Abstract
Telomerase is a ribonucleoprotein that mediates extension of the dG-rich strand of telomeres in most eukaryotes. Like telomerase derived from ciliated protozoa, yeast telomerase is found to possess a tightly associated endonuclease activity that copurifies with the polymerization activity over different affinity-chromatographic steps. As is the case for ciliate telomerase, primers containing sequences that are not complementary to the RNA template can be efficiently cleaved by the yeast enzyme. More interestingly, we found that for the yeast enzyme, cleavage site selection is not stringent, since blocking cleavage at one site by the introduction of a nonhydrolyzable linkage can lead to the utilization of other sites. In addition, the reverse transcriptase activity of yeast telomerase can extend either the 5′- or 3′-end fragment following cleavage. Two general models that are consistent with the biochemical properties of the enzyme are presented: one model postulates two distinct active sites for the nuclease and reverse transcriptase, and the other invokes a multimeric enzyme with each protomer containing a single active site capable of mediating both cleavage and extension.
Telomerase is a ribonucleoprotein that is responsible for maintaining the terminal repeats of telomeres in most organisms (1, 2, 28, 37). It acts as an unusual reverse transcriptase (RT), using a small segment of an integral RNA component as template for the synthesis of the dG-rich strand of telomeres (11, 12). DNA synthesis by telomerase in vitro is primed by oligonucleotides with telomere-like sequences. Depending on the source, telomerase in vitro can act either processively, adding many copies of a repeat without dissociating, or nonprocessively, completing only one telomeric repeat (13, 29, 31).
Telomerase activity has been detected in a wide range of organisms, including protozoa (2), yeasts (4, 17, 18, 20, 35), mice (31), Xenopus laevis (22), and humans (25). Genes encoding the RNA and RT subunit of the enzyme complex have also been cloned for many known telomerases (2, 3, 5, 8, 16, 18, 24, 26, 34). In addition, both biochemical and genetic studies point to the existence of additional protein subunits of telomerase, whose functions remain to be elucidated (7, 9, 15, 19, 27).
A telomerase-associated nuclease has been identified in Tetrahymena thermophila, Euplotes crassus, Saccharomyces cerevisiae, and Schizosaccharomyces pombe (4, 6, 10, 20, 21, 23, 29). In the case of Tetrahymena telomerase, the associated nuclease has been found to remove one or several terminal primer nucleotides prior to polymerization. Enzyme reconstituted in rabbit reticulocyte lysates with p133 (the RT subunit) and telomerase RNA retains cleavage activity, suggesting that the nuclease resides in one of these two components (5). The nuclease from E. crassus has been thoroughly characterized using a coupled cleavage-elongation assay (10, 23), which revealed the following salient features: (i) cleavage proceeds by an endonucleolytic mechanism, (ii) DNA fragments from the 3′ end can be eliminated prior to elongation of the primer by telomerase, (iii) long stretches of preferably nontelomeric sequences can be removed by the nuclease, (iv) cleavage occurs preferentially but not exclusively at the junction of match-mismatch between the primer and the RNA template, (v) the junction of match-mismatch between the primer and the RNA template can be positioned at various locations along the RNA template to effect cleavage, and (vi) primers bearing nontelomeric sequences at the 5′ end are preferentially cleaved. While not as thoroughly studied, the nuclease from other organisms exhibits properties consistent with those displayed by the Tetrahymena and E. crassus enzymes. For example, both primer-template mismatch and the presence of nontelomeric sequences at the 5′ end have been found to stimulate cleavage by the yeast telomerase-associated nuclease (21, 29).
Various functions have been suggested for the telomerase-associated endonuclease. For example, the combined cleavage and elongation activity may be useful in the de novo formation of telomeres during macronuclear development in ciliated protozoa (23). Alternatively, cleavage may serve a proofreading function given that nontelomeric sequences appear preferentially removed (10, 23). In addition, by analogy with DNA-dependent RNA polymerases, cleavages may allow an elongation-incompetent telomerase to re-engage the 3′ end of the primer prior to extension (5).
In this study, we characterized the Saccharomyces cerevisiae telomerase-associated nuclease in greater detail and found that it shares many properties that have been ascribed to the ciliate enzymes. For example, yeast cleavage activity is tightly associated with the polymerization activity. In addition, primers with sequences that are noncomplementary to the RNA template appear to be relatively efficient substrate for cleavage by yeast telomerase. The yeast nuclease also appears to act through an endonucleolytic mechanism. More surprisingly, we found that following cleavage, either one of the fragments generated by the yeast nuclease (the 5′ and the 3′ fragments) can be extended by the polymerization activity of telomerase. This result is not easily rationalized in terms of a monomeric enzyme containing a single nuclease-polymerase active site. Two models that are compatible with all of our biochemical observations are presented in the Discussion.
MATERIALS AND METHODS
Yeast strains, media, buffers, and the preparation of yeast telomerase.
JX-M3 is a haploid yeast strain identical to W303a except that the EST2 gene in the strain was fused at its C terminus to a Myc3 epitope tag using a PCR recombination method (35). JX-MH19 contains an EST2 gene whose C terminus is fused to both a Myc3 epitope tag and a His6 tag. JX-proA contains an EST2 gene with, in addition to the Myc and His tags, two copies of the immunoglobulin G (IgG) binding domain from protein A. The construction of these strains will be described in detail elsewhere.
Buffer TMG-15 contains 15% glycerol, 10 mM Tris-HCl (pH 8.0), 1.2 mM magnesium chloride, 0.1 mM EDTA, 0.1 mM EGTA, and 1.5 mM dithiothreitol (DTT). Buffer TMG-10 is identical to TMG-15 except that glycerol was included at 10%. Buffer TMG-10(500), etc., denotes buffer TMG-10 plus the millimolar concentration of sodium acetate specified by the number in parentheses. The following protease inhibitors were included in all buffers: 1 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 2 μg of pepstatin A per ml, and 1 μg of leupeptin per ml.
Purification of yeast telomerase.
For preparation of whole-cell extracts, the yeast strains DG338 (a gift of D. Garfinkel, National Cancer Institute), W303a, JX-M3, JX-MH19, or JX-proA was grown in YPD medium, lysed in TMG-15(0) buffer, and the lysates were clarified by high-speed centrifugation as previously described (4, 21). To obtain active telomerase, whole-cell extracts were processed over DEAE-agarose columns as previously described (4, 21). For Myc-tag affinity purification, DEAE fractions (10 ml) prepared from the JX-M3 strain were loaded directly onto a 0.5 ml of 9E10 (Myc antibody) column. The column was washed with TMG-10(500) and then TMG-10(500) containing 1 mg of HA.11 (hemagglutinin) peptide per ml at 4°C. Telomerase was then eluted at room temperature with 1.5 ml of TMG-10(500) containing 1 mg of 9E10 (Myc) peptide per ml. The overall recovery of activity was ∼10%, while 0.05% of the load had approximately the same amount of total protein as 50% of the purified fraction, based on the staining of a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Because the estimate of the protein concentration is not precise, we did not determine the fold enrichment for telomerase in this immunoaffinity procedure. For metal affinity purification, DEAE fractions (2 ml) prepared from JX-MH19 strain were loaded directly onto a 0.2-ml Ni-nitrilo triacetic acid column (Qiagen). The column was washed successively with TMG-10(500) and TMG-10(500) containing 5 mM imidazole. Active telomerase was then eluted with TMG-10(500) containing 200 and 500 mM imidazole. The majority of telomerase was present in the 200 mM elution. The overall recovery of activity was ∼50%, while the purified fraction contained a fraction (ca. 1/20) of the starting protein based on the Bio-Rad Protein Assay (Bio-Rad Laboratories). Thus, we estimate that telomerase is enriched by about 10-fold by the metal affinity procedure. For the protein A-tag-based purification, a DEAE fraction from JX-proA (100 μl) was directly incubated with 5 μl of IgG-Sepharose beads at 4°C with gentle rotation for 2 h. The beads were washed multiple times with TMG-10(600) and then assayed for telomerase activity along with the DEAE fraction and the supernatant. More than 95% of the starting protein remained in the supernatant, while the beads contained ∼50% of the starting activity. Thus, telomerase was purified more than 10-fold by this IgG affinity procedure.
For multistep purification, the protein A-tagged enzyme was successively fractionated over DEAE, phenyl, heparin, and IgG columns. DEAE chromatography was carried out as previously described (4, 21). Active fractions from the DEAE column were pooled and loaded directly onto a phenyl Sepharose (Pharmacia) column. The column was washed successively with two column volumes each of TMG-10(500) and TMG-10(100), and the activity was eluted with two column volumes of TMG-10(0) plus 1% Triton X-100. Active fractions were pooled and loaded onto an Affi-Gel Heparin (Bio-Rad) column. The column was washed with two column volumes of TMG-10(150), and the activity was eluted with two column volumes of TMG-10(700). Active fractions were then processed over IgG-Sepharose resin as described earlier. The specific activity and the degree of purification were calculated from primer extension activity assays and protein assays with two exceptions. First, because the activity was undetectable in whole-cell extracts, the fold purification for the DEAE column fraction was based on the degree of Est2p enrichment (as determined by Western blotting). Second, because it is not possible to elute telomerase from IgG-Sepharose, we estimated the amount of total protein bound to the beads to be the difference in protein concentration of the heparin fraction before and after binding to IgG beads. For Western analysis of protein A-tagged Est2p, proteins from extracts or DEAE fractions were separated in by SDS–8% PAGE and transferred onto nitrocellulose membrane. Primary anti-protein A antibody (Sigma) and secondary antibody were used at 1:1,000,000 and 1:5,000 dilutions, respectively. Immunoreactive species were visualized using the ProtoBlot system (Promega).
Primer preparation.
DNA primers were purchased from GeneLink (Thornwood, N.Y.) and gel purified prior to use in polymerization assays. Crude primers were dissolved in distilled H2O at 1 mg/ml and fractionated on a 16% denaturing polyacrylamide gel. Full-length DNA fragments were visualized by ethidium bromide staining and UV transillumination, isolated as small gel slices, and eluted overnight at 37°C with 400 μl of extraction buffer containing 0.1% SDS, 0.3 M sodium acetate, 10 mM magnesium acetate, and 1 mM EDTA. The DNA was recovered from the extraction buffer by ethanol precipitation in the presence of 5 μg of glycogen and resuspended in a suitable volume of water. The concentration of the purified DNA primer was again quantified by PAGE and ethidium bromide staining.
Primers bearing methylphosphonate linkages were also purchased from GeneLink and then gel purified prior to use. Resistance to nuclease was confirmed by using Escherichia coli Exonuclease III (New England Biolabs). Primers terminating in dideoxynucleotides were made by treating 500 pmol of DNA primer with 34 U of terminal deoxynucleotide transferase (USB; 17 U/μl) and 833 μM dideoxynucleotides (ddTTP or ddGTP) in 30 μl of total volume containing 1× buffer (USB) at 37°C for 3 h. After phenol-chloroform extraction, full-length DNA was recovered by ethanol precipitation and gel purified as described above.
Coupled cleavage-extension assay.
A standard cleavage-extension assay (30 μl) contained 50 mM Tris-HCl (pH 8.0), 1 mM spermidine, 1 mM DTT, 1 mM MgCl2, 130 μM dTTP, 1 to 2 μl of [α-32P]dGTP (3,000 Ci/mmol, 10 μCi/μl), and various amounts of DNA primers and telomerase fractions. Reactions were started by the addition of telomerase fraction to a premixed cocktail consisting of all the other components. Reactions were continued for 1 h at 30°C, and labeled products were processed as described previously (21).
RESULTS
Observation of a nuclease in yeast telomerase fractions.
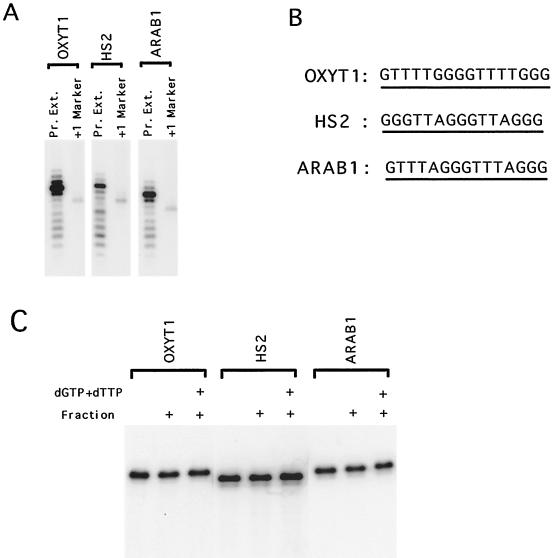
For purification and characterization of yeast telomerase, we utilized a direct primer extension assay (4, 21). Under standard reaction conditions, the yeast enzyme is nonprocessive and gives rise predominantly to a “primer + 3” product (21; Fig. 1A). Interestingly, the use of certain primers in extension assays, especially those consisting of repeats from other organisms, often yielded products that are shorter than the input primer. For example, when Oxytricha, human, and Arabidopsis repeats are utilized as primers, as much as 20% of the labeled products in the polymerization assays were shorter than the starting primer (Fig. 1A and B). To rule out the possibility that there is excess nonspecific nuclease in the partially purified yeast telomerase fractions, we assembled mock telomerase reactions using the fraction, unlabeled primer, and unlabeled nucleotide triphosphates. Also included in each reaction was a small amount of end-labeled tracer oligonucleotide used to monitor the fate of the input primer. As shown in Fig. 1C, the vast majority of the starting primers are neither shortened nor extended, even in the presence dGTP and dTTP. This result is consistent with the large molar excess of primer over active telomerase as determined by the polymerization assay. No discrete bands can be visualized in the region of the gel presumed to contain the nuclease-derived products, and quantification indicates that this region possesses <2% of the radioactivity present in the full-length bands. These results are quite consistent with earlier observations on the existence of a specific nuclease in yeast telomerase fractions (4, 21, 29).
FIG. 1.
Yeast telomerase partially purified by DEAE chromatography contains a nuclease activity. (A) Polymerization assays were performed using 5 μM concentrations of various primers and 9 μg of DEAE fractions (Pr.Ext.). Primers labeled by terminal transferase and cordycepin were run alongside the reaction products as size standards (+1 Marker). (B) The sequences of the primers used in panel A. (C) Concentrations (5 μM) of various primers (containing a small amount of labeled tracer) were incubated alone, with telomerase fractions, or with telomerase fraction and deoxynucleotide triphosphates. The DNA was recovered and analyzed by denaturing gel electrophoresis as in standard primer extension assays.
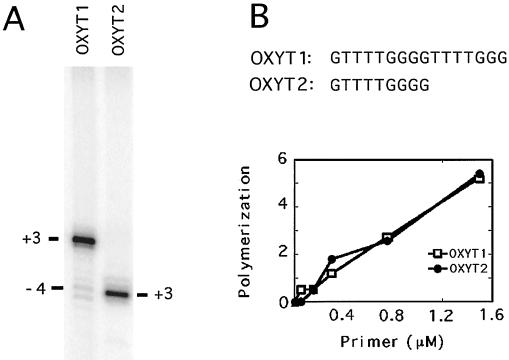
Since <2% of the input primers were cleaved yet as much as 20% of the extension products were derived from cleaved DNA, telomerase appears to preferentially extend cleaved DNA (by at least 10-fold). This preferential extension can be explained by either a coupling between the telomerase and the nuclease or by short primers being intrinsically superior substrates for telomerase. To address the latter possibility, we assessed the activity of two primers of different lengths (OXYT1 and OXYT2) bearing the Oxytricha telomeric repeats at increasing primer concentrations. The shorter primer (OXYT2) was designed to mimic the size of the cleaved but not yet extended DNA derived from the longer primer (OXYT1). As shown in Fig. 2A, the cleavage-derived products of OXYT1 are indeed similar in size to the direct extension products of OXYT2. Furthermore, at the same molar concentration, OXYT1 and OXYT2 supported a nearly identical amount of DNA synthesis, suggesting that short DNAs are not intrinsically better substrates for yeast telomerase (Fig. 2B). Therefore, a physical or functional coupling between the nuclease and telomerase appears likely.
FIG. 2.
Long and short heterologous primers are utilized by yeast telomerase at comparable efficiencies. (A) Polymerization assays were performed using 1.5 μM concentrations of either OXYT1 or OXYT2 as the DNA primer and 2.3 μg of DEAE fractions. The locations of the “+3” and “−4” products for OXYT1 and that of the “+3” product for OXYT2 are indicated by horizontal bars. (B) Polymerization assays were carried out using increasing concentrations of either OXYT1 or OXYT2, and the signals derived from direct extension of the primers were quantified and plotted.
Affinity-purified telomerase exhibits the same nuclease activity.
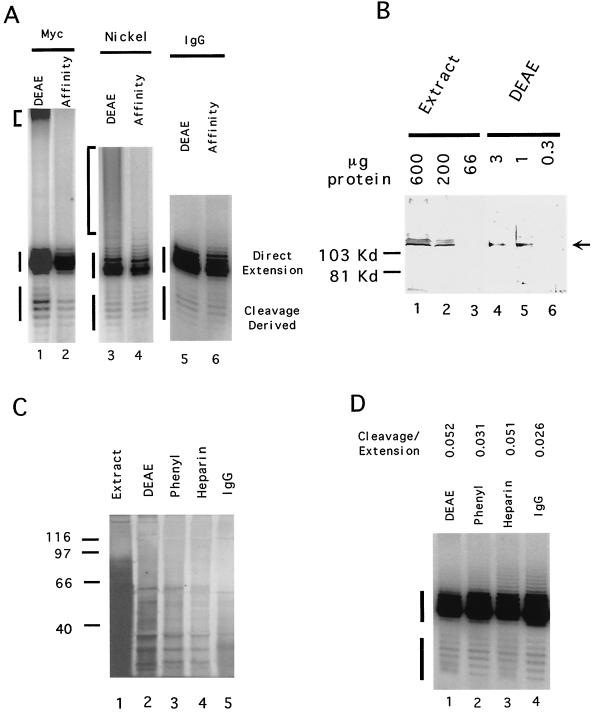
To determine if the coupling observed between the nuclease and telomerase in the DEAE fraction can be explained by physical association, we further purified yeast telomerase using three different affinity tags (a Myc3 tag, a His6 tag, and a protein A tag) and the appropriate chromatographic resins. All three tags were fused to the C terminus of Est2p and had no effect on telomere maintenance or telomerase activity (J. Xia and N. F. Lue, unpublished data). The detailed purification procedures and the estimates for the degrees of enrichment are presented in Materials and Methods. In each case, the affinity-tagged telomerase was first purified over a DEAE column (4, 21). Active enzymes were then adsorbed onto the appropriate affinity columns and either eluted with specific competitors before analysis (for Myc- and His-tagged enzymes) or directly assayed on the resin (for protein A-tagged enzymes). As shown in Fig. 3A, each of the purification procedures resulted in telomerase that was still capable of catalyzing the cleavage-extension reaction on the OXYT1 primer. Furthermore, in each case the fraction of the products that were shorter than the starting primer was similar for both the DEAE and the affinity-purified enzyme (compare lanes 1 and 2, 3 and 4, and 5 and 6). A contaminating activity (or activities) capable of generating labeled high-molecular-weight products is evident in DEAE fractions derived from the Myc- and His-tagged strains (lanes 1 and 3, indicated by brackets to the left of the panels). This activity (or activities) was successfully removed by the affinity procedures.
FIG. 3.
Affinity-purified yeast telomerase exhibits a similar cleavage activity as that found in DEAE fractions. (A) Polymerization assays were carried out using OXYT1 (1 μg) as the DNA primer and DEAE fraction (DEAE) or affinity-purified telomerase (Affinity) as the source of telomerase. The affinity resin utilized for each purification is indicated at the top. The amount of protein used for each reaction is as follows: lane 1, 3 μg; lane 2, 45 ng; lane 3, 3 μg of protein; lane 4, 0.4 μg of protein; lane 5, 3 μg of protein; lane 4, ∼0.15 μg of protein. “Direct extension” or “cleavage-derived” products are marked by vertical lines to the left of the panels. Contaminating activity or activities present in the DEAE fraction and responsible for the labeling of high-molecular-weight products (indicated by brackets to the left of the panels) can be removed by the Myc affinity or the nickel affinity chromatographic procedures. (B) Immunoblotting was used to estimate the degree of Est2p enrichment over the DEAE column. Protein A-tagged Est2p from extracts or DEAE fractions was detected using anti-protein A antibodies. The amount of protein loaded is indicated at the top. The location of the protein A-tagged Est2p is indicated by an arrow. (C) Protein compositions of fractions from successive column steps were analyzed by SDS-PAGE and silver staining. The identities and the amounts of the fractions utilized were as follows: lane 1, extract, 30 μg; lane 2, DEAE, 10 μg; lane 3, phenyl, 3 μg; lane 3, heparin, 1.2 μg; lane 4, IgG, ∼0.12 μg. (D) Polymerization assays were carried out using OXYT1 (2 μg) as the DNA primer and fractions from successive column steps. The identities of the fractions used in each reaction were as follows: lane 1, DEAE, 10 μg; lane 2, phenyl, 3 μg; lane 3, heparin, 1.2 μg; lane 4, IgG, ∼0.12 μg. The ratios of cleavage to extension products are listed at the top.
To further eliminate the possibility of an unrelated, contaminating nuclease, we purified protein A-tagged telomerase using four consecutive chromatographic steps (DEAE, phenyl, heparin, and IgG; see Table 1). The degree of purification was monitored throughout the procedure by protein and activity assays with two exceptions. First, because the activity was undetectable in whole-cell extracts, the fold purification for the DEAE column fraction was based on the degree of Est2p enrichment as determined by Western blotting using anti-protein A antibodies (Fig. 3B). Second, because it is not possible to elute telomerase from IgG-Sepharose, we estimated the amount of total protein bound to the beads to be the difference in protein of the heparin fraction before and after binding to IgG beads. Changes in the polypeptide compositions of the fractions are evident during purification (Fig. 3C). However, because of the low abundance of telomerase in yeast, telomerase-specific polypeptides cannot be identified even after this multistep purification procedure. When tested in the primer extension assay (Fig. 3D), the nuclease-derived products are evident following each chromatographic step, and PhosphorImager analysis indicates that the ratio of cleavage to extension products varied by no more than twofold. Based on these studies, we conclude that cleavage of starting primers by telomerase fractions is unlikely to be due to an unrelated contaminant.
TABLE 1.
Purification of yeast telomerase
| Fraction | Protein (mg) | Activity (U)a | Fold purification | Cleavage/ extension ratio |
|---|---|---|---|---|
| Extract | 2,100 | ND | ||
| DEAE | 5.5 | 1,000 | 200b | 0.052 |
| Phenyl | 1.8 | 1,010 | 620 | 0.031 |
| Heparin | 0.19 | 270 | 1,560 | 0.051 |
| IgG | ∼0.010 | 140 | ∼15,400 | 0.026 |
Telomerase activity was determined in primer extension assays. Incorporation of labeled dGMP into RNase-sensitive bands was quantified using a PhosphorImager. The total activity of the DEAE fraction was arbitrarily set to 1,000, and the activities of the other fractions were calculated accordingly. ND, not determined.
The fold of purification for the DEAE fraction is based on the degree of Est2p enrichment as determined by Western analysis (Fig. 3B).
The effects of reaction parameters on telomerase-mediated primer cleavage.
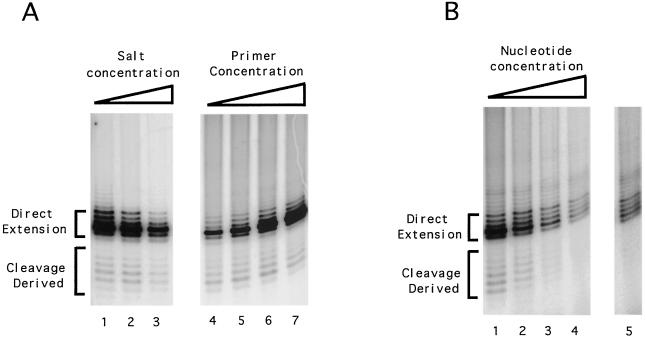
To determine if the extent of cleavage is affected by any reaction parameters, we varied the duration and the concentrations of the components of the reaction. Time course experiments indicate that the “direct extension” and “cleavage derived” products accumulate with similar kinetics (38), both being complete within ∼15 min. Prolonged incubation does not result in an increase in the relative amount of the cleavage products. Thus, there appears to be little nonspecific nuclease in the fraction that can degrade the labeled products, a finding consistent with the earlier tracer experiment (Fig. 1C). Increasing the salt concentration also did not appreciably affect the ratio of the two classes of products (Figure 4A, lanes 1 to 3). Increasing primer concentration from 3 to 24 μM reduced the relative amount of cleavage products by threefold, suggesting that the direct extension reaction pathway is more favorable at high primer concentrations (Fig. 4A, lanes 4 to 7). More interestingly, when the total dGTP concentration was increased by about 10-fold over the standard reaction (to 2 μM), the cleavage products were almost completely abolished, despite the presence of a significant amount of direct extension products (Fig. 4B, lanes 4 and 5). Thus, the cleavage-extension reaction pathway appears to be favored when the concentration of dGTP is low. Cleavage-derived products were not evident in some published studies on yeast telomerase (18, 19). This discrepancy is most likely due to the use of different primers and higher nucleotide concentrations in these other studies. The concentration of dGTP has been reported to affect the processivity and template utilization of the Euplotes aediculatus telomerase (14). Whether these effects of dGTP are related to its ability to influence cleavage remains to be determined.
FIG. 4.
Effects of salt, primer, and nucleotide concentration on the coupled cleavage and extension reactions mediated by yeast telomerase. (A) Polymerization assays were carried out using OXYT1 as the DNA primer and DEAE column fractions as the source of telomerase. For reactions 1 to 3, the DEAE fraction was first desalted using Centricon-30. Sodium acetate was then added to the following final concentrations: lane 1, 0 mM; lane 2, 150 mM; lane 3, 300 mM. For reactions 4 to 7, the following concentrations of OXYT1 primer were used: lane 4, 3 μM; lane 5, 6 μM; lane 6, 12 μM; lane 7, 24 μM. (B) Polymerization assays were carried out using OXYT1 as the DNA primer and DEAE column fractions as source of telomerase. In addition to 0.2 μM labeled dGTP (3,000 Ci/mmol; NEN), unlabeled dGTP was added to the following concentrations: lane 1, 0 μM; lane 2, 0.5 μM; lane 3, 1.0 μM; lanes 4 and 5, 2.0 μM. Lane 5 represents a longer exposure than lane 4.
Primers bearing nontelomeric cassettes are susceptible to cleavage by telomerase.
Characterization of the ciliate telomerase-associated nuclease suggests that the cleavage pathway is affected by primer-RNA interactions (10, 21, 23, 29). In general, primer-template mismatches can apparently promote cleavage. In particular, a primer containing a telomeric cassette embedded in nontelomeric sequences was an especially good substrate for cleavage by Euplotes telomerase. To determine if the yeast telomerase nuclease has similar properties, we tested yeast telomeric primers bearing nontelomeric cassettes at their 5′ or 3′ end in the polymerization reactions. As shown in Fig. 5, a primer containing either an 8- or a 14-nucleotide (nt) nontelomere cassette at its 5′ end (TEL51 and TEL52) was efficiently extended by telomerase. Such primers also gave rise to a significant amount of “cleavage-derived” products. Interestingly, the size of the cleavage-derived products was similar for these two primers (Fig. 5B, compare lanes 1 and 3). This result suggests that cleavage might have occurred predominantly near the junction of the telomeric and nontelomeric cassettes, thereby releasing telomeric fragments of similar size to be extended by the RT. If this conjecture is true, then telomerase is not only capable of extending the 5′ cleavage fragment, as previously reported, but also the 3′ fragment. This possibility was confirmed in experiments reported in the following sections.
FIG. 5.
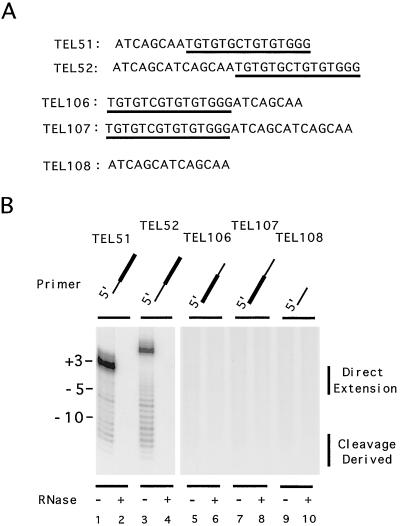
Effects of flanking nontelomeric cassettes in the DNA primer on cleavage and extension by yeast telomerase. (A) The sequences of the oligonucleotides used for the assays in panel B. The telomeric portion of the primer is underlined. (B) Polymerization assays were carried out using DEAE column fractions as a source of telomerase and various primers as indicated at the top of the panel. The reactions were carried out in the absence or presence of RNase A as indicated at the bottom of the panel. The sizes of the various products in relation to TEL51 (as determined in a separate assay) are indicated to the left, while the regions of the gel containing the direct-extension and cleavage-derived products are indicated by vertical lines to the right of the panel.
In contrast to primers with 5′ nontelomeric cassettes, primers with the same two cassettes at their 3′ end (TEL106 and TEL107) were poor substrates for telomerase-mediated extension, and few cleavage products could be observed in these reactions. As expected, the 14-nt nontelomere cassette on its own failed to yield any extension product (TEL108).
The yeast telomerase-associated nuclease acts endonucleolytically.
The cleavage-derived products for most primers had a nonrandom distribution. For example, for both HS2 and OXYT1, the cleavage-derived products were most prominent around the “primer-4” position (Fig. 6B, lanes 1 and 5). This suggests that the telomerase-associated nuclease cleaved DNA preferentially at internal locations, acting as an endonuclease. However, one can also postulate that an exonuclease was responsible and that preferential stalling of the nuclease at particular locations or preferential extension of cleavage products bearing optimal 3′-end sequences gave rise to the observed pattern of product synthesis. To distinguish between these alternatives, we carried out polymerization reactions using primers derivatized with methylphosphonate linkages (Fig. 6A). This modification was expected to render the phosphodiester bond resistant to nuclease attack. If an exonuclease was responsible for the observed primer degradation, the placement of a methylphosphonate linkage between the 3′-most two bases should inhibit the formation of all of the short products. In contrast, if an endonuclease was responsible, then the predominant short products (e.g., the “primer-4” band) should be unaffected because the cleavage that resulted in these products should have occurred far away from the modified linkages.
FIG. 6.
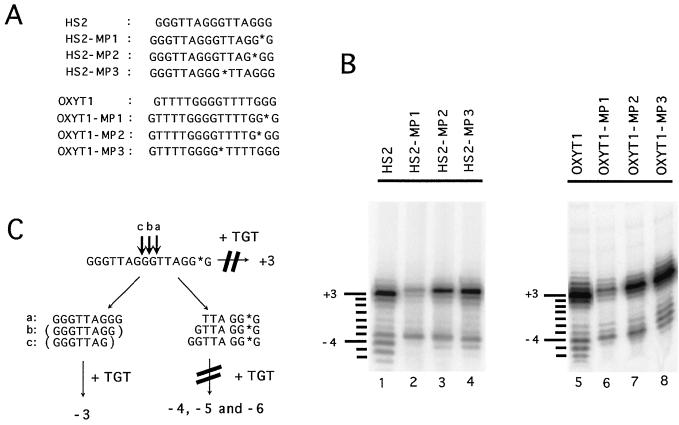
Effects of methylphosphonate linkages in the DNA primer on cleavage and extension by yeast telomerase. (A) The regular and derivatized oligonucleotides used for the reactions in panel B are listed. The location of the methylphosphonate linkage is denoted by an asterisk. (B) Polymerization assays were carried out using 0.5 μg of the various primers as indicated at the top and either 5 μg (lanes 1 to 4) or 3 μg (lanes 5 to 8) of the DEAE fraction. The lengths of the labeled products relative to the starting primers are indicated by lines and numbers to the left of the panels. (C) A schematic illustration of the cleavage-elongation pathways that can account for the reaction products visualized in lanes 1 and 2 of panel B. The nuclease is proposed to act endonucleolytically and to act predominantly in the middle G tract. As described in the text, yeast telomerase strongly prefers to extend primers that have three Gs at their 3′ end and extends these primers predominantly by 3 nt. Thus, reaction a generates two fragments that can both be efficiently extended, leading to the synthesis of the primer-3 and primer-6 products. Reactions b and c each generate only one efficient substrate, leading to the synthesis of the primer-5 and primer-4 products. The methylphosphonate substitution in MP-1 (marked by an asterisk) strongly inhibits extension of the nearby 3′ OH group by telomerase, causing the loss of the “+3” product as well as the “−4,” “−5,” and “−6” products.
As shown in Fig. 6B, some of the cleavage-derived products were retained despite the substitution of one of the two 3′-most phosphodiester linkages of the HS2 or the OXYT1 primer (the HS2-MP1, HS2-MP2, OXYT1-MP1, and OXYT1-MP2 oligonucleotides). These observations suggest that some of the short products must be generated by an endonuclease, as in the case of the Euplotes telomerase-mediated cleavage. Close inspection of the reaction products derived from modified HS2 oligonucleotides revealed two interesting features. First, the amount of “direct extension” products (as evidenced by the intensity of the “primer + 3” band) was greatly inhibited by a methylphosphonate at the 3′-most linkage (Fig. 6B, compare lane 2 with lane 1). This suggests that the last phosphodiester linkage of the primer may make a functionally important interaction with telomerase, which can be disrupted by the modification. Methylphosphonate linkages positioned near the 3′ end has also been found to inhibit extension by E. crassus telomerase (D. E. Shippen, personal communication). Second, the “primer-4” and “primer-5” products were almost completely abolished (Fig. 6B, compare lane 2 with lane 1), just like the direct elongation products. This similarity suggests that the primer-4 and primer-5 products may also be derived from primers with methylphosphonate modification near the 3′ end. In other words, these products may be due to extension of the 3′ cleavage products. In contrast, the “primer-3” product was virtually unaffected, suggesting that it may be derived from the 5′ fragment generated by the nuclease.
Extensive characterization of primer utilization by yeast telomerase indicates that the enzyme preferentially extends oligonucleotides with 3 Gs at their 3′ end. Furthermore, in our reaction condition, the enzyme has a strong tendency to pause or dissociate after adding 3 nt (TGT) (21; Xia and Lue, unpublished). Taking this property of telomerase into consideration, we can account for all of the cleavage-elongation products of HS2 by the hypothetical scheme presented in Fig. 6C. In this model, cleavage occurs preferentially in the middle G tract. Cleavage between the ninth and tenth nucleotides of HS2 (reaction a) leads to the creation of a 5′ fragment (GGGTTAGGG) that is expected to be a good substrate for telomerase and to yield a predominant 12-nt product (GGGTTAGGGTGT) at the primer-3 location, precisely as was observed. The same cleavage should also give rise to a 3′ fragment (TTAGGG) that can yield a 9-nt product (TTAGGGTGT) at the primer-6 position. This was also observed. Cleavage between the eighth and ninth nucleotides (reaction b) and between the seventh and eighth nucleotides (reaction c), on the other hand, would yield 5′ fragments that are poor substrate for telomerase but 3′ fragments that are good substrates (GTTAGGG and GGTTAGGG). These 3′ fragments are expected to give rise predominantly to the primer-5 and primer-4 products, respectively. An important prediction of this scheme is that the extension products of the 3′ fragments (primer-4, primer-5, and primer-6) should be inhibited by the MP1 modification, while the extension products of the 5′ fragment (primer-3) should not. This prediction was entirely consistent with the observation made here (Fig. 6B, compare lanes 2 and 1). A similar argument can be made to account for the cleavage-elongation products of the OXYT1 primer if cleavages occur predominantly in the middle G tract.
Flexibility of the nuclease cleavage site.
To test if the predicted cleavage sites were in fact utilized by yeast telomerase, we designed primers (HS2-MP3 and OXYT1-MP3) to specifically render one of the linkages nonhydrolyzable. Both MP3 modifications abolished some but not all of the reaction products, as expected (Fig. 6B, compare lanes 1 and 4 and lanes 5 and 8). More interestingly, the OXYT1-MP3 oligonucleotide gave rise to some products that are not observed in the case of OXYT1. Thus, for OXYT1-MP3, the primer-2 and primer-3 bands are stronger than the primer-4 band, a finding that was precisely the reverse of the pattern for OXYT1 (lanes 5 and 8). These results suggest the interesting possibility that when a preferred cleavage site is resistant to the nuclease, other sites can be utilized, leading to a different distribution of fragments.
Extension of either the 5′ or the 3′ cleavage products by yeast telomerase.
Both the nontelomeric cassette study (Fig. 5) and the methylphosphonate substitution study (Fig. 6) suggest that either the 5′ or the 3′ fragment generated by the nuclease can be extended by the RT activity of telomerase. To confirm this conjecture, DNA primers terminating in dideoxynucleotides were synthesized by using terminal transferase and the appropriate dideoxynucleotide triphosphates and then subjected to the extension assay. All products resulting from the addition of nucleotides to the 3′ cleavage fragment were expected to be abolished by this modification, while those from the addition of nucleotides to the 5′ cleavage fragment should be unaffected.
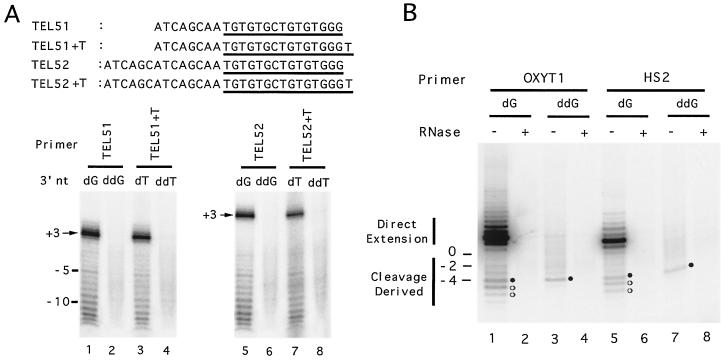
Primers containing nontelomeric cassettes (Fig. 5), as well as primers containing heterologous repeats (Fig. 1, 4, and 6), were tested in this assay. As shown Fig. 7A, when primers bearing 5′ nontelomeric and 3′ telomeric cassettes were utilized, all of the direct extension products can be abolished by substituting the last nucleotide of the primer with dideoxynucleotide, as expected. More significantly, the cleavage-derived products can also be entirely abolished by substituting the last nucleotide of the primer with dideoxynucleotide. However, when the same analysis was applied to primers bearing heterologous telomeric repeats, different results were obtained. For example, substitution of the last dG residue of OXYT1 with ddG eliminated some but not all of the cleavage-derived products (Fig. 7B, lanes 1 and 3). Substitution of the last dG residue of HS2 with ddG had similar effects (Fig. 7B, lanes 5 and 7). Most importantly, in the case of OXYT1 and HS2 the cleavage products eliminated by the ddGMP modification are precisely those eliminated by the MP1 modification, a finding consistent with the notion that both modifications abolished labeling of the 3′ cleavage fragment. For example, the OXYT1-ddG oligonucleotide yielded the prominent primer-4 product (indicated by a closed circle in lane 3) but not the primer-5 or primer-6 products evident in the case of the OXYT1 oligonucleotide (indicated by open circles in lane 3). This was precisely what was observed for the OXYT1-MP1 oligonucleotide. The same comparison can be made between the products generated by HS2-ddG and HS2-MP1 oligonucleotides (compare lane 7 of Fig. 7B and lane 2 of Fig. 6B). As expected for telomerase-mediated extension, all of the cleavage-derived products from either the native or the ddG-modified primers were sensitive to RNase A pretreatment (lanes 2, 4, 6, and 8). Taken together, the dideoxy substitution experiments suggest that in the case of primers containing nontelomeric cassettes, telomerase appears to preferentially extend the 3′ cleavage fragment, while in the case of primers containing heterologous repeats, telomerase appears to be capable of extending both the 5′ and the 3′ fragments derived from cleavage.
FIG. 7.
Effects of 3′ dideoxynucleotide substitutions in the DNA primer on cleavage and extension by yeast telomerase. (A) Polymerization reactions were carried out using 160 ng (lanes 1 to 4) or 400 ng (lanes 5 to 8) of the DNA primers (as indicated at the top of the panels) and 5 μg of the DEAE fractions. The primers bear either a deoxy- or a dideoxynucleotide at their 3′ termini. The sequences of the oligonucleotides used are shown at the top, and the GT-rich (yeast telomere-like) parts of the oligonucleotides are underlined. (B) Polymerization reactions were carried out using 0.5 μg of the DNA primers (as indicated at the top of the panel) and 5 μg of the DEAE fractions. The primers used in lane 3, 4, 7, and 8 bear dideoxynucleotides at their 3′ ends. RNase A was added to the reactions in lanes 2, 4, 6, and 8. Products derived from direct extension or cleavage followed by extension (cleavage derived) are indicated by vertical bars to the left of the panels. Bands unaffected or abolished by the dideoxynucleotide substitution are indicated by closed or open circles, respectively.
DISCUSSION
We have shown that, like ciliate telomerases, yeast telomerase has a tightly associated endonuclease activity that can cleave the starting primer prior to extension by the RT subunit. Novel aspects of this work include the demonstration (i) that the nuclease can be affinity purified along with the RT subunit of telomerase, (ii) that both the 5′ and the 3′ fragments derived from cleavage can be extended by telomerase, and (iii) that the loss of one nuclease site can lead to the preferential utilization of other sites.
The ability of yeast telomerase to extend either one of the cleaved fragments is somewhat surprising in light of earlier studies showing that extension occurs mostly on the 5′ fragment (23). This discrepancy is most likely explained by the use of primers bearing 3′ nontelomeric cassettes in these earlier studies. Such nontelomeric cassettes, once released from the rest of the primers, are probably inefficient substrates for telomerase extension. Indeed, for primers that bear a 5′ nontelomeric cassette and a 3′ telomeric cassette, the cleavage-derived products are all due to labeling of the 3′ fragments, a result consistent with the 3′ telomeric cassettes being better substrates for yeast telomerase than the 5′ nontelomeric cassettes. Similarly, the ability of yeast telomerase to extend both of the fragments derived from cleavage of heterologous repeats is explained by both fragments' ability to form a hybrid with the RNA template and serve as a substrate for extension.
Initial studies of the Tetrahymena enzyme revealed similarities between RNA polymerase-mediated transcript cleavage and telomerase-mediated primer cleavage (5, 36). Such observations raise the interesting possibility that telomerase uses the polymerization site to carry out the cleavage reaction. (The evidence that RNA polymerase mediates transcript cleavage through the polymerization active site is compelling [32]). In this model, one would expect telomerase to extend exclusively the 5′ fragment, because the 3′-OH group of this fragment would be located optimally at the polymerase active site immediately following cleavage. This expectation is clearly not met by our results.
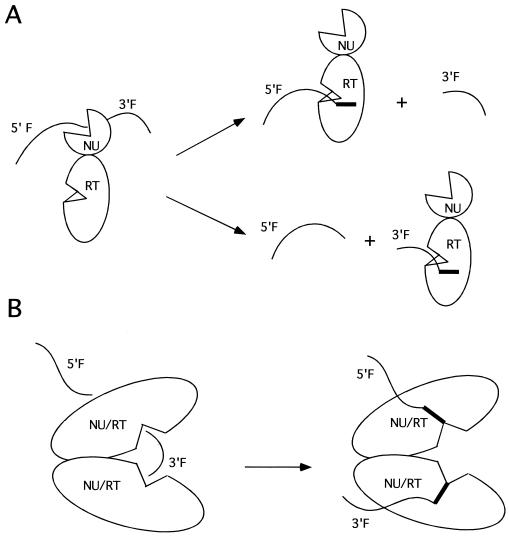
To account for the ability of yeast telomerase to extend both the 5′ and the 3′ fragments generated by cleavage, we propose two general models. The first model postulates two distinct active sites for the RT and nuclease activity in a single polypeptide. These two active sites are flexibly positioned relative to each other such that following the cleavage reaction, the RT domain can stochastically interact with and extend either the 5′- or the 3′-end fragment (Fig. 8A). Existing biochemical data suggest that the telomerase complex can interact with an extended region of the DNA primer, from the 3′ end where polymerization takes place, to approximately 25 nt upstream. Thus, both the 5′ and the 3′ cleavage products may remain associated with the complex and serve as substrates for extension. This general model is consistent with an earlier study by Greene et al. (10) showing a flexible relationship between the nuclease and RT of telomerase. An implication of this model is that a single complex cannot extend both cleavage products simultaneously.
FIG. 8.
Models for telomerase-mediated cleavage-extension reactions. (A) Telomerase is shown to possess two distinctive active sites for nuclease (NU) and RT activity. Following cleavage, the RT domain can capture stochastically either the 5′-end fragment (5′F) or the 3′-end fragment (3′F) for extension. (B) Telomerase is shown to be a dimeric enzyme containing two active sites. Each active site is bifunctional and capable of mediating both primer cleavage and extension (NU/RT). Following cleavage of the starting primer by one of the protomers, the resulting 5′-end fragment (5′F) and 3′-end fragment (3′F) can both be extended because of the presence of the two bifunctional active sites.
A second plausible model invokes a single active site that mediates both cleavage and extension but postulates that yeast telomerase is multimeric (Fig. 8B). If, for example, telomerase is a dimer, then one protomer can be acting as a nuclease. Following cleavage, this protomer would be ideally positioned to extend the 5′ fragment, while the other protomer can capture the 3′ fragment for extension. In this fashion, a single telomerase complex would be capable of elongating both cleavage products. Consistent with this second model are recently published experimental results showing that yeast telomerase may indeed be multimeric (30). Our two general models are not mutually exclusive, and features of both may be combined. For example, a multimeric telomerase containing distinct nuclease and RT active sites would also be consistent with our experimental results. Clearly, more analysis is necessary to determine the molecular coupling mechanisms between the nuclease and RT of telomerase.
The function of telomerase-associated nuclease remains to be elucidated. That a nonciliate telomerase can be shown to possess a tightly associated nuclease activity indicates that the latter is not likely to be exclusively involved in developmentally mediated chromosome fragmentation. Otherwise, our data are compatible with previously proposed functions, such as enhancing the fidelity of DNA synthesis and enhancing elongation efficiency. Another speculative function for the nuclease is raised by our finding that telomerase may be engaged with the 3′ fragment following cleavage (Fig. 8A). Cleavage in this case can result in the release of the enzyme from telomeric ends and completely abort the elongation of chromosomes. This may be one way of negatively regulating the action of the enzyme. (Extension of the released 3′ fragment would not appear to have any physiologic significance and may simply be an unintended consequence of the cleavage reaction.) Continued analysis of the telomerase-associated nuclease in a genetically tractable organism may eventually allow these proposed functions to be tested in vivo.
ACKNOWLEDGMENTS
We thank B. Futcher, B. Schneider, and B. Schwer for strains and plasmids and D. Shippen for communicating unpublished results.
This work was supported by an American Cancer Society research grant and a U.S. Army Breast Cancer Idea Award.
REFERENCES
- 1.Blackburn E H, Greider C W. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 2.Blackburn E H. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 3.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytrica trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 5.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins K, Greider C W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 7.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Funk W D, Wang S-S, Weinrich S L, Avillion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J, Le S, West M D, Harley C B, Andrews W H, Greider C W, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene E C, Bednenko J, Shippen D E. Flexible positioning of the telomerase-associated nuclease leads to preferential elimination of nontelomeric DNA. Mol Cell Biol. 1998;18:1544–1552. doi: 10.1128/mcb.18.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 12.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 13.Greider C W. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond P W, Cech T R. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 1997;25:3698–3704. doi: 10.1093/nar/25.18.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O Amgen EST Program. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 16.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J-J, Zakian V. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 18.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 19.Lingner J, Cech T R, Hughes T R, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lue N F, Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lue N F, Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. doi: 10.1093/nar/26.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantell L L, Greider C W. Telomerase activity in germline and embryonic cells of Xenopus. EMBO. 1994;13:3211–3213. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melek M, Greene E C, Shippen D E. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoof M J, Liu Q, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 25.Morin G. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama J-I, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 28.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 29.Prescott J, Blackburn E H. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal non-processivity in vivo and in vitro. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 30.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prowse K R, Avilion A A, Greider C W. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci USA. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudd M D, Izban M G, Luse D S. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 34.Singer M S, Gottschling D E. TLC1:template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 35.Steiner B R, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcription elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 37.Zakian V A. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]