Abstract
Background: The ongoing outbreak of SARS-CoV-2 represents a significant challenge to international health. Several reports have highlighted the importance of ACE2 on the pathogenesis of COVID-19. The spike protein of SARS-CoV-2 efficiently binds to the angiotensin-converting enzyme 2 (ACE2) receptors and facilitates virus entry into the host cell. In the present study, we hypothesize that a functional insertion/deletion polymorphism-rs4646994 I/D and rs4240157 T > C in the ACE gene could be associated with SARS-CoV-2 infection and mortality. Methodology: This study included 117 consecutive COVID-19 patients and 150 age matched healthy controls (ACE2-rs4646994 I/D) and 100 age matched healthy controls with ACE2 rs4240157 T > C. We used Mutation specific PCR (MSP) for ACE2-rs4646994 I/D genotyping and amplification refractory mutation system (ARMS-PCR) for ACE2 rs4240157 T > C genotyping. Results: Results indicated that there were significant differences in the genotype distributions of ACE2-rs4646994 I/D polymorphisms (p < 0.030) and ACE2 rs4240157 T > C between COVID-19 patients and controls (p-values < 0.05). Higher frequency of DD genotype (48.71%) and D allele (0.67) was reported in COVID-19 patients than controls. Our results showed that the ACE2-DD genotype was strongly associated with increased COVID-19 severity (OR 2.37 (95%) CI = (1.19–4.70), RR = 1.39 (1.09–1.77), p < 0.013) and also a strong association was seen with ACE2-ID genotype with COVID-19 severity (OR 2.20 (95%) CI = (1.08–4.46), p < 0.020) in the codominant model. In allelic comparison, the D allele was strongly associated with COVID-19 severity (OR 1.58 (95% CI) (1.11–2.27), RR 1.21 (1.05–1.41) p < 0.010). A significant correlation of ACE2-I/D genotypes was reported with Age (p < 0.035), T2D (p < 0.0013), hypertension (p < 0.0031) and coronary artery disease (p < 0.0001). Our results indicated ACE2-DD genotype was strongly associated with increased COVID-19 mortality (OR 8.25 (95%) CI = (2.40 to 28.34), p < 0.008) and also ACE2-DD + DI genotype was strongly associated with increased COVID-19 mortality with OR 4.74 (95%) CI = (1.5214 to 14.7915), p < 0.007. A significant correlation was reported between COVID-19 patients and age matched controls (p < 0.0007). Higher frequency of heterozygosity TC (40%) followed by ACE2-CC genotype (24.78%) was reported among COVID-19 patients. Using multivariate analysis, ACE2–CT genotype was strong associated with SARS-CoV-2 severity with an OR 2.18 (95% CI) (1.92–3.99), p < 0.010 and also ACE2–CC genotype was linked with COVID-19 severity with an OR 2.66 (95% CI) (1.53–4.62), p < 0.005. A significant correlation of ACE2-T > C genotypes was reported with gender (p < 0.04), T2D (p < 0.035). ACE2-CC genotype was strongly associated with increased COVID-19 mortality OR 3.66 (95%) CI = (1.34 to 9.97), p < 0.011 and also ACE2-C allele was associated with COVID-19 mortality OR 2, 01 (1.1761–3.45), p < 0.010. Conclusions: It is concluded that ACE-DD genotype and D allele was strongly associated with increased COVID-19 patient severity. In addition, ACE I/D polymorphism were strongly associated with advanced age, diabetes and ischemic heart disease in COVID-19 patients whereas ACE-II genotype was a protective factor against the development of severe COVID-19. ACE2-DD genotype was strongly associated with increased COVID-19 mortality. Additionally, ACE2–CC and CT genotypes were strongly associated with COVID-19 severity. Therefore, our study might be useful for identifying the susceptible population groups for targeted interventions and for making relevant public health policy decisions.
Keywords: gene polymorphism, infection, SARS-CoV-2, COVID-19, pathogenesis, ACE1-angiotensin-converting enzyme 1, ACE2-angiotensin-converting enzyme 2, severe acute respiratory syndrome coronavirus 2, susceptibility to SARS-CoV-2
1. Introduction
The coronavirus disease, designated as COVID-19 and declared as pandemic by the World Health Organization is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [1,2,3,4]. COVID-19 has infected around 224 million people across the globe and has resulted in more than 4.86 million deaths till date (https://www.worldometers.info/coronavirus, accessed on 12 October 2021). While as the individuals more than 50 years of age were prone to higher severity and mortality of the disease, the common comorbidities included hypertension, obesity, and diabetes [5,6]. COVID-19 has exhibited geographical variations in its spread and mortality and it has caused more morbidity and mortality in Europe and USA [7,8,9].
SARS-CoV-2 is an enveloped RNA virus with three structural protein components which are described as M (membrane), E (envelop) and S (spike) proteins [10,11]. The SARS-COV-2 makes its entry into the host cells through the S proteins. The two subunits of S protein, S1 and S2 are used by the virus to attach to the host cell membrane. The C-terminal domain (CTD) of S1 subunits is vital for the interaction and attachment of S proteins to the protein receptors of the host cell [4,12,13].
Just like the SARS-CoV, the human angiotensin converting enzyme-2 (ACE2) has emerged as the target for the SARS-CoV-2 [14,15]. ACE2 is expressed ubiquitously in different organs of the human body that include heart, lungs, kidneys, intestines and endothelium [16,17,18,19]. ACE2, a type I transmembrane zinc finger glycoprotein, is a mono-carboxypeptidase (with 805 amino acid residues and MW of 100 KD) and converts angiotensin 1 to angiotensin 1-9 [20]. The gene for ACE2, present on chromosome X (Xp22) comprises of 39.98 kb of genomic DNA and has 18 exons [20]. Many ACE2 variants are associated with several common diseases, whose incidence depends on the balance in the renin-angiotensin-aldosterone (RAAS) pathway. Hypertension is associated with rs1514283, rs2074192, rs233575, rs4646155, rs4646176, rs2285666, rs879922, rs2106809, rs4646188, rs4240157, rs4830542, rs2158083, and rs879922 [21,22]. The role of ACE2 is very critical since the inhibition of RAAS pathway leads to upregulation of ACE2 which alleviates ARDS and myocarditis symptoms in COVID-19 patients but at the same time increased ACE2 expression may increase the entry of virus into host cells, complication the overall clinical picture [23].
Geographic distribution of ACE I (insertion in intron 16) allele was summarized by Saab et al. and its frequency increases eastwards and westwards from the Middle East [24]. The genetic polymorphism of ACE2 gene is well known world over with racial and ethnic variations [25,26] having varying influences on the altered functions of RAAS pathway [27]. The polymorphism of ACE2 gene has been associated with hypertension in Chinese [28,29], Canadian [30], Indian [31] and Brazilian populations [32]. The ACE2 polymorphism has been associated with varying degree of disease severity and clinical outcomes of COVID-19, with absence of ACE D/D genotype conferring protection against severe lung injury [33]. The African Americans with high frequency of D allele have depicted higher mortality rates in USA [34,35]. Similarly, higher fatalities have been reported from many parts of Europe (https://ec.europa.eu/eurostat/statistics-explained/index.php/Main_Page, accessed on 12 October 2021). In contrast lower mortality rates have been observed in Asian patients who harbor high frequency of II genotype as compared to DD genotype [36] and (https://www.worldometers.info/coronavirus/coronavirus-death-rate, accessed on 12 October 2021). The D allele results in an increased angiotensin-2 level (as a consequence of increased ACE-1 and decreased ACE-2 levels) that leads to increased microvascular permeability resulting in pulmonary edema and other complications [37,38].
In the current scenario, the knowledge of ACE2 gene polymorphism in different populations assumes a huge significance. However, although there have been many published reports on the ACE2 gene polymorphism from different parts of the globe, we came across only one report from Saudi Arabia suggesting a link between the ACE2-rs4646994 I/D polymorphism and the obesity [39]. To the best of our knowledge, the current study on the genetic polymorphism ACE2-rs4646994 I/D and ACE2 rs4240157 C > T in the ACE2 gene is the first one from Saudi Arabia and was carried out in the COVID-19 patients with different disease severity/outcomes and healthy controls.
2. Methodology
2.1. Study Population
This population-based case-control study (prospective cohort study) was conducted on COVID-19 patients and 150 age matched healthy controls. COVID-19 laboratory confirmation was defined as a positive result on RT-qPCR (SARS-CoV-2 RT-qPCR detection kit) of nasal and oropharyngeal swab specimens. We selected COVID-19 positive cases confirmed by a real-time reverse transcription polymerase chain reaction (rRT-PCR) test.
This study was a collaborative study. Specimens from positive cases of COVID-19 were collected from different hospitals in Saudi Arabia (Bisha, Abha and Tabuk). The sample collection was conducted between September 2020 and April 2021.
2.2. Ethics Approvals
The ethical approvals were obtained from local RELOC committees of College of Medicine, University of Bisha (Ref. No. UBCOM/H-06-BH-087 (05/25)), University of Tabuk (Ref. No: KAEK2020/4/4) and College of Medicine, King Khalid University, Abha (Ref. No. KKU-H-06-B-091) in accordance with human subjects and complied with the principles of the Helsinki Declaration. Informed consent was obtained before collecting samples from all patients and control subjects.
2.3. Data Collection
All the subjects were interviewed using a structured questionnaire regarding epidemiological/demographic data, past history of CAD, T2D, and history of addiction particularly smoking, family history of any significant diseases.
2.3.1. Sample Collection from COVID-19 Patients
About 3 mL of peripheral blood sample was collected in an EDTA or Lavender top tube for all COVID-19 patients. The Blood specimens were immediately stored at −20 °C to −30 °C. The COVID-19 patients who’s who oxygen saturation was less than 60 and needed mechanical ventilatory support in ICU were classified as having severe COVID-19 disease.
2.3.2. Sample Collection from Healthy Controls
All healthy age matched controls specimens have been timed around routine blood draws that are part of routine workout, and hence did not require additional phlebotomy and all participants were provided written informed consent form. About 3 mL peripheral blood was collected in EDTA tubes. The Blood specimens were immediately stored at −20 °C to −30 °C.
2.4. Genomic DNA Extraction
Genomic DNA was extracted using DNeasy Blood K (Qiagen, Hilden, Germany) as per the manufacturer’s instructions. The extracted DNA was dissolved in nuclease-free water and stored at 4 °C until use. Quality and integrity of DNA were checked by NanoDrop™ (Thermo Scientific, Waltham, MA, USA). All DNA samples from COVID-19 and controls were screened for purity by measuring optical density (OD) at 260 nm (OD260) and 280 nm (OD280). The λ260/λ280 ratios ranged from 1.83–1.99 indicating good quality DNA.
2.5. Angiotensin-Converting Enzyme Genotyping
The Angiotensin-converting enzyme-2 genotyping (ACE2-rs4646994 I/D) was done by mutation specific PCR and amplification refractory mutation system (ARMS-PCR) PCR was used to detect ACE2 rs4240157 T > C genotyping. The ARMS primers were designed by using the Primer3 software as depicted in Table 1.
Table 1.
Primers for ACE2 gene polymorphisms.
| Direction | Primer Sequence | PCR Product | Annealing Temperature |
|---|---|---|---|
| ARMS-PCR primers of ACE2-rs4646994 I/D gene polymorphism | |||
| ACE-F | 5′-CTGGAGACCACTCCCATCCTTTCT-3′ | 490-bp (II) | 58 °C |
| ACE-R | 5′-GATGTGGCCATCACATTCGTCAGAT-3′. | 190-bp(DD) | |
| ARMS-PCR primers of ACE2 rs4240157 T > C gene polymorphism | |||
| ACE2 Fo | GCTGAGTTCTCAAAATAATGCCATAGAT | 386 bp | 60 °C |
| ACE2 Ro | GCATTTCTTTCCAATCATTAAGAGTTCA | ||
| ACE2 FI-T | GCCTCAGAACATTACAGAATCAACCT | 244 bp | |
| ACE2 RI-C | GAGGGTTGGTAAATAGTGTTCAGTGG | 194 bp | |
The PCR was done in a reaction volume of 25 µL containing template DNA (50 ng), Fo—0.25 µL, R0—0.25 µL, RI—0.25 µL, RI—0.25 µL of 25 pmol of each primer and 10 µL from GoTaq® Green Master Mix (cat no M7122) (Promega, Madison, WI, USA). The final volume of 25 µL was adjusted by adding nuclease free double distilled water (ddH2O). Finally, 2 µL of DNA was added from each patient. The thermocycling conditions used were: 95 °C for 10 min followed by 40 cycles of 95 °C for 35 s, annealing temperature ACE2-rs4646994 I/D (58 °C) and ACE2 rs4240157 C > T (60 °C) gene polymorphism for 40 s, 72 °C for 43 s followed by the final extension at 72 °C for 10 min.
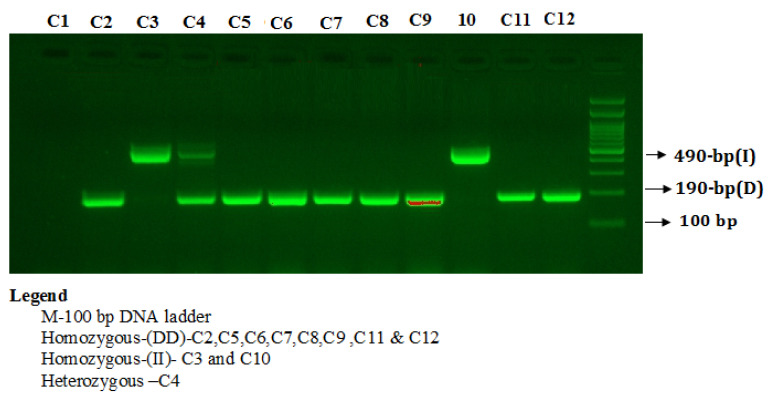
PCR products were separated on 2% agarose gel stained with 2 µL of sybre safe stain (Thermo Scientific, Waltham, MA, USA) and visualized on a UV trans illuminator from Bio-Rad (Hercules, CA, USA). The PCR products were electrophoresed on a 2% agarose gel stained with ethidium bromide to visualize three patterns: I/I (490-bp fragment), D/D (190-bp fragment), and I/D (both 490- and 190-bp fragments) as depicted in Figure 1.
Figure 1.
Detection of Angiotensin I-converting enzymes (ACE2) insertion/deletion (I/D) rs4343 gene polymorphism.
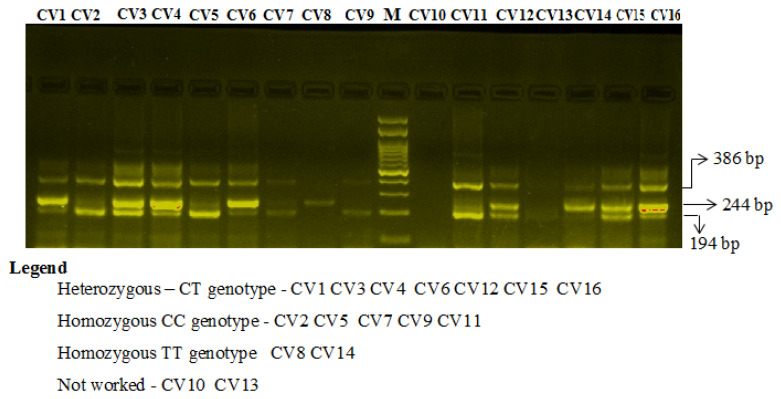
Primers FO and RO flank the exon of the ACE2 rs4240157 T > C gene, resulting in a band of 386 bp to act as a control for DNA quality and quantity. Primers FI and RO amplify a wild-type allele (T allele), generating a band of 244 bp, and primers FO and RI generate a band of 194 bp from the mutant allele (C allele) as depicted in Figure 2.
Figure 2.
Two percent gel electrophoresis of angiotensin-converting enzyme 2 (ACE2) rs4240157 C/T genotyping by the amplification-refractory mutation system (ARMS) in COVID-19 cases.
2.6. Statistical Analysis
Deviations from Hardy–Weinberg disequilibrium (HWD) were calculated by chi-square (χ2) goodness-of-fit test. Group differences were compared using Student’s two-sample t-test or one-way analysis of variance (ANOVA) for continuous variables and chi-squared for categorical variables. Differences in the ACE-rs4646994 I/D, and ACE2 rs4240157 C > T allele and genotype frequencies between groups were evaluated using Chi-square test. The associations between ACE2-rs4646994 I/D, ACE2 rs4240157 C > T genotypes and risk of COVID-19 patients were estimated by computing the odds ratios (ORs), risk ratios (RRs) and risk differences (RDs) with 95% confidence intervals (CIs). Allele frequencies among cases as well as controls were evaluated by using the chi-square Hardy–Weinberg equilibrium (HWE) test. A p-value < 0.05 was considered significant. All statistical analyses were performed SPSS 16.0.
3. Results
3.1. Demographic Features
All demographic features of the 117 consecutive COVID-19 patients are summarized in Table 2. Of 117 consecutive patients, 20 (17.10%) patients were below or equal to 40 years and 97 (82.90%) were above 40 years of age, 85 (72.64%) were males and 32 (27.36%) were females. Among 117 COVID-19 patients, 47 (40.17%) had T2DM, 37 (9.40%) were having hypertension, 11 (9.40%) having CKD and 47 (40.17%) were having low oxygen saturation (<60 mm Hg) at the time of admission (Table 2). 57 (48.71%) stayed more than 30 days in hospital. Regarding treatment of COVID-19 patients 79/117 (67.52%) were treated with antiviral therapy whereas 77/117 (65.81%) were treated with steroid therapy. Clinical outcome of the COVID-19 patients showed that 43/117 (36.75%) patients died and 74/117 (63.24%) were discharged and survived. At the time of diagnosis of COVID-19 patients, 45/117 (38.57%) had high levels of ALT, 104/117 (97.44%) had high levels of CRP and 48/117 (41.3%) had high levels of AST (Table 2).
Table 2.
Baseline characteristics of the COVID-19 patients. T2D—type2 diabetes; CKD—chronic kidney disease; CAD—coronary artery disease; SpO2—partial pressure of oxygen; ALT—alanine transaminase; CRP—C reactive protein; AST—aspartate transaminase.
| Clinical Feature | Variable | N = 117 | % |
|---|---|---|---|
| Age | >40 | 97 | 82.90 |
| <40 | 20 | 17.09 | |
| Gender | Males | 85 | 72.64 |
| Females | 32 | 27.36 | |
| T2D | Yes | 47 | 40.17 |
| No | 70 | 59.83 | |
| CKD | Yes | 11 | 9.40 |
| No | 106 | 90.60 | |
| Hypertension | Yes | 37 | 31.62 |
| No | 80 | 68.37 | |
| CAD | Yes | 17 | 14.53 |
| No | 100 | 85.47 | |
| Oxygen saturation | <60 | 47 | 40.17 |
| >80 | 70 | 59.83 | |
| Duration in hospital | >30 | 57 | 48.71 |
| <30 | 60 | 51.29 | |
| ALT | <36 | 72 | 61.53 |
| >36 | 45 | 38.57 | |
| CRP | 0.8 | 13 | 2.56 |
| 0.8 high | 104 | 97.44 | |
| AST | <40 | 69 | 58.97 |
| >40 | 48 | 41.3 | |
| Antiviral therapy | Yes | 79 | 67.52 |
| No | 38 | 32.48 | |
| Steroids therapy | Yes | 77 | 65.81 |
| No | 40 | 34.19 | |
| Survival | Yes | 43 | 36.75 |
| No | 74 | 63.24 |
3.2. Healthy Controls
Out of 150 age matched heathy controls 100 (66.67%) were males and 50 (33.33%) were females and 110 (73.33%) were above 40 years of age and 40 (26.67%) were below or equal to 40 years.
3.3. Statistical Comparisons between Patients and Controls (p Values) for ACE I/D Genotypes
We found that the ACE I/D genotype (ACE2-rs4646994 I/D) frequency among all the study participants is in compliance to the HWE. The genotype distributions and allele frequencies of the SNPs located in the ACE2-rs4646994 I/D showed no deviation in HWE (all p > 0.05) (χ2 = 2.24 p ≤ 0.13) in the COVID-19 patient group and similarly the genotype distributions and allele frequencies showed that no deviation in HWE (all p > 0.05) (χ2 = 0.52 p ≤ 0.47) in the controls. Thus, we chose 10% samples from normal control group randomly to review genotyping results, showing that the accuracy rate was more than 99%. In COVID-19 patients, the II, DI and DD genotype frequencies were 13.67%, 37.60% and 48.71% respectively whereas in healthy controls II, DI and DD genotype frequencies were 26.66%, 33.34%, and 40% respectively (Table 3). The distribution of ACE2-rs4646994 I/D genotypes observed between COVID-19 patients and healthy controls was significant (p < 0.030). Moreover, the frequency of D allele (fD) was found to be significantly higher among COVID-19 patients than in HC (0.67 vs. 0.57) (Table 3).
Table 3.
Association of ACE2-rs4646994 I/D genotypes in COVID-19 severity.
| Subjects | N | II% | DI% | DD% | fD | χ2 | I | D | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| Cases | 117 | 16 (13.67) | 44 (37.60) | 57 (48.71) | 2 | 6.77 | 0.33 | 0.67 | 0.03 |
| Controls | 150 | 40 (26.66) | 50 (33.34) | 60 (40) | 0.43 | 0.57 |
3.4. Multivariate Analysis to Estimate the Association between ACE2-rs4646994 I/D Genotypes and Risk to COVID-19 Severity
A multivariate analysis based on logistic regression like odds ratio (OD) and risk ratio (RR) with 95% confidence intervals (CI) were calculated for each group to estimate the association between ACE2-rs4646994 I/D genotypes and risk to COVID-19 and the data are summarized in Table 4. Our results indicated that in the codominant model, the ACE2-ID genotype was strongly associated with increased COVID-19 severity with OR 2.20 (95%) CI = (1.08–4.46), RR = 1.34 (1.04–1.72), p < 0.020. Similarly, ACE2-DD genotype was strongly associated with increased COVID-19 severity with OR 2.37 (95%) CI = (1.19–4.70), RR = 1.39 (1.09–1.77), p < 0.013. There is a strong association observed between ACE2–II genotype vs. ACE2-(DI + DD) genotype in dominant inheritance model and leads to increased COVID-19 severity with OR = 2.39 (95%) CI (1.21–4.35), RR = 1.37 (1.11–1.69), p < 0.010 (Table 4). In allelic comparison, the D allele was strongly associated with COVID-19 severity with an OR 1.58 (95% CI) (1.11–2.27), RR 1.21 (1.05–1.41) p-value = 0.010. No significance was observed in over dominant inheritance model. This result indicates a potential dominant effect of ACE2-DD genotype and D allele on COVID-19 severity in our study group.
Table 4.
Multivariate and ordinal regression risk factor analysis for COVID-19 severity with ACE2-rs4646994 I/D genotypes.
| Genotypes | Healthy Controls | COVID-19 Cases | Odd Ratio OR (95% CI) |
Risk Ratio RR (95% CI) |
p-Value | Status |
|---|---|---|---|---|---|---|
| (N = 150) | (N = 117) | |||||
| Codominant | ||||||
| ACE2-II | 40 | 16 | 1 (ref.) | 1 (ref.) | ||
| ACE2-ID | 50 | 44 | 2.20 (1.08–4.46) | 1.34 (1.04–1.72) | 0.02 | S |
| ACE2-DD | 60 | 57 | 2.37 (1.19–4.70) | 1.39 (1.09–1.77) | 0.013 | S |
| Dominant | ||||||
| ACE2-II | 40 | 16 | 1 (ref.) | 1 (ref.) | ||
| ACE2-(DI + DD) | 110 | 101 | 2.39 (1.21–4.35) | 1.37 (1.11–1.69) | 0.010 | S |
| Recessive | ||||||
| ACE2-(II + DI) | 90 | 60 | 1 (ref.) | 1 (ref.) | ||
| ACE2-DD | 60 | 57 | 1.42 (0.87–2.32) | 1.17 (0.93–1.45) | 0.15 | NS |
| Allele | ||||||
| ACE2-I | 130 | 76 | 1 (ref.) | 1 (ref.) | ||
| ACE2-D | 170 | 158 | 1.58 (1.11–2.27) | 1.21 (1.05–1.41) | 0.010 | S |
| Over dominant | ||||||
| ACE2-II + DD | 90 | 73 | 1 (ref.) | 1 (ref.) | ||
| ACE2-ID | 50 | 44 | 1.08 (0.65–1.80) | 1.03 (0.82–1.31) | 0.75 | NS |
3.5. Association of ACE2-rs4646994 I/D Genotypes with Comorbid Conditions by COVID-19 Severity
The statistical comparisons (p-values) of ACE2-rs4646994 I/D genotypes with comorbid conditions by COVID-19 severity was done by using a multivariate analysis based on logistic regression like odds ratio (OD) and risk ratio (RR) with 95% confidence intervals (CI) (Table 5). Results showed that the there was a significant correlation between the ACE2-rs4646994 I/D genotypes with respect to the age of the COVID-19 patients (p < 0.035), COVID-19 patients having T2D (p < 0.0013), COVID-19 patients having hypertension (p < 0.0031), COVID-19 patients having coronary artery disease (p < 0.0001) (Table 5). Similarly, a significant correlation was reported between ACE2 rs4240157 genotypes and COVID-19 cases whose oxygen saturation was less than <60 mm Hg. (p < 0.0001). A significant correlation was reported for ACE2 rs4240157 genotype distribution with respect to duration of COVID-19 cases in the hospital (p < 0.044). Similarly, in terms of treatment, a significant correlation was reported between antiviral therapy with respect to and ACE2D/I genotype (Table 5).
Table 5.
Statistical comparisons (p-values) of ACE2-rs4646994 I/D genotypes with comorbid conditions and some clinical parameters with COVID-19 disease severity. T2D—type2 diabetes; CKD—chronic kidney disease; CAD—coronary artery disease; SpO2—partial pressure of oxygen; ALT—alanine transaminase; CRP—C reactive protein; AST—aspartate transaminase.
| Clinical Feature | Variable | N = 117 | II | DI | DD | fD | χ2 | p |
|---|---|---|---|---|---|---|---|---|
| Age | >40 | 97 | 10 | 40 | 47 | 2 | 6.7 | 0.035 |
| <40 | 20 | 6 | 04 | 10 | ||||
| Gender | Males | 85 | 12 | 30 | 43 | 2 | 0.71 | 0.70 |
| Females | 32 | 04 | 14 | 14 | ||||
| T2D | Yes | 47 | 5 | 27 | 15 | 2 | 13.3 | 0.0013 |
| No | 70 | 11 | 17 | 42 | ||||
| CKD | Yes | 11 | 3 | 2 | 6 | 2 | 2.94 | 0.222 |
| No | 106 | 13 | 42 | 51 | ||||
| Hypertension | Yes | 37 | 10 | 17 | 10 | 2 | 13.28 | 0.0031 |
| No | 80 | 06 | 27 | 47 | ||||
| CAD | Yes | 17 | 10 | 02 | 05 | 2 | 34.22 | 0.0001 |
| No | 100 | 06 | 42 | 52 | ||||
| Oxygen saturation [SpO2, mm Hg] | <60 | 47 | 06 | 05 | 36 | 2 | 27.1 | 0.0001 |
| >80 | 70 | 10 | 39 | 21 | ||||
| Duration in hospital | >30 | 57 | 10 | 15 | 32 | 2 | 6.24 | 0.044 |
| <30 | 60 | 06 | 29 | 25 | ||||
| ALT | <36 | 72 | 10 | 24 | 38 | 2 | 1.55 | 0.46 |
| >36 | 45 | 06 | 20 | 19 | ||||
| CRP | <0.8 | 13 | 0 | 4 | 9 | 2 | 3.44 | 0.17 |
| >0.8 high | 104 | 16 | 40 | 48 | ||||
| AST | <40 | 69 | 10 | 19 | 40 | 2 | 7.57 | 0.022 |
| >40 | 48 | 06 | 25 | 17 | ||||
| Antiviral therapy | Yes | 79 | 08 | 21 | 50 | 2 | 20.1 | 0.0001 |
| No | 38 | 08 | 23 | 07 | ||||
| Steroids therapy | Yes | 77 | 11 | 29 | 37 | 2 | 0.08 | 0.96 |
| No | 40 | 05 | 15 | 20 | ||||
| Survival | Death occurred | 43 | 11 | 20 | 12 | 2 | 13.23 | 0.0007 |
| Survived | 74 | 05 | 24 | 45 |
3.6. Correlation of ACE2 I/D Genotypes with Age of COVID-19 Disease Severity
As can be seen in Table 6, our result indicated that in the codominant model, the ACE2-ID heterozygosity genotype (II vs. DI) was strongly associated with increased COVID-19 severity in subjects with age >40 with OR 6.0 (95%) CI = (1.41 to 25.38), RR = 4.12 (1.33 to 12.74), p < 0.014. Similarly, ACE2-DD genotype (II vs. DD + DI) was strongly associated with increased COVID-19 severity in subjects with age >40 with OR 3.72 (95%) CI = (1.17 to 11.88), RR = 2.70 (1.21–6.0), p < 0.026. In allelic comparison, the D allele was strongly associated with COVID-19 severity with an OR 1.58 (95% CI) (1.11–2.27), RR 1.21 (1.05–1.41) p-value = 0.010 (Table 6).
Table 6.
Statistical comparisons (p-values) of ACE2-rs4646994 I/D genotypes with age of COVID-19 severity.
| Age | <40 | >40 | Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-Value |
|---|---|---|---|---|---|
| N = 20 | N = 97 | ||||
| Codominant inheritance model | |||||
| II | 06 | 10 | Ref | Ref | |
| DI | 04 | 40 | 6.0 (1.41 to 25.38) | 4.12 (1.33 to 12.74) | 0.0149 |
| DD | 10 | 47 | 2.82 (0.83 to 9.55) | 2.13 (0.91 to 4.98) | 0.09 |
| Dominant inheritance model | |||||
| II | 06 | 10 | Ref | Ref | |
| DD + DI | 14 | 87 | 3.72 (1.17 to 11.88) | 2.70 (1.21 to 6.0) | 0.026 |
| Recessive inheritance model | |||||
| II + DI | 10 | 50 | Ref | Ref | |
| DD | 10 | 47 | 0.94 (0.35 to 2.46) | 0.95 (0.42 to 2.11) | 0.89 |
| Over dominant inheritance model | |||||
| II + DD | 16 | 57 | Ref | Ref | |
| DI | 04 | 40 | 2.80 (0.87 to 9.025) | 2.41 (0.86 to 6.75) | 0.08 |
3.7. Correlation of ACE2 I/D Genotypes with Mortality of COVID-19 Patients
The results are summarized in Table 7 and indicate that in the codominant model, ACE2-DD genotype (II vs. DD) was strongly associated with increased COVID-19 mortality OR 8.25 (95%) CI = (2.40 to 28.34), RR = 3.26 (1.7894 to 5.9596), p < 0.008. Also, in dominant model, ACE2-DD + DI genotype (II vs. DD + DI) was strongly associated with increased COVID-19 mortality with OR 4.74 (95%) CI = (1.5214 to 14.7915), RR = 2.16 (1.4014 to 3.3599), p < 0.007. Similarly, in recessive inheritance model, ACE2-DI + II genotype (DD vs. DI + II) was strongly associated with increased COVID-19 mortality with OR 4.0 (95%) CI = (1.7769 to 9.0432), RR = 2.45 (1.4031 to 4.2927), p < 0.0008.
Table 7.
Statistical comparisons (p-values) of ACE2-rs4646994 I/D genotypes with mortality of COVID-19 patients.
| N = 43 | N = 74 | Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-Value | |
|---|---|---|---|---|---|
| Codominant inheritance model | |||||
| II | 11 | 05 | Ref | Ref | |
| DI | 20 | 24 | 2.64 (0.7854 to 8.8737) | 1.51 (0.9524 to 2.4019) | 0.11 |
| DD | 12 | 45 | 8.25 (2.40 to 28.34) | 3.26 (1.7894 to 5.9596) | 0.008 |
| Dominant inheritance model | |||||
| II | 11 | 05 | Ref | Ref | |
| DD + DI | 32 | 69 | 4.74 (1.5214 to 14.7915) | 2.16 (1.4014 to 3.3599) | 0.007 |
| Recessive inheritance model | |||||
| II + DI | 31 | 29 | Ref | Ref | |
| DD | 12 | 45 | 4.0 (1.7769 to 9.0432) | 2.45 (1.4031 to 4.2927) | 0.0008 |
| Overdominant inheritance model | |||||
| II + DD | 23 | 50 | Ref | Ref | |
| DI | 20 | 24 | 0.55 (0.2551 to 1.1946) | 0.69 (0.4340 to 1.1070) | 0.13 |
3.8. Statistical Comparisons between Cases and Controls (p-Values) for ACE2 rs4240157 C > T Genotypes
The genotype distributions and allele frequencies of the ACE2 rs4240157 T > C showed that no deviation was detected in HWE (all p > 0.05) (χ2 = 3.38 p ≤ 0.06) in the controls. Thus, we chose 10% samples from normal control group randomly to review genotyping results, showing that the accuracy rate were more than 99%. The frequency of ACE2 rs4240157 T > C gene variation in COVID-19 cases and controls was TT (35%), TC (40.17%) and CC (24.78%) and controls TT (59%), TC (31%) and CC (10%) respectively (Table 8). The ACE2 rs4240157 T > C gene variation observed between COVID-19 patients and controls was statistically significant (p < 0.0007). Moreover, the frequency of C allele was found to be higher among COVID-19 patients than in HC (0.45 vs. 0.26).
Table 8.
ACE2 rs4240157 T > C gene polymorphism in COVID-19 patients and controls.
| Subjects | N | TT% | CT% | CC% | fD | χ2 | T | C | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| Cases | 117 | 41 (35) | 47 (40.17) | 29 (24.78) | 2 | 14.34 | 0.55 | 0.45 | 0.0007 |
| Controls | 100 | 59 (59) | 31 (31) | 10 (10) | 0.74 | 0.26 |
3.9. Multivariate Analysis to Estimate the Association of ACE2-rs4646994 I/D Genotypes with COVID-19 Severity
A multivariate analysis based on logistic regression like odds ratio (OD) and risk ratio (RR) with 95% confidence intervals (CI) were calculated for each group to estimate the association of ACE2 rs4240157 T > C genotypes with COVID-19 severity (Table 9). In the co-dominant inheritance model, ACE2–CT genotype was associated with COVID-19 severity with an OR 2.18 (95% CI) (1.92–3.99), RR 1.48 (1.07–2.04) p < 0.010 as well as ACE2–CC genotype was linked with COVID-19 severity with an OR 2.66 (95% CI) (1.53–4.62), RR 1.68 (1.25–2.26) p < 0.005. (Table 9). In case of dominant inheritance model, ACE2-(CT + CC) genotype was associated with COVID-19 severity with an OR 2.66 (95% CI) (1.53–4.62), RR 1.68 (1.25–2.26) p < 0.005. In case of recessive inheritance model, ACE2–CC vs. ACE2-(TT + CT) genotype was associated with covid-19 severity with an OR 2.96 (95% CI) (1.36–6.44), RR 1.97 (1.13–3.43) p < 0.006. In allelic comparison, the T allele was compared with the C allele. There was a no significant association of C allele with the COVID-19 severity with OR 0.86 (0.59–1.27), RR 0.92 (0.75–1.13), p-value = 0.46.
Table 9.
Multivariate analysis of ACE2 rs4240157 T > C gene polymorphism with COVID-19 severity.
| Genotypes | Healthy Controls | COVID-19 Cases | OR (95% CI) | Risk Ratio (RR) | p-Value |
|---|---|---|---|---|---|
| (N = 100) | (N = 117) | ||||
| Codominant | |||||
| ACE2-TT | 59 | 41 | 1 (ref.) | 1 (ref.) | |
| ACE2-CT | 31 | 47 | 2.18 (1.92–3.99) | 1.48(1.07–2.04) | 0.01 |
| ACE2-CC | 10 | 29 | 2.66 (1.53–4.62) | 1.68(1.25–2.26) | 0.005 |
| Dominant | |||||
| ACE2-TT | 59 | 41 | 1 (ref.) | 1 (ref.) | |
| ACE2-(CT + CC) | 41 | 76 | 2.66 (1.53–4.62) | 1.68(1.25–2.26) | 0.005 |
| Recessive | |||||
| ACE2-(TT + CT) | 90 | 88 | 1 (ref.) | 1 (ref.) | |
| ACE2-CC | 10 | 29 | 2.96(1.36–6.44) | 1.97 (1.13–3.43) | 0.006 |
| Allele | |||||
| ACE2-T | 100 | 121 | 1 (ref.) | 1 (ref.) | |
| ACE2-C | 100 | 105 | 0.86 (0.59–1.27) | 0.92 (0.75–1.13) | 0.46 |
3.10. Association of ACE2 rs4240157 T > C Genotypes with Gender and Comorbid Conditions by COVID-19 Severity
The statistical comparisons (p-values) of ACE2 rs4240157 T > C genotypes with comorbid conditions by COVID-19 severity was done by using a multivariate analysis based on logistic regression like odds ratio (OD) and risk ratio (RR) with 95% confidence intervals (CI) (Table 10). Results showed that there was a significant correlation between the ACE2 rs4240157 T > C genotypes with respect to the gender of the COVID-19 patients (p < 0.046). A significant correlation was reported between the ACE2 rs4240157 T > C genotypes and COVID-19 severity having T2D (p < 0.007), hypertension (0.006) and coronary artery disease (p < 0.049). Similarly, a significant correlation was reported between ACE2 rs4240157 T > C genotypes and COVID-19 cases whose oxygen saturation was less than <60 mm Hg. (p < 0.0001). A significant correlation was reported between steroid therapy and ACE2 rs4240157 T > C genotypes in COVID-19 patients. A significant correlation was also reported between ACE2 rs4240157 T > C genotypes in COVID-19 patients with hypertension (p < 0.006) as shown in Table 10.
Table 10.
Statistical comparisons (p-values) of ACE2 rs4240157 T > C genotypes with comorbid conditions and some clinical parameters with COVID-19 severity. T2D—type2 diabetes; CKD—chronic kidney disease; CAD—coronary artery disease; SpO2—partial pressure of oxygen; ALT—alanine transaminase; CRP—C reactive protein; AST—aspartate transaminase.
| Clinical Feature | Variable | N | TT | CT | CC | fD | χ2 | p |
|---|---|---|---|---|---|---|---|---|
| Age | <45 | 20 | 07 | 07 | 06 | 2 | 0.43 | 0.80 |
| >45 | 97 | 34 | 40 | 23 | ||||
| Gender | Males | 85 | 26 | 33 | 26 | 2 | 6.12 | 0.046 |
| Females | 32 | 15 | 14 | 03 | ||||
| T2D | Yes | 47 | 11 | 27 | 09 | 2 | 9.88 | 0.007 |
| No | 70 | 30 | 20 | 20 | ||||
| CKD | Yes | 11 | 04 | 04 | 03 | 2 | 0.08 | 0.960 |
| No | 106 | 37 | 43 | 26 | ||||
| Hypertension | Yes | 37 | 14 | 16 | 07 | 2 | 1 | 0.006 |
| No | 80 | 27 | 31 | 22 | ||||
| CAD | Yes | 17 | 05 | 11 | 01 | 2 | 6.03 | 0.049 |
| No | 100 | 36 | 36 | 28 | ||||
| Oxygen saturation | <60 mm Hg | 47 | 14 | 13 | 20 | 2 | 13.34 | 0.001 |
| >80 mm Hg | 70 | 33 | 28 | 09 | ||||
| Duration in hospital | >30 days | 57 | 17 | 26 | 14 | 2 | 1.69 | 0.420 |
| <30 days | 60 | 24 | 21 | 15 | ||||
| ALT | >36 units | 45 | 19 | 21 | 05 | 2 | 7.36 | 0.025 |
| <36 units | 72 | 22 | 26 | 24 | ||||
| CRP | >0.8 mg | 104 | 38 | 43 | 23 | 2 | 3.61 | 0.16 |
| <0.8 mg | 13 | 03 | 04 | 06 | ||||
| AST | >40 | 48 | 20 | 19 | 09 | 2 | 2.22 | 0.32 |
| <40 | 69 | 21 | 28 | 20 | ||||
| Antiviral therapy | Yes | 79 | 27 | 33 | 19 | 2 | 0.26 | 0.87 |
| No | 38 | 14 | 14 | 10 | ||||
| Steroids therapy | Yes | 77 | 31 | 23 | 23 | 2 | 10.23 | 0.006 |
| No | 40 | 10 | 24 | 06 | ||||
| Survival | Death occurred | 43 | 14 | 10 | 19 | 2 | 15.29 | 0.0005 |
| Survived | 74 | 27 | 37 | 10 |
3.11. Correlation of ACE2 rs4240157 T > C Genotypes with Mortality of COVID-19 Patients
Our results indicated that in the codominant model, the ACE2-CC genotype (TT vs. CC) was strongly associated with increased COVID-19 mortality OR 3.66 (95%) CI = (1.34 to 9.97), RR = 1.90 (1.10–3.30), p < 0.011 (Table 11). Similarly, in the recessive inheritance model, ACE2-(TT + CT) genotype was strongly associated with increased COVID-19 mortality with OR 5.0 (95%) CI = (2.0640–12.437), RR = 2.10 (1.2567 to 3.53), p < 0.004. In allelic comparison, the T allele was compared with the C allele. There was a significant association of C allele with the COVID-19 severity with OR 2, 01 (1.1761–3.45), RR 1.29 (1.05–1.59), p < 0.010 (Table 11).
Table 11.
Statistical comparisons (p-values) of ACE2 rs4240157 T > C genotypes with mortality of COVID-19 patients.
| Codominant Model | N = 74 | N = 43 | Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-Value |
|---|---|---|---|---|---|
| TT | 27 | 14 | Ref | Ref | |
| CT | 37 | 10 | 0.52 (0.20 to 1.34) | 0.83 (0.64 to 1.09) | 0.17 |
| CC | 10 | 19 | 3.66 (1.34 to 9.97) | 1.90 (1.10–3.30) | 0.011 |
| Dominant model | |||||
| TT | 27 | 14 | Ref | Ref | |
| CC + CT | 47 | 29 | 1.19 (0.5378 to 2.6332) | 1.06 (0.8028 to 1.4124) | 0.66 |
| Recessive model | |||||
| TT + CT | 64 | 24 | Ref | Ref | |
| CC | 10 | 19 | 5.06 (2.0640–12.437) | 2.10 (1.2567 to 3.53) | 0.004 |
| Allele | |||||
| T | 91 | 38 | Ref | Ref | |
| C | 57 | 48 | 2.01 (1.1761–3.45) | 1.29 (1.05 to 1.59) | 0.010 |
Table 12 summarizes the comparison of ACE2 rs4240157 T > C genotypes with COVID-19 comorbidities. As can be seen, CT genotype is highly significant in T2D OR 3.68 (95% CI 1.495 to 9.064), and RR 1.71 (1.175 to 2.515) in codominant model. In the dominant model CC + CT genotype is also significant in T2D with OR 2.45 (95% CI 1.076 to 5.599) and RR 1.39 (1.048 to 1.844). CT genotype is also very significant with reference to age with p-value of 0.014, OR 3.36 (95% CI 1.268 to 8.908) and RR 2.48 (1.155 to 5.353).
Table 12.
Statistical comparisons (p-values) of ACE2 rs4240157 T > C genotypes in COVID-19 patients with various comorbidities.
| Type 2 Diabetes | |||||
|---|---|---|---|---|---|
| Codominant Model | N = 70 | N = 47 | Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-Value |
| TT | 30 | 11 | Ref | Ref | |
| CT | 20 | 27 | 3.68 (1.495 to 9.064) | 1.71 (1.175 to 2.515) | 0.004 |
| CC | 20 | 09 | 1.22 (0.430 to 3.496) | 1.06 (0.780 to 1.441) | 0.070 |
| Dominant model | |||||
| TT | 30 | 11 | Ref | Ref | |
| CC + CT | 40 | 36 | 2.45 (1.076 to 5.599) | 1.39 (1.048 to 1.844) | 0.032 |
| Allele | |||||
| T | 70 | 47 | Ref | Ref | |
| C | 60 | 45 | 1.11 (0.654 to 1.906) | 1.04 (0.838 to 1.307) | 0.068 |
| Hypertension | |||||
| Codominant model | N = 80 | N = 37 | Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-value |
| TT | 27 | 14 | Ref | Ref | |
| CT | 31 | 16 | 0.99 (0.4114 to 2.4084) | 0.99 (0.7387 to 1.3495) | 0.99 |
| CC | 22 | 07 | 0.61 (0.2110 to 1.7850) | 0.86 (0.642 to 1.173) | 0.370 |
| Dominant model | |||||
| TT | 27 | 14 | Ref | Ref | |
| CC + CT | 53 | 23 | 0.83 (0.3723 to 1.8814) | 0.94 (0.7241 to 1.2315) | 0.67 |
| Coronary artery disease | |||||
| Codominant model | N = 100 | N = 17 | Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-value |
| TT | 36 | 05 | Ref | Ref | |
| CT | 36 | 11 | 2.20 (0.694 to 6.973) | 1.14 (0.943 to 1.393) | 0.180 |
| CC | 28 | 1 | |||
| Dominant model | |||||
| TT | 36 | 05 | Ref | Ref | |
| CC + CT | 64 | 12 | 0.25 (0.028 to 2.328) | 0.90 (0.796 to 1.039) | 0.22 |
| Age | |||||
| Codominant model | <45 N = 20 |
>45 N = 97 |
Odd Ratio (95% CI) | Risk Ratio (95% CI) | p-value |
| TT | 20 | 34 | Ref | Ref | |
| CT | 07 | 40 | 3.36 (1.268 to 8.908) | 2.48 (1.155 to 5.353) | 0.014 |
4. Discussion
Coronaviruses are divided into α, β, γ, and δ genera on the basis of the target host [1,2,3]. Out of these, mammals are infected by α and β-CoV whereas γ and δ-CoV genera tend to infect birds. The current COVID-19 pandemic is found to be caused by SARS-CoV-2 [3,4]. It is a β-CoV that is enveloped, non-segmented and positive-sense RNA virus. Genome sequencing results showed that this newly discovered virus shares 96.2% identity with bat CoV RaTG13 and 79.5% identity with SARS-CoV [10].
4.1. Role of RAAS in SARS-CoV-2 Infection
Different ACE genotypes are believed to be associated with development of acute respiratory distress syndrome and several studies have reported that the distribution frequency of ACE insertion/insertion (II) genotype might have a significant influence on COVID-19 mortality [36,40]. It has been reported that ACE2 levels correlate with susceptibility to SARS-CoV-2 infection and men have a higher ACE2 expression in lung than women and Asian people express ACE2 higher than Caucasian and African American populations [41,42].
4.2. Association of ACE2-D/I with Gender
Angiotensin converting enzyme is a metallopeptidase that converts angiotensin I (AT-I) to angiotensin II (AT-II) that acts as a vasoconstrictor in addition to its other functions. ACE also degrades bradykinin, a vasodilator substance. ACE2 is also a known SARS virus receptor [14]. Both ACE1 and ACE2 are highly polymorphic. Polymorphisms present in ACE1 are just as important as those in ACE2. As it was reported that the conversion of Ang II to Ang (1–7) by ACE2 is higher in males than females [43] because ACE2 gene is located on the X chromosome and men express more ACE2 than women that depends on the allelic expressions by women, therefore women may be considered of lower sensitivity against the SARS-CoV-2 infection and its lethal effects [43,44].
Several research reports in mice have shown the protective effect of ACE2, whereby severe lung failure is associated with ACE2 downregulation. Immediately after SARS-CoV-2 infection, there is a downregulation of ACE2 [37,38,45]. ACE gene on chromosome 17 consists of 26 exons and 25 introns. A functional insertion-deletion (I/D) polymorphism of 287 bp Alu repeat sequence has been reported in the intron 16 of the ACE gene [44,46]. Jeong et al. have reported that the deletion (D) and insertion allele (I) are associated with an increased and decreased ACE level and enzyme activity, respectively [47]. The ACE2 gene exhibits a high degree of genetic polymorphism [42]. Itoyama et al. [48] reported the correlation of ACE-D allele with the incidence of pneumonia in SARS patients and the death of subjects with acute respiratory distress syndrome. These results tempted us to investigate the role of ACE2 I/D polymorphism in susceptibility to SARS-CoV-2 infection and related mortality. Our results indicated higher overall frequencies of deletion polymorphism in COVID-19 patients (67%) than the insertion polymorphism (33%). The distribution of homozygote insertion (II) genotype is seen to be higher in the healthy population (26.66%) compared with the COVID-19 patients (13.67%). The frequency of heterozygote ID genotype is comparable between COVID-19 patients (37.60%) and control (33.34%) groups. We observed a significant difference in the distribution of homozygote deletion (DD) genotype between COVID-19 cases and controls (p < 0.03) (Table 3).
4.3. Distribution of ACE2-II, DI and DD Genotypes in the World Populations
The distribution of ACE2-II, DI and DD genotypes in the SARS-CoV-2 patients in the world populations has been summarized in the Table 13 [49]. The frequency of DD genotype in our patient population was high (48.71%) corresponding to the frequency of Brazil (49%), Finland (42.61%), Turkey (34.44%), Iran (37.50%), Spain (37.84%), Russia (39.38%), Italy (37.61%).
Table 13.
Distribution of ACE2- II, DI and DD genotypes in the SARS-CoV-2 patients in the world populations.
| Country | Sample Size (N) | DD Genotype | % DD | ID Genotype | %ID | II Genotype | %II |
|---|---|---|---|---|---|---|---|
| USA | 4320 | 1331 | 30.81 | 2145 | 49.65 | 844 | 19.54 |
| UK | 10566 | 2896 | 27.41 | 5241 | 49.60 | 2429 | 22.99 |
| Canada | 299 | 85 | 28.43 | 149 | 49.83 | 65 | 21.74 |
| Turkey | 672 | 238 | 35.42 | 303 | 45.09 | 131 | 19.49 |
| Australia | 444 | 134 | 30.18 | 216 | 48.65 | 94 | 21.17 |
| Austria | 2251 | 619 | 27.50 | 1108 | 49.22 | 524 | 23.28 |
| Belgium | 3980 | 1074 | 26.98 | 2030 | 51.01 | 876 | 22.01 |
| India | 660 | 130 | 19.70 | 316 | 47.88 | 214 | 32.42 |
| China | 3764 | 1154 | 30.66 | 1228 | 32.62 | 1382 | 36.72 |
| Czech | 443 | 116 | 26.19 | 225 | 50.79 | 102 | 23.02 |
| Germany | 5047 | 1345 | 26.65 | 2581 | 51.14 | 1121 | 22.21 |
| Japan | 379 | 68 | 17.94 | 185 | 48.81 | 126 | 33.25 |
| Netherlands | 6110 | 1705 | 27.91 | 3056 | 50.02 | 1349 | 22.08 |
| South-Africa | 300 | 46 | 15.33 | 158 | 52.67 | 96 | 32.00 |
| Israel | 87 | 9 | 10.34 | 50 | 57.47 | 28 | 32.18 |
| Korea | 785 | 256 | 32.61 | 294 | 37.45 | 235 | 29.94 |
| Denmark | 126 | 41 | 32.54 | 43 | 34.13 | 42 | 33.33 |
| Italy | 3151 | 1185 | 37.61 | 1358 | 43.10 | 608 | 19.30 |
| Russia | 320 | 126 | 39.38 | 125 | 39.06 | 69 | 21.56 |
| Spain | 1665 | 630 | 37.84 | 767 | 46.07 | 268 | 16.10 |
| Iran | 200 | 75 | 37.50 | 96 | 48.00 | 29 | 14.50 |
| Turkey | 90 | 31 | 34.44 | 14 | 15.55 | 45 | 50 |
| Finland | 115 | 49 | 42.61 | 52 | 45.22 | 14 | 12.17 |
| Brazil | 104 | 51 | 49.04 | 33 | 31.73 | 20 | 19.23 |
| Saudi Arabia [current study] |
117 | 57 | 48.71 | 44 | 37.60 | 16 | 13.67 |
Lee et al., reported that populations in France, Italy and Spain have shown high D allele frequency between 82% and 87% [50]. Similarly, African Americans in the United States have been reported to have the highest D allele frequency (89%) compared with white Americans (69%) [33]. Conversely, populations in East Asian countries such as China, Japan, Taiwan and Korea have a high frequency of the II genotype [51]. Pati et al. [52] reported that the low DD genotype frequency and high II genotype frequency in the ACE gene is strongly correlated with the relatively low mortality rate of COVID-19 among these populations [52]. A higher mortality rate has been shown among the European populations as in the case of black ethnicity in the United States [34]. In the present study, we observed a positive correlation of allele D with infection and mortality rate of COVID-19.
Recent studies have also shown that the ethnic variations of the ACE I/D genotype tend to correlate with the variations in outcomes where populations with a high D genotype frequency tend to experience higher mortality rates [23]. Our results indicated that in the codominant model, the ACE2-DD genotype was strongly associated with increased COVID-19 mortality OR 8.25 (95%) CI = (2.40 to 28.34), p < 0.008 (Table 7). Mortality of SARS-CoV-2 subjects is dependent on a wide range of complex phenomena and genetic mutations are one among them [53,54,55,56]. The prevalence of the DD genotype is higher in patients with severe lung infections and is significantly correlated with a high death rate. Mortality related to SARS-CoV-2 infections has been associated to various comorbid conditions such as hypertension, diabetes, hyperlipidemia, coronary artery disease and renal disease [23,57,58,59,60,61,62,63,64]. High producer of ACE, the DD genotype has been associated with susceptibility to hypertension. [34,35,55,62], Type 2 diabetes [59,60], coronary artery disease [57,59] indicating a possible role of ACE I/D polymorphism with SARS-CoV-2 related mortality. Our results reported a significant correlation of ACE2-DD genotypes with advanced age of the COVID-19 patients (p < 0.035), with T2D (p < 0.0013), hypertension (p < 0.0031) and coronary artery disease (p < 0.0001) [Table 5]. Our study revealed that age is a significant risk factor for severe disease. Results showed that the there was a significant correlation between the ACE2-rs4646994 I and D genotypes with respect to the Age of the COVID-19 patients (p < 0.035). Advanced age is a significant risk factor for mortality from COVID-19, the important reason being the associated increased frequency of comorbid conditions with advancing age [61,63].
4.4. Role of ACE2 rs4240157 T > C Gene Polymorphism in COVID-19 Severity
Our results indicated a significant correlation of ACE2 rs4240157 T > C genotypes with the COVID-19 severity among the COVID-19 patients with T2D (p < 0.007), with coronary artery disease (p < 0.049) and hypertension (p < 0.006) (Table 10). Recent studies identified ACE2 polymorphisms that might influence disease severity and indicated that out of 10 studied SNPs, 5 polymorphisms (rs6632680, rs4830965, rs1476524, rs4240157 and rs2048683) indicated an association with higher tissue specific expression of ACE2 resulting in hospitalization whereas rs1548474 polymorphism showed correlation with low tissue expression and lesser severity [23,37,38,39,40]. Variation in circulating ACE2 levels was speculated to be controlled by genetic factors including rs2106809 polymorphism [39,63]. Xiao et al., [64] have earlier reported that a point mutation in the ACE2 gene (Leu584Ala) facilitates entry of SARS-CoV-1 into host cells. Recent studies have proven that several amino acid variants can potentially affect the interaction between the viral S1 protein and ACE2 receptors and thus the level of infection [65]. Different amino acid residues expressed within the ACE2 receptor were observed to be very relevant either by promoting or preventing viral infection. A total of 13 ACE2 polymorphisms enhanced ACE2/S1 recognition, thereby facilitating SARS-CoV-2 infection while as in contrast, 18 SNPs hindered interactions between ACE2 and S1, thereby reduces the infection rate [62]. Patel et al., [59] reported that ACE 2 rs2074192, rs4240157 and rs4646188 variations in T2D exhibited higher risk with hypertension among persons of Australian descent. ACE2 s4240157 (p < 0.001) and rs4646156 (p < 0.037) are correlated with increased blood pressure [64]. Our results reported higher frequency of heterozygosity among SARS-CoV-2 cases of ACE2-rs4240157 TC genotypes (40.17%) followed by TT genotypes (35%), and CC genotypes (24.78%) respectively [Table 8].
In our study, the ACE2 rs4240157 T > C gene polymorphism observed between COVID-19 patients and controls was statistically significant (p < 0.0007). Wooster et al., [55] reported that ACE2 rs4240157 polymorphisms is associated with COVID-19 disease severity as it might be inducing higher tissue specific expression of ACE2 resulting in the hospitalization of COVID-19 patients. Pouladi et al., [62] reported the association ACE2 rs4240157 T > C gene polymorphism with hypertension and other related heart diseases. Our results indicated ACE2-CT genotype association with COVID-19 severity with an OR 2.18 (95% CI) (1.92–3.99), p < 0.010 and also ACE2-CC genotype was linked with COVID-19 severity with an OR 2.66 (95% CI) (1.53–4.62), p < 0.005 (Table 4). We observed that ACE2-CC genotype was associated with increased COVID-19 mortality with OR 3.66 (95%) CI = (1.34 to 9.97), p < 0.011 (Table 10). Similarly, in recessive inheritance model, ACE2-(TT + CT) genotype was associated with increased COVID-19 mortality with OR 5.0 (95%) CI = (2.0640–12.437), p < 0.004.
Despite the relatively small sample size, our results indicate that there is a close association between the ACE I/D gene polymorphism with clinical severity of COVID-19 disease. In addition, advanced age, coronary artery disease and diabetes were independent risk factors for the development of severe COVID-19 disease and mortality. The ACE-II genotype was a protective factor against the development of severe COVID-19 disease. Since this study is a first of its kind from Saudi Arabia, we believe that the relation of ACE2 DD genotype with the disease severity and clinical outcome in COVID-19 patients should be further investigated with more extensive studies. However, none of the studied SNPs are common in the general population. This finding might help in the recognition of people less and more prone to COVID-19.
5. Conclusions
It is concluded that ACE-DD genotype and D allele was strongly associated with increased COVID-19 disease severity. In addition, ACE I/D polymorphisms were strongly associated with advanced age, diabetes and coronary artery disease in COVID-19 patients whereas ACE-II genotype was a protective factor against the development of severe COVID-19. ACE2-DD genotype was strongly associated with increased SARS-CoV-2 mortality. Additionally, ACE2-CC and CT genotypes were strongly associated with SARS-CoV-2 severity. Therefore, our study might be useful for identifying the susceptible population groups for targeted therapeutic interventions and for making relevant public health policy decisions. The limitations of the study include a relatively smaller sample size of 117 patients and the fact that COVID-19 patients without symptoms were not included in the study.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at University of Bisha Saudi Arabia for funding this work through “COVID-19 Initiative Project’” under Grant Number (UB-COVID-02-1441). The authors highly appreciate the COVID-19 patients and healthy controls for their participation in this study. The facilities extended by universities of Bisha, Tabuk and King Khalid, Abha, KSA are gratefully acknowledged. Special thanks to Mohammad Jeelani and Rabia Farooq for their technical assistance.
Author Contributions
Conceptualization, M.M.M., R.M., B.A.A. and M.A.A.A.; Data Curation, R.M., M.M.M., B.A.A., J.I.W.; Formal Analysis, M.M.M., R.M., M.A.A.A. and A.M.A.-S.; Investigation, M.M.M., R.M., B.A.A. and J.I.W.; Methodology, M.M.M., R.M. and M.A.A.A.; Project Administration, M.M.M. and M.A.A.A.; Resources, M.M.M., R.M., B.A.A., J.I.W., M.H.A. and M.A.A.A.; Validation, M.M.M., R.M. and M.A.A.A.; Visualization, M.M.M., B.A.A., M.A.A.A. and A.M.A.-S.; Writing—Original Draft, M.M.M. and R.M.; Writing—Review and Editing, M.M.M., R.M., B.A.A., J.I.W., M.A.A.A., M.H.A. and A.M.A.-S. All authors have read and agreed to the published version of the manuscript.
Funding
COVID-19 initiative project from Deanship of Scientific Research at University of Bisha Saudi Arabia.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Ethics Committees of College of Medicine, University of Bisha, Bisha (Ref No. UBCOM/H-06-BH-087(05/25) Dated: 21/12/2020, University of Tabuk, Tabuk (Ref. No. UT-128-25-2020, Dated: 07/11/2020) and King Khalid University, Abha (Ref No. KKU-H-06-B-091, Dated: 12/03/2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajgain K.T., Badal S., Bajgain B.B., Santana M.J. Prevalence of comorbidities among individuals with COVID-19: A rapid review of current literature. Am. J. Infect. Control. 2020;49:238–246. doi: 10.1016/j.ajic.2020.06.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasmi A., Peana M., Pivina L., Srinath S., Benahmed A.G., Semenova Y., Menzel A., Dadar M., Bjørklund G. Interrelations between COVID-19 and other disorders. Clin. Immunol. 2021;224:108651. doi: 10.1016/j.clim.2020.108651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poorolajal J. Geographical Distribution of COVID-19 Cases and Deaths Worldwide. J. Res. Heal. Sci. 2020;20:e00483. doi: 10.34172/jrhs.2020.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC COVID-19 Response Team. Bialek S., Bowen V., Chow N., Curns A., Gierke R., Hall A., Hughes M., Pilishvili T., Rithey M., et al. Geographic Differences in COVID-19 Cases, Deaths, and Incidence—United States, February 12–April 7, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:465–471. doi: 10.15585/mmwr.mm6915e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorci G., Faivre B., Morand S. Explaining among-country variation in COVID-19 case fatality rate. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-75848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss S.R., Navas-Martin S. Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadi-Alijanvand H., Rouhani M. Studying the Effects of ACE2 Mutations on the Stability, Dynamics, and Dissociation Process of SARS-CoV-2 S1/hACE2 Complexes. J. Proteome Res. 2020;19:4609–4623. doi: 10.1021/acs.jproteome.0c00348. [DOI] [PubMed] [Google Scholar]
- 12.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-Dos-Santos A.J., Da Costa J., Zhang L., Pei Y., et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nat. Cell Biol. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 18.Keidar S., Kaplan M., Gamliel-Lazarovich A. ACE2 of the heart: From angiotensin I to angiotensin (1–7) Cardiovasc. Res. 2007;73:463–469. doi: 10.1016/j.cardiores.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Danilczyk U., Sarao R., Remy C., Benabbas C., Stange G., Richter A., Arya S., Pospisilik J.A., Singer D., Camargo S., et al. Essential role for collectrin in renal amino acid transport. Nat. Cell Biol. 2006;444:1088–1091. doi: 10.1038/nature05475. [DOI] [PubMed] [Google Scholar]
- 20.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 21.Pan Y., Wang T., Li Y., Guan T., Lai Y., Shen Y., Zeyaweiding A., Maimaiti T., Li F., Zhao H., et al. Association of ACE2 polymorphisms with susceptibility to essential hypertension and dyslipidemia in Xinjiang, China. Lipids Heal. Dis. 2018;17:1–9. doi: 10.1186/s12944-018-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu N., Yang Y., Wang Y., Liu Y., Fu G., Chen D., Dai H., Fan X., Hui R., Zheng Y. ACE2 gene polymorphism and essential hypertension: An updated meta-analysis involving 11,051 subjects. Mol. Biol. Rep. 2012;39:6581–6589. doi: 10.1007/s11033-012-1487-1. [DOI] [PubMed] [Google Scholar]
- 23.Magrone T., Magrone M., Jirillo E. Focus on Receptors for Coronaviruses with Special Reference to Angiotensin-Converting Enzyme 2 as a Potential Drug Target—A Perspective. Endocr. Metab. Immune Disord. Drug Targets. 2020;20:807–811. doi: 10.2174/1871530320666200427112902. [DOI] [PubMed] [Google Scholar]
- 24.Saab Y.B., Gard P.R., Overall A.D.J. The geographic distribution of the ACE II genotype: A novel finding. Genet. Res. 2007;89:259–267. doi: 10.1017/S0016672307009019. [DOI] [PubMed] [Google Scholar]
- 25.Singh H., Choudhari R., Nema V., Khan A.A. ACE2 and TMPRSS2 polymorphisms in various diseases with special reference to its impact on COVID-19 disease. Microb. Pathog. 2021;150:104621. doi: 10.1016/j.micpath.2020.104621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi L., Gu Y.H., Wang X.L., An L.Z., Xie X.D., Shao W., Ma L.Y., Fang J.R., An Y.D., Wang F., et al. Association of ACE, ACE2 and UTS2 Polymorphisms with Essential Hypertension in Han and Dongxiang Populations from North-western China. J. Int. Med Res. 2006;34:272–283. doi: 10.1177/147323000603400306. [DOI] [PubMed] [Google Scholar]
- 28.Niu W., Qi Y., Hou S., Zhou W., Qiu C. Correlation of angiotensin-converting enzyme 2 gene polymorphisms with stage 2 hypertension in Han Chinese. Transl. Res. 2007;150:374–380. doi: 10.1016/j.trsl.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Malard L., Kakinami L., O’Loughlin J., Roy-Gagnon M.H., Labbe A., Pilote L., Hamet P., Tremblay J., Paradis G. The association between the angioten-sin-converting enzyme-2 gene and blood pressure in a cohort study of adolescents. BMC Med. Genet. 2013;14:117. doi: 10.1186/1471-2350-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patnaik M., Pati P., Swain S.N., Mohapatra M.K., Dwibedi B., Kar S.K., Ranjit M. Association of angiotensin-converting enzyme and angiotensin-converting enzyme-2 gene polymorphisms withessential hypertension in the population of Odisha, India. Ann. Hum. Biol. 2014;41:145–152. doi: 10.3109/03014460.2013.837195. [DOI] [PubMed] [Google Scholar]
- 31.Pinheiro D.S., Santos R.D.S., Jardim P.C.B.V., Silva E.G., Reis A., Pedrino G.R., Ulhoa C.J. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: A genetic association study in Brazilian patients. PLoS ONE. 2019;14:e0221248. doi: 10.1371/journal.pone.0221248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H., Cao J.J. Angiotensin-Converting Enzyme Gene Polymorphism and Severe Lung Injury in Patients with Coronavirus Disease 2019. Am. J. Pathol. 2020;190:2013–2017. doi: 10.1016/j.ajpath.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yancy C.W. COVID-19 and African Americans. JAMA. 2020;323:1891. doi: 10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
- 34.Dyer O. COVID-19: Black people and other minorities are hardest hit in US. BMJ. 2020;369:m1483. doi: 10.1136/bmj.m1483. [DOI] [PubMed] [Google Scholar]
- 35.Sabir J.S., El Omri A., Khan I.A., Banaganapalli B., Hajrah N.H., Zrelli H., Omar A.M.S., Alharbi M.G., Alhebshi A.M., Jansen R.K., et al. ACE insertion/deletion genetic polymorphism, serum ACE levels and high dietary salt intake influence the risk of obesity development among the Saudi adult population. J. Renin-Angiotensin-Aldosterone Syst. 2019;20:1470320319870945. doi: 10.1177/1470320319870945. [DOI] [Google Scholar]
- 36.Aung A.K., Aitken T., Teh B.M., Yu C., Ofori-Asenso R., Chin K.L., Liew D. Angiotensin converting enzyme genotypes and mortality from COVID-19: An ecological study. J. Infect. 2020;81:961–965. doi: 10.1016/j.jinf.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall R.P., Webb S., Bellingan G.J., Montgomery H.E., Chaudhari B., McAnulty R.J., Humphries S.E., Hill M.R., Laurent G.J. An-giotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute res-piratory distress syndrome. Am. J. Respir. Crit. Care Med. 2002;166:646–650. doi: 10.1164/rccm.2108086. [DOI] [PubMed] [Google Scholar]
- 38.Gómez J., Albaiceta G.M., García-Clemente M., López-Larrea C., Amado-Rodríguez L., Lopez-Alonso I., Hermida T., Enriquez A.I., Herrero P., Melón S., et al. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. 2020;762:145102. doi: 10.1016/j.gene.2020.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khamlaoui W., Mehri S., Hammami S., Elosua R., Hammami M. Association of angiotensin-converting enzyme insertion/deletion (ACE I/D) and angiotensinogen (AGT M235T) polymorphisms with the risk of obesity in a Tunisian population. J. Renin-Angiotensin-Aldosterone Syst. 2020;21:1470320320907820. doi: 10.1177/1470320320907820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto N., Ariumi Y., Nishida N., Yamamoto R., Bauer G., Gojobori T., Shimotohno K., Mizokami M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944. doi: 10.1016/j.gene.2020.144944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun P., Lu X., Xu C., Sun W., Pan B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020;92:548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatami N., Ahi S., Sadeghinikoo A., Foroughian M., Javdani F., Kalani N., Fereydoni M., Keshavarz P. Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: An ecological meta-regression. Endocrine. 2020;68:479–484. doi: 10.1007/s12020-020-02381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gwathmey T.M., Shaltout H.A., Nixon P.A., O’Shea T.M., Rose J.C., Washburn L.K. Gender differences in urinary ACE and ACE2 activities in adolescents. FASEB J. 2008;22:940. doi: 10.1096/fasebj.22.1_supplement.940.6. [DOI] [Google Scholar]
- 44.Burrell L.M., Harrap S.B., Velkoska E., Patel S.K. The ACE2 gene: Its potential as a functional candidate for cardiovascular disease. Clin. Sci. (Lond.) 2013;124:65–76. doi: 10.1042/CS20120269. [DOI] [PubMed] [Google Scholar]
- 45.Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: Role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci. 2020;21:3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villard E., Soubrier F. Molecular biology and genetics of the angiotensin-I-converting enzyme: Potential implications in cardiovascular diseases. Cardiovasc. Res. 1996;32:999–1007. doi: 10.1016/S0008-6363(96)00170-8. [DOI] [PubMed] [Google Scholar]
- 47.Jeong K.-H., Lee T.-W., Ihm C.-G., Lee S.-H., Moon J.-Y. Polymorphisms in two genes, IL-1B and ACE, are associated with erythropoietin resistance in Korean patients on maintenance hemodialysis. Exp. Mol. Med. 2008;40:161–166. doi: 10.3858/emm.2008.40.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoyama S., Keicho N., Quy T., Phi N.C., Long H.T., Ha L.D., Van Ban V., Ohashi J., Hijikata M., Matsushita I., et al. ACE1 polymorphism and progression of SARS. Biochem. Biophys. Res. Commun. 2004;323:1124–1129. doi: 10.1016/j.bbrc.2004.08.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathew J., Basheeruddin K., Prabhakar S. Differences in frequency of the deletion polymorphism of the angiotensin-converting enzyme gene in different ethnic groups. Angiology. 2001;52:375–379. doi: 10.1177/000331970105200602. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y.-J., Tsai J.C. ACE gene insertion/deletion polymorphism associated with 1998 World Health Organization definition of metabolic syndrome in Chinese type 2 diabetic patients. Diabetes Care. 2002;25:1002–1008. doi: 10.2337/diacare.25.6.1002. [DOI] [PubMed] [Google Scholar]
- 51.Huang T., Shu Y., Cai Y.-D. Genetic differences among ethnic groups. BMC Genom. 2015;16:1093. doi: 10.1186/s12864-015-2328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pati A., Mahto H., Padhi S., Panda A.K. ACE deletion allele is associated with susceptibility to SARS-CoV-2 infection and mortality rate: An epidemiological study in the Asian population. Clin. Chim. Acta. 2020;510:455–458. doi: 10.1016/j.cca.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., Hosein Z., Padda I., Mangat J., Altaf M. Comorbidity and its Impact on Patients with COVID-19. SN Compr. Clin. Med. 2020:1–8. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnan R., Sekar D., Karunanithy S., Subramanium S. Association of angiotensin converting enzyme gene insertion/deletion polymorphism with essential hypertension in south Indian population. Genes Dis. 2016;3:159–163. doi: 10.1016/j.gendis.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wooster L., Nicholson C.J., Sigurslid H.H., Cardenas C.L.L., Malhotra R. Polymorphisms in the ACE2 locus associate with severity of COVID-19 infection. medRxiv. 2020 doi: 10.1101/2020.06.18.20135152. [DOI] [Google Scholar]
- 56.Feng Y., Niu T., Xu X., Chen C., Li Q., Qian R., Wang G., Xu X. Insertion/deletion polymorphism of the ACE gene is associated with type 2 diabetes. Diabetes. 2002;51:1986–1988. doi: 10.2337/diabetes.51.6.1986. [DOI] [PubMed] [Google Scholar]
- 57.Amara A., Mrad M., Sayeh A., Lahideb D., Layouni S., Haggui A., Fekih-Mrissa N., Haouala H., Nsiri B. The Effect of ACE I/D Polymorphisms Alone and With Concomitant Risk Factors on Coronary Artery Disease. Clin. Appl. Thromb. 2018;24:157–163. doi: 10.1177/1076029616679505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: Summary of a report of 72,314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 59.Patel S.K., Wai B., Ord M., MacIsaac R.J., Grant S., Velkoska E., Panagiotopoulos S., Jerums G., Srivastava P.M., Burrell L.M. Association of ACE2 genetic variants with blood pressure, left ventricular mass, and cardiac function in Caucasians with type 2 diabetes. Am. J. Hypertens. 2012;25:216–222. doi: 10.1038/ajh.2011.188. [DOI] [PubMed] [Google Scholar]
- 60.Liu C., Li Y., Guan T., Lai Y., Shen Y., Zeyaweiding A., Zhao H., Li F., Maimaiti T. ACE2 polymorphisms associated with cardiovascular risk in Uygurs with type 2 diabetes mellitus. Cardiovasc. Diabetol. 2018;17:1–11. doi: 10.1186/s12933-018-0771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cangiano B., Fatti L.M., Danesi L., Gazzano G., Croci M., Vitale G., Bonomi M. Mortality in an Italian nursing home during COVID-19 pandemic: Correlation with gender, age, ADL, vitamin D supplementation, and limitations of the diagnostic tests. Aging (Albany NY) 2020;12:24522. doi: 10.18632/aging.202307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pouladi N., Abdolahi S. Investigating the ACE2 polymorphisms in COVID-19 susceptibility: An in silico analysis. Mol. Genet. Genom. Med. 2021;9:e1672. doi: 10.1002/mgg3.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darbani B. The Expression and Polymorphism of Entry Machinery for COVID-19 in Human: Juxtaposing Population Groups, Gender, and Different Tissues. Int. J. Environ. Res. Public Heal. 2020;17:3433. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao F., Zimpelmann J., Agaybi S., Gurley S.B., Puente L., Burns K.D. Characterization of angiotensin-converting enzyme 2 ectodomain shedding from mouse proximal tubular cells. PLoS ONE. 2014;9:e85958. doi: 10.1371/journal.pone.0085958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. arXiv. 2020 doi: 10.1016/j.jmb.2020.07.009.2005.14669 [DOI] [PMC free article] [PubMed] [Google Scholar]