole moiety connected by a phenoxide linker. The newly synthesized compounds were tested for anti-proliferative efficacy against four different cancer cell lines. The most active ones were investigated further for their possible apoptotic activity utilizing extrinsic and intrinsic apoptotic indicators.
2. Results
2.1. Chemistry Section
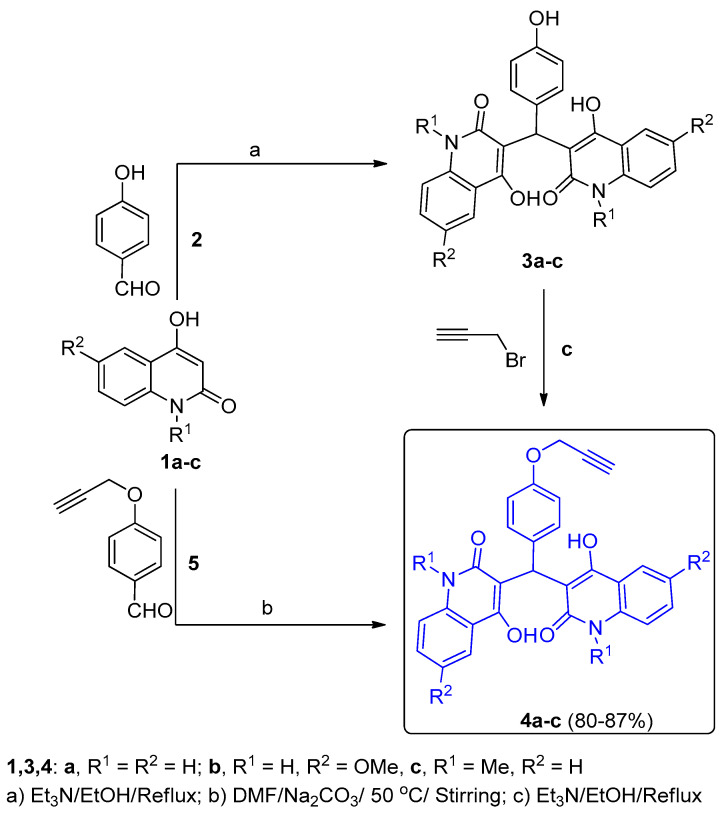
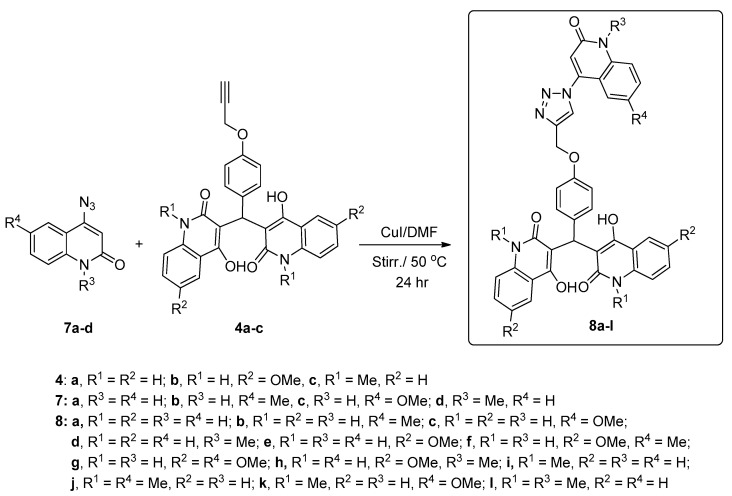
Here, in our present work, we synthesized a new various classes of quinolones/triazoles (Scheme 1 and Scheme 2) as 3,3’-((4-((1-(2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-ones) 8a–l from 4-azido-2-quinolinones 7a–d and 3,3’-((4-(prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxy-quinolin-2(1H)-ones) 4a–c by a Cu-catalyzed [3+2] cycloaddition, as shown in Scheme 2. We initially examined the synthesis of novel terminal alkynes 4a–c via interaction between 4-hydroxy-quinoline-2(1H)-ones 1a–c [20,21] and p-hydroxybenzaldehyde (2) (molar ratio 2:1) in absolute ethanol catalyzed with triethyl amine under refluxing conditions to yield 3,3’-((4-hydroxy-phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-ones) 3a–c. In DMF, the resulting phenol compound interacts with propargyl bromide to produce our terminal alkynes 4a–c in good yield.
Scheme 1.
Synthesis of bis(4-hydroxy-quinolin-2(1H)-ones) 4a–c.
Scheme 2.
Synthesis of the new target compounds 8a–l.
4-hydroxy-2-quinolinones 1a–c, on the other hand, reacted with 4-(prop-2-yn-1-yloxy)benzaldehyde (5) in a molar ratio (2:1) to generate our desired terminal alkynes 4a–c, as indicated in Scheme 1.
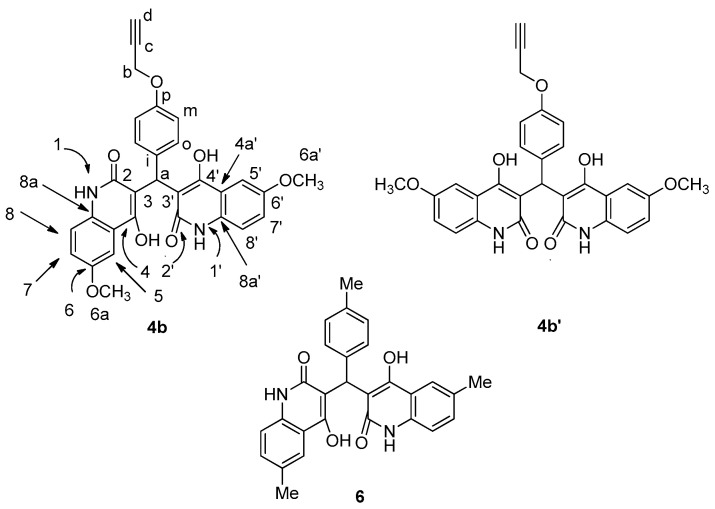
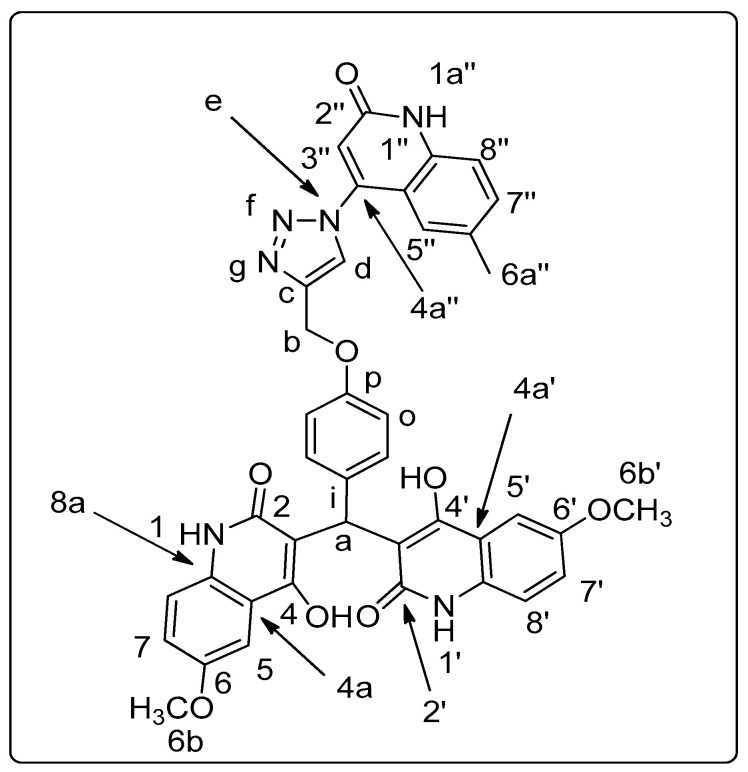
To confirm the structures of the obtained products 3,3’-(4-(prop-2-yn-1-yloxy)phenyl)methylene)-bis(4-hydroxy-quinolin-2(1H)-ones) 4a–c, the elemental analysis declares that the acquired molecular formula for 4a–c is generated from two molecules of compounds 1a–c and one molecule of aldehyde 5 with the elimination of H2O molecule or one molecule of 3a–c and one molecule of propargyl bromide with HBr molecule elimination. We discuss here the assignment of compound 4b, which was expected to be 3,3’-((4-(prop-2-yn-1-yloxy)phenyl)methylene)bis-(4-hydroxy-6-methoxyquinolin-2(1H)-one) as an example for the new terminal alkynes 4a–c (Figure 2).
Figure 2.
The structure of compound 4b, its tautomeric structure 4b’, and analog compounds 6.
The 1H NMR spectrum showed a characteristic four broad singlet signals at δH = 13.39, 12.90, 12.19, and 12.08 ppm, integrating four D2O exchangeable protons assigned as OH-4/4’, OH-4’/4, NH-1/1’, and NH-1’/1, respectively. Additionally, a singlet at δH = 6.14 ppm (1H) corresponded to q-CH-(H-a), which further confirmed from 13C NMR with a characteristic singlet at δC = 34.69 ppm (Table 1). The two methoxy groups with six protons exhibit a singlet at δH = 3.77 ppm, which further confirmed from 13C NMR with a characteristic singlet at δC = 55.41 ppm (C-6a, 6a’). Furthermore, the 1,4-disubstituted system was observed for the phenyl ring as doublet–doublet at δH = 7.02 (d, J = 8.5 Hz; 2H, H-o) and δH = 6.89 (d, J = 8.8 Hz; 2H, H-m), and both of them give 1H-1H-COSY correlation with each other and HMBC correlation with H-a and H-o (Table 1). In its 13C NMR spectra, C-8/8’ also gives HMBC correlation with a 2H doublet at δH 7.26 ppm, assigned as H-6/6’; its attached carbon appears at δC = 154.80 ppm, grounds as C-2,2’,4,4’. The 6H methyl singlet at δH = 3.77 must be H-6a/6a’ and gives HSQC correlation to carbon at δC = 55.41 ppm. C-a gives HMBC correlation to a 1H singlet at δH = 6.14, assigned as H-a; its attached carbon appears at δC = 34.69 ppm. Additionally, the four lines between δC = 166–160 give HMBC correlation with H-a and are assigned on chemical shift. Additionally, compound 4b has a tautomeric form 4b’ in which the two quinolinone rings are in the same direction. However, from 1H NMR spectra, it is clear that the two OH groups, the two NH groups, and H-5 and H-5’ are not equivalent: that means, the two quinolinone rings are spectroscopically non-equivalent because their rotation is hindered, so that the compound does not possess a σ-plane. The simplest rationale is that the tautomer populations of the two quinolinones rings differ (tautomeric equilibration being relatively fast). Additionally, the two quinolinone-NH, which were assigned as N-1/1’, give HSQC with a proton at δH 12.08 ppm, and the other N-1’/1 give HSQC with a proton at δH 12.19 ppm, which confirms that the two quinolone rings are not equivalent in comparison with the analogous compound 6 prepared previously [22]. Although the two quinolinone rings are stated to be spectroscopically non-equivalent, some of their signals co-resonate because their rotation is hindered, so that the compound does not possess a σ plane and must be assigned as 4b, not 4b’.
Table 1.
Spectrum data for compound 4b.
| 1H NMR | 1H-1HCOSY | Assignment | |
| 13.39 (bs; 1H) | OH-4/4’ | ||
| 12.90 (bs; 1H) | OH-4’/4 | ||
| 12.19 (bs; 1H) | NH-1/1’ | ||
| 12.08 (bs; 1H) | NH-1’/1 | ||
| 7.40 (d, J = 9.0; 2H) | 7.26 | H-8, 8’ | |
| 7.40 (bs; 1H) | H-5/5’ | ||
| 7.33 (bs; 1H) | H-5’/5 | ||
| 7.26 (bd, J = 8.6; 2H) | 7.40 | H-7, 7’ | |
| 7.02 (d, J = 8.5; 2H) | 6.89 | H-o | |
| 6.89 (d, J = 8.8; 2H) | 7.02 | H-m | |
| 6.14 (s; 1H) | H-a | ||
| 4.76 (d, J = 2.0; 2H) | 3.55 | H-b | |
| 3.77 (s; 6H) | H-6a, 6a’ | ||
| 3.55 (bs; 1H) | 4.76 | H-d | |
| 15N NMR | HSQC | HMBC | Assignment |
| 144.2 | 12.08 | 7.40 | N-1/1’ |
| 142.9 | 12.19 | 7.40 | N-1’/1 |
| 13C NMR | HSQC | HMBC | Assignment |
| 165.76, 164.10 | 6.14 | C-2, 2’ | |
| 161.84, 160.91 | 6.14 | C-4, 4’ | |
| 155.31 | 7.02, 6.89, 4.76 | C-p | |
| 154.80 | 7.40, 7.26, 3.77 | C-6, 6’ | |
| 131.52 | 7.40, 7.26 | C-8a, 8a’ | |
| 130.29 | 6.89, 6.14 | C-i | |
| 127.29 | 7.02 | 7.02, 6.14 | C-o |
| 120.63 | 7.26 | C-7, 7’ | |
| 117.46 | 7.40 | 7.40 | C-8, 8’ |
| 117.21, 116.70 | 7.40, 7.33 | C-4a, 4a’ | |
| 114.35 | 6.89 | 6.89, 6.89 | C-m |
| 111.74, 111.25 | 6.14 | C-3, 3’ | |
| 103.82 | 7.40, 7.33 | C-5, 5’ | |
| 79.43 | 4.76 | C-c | |
| 78.00 | 3.55 | 3.55 | C-d |
| 55.41 | 3.77 | C-6a, 6a’ | |
| 55.34 | 4.76 | C-b | |
| 34.69 | 6.14 | 7.02, 6.14 | C-a |
Secondly, 4-azido-2-quinolinones 7a–d reacted with 3,3’-((4-(prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-ones) 4a–c in DMF as a solvent in molar ration (1.2:1) and catalyzed by 10 mol% of CuI to give 3,3’-((4-((1-(2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)-bis(4-hydroxyquinolin-2(1H)-one) 8a–l in an excellent yields (Scheme 2).
Different spectral data (1H NMR, 13C NMR, 2D NMR, and 15N NMR; see Supplementary Materials) addition to mass spectrometry, as well as elemental analyses, were used to confirm the structures of all new compounds 8a–l. For example, compound 8f was assigned as 3,3’-((4-((1-(6-methyl-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)-bis(4-hydroxy-6-methoxyquinolin-2(1H)-one). It exhibited a molecular formula of C40H32N6O8, representing a product from one molecule of 4b and one molecule of 7b without elimination. Compound 8f was further confirmed by mass spectrometry with m/z = 724 and elemental analysis (Figure 3), which confirm that compound 8f is resulting from the reaction of compound 4b with 4-azido-6-methyl-2-quinolinone 7b in molar ratio (1:1) without elimination.
Figure 3.
Structure of compound 8f.
The 6-methyl-4-triazolo-2-quinolinone substructure (including the triazole ring) is assigned by analogy with compound 8b. In both compounds, the two OH groups, the two NH groups, and H-5 and H-5’ are not equivalent: the simplest rationale is that the tautomer populations of the two quinolinone rings differ (tautomeric equilibration being relatively fast). Consistent with this idea, N-1 and N-1’, C-2 and C-2’, C-3 and C-3’, and C-4 and C-4’ are also non-equivalent in both samples; C-4a and C-4a’ are non-equivalent in compound 4b, and C-8a and C-8a’ are non-equivalent in compound 8f (Table 2, Exp. Section). The 1H-NMR spectrum of 8f showed five protons that appeared as broad signals at δH 13.41, 12.91, 12.20, and 12.09 ppm for the quinolinone rings as two hydroxyl groups OH-4/4’, OH-4’/4, quinolinone-N-1/1’,1″ and quinolinone-N-1’/1, respectively. These two protons were exactly assigned by analogy with 4b (Figure 2) and 2D NMR (Table 2). Additionally, there were five doublets at δH 7.47 (d, J = 8.3 Hz; H-7″), 7.40 (d, J = 9.2 Hz; 2H, H-8,8’), and 7.38 (d, J = 8.4 Hz; 2H, H-5/5’, 8″), in addition to a 1,4-disubstituted doublet-doublet at δH 7.05 (d, J = 8.2, Hz; 2H, H-o) and 7.01 (d, J = 8.2 Hz; 2H, H-m), and four singlets at δH 7.22, 6.81, 5.27, 3.82, and 2.29 ppm, which were assigned as H-5’’, H-a, H-3″, H-a, H-b, H-6b, 6b’, and H-6a″, respectively. The 13C NMR spectrum for 8f showed that four signals appeared at δC 165.85, 164.13, 161.84, and 160.84 ppm, assigned as C-2,2’, C-4,4’, and C-2″,4″, respectively. Additionally, the signal at δC 60.85, which gives HSQC correlation with the proton at δH 5.27, was assigned as C-b (-O-CH2), in addition to two signals at 55.40 and 20.50 ppm, which was assigned for two methoxy groups (C-6b, 6b’) and methyl group (C-6a″), respectively. Furthermore, we see four types of nitrogen in the 15N NMR spectrum; one of them appears at δN 247.4, which indicates an sp3 nitrogen; it was assigned as N-e and gave HMBC correlation with a proton at δH 8.84 and 6.81 (H-d, H-3″) but gives no HSQC correlation. The second nitrogen atom resonated at δN 151.6 ppm, which assigned as N-1″ and also gives HMBC correlation with a proton at δH 7.38 and 6.81 (H-5/5’, 8″ and H-3″). The last two nitrogen at δN 144.2 and 142.8 ppm., which they assigned as N-1’/1 and N-1/1’, and they give HSQC correlation with the attached protons at δH 12.09 and 12.20, respectively.
Table 2.
Spectrum data for compound 8f.
| 1H NMR | 1H-1HCOSY | Assignment | |
| 13.41 (bs; 1H) | OH-4/4’ | ||
| 12.91 (bs; 1H) | OH-4’/4 | ||
| 12.20 (s; 2H) | 6.81 | NH-1/1’, 1″ | |
| 12.09 (bs; 1H) | NH-1’/1 | ||
| 8.84 (s; 1H) | H-d | ||
| 7.47 (d, J = 8.3; 1H) | 7.38, 7.22 | H-7″ | |
| 7.40 (d, J = 9.2; 2H) | 7.26 | H-8, 8’ | |
| 7.38 (d, J = 8.4; 2H) | 7.47, 7.26, 2.29 | H-5/5’, 8″ | |
| 7.33 (m; 1H) | 7.26 | H-5’/5 | |
| 7.26 (d, J = 8.1; 2H) | 7.40, 7.38, 7.33 | H-7, 7’ | |
| 7.22 (s; 1H) | 7.47, 2.29 | H-5″ | |
| 7.05 (d, J = 8.2; 2H) | 7.01, 6.15 | H-o | |
| 7.01 (d, J = 8.2; 2H) | 7.05 | H-m | |
| 6.81 (s; 1H) | 12.20 | H-3″ | |
| 6.15 (bs; 1H) | 7.05 | H-a | |
| 5.27 (s; 2H) | H-b | ||
| 3.82 (s; 6H) | H-6b, 6b’ | ||
| 2.29 (s; 3H) | 7.40, 7.38, 7.26 | H-6a″ | |
| 13C NMR | HSQC | HMBC | Assignment |
| 165.85, 164.13 | 6.15 | C-2, 2’ | |
| 161.84 | C-4, 4’ | ||
| 160.84 | C-2″, 4″ | ||
| 156.06 | 7.05, 5.27 | C-p | |
| 154.81 | 7.40, 3.82 | C-6, 6’ | |
| 143.42 | 8.84, 7.40, 7.22, 6.81, 5.27 | C-c | |
| 137.50 | 7.47, 7.22 | C-8a″ | |
| 133.15 | 7.47 | 7.47, 7.22 | C-7″ |
| 131.73, 131.51 | 7.38, 7.26, 2.29 | C-8a, 8a’ | |
| 130.07 | C-i | ||
| 127.40 | 7.05 | C-o, 6″ | |
| 126.50 | 8.84 | 8.84,3 7.01, 5.27 | C-d |
| 123.15 | 7.22 | 7.47, 7.05, 2.29 | C-5″ |
| 120.82 | 7.26 | 7.01 | C-7, 7’ |
| 117.69 | 6.81 | 7.05, 6.813 | C-3″ |
| 117.45 | 7.40 | 7.403 | C-8, 8’ |
| 116.69 | 7.38, 7.33 | C-4a, 4a’ | |
| 115.89 | 7.38 | C-8″ | |
| 114.38 | 7.01 | 7.38, 6.81 | C-m, 4a″ |
| 111.86, 110.18 | 6.15 | C-3, 3’ | |
| 103.81 | 7.38, 7.33 | C-5, 5’ | |
| 60.85 | 5.27 | C-b | |
| 55.40 | 3.82 | 3.823 | C-6b, 6b’ |
| 34.71 | 6.15 | C-a | |
| 20.50 | 2.29 | 7.47, 7.22, 2.293 | C-6a″ |
| 15N NMR | HSQC | HMBC | Assignment |
| 247.4 | 8.84, 6.81 | N-e | |
| 151.6 | 12.20 | 7.38, 6.81 | N-1″ |
| 144.2 | 12.09 | N-1’/1 | |
| 142. | 12.20 | N-1/1‘ | |
| N-f, g n/o. | |||
2.2. Pharmacological Assays
2.2.1. Cell Viability Assay
A human mammary gland epithelial cell line was used in the cell viability assay (MCF-10A). MCF-10A cells were incubated with compounds 8a–l for four days, and cell viability was determined using an MTT assay [23,24]. There were no cytotoxic effects in any of the compounds tested, and most of the compounds tested had more than 84% viability at 50 µM; Table 3 shows the results.
Table 3.
Anti-proliferative activity of compounds 8a–l and doxorubicin.
| Comp. | Cell Viability% (50 µM) | Anti-Proliferative Activity IC50 ± SEM (µM) | ||||
|---|---|---|---|---|---|---|
| A-549 | MCF-7 | Panc-1 | HT-29 | Average (IC50) |
||
| 8a | 89 | 5.7 ± 0.4 | 5.3 ± 0.8 | 5.6 ± 0.6 | 5.7 ± 0.4 | 5.575 |
| 8b | 87 | 7.6 ± 0.8 | 6.9 ± 0.8 | 7.6 ± 0.6 | 7.5 ± 0.4 | 7.400 |
| 8c | 91 | 3.9 ± 0.5 | 3.2 ± 0.08 | 4.1 ± 0.2 | 4.2 ± 0.2 | 3.850 |
| 8d | 89 | 12.9 ± 0.8 | 11.5 ± 1.1 | 11.6 ± 0.8 | 11.4 ± 1.2 | 11.850 |
| 8e | 85 | 2.1 ± 0.2 | 1.5 ± 0.7 | 1.7 ± 0.1 | 2.2 ± 0.4 | 1.875 |
| 8f | 87 | 2.9 ± 0.5 | 2.6 ± 0.8 | 3.1 ± 0.2 | 3.4 ± 0.2 | 3.000 |
| 8g | 84 | 1.9 ± 0.2 | 1.2 ± 0.2 | 1.4 ± 0.2 | 1.8 ± 0.4 | 1.575 |
| 8h | 90 | 10.7 ± 2.5 | 10.6 ± 2.9 | 10.8 ± 1.9 | 10.8 ± 1.6 | 10.725 |
| 8i | 90 | 17.9 ± 0.3 | 17.8 ± 1.6 | 17.6 ± 1.5 | 17.9 ± 1.1 | 17.800 |
| 8j | 81 | 22.5 ± 0.2 | 22.1 ± 0.1 | 22.4 ± 0.2 | 22.9 ± 0.6 | 22.474 |
| 8k | 90 | 14.5 ± 2.6 | 13.6 ± 2.2 | 14.6 ± 2.9 | 14.8 ± 1.4 | 14.375 |
| 8l | 96 | 25.3 ± 2.5 | 24.9 ± 1.8 | 24.5 ± 2.3 | 24.2 ± 1.4 | 24.725 |
| Doxorubicin | -- | 1.21 ± 0.80 | 0.90 ± 0.62 | 1.41 ± 0.58 | 1.01 ± 0.82 | 1.136 |
2.2.2. Cytotoxic Activity and Evaluation of IC50
Tested 8a–l compounds were assessed for their anti-proliferative activity against four human cancer cell lines, including the pancreatic cancer cell line (Panc-1), breast cancer cell line (MCF-7), colon cancer cell line (HT-29), and epithelial cancer cell line (A-549) using propidium iodide fluorescence assay [25,26] and the reference doxorubicin. Results (IC50) are illustrated in Table 3.
From the results, it is clear that the electronic effect of the substitution (R2 and/or R4) on the phenyl groups of quinoline moieties is important for the antiproliferative activity. Compounds 8c, 8e, 8f, and 8g (R2 and/or R4 = OCH3) were the most active compounds against the tested four cell lines with average IC50 in the range of 1.575 to 3.850 μM. Compound 8g (R1 = R3 = H, R2 = R4 = OCH3) showed the utmost antiproliferative activity with average IC50 = 1.575 μM compared to the reference doxorubicin with average IC50 = 1.136 μM. Compounds 8e (R1 = R3 = R4 = H, R2 = OCH3, average IC50 = 1.875) and 8c (R1 = R2 = R3 = H, R4 = OCH3, average IC50 = 3.850) showed higher IC50 (less potent) than 8g. Replacement of OCH3 group either by H or CH3 resulted in a decrease in the antiproliferative activity. For example, compounds 8a (R1 = R2 = R3 = H, R4 = H, average IC50 = 5.575 μM) and 8b (R1 = R2 = R3 = H, R4 = CH3, average IC50 = 7.40 μM) showed a 1.5-fold and 2-fold decrease in activity relative to 8c, respectively.
Another significant factor influencing the efficacy of these compounds is the position of substituent on the scaffold studied, which had a major impact on its cytotoxic activity. Generally, triazoles 8a–c and 8e–g (R1 and/or R3 = H) showed superior anti-proliferative activity compared to their N-methyl counterparts 8d, 8h, 8i, and 8j–l ((R1 and/or R3 = CH3), Table 3. For example, the unsubstituted derivative 8a (R1 = R2 = R3 = R4 = H) showed an average IC50 of 5.575 µM against the tested cell lines. Replacement of the NH group by N-CH3, 8l (R1 = R3 = CH3, R2 = R4 = H, average IC50 = 24.725 μM) resulted in at least 4-fold reduction of the average IC50 value. In addition, the average IC50 values of 8i–k (R1 and/or R3 = CH3) were the lowest among the compounds tested, indicating the significance of the NH group for anti-proliferative action.
2.2.3. Apoptosis Assay
To discover the pro-apoptotic capability of our target compounds, the utmost active compounds 8e, 8f, and 8g were tested for their potential ability to induce apoptosis in a MCF-7 breast cancer cell line.
Activation of Proteolytic Caspases Cascade
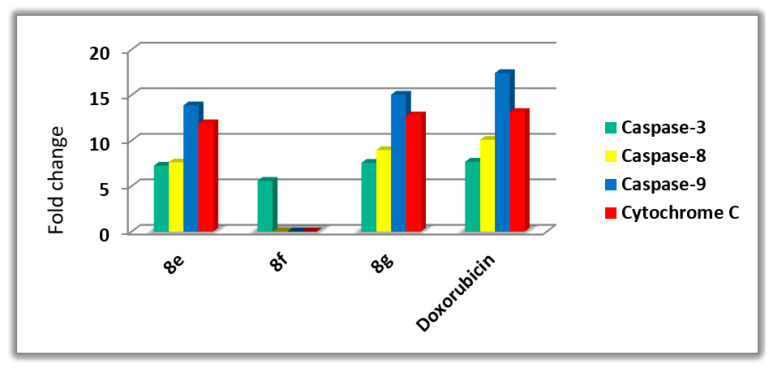
Caspases play an important role in initiating and completing the apoptotic process [27]. Caspase 3 is an essential caspase that cleaves multiple proteins in cells, resulting in apoptotic cell death [28]. The effects of compounds 8e, 8f, and 8g on Caspase 3 were investigated and compared to that of doxorubicin. The findings revealed that when compared to control cells, the studied compounds increased the level of active Caspase 3 by 5.5–7.5-fold and that 8e, 8f, and 8g had unprecedented control over Caspase 3 protein level expression (475.20 ± 4.27, and 365.60 ± 3.20, and 489.2 ± 4.13 pg/mL, respectively) compared to doxorubicin (503.2 ± 4.22 pg/mL). Compared to control untreated cells, the most active compound 8g showed a 7.45-fold increase in active Caspase 3 levels, Figure 4.
Figure 4.
Effects of compounds 8e, 8f, and 8g, and doxorubicin on active Caspases 3, 8, 9, and Cytochrome C in MCF-7 breast cancer cell line.
Compounds 8e and 8g were also tested for their effects on Caspases 8 and 9. The results showed that compound 8g raises Caspase 8 and 9 levels by 8.94- and 15.03-fold, respectively. When compared to control cells, compound 8e showed a 7.58- and 13.86-fold increase in Caspase 8 and 9, respectively, indicating activation of both the intrinsic and extrinsic pathways, with the intrinsic pathway having a stronger effect since Caspase 9 levels were higher [29].
Cytochrome C Assay
The amount of cytochrome C in a cell plays an important role in caspase activation and the initiation of the intrinsic apoptosis pathway [29,30]. Table 4 shows the effects of testing triazole derivatives 8e and 8g as cytochrome C activators in the MCF-7 human breast cancer cell line. 8e and 8g resulted in around 11.90- and 12.76-fold higher levels of cytochrome C expression than untreated control cells. The findings add to the proof that apoptosis can be attributed to cytochrome C overexpression and activation of the intrinsic apoptotic pathway triggered by the tested compounds.
Table 4.
Effects of compounds 8e, 8f, 8g, and doxorubicin on active Caspases 3, 8, 9, and cytochrome C in MCF-7 breast cancer cell line.
| Compound No. | Caspase 3 | Caspase 8 | Caspase 9 | Cytochrome C | ||||
|---|---|---|---|---|---|---|---|---|
| Conc (pg/mL) |
Fold Change | Conc (ng/mL) | Fold Change | Conc (ng/mL) | Fold Change | Conc (ng/mL) | Fold Change | |
| 8e | 475.20 ± 4.27 | 7.23 | 1.29 | 7.58 | 12.89 | 13.86 | 0.548 | 11.90 |
| 8f | 365.60 ± 3.20 | 5.57 | ND | ND | ND | ND | ND | ND |
| 8g | 489.20 ± 4.13 | 7.54 | 1.52 | 8.94 | 13.98 | 15.03 | 0.587 | 12.76 |
| Doxorubicin | 503.20 ± 4.22 | 7.66 | 1.75 | 10.07 | 16.23 | 17.40 | 0.604 | 13.13 |
| Control | 65.64 | 1 | 0.17 | 1 | 0.93 | 1 | 0.046 | 1 |
ND: Not determined.
Bax and Bcl-2 Levels Assay
Using doxorubicin as a control, the most active caspase activators 8e and 8g were further investigated for their impact on Bax and Bcl-2 levels in the MCF-7 breast cancer cell line [31,32]. Table 5 shows that 8e and 8g elicited a significant increase in Bax levels as compared to doxorubicin. 8g demonstrated a comparable induction of Bax (264.90 pg/mL) relative to doxorubicin (276 pg/mL), 32-fold higher than the control untreated breast cancer cell, followed by compound 8e (247.65 pg/mL and 30-fold change). Finally, compound 8g reduced Bcl-2 protein levels in MCF-7 cells to 1.085 ng/mL, followed by compound 8e (1.278 ng/mL) and doxorubicin (0.98 ng/mL).
Table 5.
Bax and Bcl-2 levels for compounds 8e, 8g, and Doxorubicin in MCF-7 breast cancer cell line.
| Compound No. | Bax | Bcl-2 | ||
|---|---|---|---|---|
| Conc (pg/mL) | Fold Change | Conc (ng/mL) | Fold Reduction | |
| 8e | 247.65 | 29.98 | 1.278 | 3.97 |
| 8g | 264.90 | 32.07 | 1.085 | 4.68 |
| Doxorubicin | 276.19 | 33.42 | 0.983 | 5.17 |
| Cont. | 8.26 | 1 | 5.086 | 1.00 |
Flow Cytometric Cell Cycle Analysis
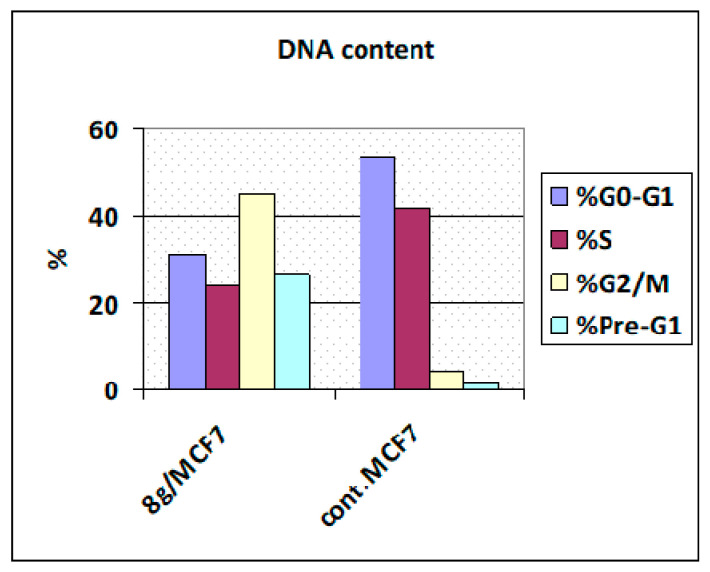
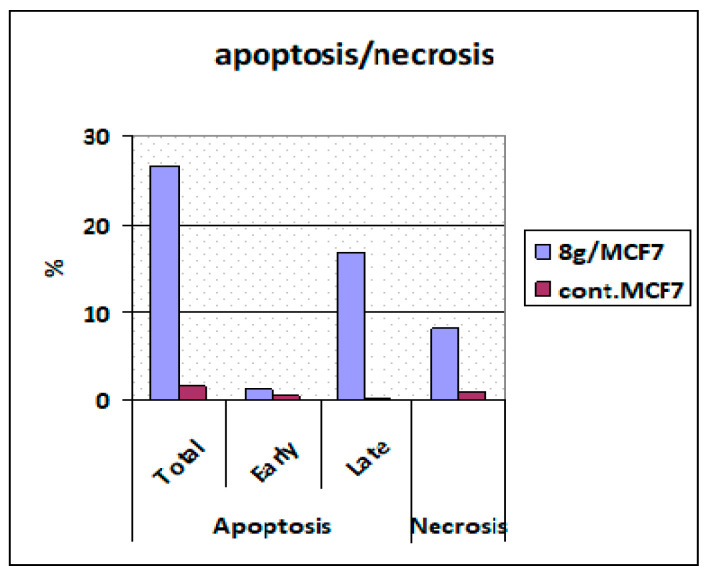
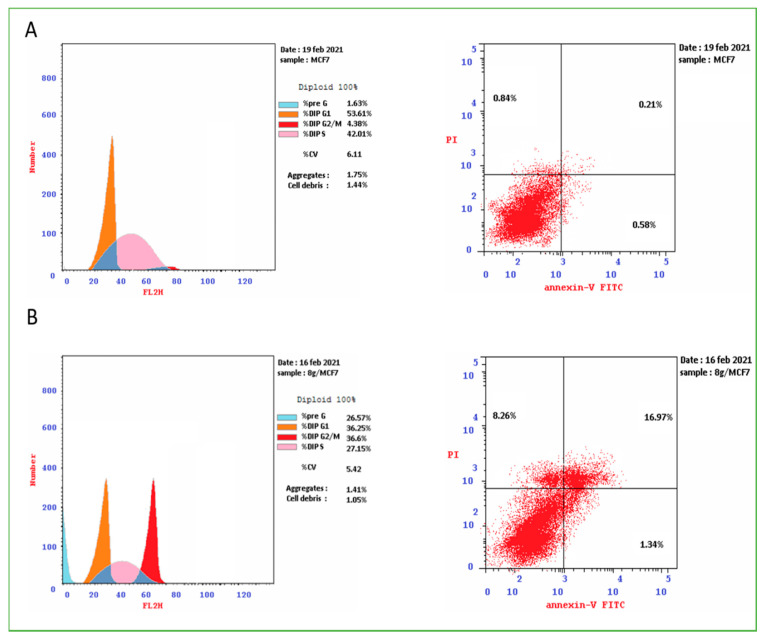
Following treatment with the most active compound, 8g, a cell cycle analysis was performed on the MCF-7 cancer cell line [33,34]. Figure 5 shows that the percentage of MCF-7 cell line cells in the G0/G1 phase was 53.61% in control and 36.25% after treatment with compound 8g, whereas the percentage of cells in the S phase was reduced with compound 8g (27.15%) compared to the control (42.01%). Compared to the control, the percentage of MCF-7 cell line in the G2/M process increased to 36.6% after treatment with 8g (4.38%). Furthermore, the percentage of apoptotic cells in the Pre-G1 process increased from 1.63% in control untreated MCF-7 cells to 26.57% and 22.17% in cells treated with 8g and doxorubicin, respectively (Figure 6 and Figure 7). According to the findings, 8g primarily displayed cell cycle arrest at the G2/M phases. Furthermore, the investigated compound is anti-proliferative rather than cytotoxic, resulting in programmed cell death and cell cycle arrest.
Figure 5.
Cell cycle analysis in MCF-7 cell line treated with compound 8g.
Figure 6.
Percentage of apoptosis and necrosis for compound 8g in MCF-7 cell line.
Figure 7.
Cell cycle analysis and Apoptosis induction analysis using Annexin V/PI of control untreated MCF-7cell line (A) and 8g (B).
3. Conclusions
A series of novel conjugated heterocycles containing three quinolinone moieties could be synthesized straightforwardly. The propargylated core of the phenoxide linker with two molecules of quinolinone could be equipped with another tethered molecule by reacting with 4-azido-2-quinolones via a Meldal–Sharpless click reaction. The target products 3,3’-((4-(prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxy-quinolin-2(1H)-ones) were obtained in good to very good yields and were characterized by different spectroscopic and elemental methods including 1H NMR, 13C NMR, 2D NMR, MS, and elemental microanalyses.
The antiproliferative activity of the new compounds was tested against a panel of cancer cell lines. Compound 8g demonstrated the highest antiproliferative activity with an average IC50 = 1.575 M compared to doxorubicin (IC50 = 1.136 M). The results show that the electronic effect of the substitution, as well as its position on the scaffold studied, have a significant impact on the activity of these compounds.
The most active compounds were further investigated for caspase, cytochrome C activation, Bax activation, and Bcl-2 downregulation compared to doxorubicin. Compounds 8e and 8g increased the levels of active Caspase 3, 8, and 9, indicating that both the intrinsic and extrinsic pathways were activated, with the intrinsic pathway having a stronger effect due to higher Caspase 9 levels. Compound 8g exhibited cell cycle arrest at the G2/M phase with a significant pro-apoptotic activity.
4. Experimental
4.1. Chemistry
Melting points were determined on Stuart electrothermal melting point apparatus and were uncorrected. All reactions were monitored with thin-layer chromatography (TLC) on Merck alumina-backed TLC plates and visualized under UV light. NMR spectra were measured in DMSO-d6 on a Bruker AV-400 spectrometer (400 MHz for 1H, 100 MHz for 13C, and 40.54 MHz for 15N), at the Chemistry Department, Florida Institute of Technology, 150 W University Blvd, Melbourne, FL 32901, USA, and Chemistry Department, Faculty of Science, Cairo University, Cairo, Egypt. Chemical shifts are expressed in δ (ppm) versus internal Tetramethylsilane (TMS) = 0 ppm for 1H and 13C, and external liquid ammonia = 0 ppm for 15N. Coupling constants are stated in Hz. Correlations were established using 1H-1H COSY, 1H-13C, and 1H-15N HSQC and HMBC experiments. Chemical shifts (δ) are reported in parts per million (ppm) relative to Tetramethylsilane (TMS) as an internal standard, and the coupling constants (J) are reported in Hertz (Hz). Splitting patterns are denoted as follows: singlet (s), broad (b), doublet (d), multiplet (m), triplet (t), quartet (q), broad of singlet (bs), doublet of doublets (dd), doublet of triplets (dt), triplet of doublets (td), and doublet of a quartet (dq). Mass spectra were recorded on a Finnigan Fab 70 eV at Al-Azhar University, Egypt. Elemental analyses were carried out on a Perkin Elmer device at the Microanalytical Institute of Organic Chemistry, Karlsruhe Institute of Technology, Karlsruhe, Germany.
4.1.1. Starting Materials
4-Hydroxy-quinoline-2(1H)-ones 1a–c [20,21], 3,3’-((4-hydroxy-phenyl)methylene) bis(4-hydroxyquinolin-2(1H)-ones) 3a–c [22], 4-(prop-2-yn-1-yloxy)benzaldehyde 5 [35], and 4-azido-2-quinolinones 7a–d [36] were synthesized according to the literature. 4-hydroxybenzaldehyde (5) and propargyl bromide (Aldrich) were used as received.
4.1.2. General Procedure for the Synthesis of Compounds 4a–c
Method A: A mixture of 4-hydroxy-2-quinolinones 1a–c (2 mmol), p-hydroxy-benzaldehyde (2) (1 mmol), and 0.5 mL of Et3N in 20 mL absolute ethanol was refluxed for 6 h. The resulting white precipitate was left to cool and then poured into ice-cold water to give 3a–c. The precipitate collected by filtration was dried well and then reacted with propargyl bromide in DMF in a molar ratio (1:2), and the reaction mixture was monitored by TLC and then poured into about 200 g ice to give 4a–c, which was collected by filtration.
Method B: A mixture of 4-hydroxy-2-quinolinones 1a–c (2 mmol), 4-(prop-2-yn-1-yloxy)benzaldehyde (5) (1 mmol), and 0.5 mL of Et3N in 20 mL absolute ethanol was refluxed for 4h. The resulting white precipitate was left to cool and then filtered off to give 4a–c in excellent yields.
3,3’-((4-(Prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-one) 4a. Colorless powder (85%), m.p. 296–98 °C; 1H NMR (DMSO-d6): δH = 13.12 (bs; 1H, OH-4/4’), 12.67 (bs; 1H, OH-4’/4), 12.24 (bs; 1H, NH-1/1’), 12.21 (bs; 1H, NH-1’/1), 7.98 (bs; 1H, H-5/5’), 7.95 (bd, J = 6.6 Hz; 1H, H-5’/5), 7.61 (dd, J = 7.6, 7.6 Hz; 2H, H-7, 7’), 7.45 (d, J = 8.2 Hz; 2H, H-8, 8’), 7.30 (bs; 2H, H-6, 6’), 7.03 (d, J = 8.5 Hz; 2H, H-o), 6.88 (d, J = 8.7 Hz; 2H, H-m), 6.14 (s; 1H, H-a), 4.76 (d, J = 1.8 Hz; 2H, H-b), 3.55 ppm (bs; 1H, H-d), 13C NMR (DMSO-d6): δC = 165.90 (C-2/2’), 163.40 (C-2’/2), 161.00 (C-4, 4’), 155.35 (C-p), 137.00 (C-8a, 8a’), 131.10 (C-7, 7’), 130.16 (C-i), 127.31 (C-o), 123.15 (C-5, 5’), 122.52 (C-6, 6’), 115.87 (C-8, 8’), 114.36 (C-m), 110.80 (C-4a, 4a’), 79.41 (C-c), 78.01 (C-d), 55.34 (C-b), 34.50 ppm (C-a), 15N NMR (DMSO-d6): δN = 144.8 ppm (N-1, 1’). Anal. Calcd for C28H20N2O5: C, 72.41; H, 4.34; N, 6.03; Found: C, 72.59; H, 4.28; N, 6.18.
3,3’-((4-(Prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxy-6-methoxy-quinolin-2(1H)-one)4b. Colorless powder (90%), m.p. 283–85 °C; NMR (DMSO-d6) (Table 1). Anal. Calcd for C30H24N2O7: C, 68.70; H, 4.61; N, 5.34; Found: C, 68.63; H, 4.55; N, 5.49.
3,3’-((4-(Prop-2-yn-1-yloxy)phenyl)methylene)bis(4-hydroxy-1-methylquinolin-2(1H)-one)4c. Colorless powder (78%), m.p. 280–82 °C; 1H NMR (DMSO-d6): δH = 12.86 (bs, 1H; OH-4/4’), 12.52 (bs, 1H; OH-4’/4), 8.05 (bs, 2H; H-5, 5’), 7.71 (ddd, J = 7.3, 7.3, 0.9 Hz, 2H; H-7, 7’), 7.66 (d, J = 8.3 Hz, 2H; H-8, 8’), 7.37 (dd, J = 7.3, 7.2 Hz, 2H; H-6, 6’), 6.98 (d, J = 8.6 Hz, 2H; H-o), 6.85 (d, J = 8.8 Hz, 2H; H-m), 6.23 (s, 1H; H-a), 4.75 (d, J = 2.2 Hz, 2H; H-b), 3.72 (bs, 6H; H-1a, 1a’), 3.56 (t, J = 2.2 Hz, 1H; H-d), 13C NMR (DMSO-d6): δC = 164.75 (b; C-2, 2’), 160.58 (b; C-4, 4’), 155.32 (C-p), 138.10 (C-8a, 8a’), 131.54 (C-7, 7’), 130.12 (C-i), 127.28 (C-o), 123.60 (C-5, 5’), 122.70 (C-6, 6’), 116.90 (C-4a, 4a’), 115.29 (C-8, 8’), 114.35 (C-m), 110.12 (b; C-3, 3’), 79.42 (C-c), 78.02 (C-d), 55.31 (C-b), 35.98 (C-a), 30.12 ppm (b; C-1a, 1a’), 15N NMR (DMSO-d6): δN = 138.7 ppm (N-1, 1’). Anal. Calcd for C30H24N2O5: C, 73.16; H, 4.91; N, 5.69; Found: C, 73.25; H, 4.81; N, 5.55.
4.1.3. General Procedure for the Synthesis of Compounds 8a–l
A mixture of terminal alkynes 4a–c (1.1 mmol) in 20 mL dimethylformamide (DMF) and CuI (10 mmol%) was stirred for 10 min at room temperature. Then, 4-azido compounds 7a–d (1.0 mmol) were added to the mixture. The reaction mixture was allowed to stir at 50 °C for 24 h, and TLC was used to monitor the reaction. After completion, the mixture was diluted with 50 mL H2O. The precipitate was filtered off to give 8a–l in excellent yields.
3,3’-((4-((1-(2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-methylene)bis(4-hydroxyquinolin-2(1H)-one)8a. Colorless powder, (75%), m.p. 280–82 °C; 1H NMR (DMSO-d6): δH = 13.18 (bs; 1H, OH-4/4’), 12.69 (bs; 1H, OH-4’/4), 12.29 (bs; 3H, NH-1, 1’, 1″), 8.86 (s; 1H, H-d), 7.96 (m; 2H, H-5, 5’), 7.63 (bd; 3H, H-7, 7’, 7″), 7.46 (bs; 4H, H-5″, 8, 8’, 8″), 7.26 (m; 2H, H-6, 6’), 7.24 (dd, J = 7.4, 7.4 Hz; 1H, H-6″), 7.05 (m; 2H, H-o), 6.86 (m; 2H, H-m), 6.16 (s; 1H, H-a), 5.27 ppm (s; 2H, H-b), 13C NMR (DMSO-d6): δC = 161.13, 160.77 (C-2, 2’, 2″, 4, 4’, 4″), 155.89 (C-p), 143.41, 143.25 (C-c), 139.18, 136.82 (C-8a, 8a’, 8a″), 131.61 (C-7, 7’, 7″), 130.83 (C-i), 127.22 (C-o), 123.74 (C-5″), 122.89 (C-5,5’), 122.34 (C-6, 6’, 6″), 117.48 (C-m), 116.37 (C-3, 3’, 3″), 115.69 (C-8, 8’, 8″), 114.22 (C-4a, 4a’, 4a″), 60.63 (C-b), 35.10 ppm (C-a), 15N NMR (DMSO-d6): δN = 247.50 (N-e or g), 152.50 (N-1″), 144.90 ppm (N-1,1’). EI-MS (m/z, %): 650 (M+, 60). Anal. Calcd for C37H26N6O6: C, 68.30; H, 4.03; N, 12.92. Found: C, 68.44; H, 3.98; N, 13.02.
3,3’-((4-((1-(6-Methyl-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-one)8b. Colorless powder, (80%), m.p. 230–32 °C; 1H NMR (DMSO-d6): δH = 13.17 (bs, 1H; OH-4/4’), 12.68 (bs, 1H; OH-4’/4), 12.20 (bs, 3H; NH-1, 1’, 1″), 8.83 (s, 1H; H-d), 7.96 (m, 2H; H-5, 5’), 7.60 (t, J = 7.3, 2H; H-7, 7’), 7.46 (t, J = 6.6, 3H; H-8, 8’, 7″), 7.38 (d, J = 8.4, 1H; H-8″), 7.30 (b, 2H; H-6, 6’), 7.22 (s, 1H; H-5″), 7.06 (d, J = 8.3, 2H; H-o), 7.01 (d, J = 8.4, 2H; H-m), 6.80 (s, 1H; H-3″), 6.16 (s, 1H; H-a), 5.26 (s, 2H; H-b), 2.28 ppm (s, 3H; H-6a″), 13C NMR (DMSO-d6): δC = 160.84 (C-2, 2’, 2″, 4, 4’, 4″), 156.09 (C-p), 143.42, 143.35 (C-c, 4″), 137.50 (C-8a″), 137.01 (C-8a, 8a’), 133.14 (C-7″), 131.73 (C-7, 7’), 131.09 (C-i), 127.43 (C-6″), 126.50 (C-d), 123.15 (C-5, 5’, 5″), 122.49 (C-6, 6’), 117.70 (C-3, 3’, 3″), 115.88 (C-8, 8’, 8″), 114.38 (C-o, m, 4a, 4a’, 4a″), 60.85 (C-b), 34.51 (C-a), 20.50 ppm (C-6a″), 15N NMR (DMSO-d6): δN = 247.2 (N-e), 151.8 (N-1″), 144.96 ppm (N-1, 1’), N-f, g n/o. EI-MS (m/z, %): 664 (M+, 28). Anal. Calcd for C38H28N6O6: C, 68.67; H, 4.25; N, 12.64. Found: C, 68.55; H, 4.33; N, 12.71.
3,3’-((4-((1-(6-Methoxy-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-one)8c. Colorless powder, (82%), m.p. 255–57 °C; 1H NMR (DMSO-d6): δH = 13.17 (bs, 1H; OH-4/4’), 12.68 (bs, 1H; OH-4’/4), 12.19 (bs, 3H; NH-1, 1’, 1″), 8.88 (s, 1H; H-d), 7.96 (m, 2H; H-5, 5’), 7.61 (m, 2H; H-7, 7’), 7.43 (bd, J = 9.0, 3H; H-8, 8’, 8″), 7.32 (bd, J = 8.3, 3H; H-6, 6’, 7″), 7.03 (bd, J = 9.7, 4H; H-o,m), 6.92 (bs, 1H; H-5″), 6.85 (s, 1H; H-3″), 6.15 (bs, 1H; H-a), 5.27 (s, 2H; H-b), 3.67 ppm (s, 3H; H-6b″), 13C NMR(DMSO-d6): δC = 166.40 (b), 164.85 (b; C-2, 2’), 161.38 (b; C-4, 4’), 160.54 (C-2″), 156.09 (C-p), 154.50 (C-6″), 143.50 (C-c), 143.10 (C-4″), 136.98 (C-8a, 8a’), 134.07 (C-8a″), 131.08 (C-7, 7’), 130.01 (b; C-i), 127.43 (C-o), 126.41 (C-d), 123.14 (C-5, 5’), 122.53 (C-6, 6’), 121.03 (C-7″), 117.95 (C-3″), 117.43 (C-8″), 115.88 (C-8, 8’), 114.84 (C-4a″), 114.38 (C-m), 111.38 (b; C-4a, 4a’), 110.15 (b; C-3), 105.51 (C-5″), 60.84 (C-b), 55.35 (C-6b″), 34.51 ppm (b; C-a), 15NNMR (DMSO-d6): δN = 248.1 (N-e), 151.0 (N-1″), 144.6 ppm (N-1, 1’), N-f, g n/o. EI-MS (m/z, %): 680 (M+, 27). Anal. Calcd for C38H28N6O7: C, 67.05; H, 4.15; N, 12.35. Found: C, 66.97; H, 4.01; N, 12.43.
3,3’-((4-((1-(1-Methly-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxyquinolin-2(1H)-one)8d. Colorless powder, (70%), m.p. 259–61 °C; 1H NMR (DMSO-d6): δH = 13.18 (bs, 1H; OH-4/4’), 12.68 (bs, 1H; OH-4’/4), 12.25 (bs, 2H; NH-1, 1’), 8.84 (s, 1H; H-d), 7.96 (m, 2H; H-5, 5’), 7.75 (t, J = 7.2, 1H; H-7″), 7.70 (d, J = 8.2, 1H; H-8″), 7.61 (m, 2H; H-7, 7’), 7.44 (m, 3H; H-8, 8’, 5″), 7.32 (m, 3H; H-6, 6’, 6″), 7.04 (m, 4H; H-o,m), 6.89 (s, 1H; H-3″), 6.16 (bs, 1H; H-a), 5.27 (s, 2H; H-b), 3.71 ppm (s, 3H; H-1a″), 13C NMR (DMSO-d6): δC = 161.27 (b; C-4, 4’), 160.29 (C-2″), 156.14 (C-p), 143.46 (C-c), 142.62 (C-4″), 140.10 (C-8a″), 136.98 (b; C-8a, 8a’), 132.27 (C-7″), 131.08 (C-7, 7’), 129.97 (b; C-i), 127.50 (C-o), 126.72 (C-d), 124.45 (C-5″), 123.11 (C-5, 5’), 122.71 (C-6, 6’, 6″), 117.18 (C-3″), 115.86 (C-8, 8’), 115.52 (C-8″), 114.43 (C-m), 114.84 (C-4a″), 60.87 (C-b), 34.60 (b; C-a), 29.55 ppm (C-1a″). C-2, 2’, 3, 3’, 4a, 4a’ n/o; 15N NMR (DMSO-d6): δN = 246.7 (N-e), 147.1 (N-1″), 144.7 ppm (N-1, 1’), N-f, g n/o. EI-MS (m/z, %): 664 (M+, 28). Anal. Calcd for C38H28N6O6: C, 68.67; H, 4.25; N, 12.64. Found: C, 68.58; H, 4.19; N, 12.77.
3,3’-((4-((1-(2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-methylene)bis(4-hydroxy-6-methoxyquinolin-2(1H)-one)8e. Colorless powder, (77%), m.p. 269–71 °C; 1H NMR (DMSO-d6): δH = 13.41 (bs, 1H; OH-4/4’), 12.90 (bs, 1H; OH-4’/4), 12.27 (s, 1H; NH-1″), 12.20 (bs, 1H; NH-1/1’), 12.09 (bs, 1H; NH-1’/1), 8.85 (s, 1H; H-d), 7.64 (dd, J = 7.5, 7.5, 1H; H-7″), 7.47 (d, J = 8.0, 1H; H-8″), 7.45 (d, J = 7.8, 1H; H-5″), 7.40 (m, 3H; H-8, 8’, 5/5’), 7.33 (m, 1H; H-5’/5), 7.25 (d, J = 7.3, 2H; H-7, 7’), 7.23 (m, 1H; H-6″), 7.03 (m, 4H; H-o,m), 6.85 (s, 1H; H-3″), 6.15 (s, 1H; H-a), 5.26 (s, 2H; H-b), 3.82 ppm (s, 6H; H-6b, 6b’), 13C NMR(DMSO-d6): δC = 164.13 (b; C-2,2’), 161.86 (b; C-4,4’), 160.99 (C-2″), 156.09 (C-p), 154.82 (C-6,6’), 143.64 (C-c), 143.42 (C-4″), 139.41 (C-8a″), 131.85 (C-7″), 131.51 (b; C-8a, 8a’), 130.08 (b; C-i), 127.40 (C-o), 126.50 (C-d), 123.98 (C-5″), 122.58 (C-6″), 120.83 (C-7,7’), 117.69 (C-8,8’), 117.47 (C-3″), 116.71 (b; C-4a, 4a’), 115.92 (C-8″), 114.45 (C-m,4a″), 111.77 (b; C-3, 3’), 103.82 (C-5, 5’), 60.87 (C-b), 55.41 (C-6b, 6b’), 34.72 ppm (C-a), 15N NMR (DMSO-d6): δN = 247.5 (N-e), 152.2 (N-1″), 144.1 (N-1’/1), 143.0 ppm (N-1/1‘), N-f, g n/o. EI-MS (m/z, %): 710 (M+, 28). Anal. Calcd for C39H30N6O8: C, 65.91; H, 4.25; N, 11.83. Found: C, 66.09; H, 4.33; N, 11.79.
3,3’-((4-((1-(6-Methyl-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxy-6-methoxyquinolin-2(1H)-one)8f. Colorless powder, (82%), m.p. 264–66 °C; NMR (DMSO-d6) (Table 2). EI-MS (m/z, %): 724 (M+, 22). Anal. Calcd for C40H32N6O8: C, 66.29; H, 4.45; N, 11.60. Found: C, 66.33; H, 4.42; N, 11.57.
3,3’-((4-((1-(6-Methoxy-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxy-6-methoxyquinolin-2(1H)-one)8g. Colorless powder, (83%), m.p. 270–72 °C; 1H NMR (DMSO-d6): δH = 13.41 (bs, 1H; OH-4/4’), 12.91 (bs, 1H; OH-4’/4), 12.21 (bs, 2H; NH-1/1’, 1″), 12.11 (bs, 1H; NH-1’/1), 8.89 (s, 1H; H-d), 7.42 (m, 4H; H-5/5’, 8, 8’, 8″), 7.33 (m, 2H; H-5’/5, 7″), 7.26 (m, 2H; H-7, 7’), 7.03 (m, 4H; H-o,m), 6.92 (s, 1H; H-5″), 6.85 (s, 1H; H-3″), 6.15 (bs, 1H; H-a), 5.27 (s, 2H; H-b), 3.82 (s, 6H; H-6b, 6b’), 3.67 ppm (s, 3H; H-6b″), 13C NMR (DMSO-d6): δC = 165.84 (b), 164.16 (b; C-2, 2’), 161.83 (b), 160.93 (b; C-4, 4’), 160.52 (C-2″), 156.05 (C-p), 154.81 (C-6, 6’), 154.50 (C-6″), 143.48 (C-c), 143.09 (C-4″), 134.07 (C-8a″), 131.58 (b; C-8a, 8a’), 130.09 (b; C-i), 127.40 (C-o), 126.41 (C-d), 121.04 (C-7″), 120.84 (C-7, 7’), 117.95 (C-3″), 117.44, 116.69 (b; C-8, 8’, 8″), 114.83 (C-4a″), 114.38 (C-m), 111.75 (b; C-3, 3’), 110.23 (b; C-4a, 4a’), 105.50 (C-5″), 103.80 (C-5, 5’), 60.84 (C-b), 55.35, 55.11 (b; C-6b, 6b’, 6b″), 34.71 ppm (b; C-a), 15N NMR (DMSO-d6): δN = 247.3 (N-e), 151.0 (N-1″), 144.4 (N-1’/1), 143.0 ppm (N-1/1‘), N-f, g n/o. EI-MS (m/z, %): 740 (M+, 26). Anal. Calcd for C40H32N6O9: C, 64.86; H, 4.35; N, 11.35. Found: C, 64.80; H, 4.44; N, 11.21.
3,3’-((4-((1-(1-Methly-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxy-6-methoxyquinolin-2(1H)-one)8h. Colorless powder, (69%), m.p. 260–62 °C; 1H NMR (DMSO-d6): δH = 13.41 (bs, 1H; OH-4/4’), 12.89 (bs, 1H; OH-4’/4), 12.19 (b, 1H; NH-1/1’), 12.08 (b, 1H; NH-1’/1), 8.84 (s, 1H; H-d), 7.77 (dd, J = 7.5, 7.5, 1H; H-7″), 7.71 (d, J = 8.3, 1H; H-8″), 7.41 (m, 4H; H-5″,8,8’,5/5’), 7.33 (m, 2H; H-6″,5’/5), 7.26 (bd, J = 6.8, 2H; H-7, 7’), 7.02 (m, 4H; H-o,m), 6.94 (s, 1H; H-3″), 6.15 (s, 1H; H-a), 5.27 (s, 2H; H-b), 3.82 (bs, 6H; H-6b, 6b’), 3.72 ppm (s, 3H; H-1a″), 13C NMR (DMSO-d6): δC = 164.14 (C-2,2’), 162.29 (C-4, 4’), 160.29 (C-2″), 156.09 (C-p), 154.78 (C-6, 6’), 143.42 (C-c), 142.62 (C-4″), 140.10 (C-8a″), 132.26 (C-7″), 131.42 (C-8a, 8a’), 130.09 (C-i), 127.39 (C-o), 126.70 (C-d), 124.44 (C-5″), 122.71 (C-6″), 120.83 (C-7, 7’), 117.47 (C-8, 8’), 117.18 (C-3″, 4a, 4a’), 115.52 (C-8″), 114.41 (C-m, 4a″), 103.82 (C-5, 5’), 60.89 (C-b), 55.41 (C-6b, 6b’), 34.64 (C-a), 29.56 ppm (C-1a″). C-3, 3’ n/o, 15N NMR (DMSO-d6): δN = 246.5 (N-e), 147.1 (N-1″), 144.2 ppm (N-1, 1’), N-f, g n/o. EI-MS (m/z, %): 724 (M+, 37). Anal. Calcd for C40H32N6O8: C, 66.29; H, 4.45; N, 11.60. Found: C, 66.22; H, 4.33; N, 11.75.
3,3’-((4-((1-(2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-methylene)bis(4-hydroxy-1-methylquinolin-2(1H)-one)8i. Colorless powder, (72%), m.p. 265–67 °C; 1H NMR (DMSO-d6): δH = 12.87 (bs, 1H; OH-4/4’), 12.54 (bs, 1H; OH-4’/4), 12.27 (bs, 1H; NH-1″), 8.85 (s, 1H; H-d), 8.07 (bs, 2H; H-5, 5’), 7.71 (m, 2H; H-7, 7’), 7.64 (m, 3H; H-8, 8’, 7″), 7.46 (m, 2H; H-5″, 8″), 7.39 (b, 2H; H-6, 6’), 7.23 (t, J = 7.6, 1H; H-6″), 7.01 (m, 4H; H-o,m), 6.84 (s, 1H; H-3″), 6.25 (bs, 1H; H-a), 5.25 (s, 2H; H-b), 3.73 ppm (bs, 6H; H-1a, 1a’), 13C NMR (DMSO-d6): δC = 165.77 (b), 164.20 (b; C-2, 2’), 160.97 (C-4, 4’, 2″), 156.11 (C-p), 143.62 (C-4″), 143.40 (C-c), 139.41 (C-8a″), 138.14 (C-8a, 8a’), 131.84 (C-7, 7’, 7″), 131.58 (C-i), 127.41 (C-o), 126.51 (C-d), 123.97 (C-5″), 123.61 (C-5, 5’), 122.74 (C-6, 6’), 122.56 (C-6″), 117.69 (C-3″), 116.94 (b; C-4a, 4a’), 115.90 (C-8″), 115.34 (C-8, 8’), 114.44 (C-m), 110.92 (b; C-3, 3’), 60.84 (C-b), 36.00 (C-a), 30.18 ppm (C-1a, 1a’), 15N NMR (DMSO-d6): δN = 247.5 (N-e), 152.2 ppm (N-1″), N-1, 1’, f, g n/o. EI-MS (m/z, %): 678 (M+, 35). Anal. Calcd for C39H30N6O6: C, 69.02; H, 4.46; N, 12.38. Found: C, 68.99; H, 4.35; N, 12.55.
3,3’-((4-((1-(6-Methyl-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxy-1-methylquinolin-2(1H)-one)8j. Colorless powder, (77%), m.p. 222–24 °C; 1H NMR (DMSO-d6): δH = 12.92 (bs, 1H; OH-4/4’), 12.50 (bs, 1H; OH-4’/4), 12.22 (bs, 1H; NH-1″), 8.83 (s, 1H; H-d), 8.03 (b, 2H; H-5, 5’), 7.65 (m, 4H; H-7, 7’, 8, 8’), 7.42 (bd, J = 8.6 Hz, 1H; H-7″), 7.35 (d, J = 8.2 Hz, 3H; H-6, 6’, 8″), 7.20 (s, 1H; H-5″), 6.99 (m, 4H; H-o, m), 6.80 (s, 1H; H-3″), 6.23 (bs, 1H; H-a), 5.25 (s, 2H; H-b), 3.71 (bs, 6H; H-1a, 1a’), 2.25 ppm (s, 3H; H-6a″), 13C NMR (DMSO-d6): δC = 165.58 (b), 164.36 (b; C-2, 2’, 2″), 160.84 (C-4, 4’, 4″), 158.06 (C-p), 143.39 (C-c), 138.08 (C-8a, 8a’), 137.50 (C-8a″), 133.08 (C-7″), 131.68, 131.47 (C-7, 7’), 129.91 (b; C-i), 127.39 (C-o, 6″), 126.48 (C-d), 123.57 (C-5, 5’), 123.12 (C-5″), 122.64 (C-6, 6’), 117.67 (C-3″), 116.90 (b; C-4a, 4a’), 115.87 (C-8″), 115.20 (C-8, 8’), 114.36, 114.32 (C-m, 4a″), 110.49 (b), 109.92 (b; C-3, 3’), 60.81 (C-b), 36.00 (C-a), 30.07 (b; C-1a, 1a’), 20.47 ppm (C-6a″), 15N NMR (DMSO-d6): δN = 247.9 (N-e), 151.9 ppm (N-1″), N-1, 1’, f, g n/o. EI-MS (m/z, %): 692 (M+, 10). Anal. Calcd for C40H32N6O6: C, 69.35; H, 4.66; N, 12.13. Found: C, 69.44; H, 4.55; N, 12.22.
3,3’-((4-((1-(6-Methoxy-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-tri-azol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxy-1-methylquinolin-2(1H)-one))8k. Colorless powder, (71%), m.p. 235–37 °C; 1H NMR (DMSO-d6): δH = 12.84 (bs, 1H; OH-4/4’) 12.55 (bs, 1H; OH-4’/4), 12.44 (bs, 1H; NH-1″), 8.86 (s, 1H; H-d), 8.02 (bs, 2H; H-5, 5’), 8.87(m, 4H; H-7, 7’, 8, 8’), 7.85 (d, 1H; H-7″), 7.39 (d, 3H; H-6, 6’, 8″), 7.31 (s, 1H; H-5″), 6.92 (m, 4H; H-o, m), 6.65 (s, 1H; H-3″), 6.23 (bs, 1H; H-a), 5.42 (s, 2H; H-b), 3.09 (bs, 6H; H-1a, 1a’), 2.49 ppm (s, 3H; H-6a″), 13C NMR (DMSO-d6): δC = 160.27 (C-4, 4’, 4″), 156.12 (C-p), 143.52 (C-c), 138.08 (C-8a, 8a’), 132.20 (C-8a″), 131.51 (C-7″), 130.43, 130.07 (C-7, 7’), 127.44 (b; C-i), 126.73 (C-o, 6″), 124.45 (C-5, 5’), 123.60 (C-5″), 122.68 (C-6, 6’), 117.11 (C-3″), 117.51 (b; C-4a, 4a’), 115.42 (C-8″), 115.23 (C-8, 8’), 114.36 (C-m), 60.81 (C-b), 36.05 (C-a), 30.09 ppm (b; C-1a, 1a’). EI-MS (m/z, %): 708 (M+, 21). Anal. Calcd for C40H32N6O7: C, 67.79; H, 4.55; N, 11.86. Found: C, 67.88; H, 4.66; N, 12.01.
3,3’-((4-((1-(1-Methyl-2-oxo-1,2-dihydroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)methylene)bis(4-hydroxy-1-methylquinolin-2(1H)-one)8l. Colorless powder, (66%), m.p. 220–22 °C; 1H NMR (DMSO-d6): δH = 12.15 (bs, 2H; OH-4/4’, OH-4’/4), 12.21 (bs, 1H; NH-1″), 8.86 (s, 1H; H-d), 8.02 (bs, 2H; H-5, 5’), 7.61 (m, 4H; H-7, 7’, 8, 8’), 7.39 (d, 1H; H-7″), 7.31 (d, 3H; H-6, 6’, 8″), 7.28 (s, 1H; H-5″), 6.98 (m, 4H; H-o, m), 6.90 (s, 1H; H-3″), 6.23 (bs, 1H; H-a), 5.24 (s, 2H; H-b), 2.31 (bs, 6H; H-1a, 1a’), 2.29 ppm (s, 3H; H-6a″), 13C NMR (DMSO-d6): δC = 160.65 (C-4, 4’, 4″), 156.16 (C-p), 143.17 (C-c), 138.16 (C-8a, 8a’), 134.16 (C-8a″), 131.58 (C-7″), 131.58, 130.07 (C-7, 7’), 127.51 (b; C-i), 126.50 (C-o, 6″), 123.68 (C-5, 5’), 122.75 (C-5″), 121.06 (C-6, 6’), 117.98 (C-3″), 117.51 (b; C-4a, 4a’), 115.30 (C-8″), 115.20 (C-8, 8’), 114.88, 114.40 (C-m, 4a″), 105.56 (b; C-3, 3’), 60.88 (C-b), 55.40 (C-6b″), 36.10 (C-a), 30.19 ppm (b; C-1a, 1a’. EI-MS (m/z, %): 692 (M+, 18). Anal. Calcd for C40H32N6O6: C, 69.35; H, 4.66; N, 12.13. Found: C, 69.43; H, 4.51; N, 12.21.
4.2. Pharmacological Assays
4.2.1. Cytotoxic Activity Using MTT Assay and Evaluation of IC50
MTT Assay
An MTT assay was performed to investigate the effect of the synthesized compounds on mammary epithelial cells (MCF-10A) at a concentration of 50 µM for the tested compound. The cells were propagated in a medium consisting of Ham’s F-12 medium/Dulbecco’s modified Eagle’s medium (DMEM) (1:1) supplemented with 10% fetal calf serum, 2 mM glutamine, insulin (10 μg/mL), hydrocortisone (500 ng/mL), and epidermal growth factor (20 ng/mL). Trypsin ethylenediamine tetra acetic acid (EDTA) was used to pass the cells every 2–3 days. Ninety-six-well flat-bottomed cell culture plates were used to seed the cells at a density of 104 cells mL−1. The medium was aspirated from all the wells of culture plates after 24 h followed by the addition of synthesized compounds (in 200 μL medium to yield a final concentration of 0.1% (v/v) dimethylsulfoxide) into individual wells of the plates. Four wells were designated to a single compound. The plates were allowed to incubate at 37 °C for 96 h. Afterward, the medium was aspirated, and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) (0.4 mg/mL) in the medium was added to each well and subsequently incubated for 3 h. The medium was aspirated, and 150 μL dimethyl sulfoxide (DMSO) was added to each well. The plates were vortexed, followed by the measurement of absorbance at 540 nm on a microplate reader. The results were presented as inhibition (%) of proliferation compared to controls comprising 0.1% DMSO.
Assay for the Anti-Proliferative Effect
To explore the anti-proliferative potential of compounds, propidium iodide fluorescence assay was performed using different cell lines such as Panc-1 (pancreas cancer cell line), MCF-7 (breast cancer cell line), HT-29 (colon cancer cell line), and A-549 (epithelial cancer cell line), respectively. To calculate the total nuclear DNA, a fluorescent dye (propidium iodide, PI) is used to attach to the DNA, thus offering a quick and precise technique. PI cannot pass through the cell membrane, and its signal intensity can be considered as directly proportional to the quantity of cellular DNA. Cells whose cell membranes are damaged or have changed permeability are counted as dead ones. The assay was performed by seeding the cells of different cell lines at a density of 3000–7500 cells/well (in 200 μL medium) in culture plates followed by incubation for 24 h at 37 °C in humidified 5%CO2/95% air atmospheric conditions. The medium was removed; the compounds were added to the plates at 10 μM concentrations (in 0.1% DMSO) in triplicates, followed by incubation for 48h. DMSO (0.1%) was used as a control. After incubation, the medium was removed, followed by the addition of PI (25 μL, 50 μg/mL in water/medium) to each well of the plates. At −80 °C, the plates were allowed to freeze for 24 h, followed by thawing at 25 °C. A fluorometer (Polar-Star BMG Tech, Beijing, China) was used to record the readings at excitation and emission wavelengths of 530 and 620 nm for each well. The percentage cytotoxicity of compounds was calculated using the following formula:
where ATC = absorbance of treated cells and AC = absorbance of control. Doxorubicin was used as a positive control in the assay.
4.2.2. Caspase 3 Activation Assay
All reagents were allowed to reach room temperature before use. All liquid reagents were gently mixed before use. The number of 8-well strips needed for the assay was determined. These were inserted into the frame(s) for current use. A total of 100 μL of the Standard Diluent Buffer was added to the zero standard wells. Well(s) reserved for chromogen blank were left empty. A total of 100 μL of standards and controls or diluted samples was added to the appropriate microtiter wells. The sample dilution chosen was optimized for each experimental system. The side of the plate was gently tapped on to mix. Wells were covered with a plate cover and incubated for 2 h at room temperature. The solution was thoroughly aspirated or decanted from the wells and the liquid was discarded; wells were washed four times. A total of 100 μL of Caspase 3 (Active) Detection Antibody solution was pipetted into each well except the chromogen blank(s). The side of the plate was gently tapped on to mix. The plate was covered with a plate cover and incubated for 1 h at room temperature. The solution was thoroughly aspirated or decanted from wells and the liquid was discarded; wells were washed four times. A total of 100 μL Anti-Rabbit IgG HRP Working Solution was added to each well except the chromogen blank(s).
The working dilution was prepared as described in Preparing IgG HRP. Wells were covered with the plate cover and incubated for 30 min at room temperature. The solution was thoroughly aspirated or decanted from the wells and the liquid was discarded. Wells were washed four times. A total of 100 μL of Stabilized Chromogen was added to each well. The liquid in the wells began to turn blue. It was incubated for 30 min at room temperature and in the dark. The incubation time for chromogen substrate was determined by the microtiter plate reader used.
Many plate readers have the capacity to record a maximum optical density (O.D.) of 2.0. The O.D. values were monitored, and the substrate reaction was stopped before the O.D. of the positive wells exceeds the limits of the instrument. The O.D. values at 450 nm could only be read after the Stop Solution had been added to each well. A total of 100 μL of Stop Solution was added to each well. The side of the plate was gently tapped to mix. The solution in the wells changed from blue to yellow. The absorbance of each well was read at 450 nm, having blanked the plate reader against a chromogen blank composed of 100 μL each of Stabilized Chromogen and Stop Solution. The plate was read within 2 h after adding the Stop Solution. A curve fitting software was used to generate the standard curve. A four-parameter algorithm provided the best standard curve fit. The concentrations for unknown samples and controls from the standard curve were read. Value(s) obtained for sample(s) by the appropriate dilution factor were multiplied to correct for the dilution in step 3. Samples producing signals greater than that of the highest standard were diluted in Standard Diluent Buffer and reanalyzed.
4.2.3. Caspase 8 Activation Assay
Cells were obtained from American Type Culture Collection; cells were grown in RPMI 1640 containing 10% fetal bovine serum at 37 °C, stimulated with the compounds to be tested for caspase8, and lysed with Cell Extraction Buffer. This lysate was diluted in Standard Diluent Buffer over the range of the assay and measured for human active Caspase 8 content. (cells were plated in a density of 1.2–1.8 × 10,000 cells/well in a volume of 100 µL complete growth medium + 100 µL of the tested compound per well in a 96-well plate for 24 h before the enzyme assay for Tubulin). The absorbance of each microwell was read on a spectro-photometer at 450 nm. A standard curve was prepared from seven human Caspase 8 standard dilutions, and human Caspase 8 concentration was determined.
4.2.4. Bax Activation Assay
All reagents, except the human Bax-α Standard, were brought to room temperature for at least 30 min before opening. The human Bax-α Standard solution was not be left at room temperature for more than 10 min. All standards, controls, and samples were run in duplicate. The Assay Layout Sheet was used to determine the number of wells to be used, and any remaining wells were put with the desiccant back into the pouch, and the ziplock was sealed. Unused wells were stored at 4 °C. A total of 100 μL of Assay Buffer was pipetted into the S0 (0 pg/mL standard) wells. A total of 100 μL of Standards #1 through #6 was pipetted into the appropriate wells. A total of 100 μL of the samples was pipetted into the appropriate wells. The plate was gently tapped to mix the contents. The plate was sealed and incubated at room temperature on a plate shaker for 1 h at ~500 rpm. The contents of the wells were emptied and washed by adding 400 μL of wash solution to every well. The wash was repeated four more times for a total of five washes. After the final wash, the wells were emptied or aspirated and firmly the plate on a lint-free paper towel was tapped to remove any remaining wash buffer. A total of 100 μL of yellow antibody was pipetted into each well, except the Blank. The plate was sealed and incubated at room temperature on a plate shaker for 1 h at ~500 rpm. The contents of the wells were emptied and washed by adding 400 μL of wash solution to every well. The wash was repeated four more times for a total of five washes. After the final wash, the wells were emptied or aspirated, and the plate was firmly tapped on a lint-free paper towel to remove any remaining wash buffer. A total of 100 μL of blue Conjugate was added to each well, except the Blank. The plate was sealed and incubated at room temperature on a plate shaker for 30 min at ~500 rpm. The contents of the wells were emptied and washed by adding 400 μL of wash solution to every well. The wash was repeated four more times for a total of five washes. After the final wash, the wells were emptied or aspirated and the plate was firmly tapped on a lint-free paper towel to remove any remaining wash buffer. A total of 100 μL of Substrate Solution was pipetted into each well. It was incubated for 30 min at room temperature on a plate shaker at ~500 rpm. A total of 100 μL Stop Solution was pipetted into to each well. The plate reader was blanked against the Blank wells, and the optical density was read at 450 nm. The average net optical density (OD) bound for each standard and sample was calculated by subtracting the average Blank OD from the average OD for each standard and sample. Using linear graph paper, the Average Net OD for each standard versus Bax concentration in each standard were plotted. A straight line was approximated through the points. The concentration of Bax in the unknowns could be determined by interpolation.
4.2.5. Bcl-2 Inhibition Assay
All reagents were thoroughly mixed without foaming before use. The microwells were washed twice with approximately 300 μL Wash Buffer per well with the thorough aspiration of microwell contents between washes. Care was taken not to scratch the surface of the microwells. After the last wash, the wells were emptied and microwell strips were tapped on an absorbent pad or paper towel to remove excess Wash Buffer. The microwell strips were used immediately after washing or placed upside down on a wet absorbent paper for not longer than 15 min. Wells were not allowed to dry. A total of 100 μL of Sample Diluent was added in duplicate to all standard wells and the blank wells. Standard (1:2 dilution) in duplicate was prepared ranging from 32 to 0.5 ng/mL. A total of 100 μL of Sample Diluent, in duplicate, was added to the blank wells. A total of 80 μL of Sample Diluent, in duplicate, was added to the sample wells. A total of 20 μL of each sample, in duplicate, was added to the designated wells. A total of 50 μL of diluted biotin was added, conjugated to all wells, including the blank wells. It was covered with a plate cover and incubated at room temperature on a microplate shaker at 100 rpm for 2 h. The plate cover was removed and the wells were emptied. Microwell strips were washed times as described in Step 2. A total of 100 μL of diluted Streptavidin-HRP was added to all wells, including the blank wells. It was covered with a plate cover and incubated at room temperature on a microplate shaker at 100 rpm for 1 h. The plate cover was removed and the wells were emptied. Microwell strips were washed three times, as described in Step 2. A total of 100 μL of mixed TMB Substrate Solution was pipetted into all wells, including the blanks. The microwell strips were incubated at room temperature (18 °C to 25 °C) for about 15 min, on a rotator set at 100 rpm. Direct exposure to intense light was avoided. The point at which the substrate reaction is stopped was determined by the ELISA reader. Many ELISA readers record absorbance only up to 2.0 O.D. Therefore, the color development within individual microwells was watched by the person running the assay, and the substrate reaction was stopped before positive wells were no longer properly detectable. The enzyme reaction was stopped by quickly pipetting 100 μL of Stop Solution into each well, including the blank wells. It was important that the Stop Solution was spread quickly and uniformly throughout the microwells to inactivate the enzyme completely. Results were read immediately after the Stop Solution was added or within one hour if the microwell strips were stored at 2–8 °C in the dark. The absorbance of each microwell was read on a spectrophotometer using 450 nm as the primary wavelength.
4.2.6. Cytochrome C Assay
Cells were obtained from American Type Culture Collection, grown in RPMI 1640 containing 10% fetal bovine serum at 37 °C, stimulated with the compounds to be tested for cytochrome C, and lysed with Cell Extraction Buffer. This lysate was diluted in Standard Diluent Buffer over the range of the assay and measured for cytochrome C content. (cells were plated in cells/well in a volume of 100 µL complete growth medium + 100 µL of the tested compound + 50 µL of 1× biotin-conjugated antibody + 100 µL of 1× streptavidin-HRP + 100 µL TMB substrate soln of per well in a 96-well plate for 24 h before assay).
4.2.7. Cell Apoptosis Assay
Apoptosis was determined by flow cytometry based on the annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining kit (BD Pharmingen, San Diego, USA). Apoptotic cells were defined as Annexin-V-positive. Cells were grown to approximately ~70% confluency and exposed to the tested compounds (8 μmol/L) for 24 h. Treated cells were trypsinized, washed twice with PBS, and transferred into microcentrifuge tubes for centrifugation at 1000 rpm for 5 min at room temperature, and then resuspended in binding buffer; 5 μL of FITC and PI were added to per Eppendorf tube, and cells were vortexed and incubated for 15 min at room temperature in the dark. Subsequently, cells were analyzed by flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA).
Acknowledgments
The authors acknowledge support by the KIT-Publication Fund of the Karlsruhe Institute of Technology. The NMR spectrometer at Florida Institute of Technology was purchased with assistance from the U.S. National Science Foundation (CHE 03-42251).
Supplementary Materials
The following are available online, Spectral data of all newly synthesized compounds.
Author Contributions
E.M.E.-S. conceptualization, methodology and writing; B.G.M.Y. methodology, conceptualization, writing, revision and submission; A.B.B. NMR, writing and revision; M.A.I.E. revision; H.A.M.G. pharmacology and revision; S.B. revision; A.M.S. writing and revision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The authors declare that they have no known competing interests.
Sample Availability
Samples of the compounds are not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Batista V.F., Pinto D.C., Silva A.M. Synthesis of quinolines: A green perspective. ACS Sustain. Chem. Eng. 2016;4:4064. doi: 10.1021/acssuschemeng.6b01010. [DOI] [Google Scholar]
- 2.Dhiman P., Arora N., Thanikachalam P.V., Monga V. Recent advances in the synthetic and medicinal perspective of quinolones. A review. Bioorg. Chem. 2019;92:103291. doi: 10.1016/j.bioorg.2019.103291. [DOI] [PubMed] [Google Scholar]
- 3.Aly A.A., Ramadan M., Abuo-Rahma G.E.-D.A., Elshaier Y.A., Elbastawesy M.A., Brown A.B., Bräse S. Advances in Heterocyclic Chemistry. Academic Press; Cambridge, MA, USA: 2020. Quinolones as prospective drugs: Their syntheses and biological applications. [Google Scholar]
- 4.Bush N.G., Diez-Santos I., Abbott L.R., Maxwell A. Quinolones: Mechanism, Lethality and Their Contributions to Antibiotic Resistance. Molecules. 2020;25:5662. doi: 10.3390/molecules25235662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao F., Zhang X., Wang T., Xiao J. Quinolone hybrids and their anti-cancer activities: An overview. Eur. J. Med. Chem. 2019;165:59. doi: 10.1016/j.ejmech.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Ganguly A., Banerjee K., Chakraborty P., Das S., Sarkar A., Hazra A., Banerjee M., Maity A., Chatterjee M., Mondal N.B. Overcoming multidrug resistance (MDR) in cancer in vitro and in vivo by a quinoline derivative. Biomed. Pharmacother. 2011;65:387. doi: 10.1016/j.biopha.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 7.Elbastawesy M.A., Aly A.A., Ramadan M., Elshaier Y.A., Youssif B.G., Brown A.B., Abuo-Rahma G.E.-D.A. Novel Pyrazoloquinolin-2-ones: Design, synthesis, docking studies, and biological evaluation as anti-proliferative EGFR-TK inhibitors. Bioorg. Chem. 2019;90:103045. doi: 10.1016/j.bioorg.2019.103045. [DOI] [PubMed] [Google Scholar]
- 8.Elbastawesy M.A., Ramadan M., El-Shaier Y.A., Aly A.A., Abuo-Rahma G.E.-D.A. Arylidenes of Quinolin-2-one scaffold as Erlotinib analogues with activities against leukemia through inhibition of EGFR TK/STAT-3 pathways. Bioorg. Chem. 2020;96:103628. doi: 10.1016/j.bioorg.2020.103628. [DOI] [PubMed] [Google Scholar]
- 9.Ramadan M., Abd El-Aziz M., Elshaier Y.A., Youssif B.G., Brown A.B., Fathy H.M., Aly A.A. Design and synthesis of new pyranoquinolinone heteroannulated to triazolopyrimidine of potential apoptotic anti-proliferative activity. Bioorg. Chem. 2020;105:104392. doi: 10.1016/j.bioorg.2020.104392. [DOI] [PubMed] [Google Scholar]
- 10.Sathish Kumar S., Kavitha P.H. Synthesis and biological applications of triazole derivatives: A review. Mini. Rev. Org. Chem. 2013;10:40. doi: 10.2174/1570193X11310010004. [DOI] [Google Scholar]
- 11.Huang M., Deng Z., Tian J., Liu T. Synthesis and biological evaluation of salinomycin triazole analogues as anti-cancer agents. Eur. J. Med. Chem. 2017;127:900. doi: 10.1016/j.ejmech.2016.10.067. [DOI] [PubMed] [Google Scholar]
- 12.Dheer D., Singh V., Shankar R. Medicinal attributes of 1,2,3-triazoles: Current developments. Bioorg. Chem. 2017;71:30. doi: 10.1016/j.bioorg.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Xu J.H., Fan Y.L., Zhou J. Quinolone-Triazole Hybrids and their Biological Activities. Heterocycl. Chem. 2018;55:1854. doi: 10.1002/jhet.3234. [DOI] [Google Scholar]
- 14.Zhang J., Wang S., Ba Y., Xu Z. 1,2,4-Triazole-quinoline/quinolone hybrids as potential antibacterial agents. Eur. J. Med. Chem. 2019;174:1–8. doi: 10.1016/j.ejmech.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Nandivada H., Jiang X., Lahann J. Click chemistry: Versatility and control in the hands of materials scientists. Adv. Mater. 2007;19:2197. doi: 10.1002/adma.200602739. [DOI] [Google Scholar]
- 16.Gehringer M., Laufer S.A. Click chemistry: Novel applications in cell biology and drug discovery. Angew. Chem. Int. Ed. 2017;56:15504. doi: 10.1002/anie.201710195. [DOI] [PubMed] [Google Scholar]
- 17.Aly A.A., El-Sheref E.M., Bakheet M.E., Mourad M.A., Bräse S., Ibrahim M.A., Nieger M., Garvalov B.K., Dalby K.N., Kaoud T.S. Design, synthesis and biological evaluation of fused naphthofuro[3,2-c]quinoline-6,7,12-triones and pyrano[3,2-c]quinoline-6,7,8,13-tetraones derivatives as ERK inhibitors with efficacy in BRAF-mutant melanoma. Bioorg. Chem. 2019;82:290. doi: 10.1016/j.bioorg.2018.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aly A.A., El-Sheref E.M., Bakheet M.E., Mourad M.A., Brown A.B., Bräse S., Nieger M., Ibrahim M.A. Synthesis of novel 1,2-bis-quinolinyl-1,4-naphthoquinones: ERK2 inhibition, cytotoxicity and molecular docking studies. Bioorg. Chem. 2018;81:700. doi: 10.1016/j.bioorg.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Elbastawesy M.A., El-Shaier Y.A., Ramadan M., Brown A.B., Aly A.A., Abuo-Rahma G.E.-D.A. Identification and molecular modeling of new quinolin-2-one thiosemicarbazide scaffold with antimicrobial urease inhibitory activity. Mol. Divers. 2020;25:13. doi: 10.1007/s11030-019-10021-0. [DOI] [PubMed] [Google Scholar]
- 20.Abass M. Chemistry of substituted quinolinones. Part II synthesis of novel 4-pyrazolylquinolinone derivatives. Synth. Commun. 2000;30:2735. doi: 10.1080/00397910008086898. [DOI] [Google Scholar]
- 21.Ismail M.M., Abass M., Hassan M.M. Chemistry of substituted quinolinones. Part VI. Synthesis and nucleophilic reactions of 4-chloro-8-methylquinolin-2(1H)-one and its thione analogue. Molecules. 2000;5:1224. doi: 10.3390/51201224. [DOI] [Google Scholar]
- 22.Aly A.A., El-Sheref E.M., Mourad A.-F.E., Bakheet M.E., Bräse S., Nieger M. One-pot synthesis of 2,3-bis-(4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl) succinates and arylmethylene-bis(3,3’-quinoline-2-ones) Chem. Pap. 2019;73:27. doi: 10.1007/s11696-018-0561-0. [DOI] [Google Scholar]
- 23.Hisham M., Youssif B.G., Osman E.E.A., Hayallah A.M., Abdel-Aziz M. Synthesis and biological evaluation of novel xanthine derivatives as potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019;176:117. doi: 10.1016/j.ejmech.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Al-Wahaibi L.H., Gouda A.M., Abou-Ghadir O.F., Salem O.I., Ali A.T., Farghaly H.S., Abdelrahman M.H., Trembleau L., Abdu-Allah H.H., Youssif B.G. Design and synthesis of novel 2,3-dihydropyrazino[1,2-a]indole-1,4-dione derivatives as anti-proliferative EGFR and BRAFV600E dual inhibitors. Bioorg. Chem. 2020;104:104260. doi: 10.1016/j.bioorg.2020.104260. [DOI] [PubMed] [Google Scholar]
- 25.Youssif B.G., Mohamed A.M., Osman E.E.A., Abou-Ghadir O.F., Elnaggar D.H., Abdelrahman M.H., Treamblu L., Gomaa H.A. 5-Chlorobenzofuran-2-carboxamides: From allosteric CB1 modulators to potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019;177:1–11. doi: 10.1016/j.ejmech.2019.05.040. [DOI] [PubMed] [Google Scholar]
- 26.Abdelazeem A.H., El-Saadi M.T., Said E.G., Youssif B.G., Omar H.A., El-Moghazy S.M. Novel diphenylthiazole derivatives with multi-target mechanism: Synthesis, docking study, anti-cancer and anti-inflammatory activities. Bioorg. Chem. 2017;75:127. doi: 10.1016/j.bioorg.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen T., Li J.X., Thomas B.F., Wiley J.L., Kenakin T.P., Zhang Y. Allosteric modulation: An alternate approach targeting the cannabinoid CB1 receptor. Med. Res. Rev. 2017;37:441. doi: 10.1002/med.21418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen G.M. Caspases: The executioners of apoptosis. Biochem. J. 1997;326:1. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slee E.A., Adrain C., Martin S.J. Executioner caspase-3, -6, and-7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. 2001;276:7320. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 30.Fulda S., Debatin K.-M. Extrinsic versus intrinsic apoptosis pathways in anti-cancer chemotherapy. Oncogene. 2006;25:4798. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 31.Abdelbaset M.S., Abuo-Rahma G.E., Abdelrahman M.H., Ramadan M., Youssif B.G., Bukhari S.N.A., Mohamed M.F., Abdel-Aziz M. Novel pyrrol-2(3H)-ones and pyridazin-3(2H)-ones carrying quinoline scaffold as anti-proliferative tubulin polymerization inhibitors. Bioorg. Chem. 2018;80:151. doi: 10.1016/j.bioorg.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Abou-Zied H.A., Youssif B.G., Mohamed M.F., Hayallah A.M., Abdel-Aziz M. EGFR inhibitors and apoptotic inducers: Design, synthesis, anti-cancer activity and docking studies of novel xanthine derivatives carrying chalcone moiety as hybrid molecules. Bioorg. Chem. 2019;89:102997. doi: 10.1016/j.bioorg.2019.102997. [DOI] [PubMed] [Google Scholar]
- 33.Abdelrahman M.H., Aboraia A.S., Youssif B.G., Elsadek B.M. Design, synthesis and pharmacophoric model building of new 3-alkoxymethyl/3-phenyl indole-2-carboxamides with potential anti-proliferative activity. Chem. Biol. Drug Des. 2017;90:64. doi: 10.1111/cbdd.12928. [DOI] [PubMed] [Google Scholar]
- 34.Abdelbaset M.S., Abdel-Aziz M., Abuo-Rahma G.E.-D.A., Abdelrahman M.H., Ramadan M., Youssif B.G. Novel quinoline derivatives carrying nitrones/oximes nitric oxide donors: Design, synthesis, anti-proliferative and caspase-3 activation activities. Arch. Pharm. 2019;352:1800270. doi: 10.1002/ardp.201800270. [DOI] [PubMed] [Google Scholar]
- 35.Okuno T. 4-(Prop-2-yn-1-yloxy) benzaldehyde. Acta Crystall. Sect. E Rep. Online. 2013;69:o125. doi: 10.1107/S1600536812050866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aizikovich A., Kuznetsov V., Gorohovsky S., Levy A., Meir S., Byk G., Gellerman G. A new application of diphenylphosphorylazide (DPPA) reagent: Convenient transformations of quinolin-4-one, pyridin-4-one and quinazolin-4-one derivatives into the 4-azido and 4-amino counterparts. Tetrahedron Lett. 2004;45:4241. doi: 10.1016/j.tetlet.2004.04.032. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.