Abstract
Mammalian genes are characterized by relatively small exons surrounded by variable lengths of intronic sequence. Sequences similar to the splice signals that define the 5′ and 3′ boundaries of these exons are also present in abundance throughout the surrounding introns. What causes the real sites to be distinguished from the multitude of pseudosites in pre-mRNA is unclear. Much progress has been made in defining additional sequence elements that enhance the use of particular sites. Less work has been done on sequences that repress the use of particular splice sites. To find additional examples of sequences that inhibit splicing, we searched human genomic DNA libraries for sequences that would inhibit the inclusion of a constitutively spliced exon. Genetic selection experiments suggested that such sequences were common, and we subsequently tested randomly chosen restriction fragments of about 100 bp. When inserted into the central exon of a three-exon minigene, about one in three inhibited inclusion, revealing a high frequency of inhibitory elements in human DNA. In contrast, only 1 in 27 Escherichia coli DNA fragments was inhibitory. Several previously identified silencing elements derived from alternatively spliced exons functioned weakly in this constitutively spliced exon. In contrast, a high-affinity site for U2AF65 strongly inhibited exon inclusion. Together, our results suggest that splicing occurs in a background of repression and, since many of our inhibitors contain splice like signals, we suggest that repression of some pseudosites may occur through an inhibitory arrangement of these sites.
The removal of introns during pre-mRNA splicing represents a fundamental step in the transfer of information from DNA to protein. Introns are defined by at least three discrete elements: the 5′ splice site, the branch point, and the 3′ splice site. The spliceosome assembles around these three elements and excises introns with high fidelity in an ordered, stepwise process. In the yeast genes that contain introns, the almost perfect agreement of these elements with three consensus sequences appears sufficient to account for a high fidelity of splicing. Higher eukaryotes, on the other hand, have larger introns defined by more-degenerate splicing signals. In general, these large introns contain many close matches to the 5′ and 3′ splice site consensus sequences (54). These pseudosites often compare favorably to real splice sites yet are not used in the splicing reaction. Discovering what causes real sites to be used in favor of these sometimes stronger pseudosites is fundamental to our understanding of pre-mRNA processing.
Since consensus sequences for splice sites were first compiled (43), there has been much effort devoted to finding additional requirements that predict whether a site will be used. It has been proposed that a branch point, a 3′ splice site, and a 5′ splice site are recognized as a tripartite signal defining an exon rather than an intron (52). Several lines of experimental evidence support this exon definition model. (i) Downstream 5′ splice sites have a stimulatory effect on splicing at an upstream 3′ splice sites in single intron constructs (20, 52). (ii) The predominant phenotype of a mutation in a splice site of an internal exon is exon skipping rather than intron retention or the activation of a cryptic site (12, 36, 48). (iii) Mutations at a 5′ splice site that eliminate splicing can be suppressed by additional mutations that strengthen the upstream 3′ splice site across the exon (12).
The exon definition model implies a bridging activity across an exon, possibly leading to steric constraints on the size of an exon. Consistent with this idea, most human exons are between 50 and 200 nucleotides (nt) long (30, 67). Reducing exon size to <50 nt can impose a requirement for a splicing enhancer element (25, 32) or require the transient definition of a larger exon intermediate (59). However, a constraint based on maximum length is less clear (15, 29, 59, 61). A substantial minority of true exons fall outside this size range, and the close grouping of false splice sites within introns is such that many of them define pseudoexons that fall within the permissive size range (54). Thus, it seems unlikely that exon size could be a general factor in the distinction between true and false splice sites.
It has been known for some time that many alternatively spliced exons, small exons, or exons with weak splice sites rely upon the activity of enhancers for their inclusion in mRNA (65). These enhancers are often purine rich and in some cases have been shown to function by binding SR proteins (24, 39, 60). The SR proteins, in turn, are thought to act by recruiting additional splicing factors. For instance, enhancers of splicing at 3′ sites have been shown to promote binding of U2AF65 to the polypyrimidine tract (64). However, a recent analysis of alternative splicing of immunoglobulin transcripts showed that enhancers can also act by countering the effect of an inhibitory element (33), rather than by a positive recruitment mechanism. Enhancers have also been proposed to act in the definition of constitutively spliced exons. The constitutively spliced β-globin exon 2 has been shown to contain enhancer elements, with several discrete sequences interacting with specific SR proteins (40, 53). The presence of enhancers in β-globin, however, do not seem to reflect a general requirement for enhancers in all exons since only a small minority of hundreds of known mammalian splicing mutants lie outside the splice sites themselves (12, 36, 44, 48, 49, 62).
In addition to sequences that promote exon inclusion, there are sequences that inhibit splicing, so-called exonic or intronic splicing silencers. The silencers are less well characterized; they can be purine or pyrimidine rich and bind a diverse array of proteins. Perhaps the best understood example of negative regulation is the role of the Drosophila sxl protein in the sex-specific processing of tra pre-mRNA, where sxl is thought to block access of U2AF65 to the 3′ splice site by binding to a cis element (63). Polypyrimidine tract binding protein (PTB), also known as hnRNP I, can function by antagonizing U2AF65 action, as has been shown in the processing of alpha-tropomyosin and GABA(A) receptor gamma2 pre-mRNAs (4, 37). Other silencers have been shown to function through interactions with hnRNP A1 (7, 11, 21), hrp48 (a Drosophila protein in the hnRNP A family) (55), and hnRNP H (13).
Our incomplete understanding of splicing regulatory elements prevents us from predicting the presence of exons in a typically long vertebrate transcript, at least not without searching for open reading frames. Splicing at some true sites may be stimulated by enhancers, splicing at some pseudosites may be inhibited by silencers, and some sites may be defined by the interplay between these two types of elements. Due to the multitude of pseudo-splice sites within introns, we decided to search for negatively acting sequences that may be repressing intronic pseudosites. Early work identified several exonic and intronic sequences that inhibited splicing when ligated downstream of a 3′ splice site (25, 51).
To look for intron sequences that inhibit splicing, we constructed several libraries and genetically selected for sequences that could disrupt splicing when inserted into the central exon of a three-exon minigene. We used human genomic DNA as a source rich in intron sequences. A library of human DNA inserts readily yielded sequences that inhibited exon splicing. A comparative screen of human and bacterial DNA sequences revealed that over a third of 19 randomly chosen human inserts caused aberrant processing of the pre-mRNA, whereas none of 27 bacterial inserts did so. We concluded that the human genome is rich in specific sequences that inhibit splicing, and we suggest that such sequences may play a role in silencing inappropriate splice sites.
MATERIALS AND METHODS
Constructs.
All libraries were constructed by cloning restriction fragments into a pD2B-based minigene. pD2B is a three-exon dhfr minigene (for dihydrofolate reductase [DHFR]) driven from its own promoter in the vector pSP72 (Promega) and containing an extra copy of dhfr exon 2 as the internal exon (see Fig. 1, line 2). The upstream portion of the minigene, from the full promoter to 91 bp into the second intron, is genomic in sequence. The remaining 181 bp of the second intron is a repeat of genomic dhfr intron 1. The third exon is made up of exons 2 to 6 from cDNA, followed by genomic dhfr polyadenylation signals. The construction of pD2B has been described previously (15). pD2C3SfuI is a derivative of pD2B that contains XhoI and ApaI sites on either side of the SfuI site in the central exon.
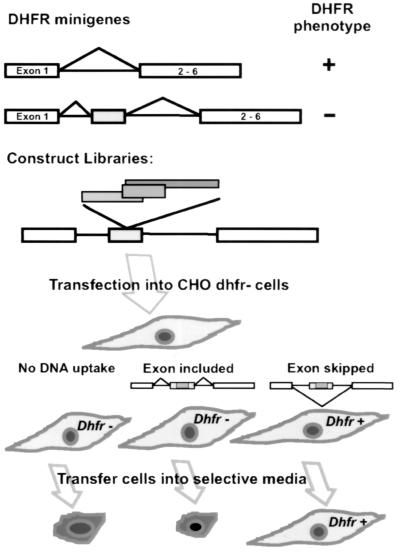
FIG. 1.
Selection scheme for sequences that inhibit splicing. The top diagram depicts a basic dhfr minigene (in pDCH1P) retaining the 300-nt dhfr intron 1 as the sole intron, with exons 2 through 6 originating from cDNA. This minigene confers a DHFR+ growth phenotype (ability to grow in the absence of purines and thymidine) when transfected into DHFR− recipient cells. pDCH1P was modified to produce pD2B, carrying the minigene shown in the second diagram. This gene has an extra copy of exon 2 inserted into the intron. This extra 50-nt exon is efficiently included in the mRNA, resulting in a message that cannot code for a functional DHFR enzyme. Libraries were constructed by cloning DNA fragments into the unique SfuI restriction site engineered into the central exon. Inserts that reduce splicing result in partial or complete skipping of the central exon, thus restoring the production of functional mRNA.
Human libraries.
Human placental DNA was fragmented with a cocktail of HinPI, TaqI, and MspI restriction enzymes; the 50- to 250-bp fraction was purified by gel electrophoresis. These fragments were ligated into the SfuI site of pD2B, whose 6-bp recognition site begins at position +16 in the 50-bp internal exon of pD2B. The products of ligation were used to transform the chemically competent SURE strain of Escherichia coli (Stratagene). After selection on ampicillin plates, the resistant colonies were pooled and the DNA was extracted. Redigestion with SfuI was used to eliminate residual uninserted vector.
A second human library was constructed in the plasmid pD2C3SfuI. Human placental DNA was fragmented as described above, and the fragments of 60 to 120 bp were purified by gel electrophoresis. The fragments were ligated into pD2C3SfuI and used to transform the XL-1 Blue strain of E. coli (Stratagene). In this case, the transformed bacteria were plated directly after heat shock to reduce the chance of sister colonies contributing duplicate sequences in the subsequent screen.
A third library was constructed in pCMVD2C3, which carries a dhfr minigene driven by the cytomegalovirus (CMV) promoter. pCMVD2C3 was constructed by cloning the dhfr minigene from pD2C3, which is described below. The region from the major transcriptional start site (42) to a position 88 bp downstream of the translation termination site in the 3′ untranslated region was cloned between the NheI site and the NotI sites of pEGFP-N1 (Clontech), eliminating the green fluorescent protein (GFP) sequence. The resulting construct, pCMVD2C3, should produce a transcript starting 14 nt upstream of the 5′ end of dhfr mRNA and terminating with a simian virus 40 poly(A) site provided by the vector. Human DNA was fragmented with a cocktail of XhoI and the blunt cutters AluI, RsaI, and HaeIII. The 50- to 150-bp fraction of the restricted DNA was purified and isolated as described above. These fragments were ligated into the SmaI-XhoI sites of pCMVD2C3. The SmaI site overlaps the ApaI site in the clamp region of the synthetic inserts described below.
Synthetic library.
The two single stranded 5′ phosphorylated oligomers CGCGGGCCCGGGCTGTGN20TTTATGCTCTCGAGTA and CGTACTCGAGAGCATAAAN20CACAGCCCGGGCCCG were allowed to anneal. The resulting double-stranded DNA contained a randomized 20-nt core flanked by clamps containing the unique restriction sites XhoI and ApaI and capped by two SfuI-compatible sticky ends. The 51-bp fragments were ligated into the SfuI site of exon 2 of pD2B as described above and transformed into the XL-1 Blue strain of E. coli. About 10,000 transformed colonies were pooled for DNA extraction.
Bacterial library.
E. coli genomic DNA was fragmented with a cocktail of HinPI, TaqI, and MspI restriction enzymes, and the 80- to 120-bp fraction was gel purified. The fragments were ligated into pD2C3SfuI and used to transform the XL-1 strain of E. coli. The transformed bacteria were plated directly after heat shock to reduce the chance of sister colonies contributing duplicate sequences in the subsequent screen.
Recloning inserts.
Two isolates (pD2C2 and pD2C3) of the synthetic library were used for recloning various inserts. The restriction sites XhoI and ApaI were used for the recloning. In pD2C3 the XhoI site is upstream of the ApaI sites, whereas in pD2C2 the ApaI site is upstream of the XhoI site. pD2C3 was used as the vector to remake the splicing constructs from the inserts found in dhfr+ recipients of the pD2B human library. The inhibitory inserts were amplified from genomic DNA and cloned via XhoI and ApaI sites introduced within the primers. The 5′ primer was GAACGAACTCGAGTACTTC, and the 3′ primer was TGAGGAGGTGGTGGGCCCTCTTT. pD2C2 was used as a host to invert inserts by recloning XhoI-ApaI fragments that had been originally cloned in pD2C3. To insert sequences B11 and B36 into the aprt gene, we cleaved plasmids carrying these inserts (pB11 and pB36) at Acc113I and HaeIII restriction sites in the flanking dhfr sequence and cloned the small fragment into the EcoRV site in exon 2 of pAPRTWT (34).
Constructs with known binding sites.
The following double-stranded oligomers were cloned between the XhoI and the ApaI sites of pD2C2: hnRNP A1, TATGATAGGGACTTAGGGT; hnRNP H, TAAATGTGGGACCTAGA; PTB, CTGCAGCCTGGAGCTCCTCTCGTGGCC; and U2AF65, TTTTTTTTTCCTTTTTTTTTCCTTTTTTTTT. The hnRNPA 1 and PTB sequences were derived from SELEX experiments (9, 58), the hnRNP H sequence was identified as the hnRNP H binding site in the rat beta-tropomyosin gene (13), and the U2AF65 sequence contains the consensus U2AF binding site derived from a panel of SELEX winners (58).
Transfection.
All transfections were performed with Lipofectamine (Life Technologies) using the conditions recommended by the manufacturer for CHO K1 cells, except for the DNA concentrations. Near-confluent 100-mm plates of dhfr− null CHO DG44 cells (3 × 106 cells) were transfected with various amounts of plasmid DNA and enough human genomic DNA to bring the total to 7 μg. In experiments in which transfectants were selected for a DHFR+ phenotype, DNA from the pD2B human library or synthetic library was used in a series of 1:10 dilutions. In the case of the pD2B human library, the starting plasmid concentration was 1.38 μg/dish. After a 24-h recovery period, the transfected cells were trypsinized and transferred to a larger dish for selection in F-12 medium lacking glycine, hypoxanthine, and thymidine and supplemented with 7% dialyzed fetal calf serum. In experiments in which no selection for DHFR was applied, transfectants were selected for G418 resistance conferred by a cotransfecting neo plasmid, pEGFP-N1 (Clontech). DG44 cells (5 × 105) in a 35-mm dish were cotransfected with 0.5 μg of pEGFP-N1 and approximately 0.5 μg of a plasmid bearing an insert in the dhfr minigene. After a 24-h recovery period, cells were transferred to a 100-mm dish and challenged in F-12 medium containing G418 (400 μg of active compound/ml). Surviving colonies were pooled for further analysis.
RNA and DNA extraction.
RNA was extracted by the method of Huang and High (31) for the experiments of Fig. 2. All other analyses were performed on total RNA isolated with s.n.a.p columns (Invitrogen) using the manufacturer's recommended procedure. DNA was extracted with DNAzol (MRC) using the protocol supplied by the manufacturer.
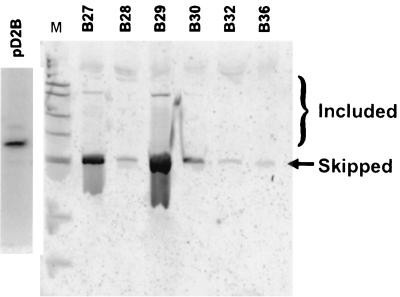
FIG. 2.
Inhibition of splicing in transfectants selected for a DHFR+ growth phenotype. CHO dhfr-deficient cells were transfected with a plasmid library containing human DNA insertions in the central exon of a dhfr test minigene, as described in the text. Colonies that exhibited a DHFR+ growth phenotype (growth in −GHT medium) were expanded and assayed for their splicing phenotype by RT-PCR, using primers located in exons 1 and 4. A phosphorimage of an electrophoretic gel is shown. The bracket indicates the positions of bands corresponding to inclusion of the exons with inserts of various sizes. Splicing in cells transfected with the uninserted parental plasmid (pD2B) is also shown.
RT-PCR.
Reverse transcriptase (RT) reactions were performed using approximately 1 μg of total RNA by the random primer protocol supplied by the manufacturer (Life Technologies). One-fifth of the 20-μl reaction mixture was used in the subsequent PCR. PCR was performed with HotWax Mg++ beads (Invitrogen) using the conditions recommended by the manufacturer, except for the inclusion of radioactive substrate (14). Unless otherwise specified, all reactions were performed with an annealing temperature of 55°C for 27 cycles. In the case of genomic PCR shown in Fig. 3, 1 μg of DNA was amplified for 35 cycles. PCR products were separated by electrophoresis in a 2% modified agarose gel (Trevigel 500; Trevigen). ImageQuant software was used for the quantitative comparison of spliced products after phosphorimaging.
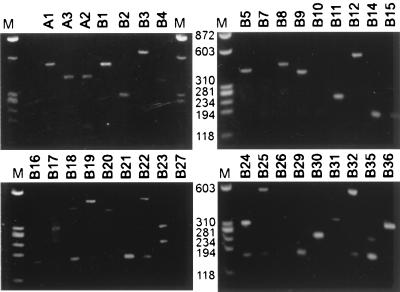
FIG. 3.
Distribution of insert sizes in transfectants selected for dhfr splicing inhibition. Genomic DNA samples from clones selected for a DHFR+ growth phenotype were PCR amplified with primers flanking the insert site. The PCR products were stained with ethidium bromide after electrophoresis in 2% modified agarose gel. A 175-bp PCR product was sometimes seen. This size corresponds to a product containing exon 2 without an insert. Since the template contains a duplication of the intron 2 region (to which the 5′ primer can anneal), such an artifactual product could have arisen by DNA recombination during PCR.
RESULTS
Genetic selection of genomic sequences that inhibit splicing.
Most splicing mutations result in exon skipping (12, 35, 47). We took advantage of that fact to select for sequences that inhibit splicing. Our strategy was to insert fragments of human genomic DNA into the central exon of a three-exon dhfr minigene. The central exon in this minigene is an extra exon; its inclusion into dhfr mRNA results in a 50-nt insertion that disrupts DHFR enzymatic activity. If an insert inhibits the splicing of this central exon, the two terminal exons are joined to form dhfr mRNA that codes for the functional enzyme. We have previously used this minigene, carried in pD2B, to select for a large number of base substitution mutants deficient in splicing of the central exon (14).
To create a library of human genomic fragments from which to select splicing inhibitory sequences, we fragmented human placental DNA with a mixture of the three restriction enzymes, TaqI, MaeII, and HpaI, all of which produce overhangs complementary to those produced by SfuI. Fragments in the 50- to 250-bp range were size selected and cloned into the unique SfuI site in the central exon of the pD2B minigene. After transfection of pooled plasmid DNA into a CHO DHFR-deficient mutant, the cells were challenged in a selective medium lacking glycine, a source of purines (e.g., hypoxanthine), and thymidine (−GHT medium). DHFR-deficient cells cannot grow in this medium, nor can cells that receive the minigene that are proficient in splicing in the central “killer” exon. CHO cells can grow with <2% of endogenous, wild-type DHFR activity (12), so even a small proportion of exon skipping yields transfectants that form colonies in −GHT (14). This scheme is depicted in Fig. 1.
Four transfections were carried out with 10-fold serial dilutions of the insert library. The frequency of colonies in −GHT medium decreased in approximate proportion to the dilution: 1.4 μg yielded approximately 1,500 colonies, 140 ng yielded 146 colonies, 14 ng yielded 38 colonies, and 1.4 ng yielded 3 colonies. We isolated colonies from the two lowest dilutions of plasmid DNA to minimize the probability of isolating transfectants with more than a single copy of the dhfr minigene. Forty colonies were expanded, and RNA was extracted from five of these for a preliminary check of the splicing phenotype. All five expressed dhfr mRNA that lacked the central exon, along with various amounts of mRNA that included this exon (Fig. 2). In contrast, transfectants that carry the uninserted pD2B minigene exhibit very little (<5%) exon skipping (14). The region encompassing the central exon in 34 transfectants was amplified from genomic DNA by PCR, and the PCR products were analyzed by gel electrophoresis (Fig. 3). Three yielded no central exon band and were assumed to have arisen from the elimination of the killer exon by intragenic recombination. Six of the PCR reactions yielded an artifactual 175-bp band. Seven had inserts larger than 250 bp. Of the 18 remaining products, 12 with small inserts were chosen for further study.
Sequencing of the PCR products revealed that some cell lines contained identical inserts. B11 and B30, for instance, both carried a 61-bp fragment of an Alu repeat. A2 and A3 were also identical. Human DNA is rich in repeated sequences so, since no effort was made to remove repetitive sequences, it is not surprising that we obtained multiple hits with highly repeated sequences such as Alu. Moreover, since the transfected cells were replated for selection, some sister colonies may also have been present.
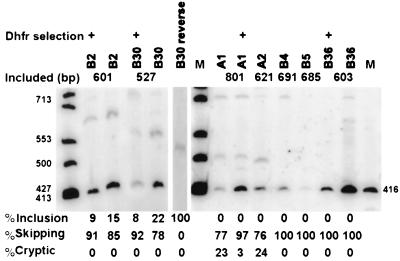
It is possible that the selected transfectants could have been skipping the internal exon for reasons unrelated to the activity of the insert. For instance, the inserted plasmid could have suffered a spontaneous splice site mutation that caused this exon to be skipped. Alternatively, the selected transfectants may have arisen from cell clones with increased central exon skipping caused by heritable changes in the expression of trans-acting factors in the host cell rather than being caused by the insert. To test these possibilities, eight of the inserts were PCR amplified from the DNA of the transfectant clones and recloned into the central exon of the dhfr minigene vector. The splicing phenotype was then determined after a secondary transfection into DHFR-deficient cells. DHFR-deficient DG44 cells were cotransfected with plasmids containing the recloned inserts and a plasmid carrying the neo gene; transfectants were selected for resistance to G418 rather than for DHFR activity, thereby avoiding any selective pressure being placed on the splicing phenotype. G418-resistant colonies were pooled, expanded, and tested for dhfr splicing by RT-PCR. As can be seen in Fig. 4, all of the constructs retested in this way displayed a predominant skipping phenotype, verifying that splicing inhibition caused by the insert sequences per se had been the basis of the original selection.
FIG. 4.
Inhibition of splicing in secondary transfectants carrying minigenes with recloned inserts. Inserts were PCR amplified from the DNA of eight selected primary transfectants and recloned into the central exon of the minigene vector. CHO dhfr null cells were retransfected with each of these plasmids, along with a neo vector. G418-resistant colonies were pooled and tested for their dhfr splicing phenotype by RT-PCR as described in the legend to Fig. 2. In some cases, as indicated, the transfected populations were selected for a DHFR+ phenotype in parallel with the G418 selection. The number above the lanes indicates the size of the RT-PCR product expected for exon inclusion. Below each lane is shown the proportion of exon skipping versus exon inclusion or cryptic splicing, as determined by PhosphorImager analysis.
The inhibitory sequences function in a heterologous context.
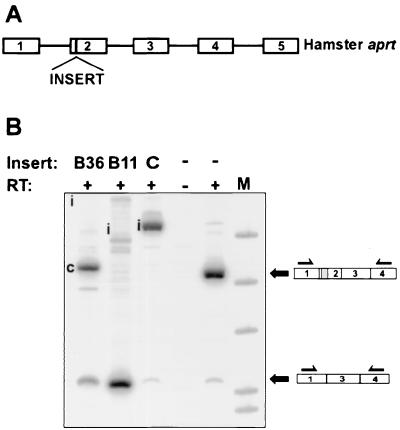
It is possible that the insertion of a foreign sequence into dhfr exon 2 produced an inhibitory effect specific to dhfr exon 2. For instance, the insert might create an inhibitory secondary structure, either by sequestering positive signals or by disrupting the positive secondary structure. If this were the case, we would expect the splicing inhibitory sequences to be context specific and not to affect splicing if inserted into an unrelated pre-mRNA. To test this idea, we inserted the B36 and B11 sequences into position +4 of exon 2 of the cloned genomic aprt gene. Both B11 and B36 inhibited the correct splicing of aprt exon 2 (Fig. 5). B11 resulted in almost complete skipping of the exon. B36 also inhibited normal exon 2 splicing but spliced predominantly to a cryptic site (possibly the 3′ splice site-like sequence, CTTCTCTCCCAACTCCCCGCAG/C) instead of the aprt 3′ splice site. In contrast, insertion of an arbitrarily chosen 77-bp fragment (starting from position +8 in dhfr intron 1) at the same position had no effect on the splicing of this exon (Fig. 5). These results suggest that these inhibitory sequences can act autonomously.
FIG. 5.
Sequences selected for inhibition of dhfr splicing also inhibit aprt splicing. The inserts B11 and B36 were cloned into position +4 in aprt exon 2 in the plasmid pAPRTWT, which carries the full genomic hamster aprt gene. The control construct, shown in lane C, is identical to B11 and B36 except that it carries a 77-bp insert derived from dhfr intron 1 (positions +8 to +84) instead of an inhibitory insert in position +4 of exon 2. The inserted aprt plasmids, along with a neo plasmid, were used to transfect CHO U1S, an aprt double-deletion mutant. Transfectants resistant to G418 were pooled, expanded, and analyzed for aprt splicing by RT-PCR using primers located in exons 1 and 4. The letter “i” in the B36 and B11 lanes indicate the expected positions for bands corresponding to the inclusion of aprt exon 2. The c in lane B36 indicates the product of cryptic splicing.
Screening for genomic sequences that inhibit splicing.
The ease with which inhibitory inserts were isolated from this pool of human genomic DNA fragments raised the possibility that many sequences may be inhibitory. We therefore attempted to select for such inhibitory sequences from a library of random synthetic 20-mers. However, an experiment carried out on the same scale as that described above for human genomic fragments yielded no insert-dependent DHFR-positive colonies. Although some colonies were produced in transfections carried out with very high concentrations of DNA, all the dhfr-positive cell clones were carrying minigenes that had eliminated their central exon (data not shown). They presumably originated by homologous recombination between the duplicated regions encompassing dhfr exon 2 in this minigene (14). We concluded that either the 20-mer was not long enough to specify an inhibitory sequence or the human genome was a richer source of such sequences than a random library.
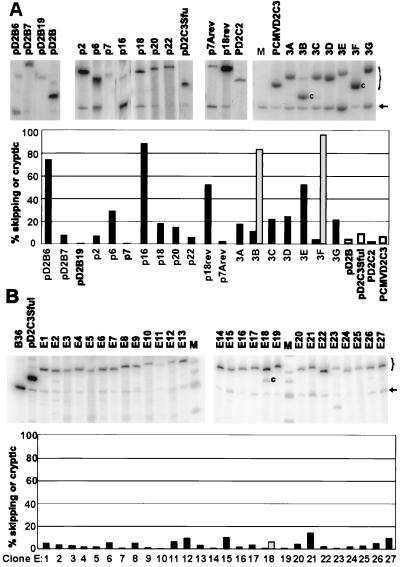
To explore this latter possibility, we screened human genomic libraries for the skipping phenotype, rather than selecting for it. The 80- to 120-bp size fraction of restriction enzyme-digested human DNA was cloned into the central exon of the pD2B minigene as described above, and the plasmids from individual E. coli colonies were characterized by sequencing their inserts. Several constructs contained inserts derived from highly repeated DNA that were nearly identical to each other; only one representative of each insert was examined. Nineteen different insert sequences were studied further, including two that were purposely produced by inversion of a given insert. Each plasmid was transfected into CHO dhfr-deficient cells, together with a plasmid bearing the neo gene, and pooled permanent transfectants were selected on the basis of their resistance to G418. The dhfr splicing phenotype in these populations was then determined by RT-PCR. As can be seen in Fig. 6A, 12 of the 19 clones predominantly included the central exon (<25% skipping), 5 exhibited substantial (25 to 100%) exon skipping, and 2 exhibited cryptic splicing to a site within the insert. Thus, 7 of 19 inserts (37%) caused an inhibition of normal splicing of the central exon; these 7 inhibitory sequences were found by scanning our set totaling 1.9 kb of unrelated human DNA sequences.
FIG. 6.
Inhibitory sequences occur more frequently in human than in E. coli genomic DNA. (A) Pooled permanent transfectants carrying dhfr minigenes with unique inserts of human DNA were assayed for splicing by RT-PCR and PAGE. The additional lanes marked pD2B, pD2CSfu, pD2C2, and pCMVD2C3 are vector controls for the sample lanes immediately preceding or following them. M is a size marker (416 bp) for skipped mRNA. The size of this band is also marked with an arrow. The letter “c” indicates an mRNA produced by splicing to a cryptic site. The size range of mRNAs that included the inserted exon is indicated by a bracket in the right margin. Each lane was quantified using a PhosphorImager and ImageQuant software. The bar chart shows the percent skipping in black and percent cryptic splicing in gray. Open bars represent control constructs. (B) Pooled permanent transfectants carrying dhfr minigenes with unique E. coli DNA inserts were assayed by RT-PCR and PAGE. All inserts were introduced into pD2C3SfuI, whose splicing phenotype can be seen in both panels A and B. B36 and pD2C3SfuI provide size controls for skipped mRNA and included mRNA, respectively. M indicates size markers. Other features are labeled as for panel A.
Is this high frequency of inhibition due to the ease with which splicing can be affected by random sequences of about 100 nt, or are inhibitory sequences especially prevalent in the human genome? To answer this question we repeated the experiment using E. coli DNA as a source of the inserts. Twenty-seven plasmids with an average insert size of 120 nt were isolated, stably transfected into CHO dhfr-deficient cells, and analyzed by RT-PCR. All but 1 of the 27 plasmids spliced normally, with more than 90% exon inclusion; the exception spliced normally 85% of the time (Fig. 6B). Thus, fewer than 1 in 27 of the E. coli DNA inserts was inhibitory at the 25% or greater level, or less than 1 in about 3.2 kb of bacterial DNA examined. The difference in the frequency of inhibitory inserts between the human and bacterial human library is significant at the P = 0.01 level (chi-square test with Yates correction). We conclude that sequences that can inhibit splicing are enriched in the human genome.
Effect of recognized inhibitory sequences on minigene transcript splicing.
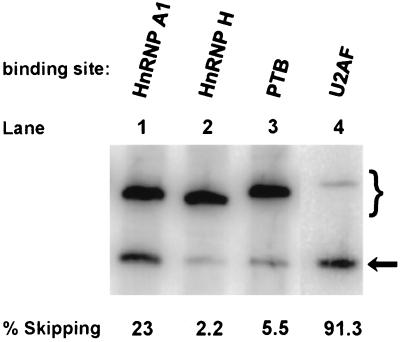
The high frequency in the human genome of sequences that inhibited splicing focused our attention on sequences that are the targets of abundant RNA-binding proteins. We wondered whether much shorter sequences of this type could also act as inhibitors of constitutive splicing in this system. We chose sequences known to be bound by hnRNP A1, hnRNP H, PTB (hnRNP I), and U2AF65 to test the ability of those factors to inhibit the inclusion of our exon. hnRNP A1 has been shown to inhibit splicing in hnRNP A1 (7), human immunodeficiency virus (HIV) tat (11), and human fibroblast growth factor receptor 2 (21) transcripts. The insertion of a 19-nt sequence selected for tight binding to hnRNP A1 (1, 9) showed some inhibitory activity in dhfr minigene transcripts but still allowed over 75% exon inclusion (Fig. 7, lane 1). The hnRNP H sequence was that of the alternatively spliced exon 7 of the rat beta-tropomyosin transcript; binding of hnRNP H to this sequence correlates with its exclusion in favor of exon 6 (13). As shown in Fig. 7, lane 2, the insertion of this 17-nt sequence did not inhibit splicing of the central dhfr exon in pooled permanent transfectants. PTB and U2AF65 both can bind to polypyrimidine tracts (PPTs), although their preferred sequences emerge as different from iterative binding selection procedures (58, 66). To increase the chance of distinguishing these two activities, we chose a selected version of the PTB binding sequence (p53.6 in reference 56) that appeared to have the least overlap with the U2AF65 binding site consensus (Fig. 7). This 27-nt PTB element was without effect on splicing in dhfr minigene transcripts (Fig. 7, lane 3). The 31-nt U2AF65 sequence was chosen as a combination of sequences found by iterative selection for binding (58, 66) and sequences shown to enhance splicing in a functional assay (19). This sequence inhibited splicing strongly (91% skipping) (Fig. 7, lane 4). It should be noted that this sequence is not directly followed by an AG dinucleotide and so does not constitute a 3′ splice site. This last result indicates that a sequence as short as 31 nt can strongly inhibit inclusion of this constitutive dhfr exon and suggested polypyrimidine tracts as potentially active components of our inhibitory inserts.
FIG. 7.
Effect of recognized protein binding sequences on splicing. Four oligomers corresponding to sequences known to bind the proteins hnRNP A1, hnRNP H, PTB, and U2AF65 were inserted between the XhoI and ApaI sites near the center of the central exon of pD2C2. The resulting plasmids were transfected into CHO cells and pooled permanent transfectants were assayed for splicing by RT-PCR as indicated. The oligomers were hnRNP A1 TATGATAGGGACTTAGGGT), hnRNP H (TAAATGTGGGACCTAGA), PTB (CTGCAGCCTGGAGCTCCTCTCGTGGCC), and U2AF65 (TTTTTTTTTCCTTTTTTTTTCCTTTTTTTTT). The numbers beneath the lanes show the percent skipped.
Characteristics of sequences.
We sequenced 10 inhibitory inserts, obtained either by selection or screening. In a search for possible commonalities, all possible pairs of the 10 sequences were aligned. FastA was used to search each insert against a local database of all 10 inserts. The frequency and quality of the matches found was not different from that expected from a set of random sequences, suggesting that the inhibitory regions were too small and/or too degenerate to be detected within such a small group. However, the motif discovery program MEME (5) found several 10-mer motifs, the top three being the G-rich sequence GGCAGGGUGG and two pyrimidine-rich sequences (CUUACUCUUC and UCUUUCACCG). As can be seen at the bottom of Fig. 8, 6 of the 10 sequences contained good matches to the G-rich consensus sequence. An examination of the sequences that encompass this motif in the 6 inserts revealed an abundance of G triplet repeats (Fig. 8). Multiple sequence alignments with low gap and extension penalties confirmed the presence of clusters of G triplets, principally within the 5′ region of B2, B36, P16, and B5. G triplets have been proposed as a distinguishing characteristic of the 5′ and 3′ edges of many introns (41, 46).
FIG. 8.
Human genomic sequences that inhibit splicing. Each of the 10 sequences inhibits splicing of the test exon by at least 25% (Fig. 6). Polypyrimidine tracts (length of 11 or more with a pyrimidine content of 85% or more) are underlined. Runs of three or more G's are in boldface. A G-rich consensus sequence found in 6 of the 10 sequences is double underlined; the agreements to this consensus are shown at the bottom of the figure.
The two pyrimidine-rich motifs are less well conserved and probably reflect the multiple PPTs found in 8 of the 10 sequences. Tracts of at least 11 nt containing at least 85% pyrimidines are underlined in Fig. 8. Most of these PPTs are U-rich, and five of the insert sequences have an overall U content in the range of 33 to 43%. It is possible that these tracts represent binding sites for the 3′ splicing factor U2AF65. The presence of the G triplets and PPTs in an insert was not mutually exclusive, since four of the sequences contained both of these elements.
A global comparison of intron and exon sequences for all possible hexamers has shown that certain hexamers are highly discriminate for introns (67). We wished to examine the inhibitory inserts for such intron discriminate sequences, but the occurrence of specific hexamers was too low to be statistically significant. We therefore generated similar data for tetramers, scanning an annotated database of over 2,000 exons and introns from nonredundant human genes (M. Reese, U. Ohler, D. Kulp, and A. Gentles, Human Gene Database [http://www.fruitfly.org/sequence/human-datasets.html: GENIE gene finding data set, with data taken from http://whitefly.lbl.gov/seq_tools/datasets/Human/exons_v105 and /introns_v105]). The 25 most discriminate of the 256 tetramers were highly T rich (58%) and AT rich (91%). The average frequency of these top 25 tetramers among the inhibitory sequences shown in Fig. 8 was 0.49/100 nt but was considerably lower (0.28/100 nt) in a set of noninhibitory sequences of comparable size (data not shown). Although these data sets are too small (ca. 1,000 nt) to be conclusive, the result is consistent with the idea that sequences that inhibit splicing are enriched for sequences that are more frequent in introns than in exons.
We used BLAST 2.0 to search human genomic and EST databases for matches to the 10 sequences. The results are shown in Table 1 for sequences that showed identity or near identity to a database sequence. Four of the 10 sequences (7Arev, B11, P6, and B9) contained matches to highly repeated sequences (Alu, PTR5, and human satellite III). This frequency is not unexpected given the fact that more than 30% of the human genome is made up of such repeated sequences. Several sequences matched uncharacterized cosmids in the database. One sequence, P16, corresponded to an intron in a recognized gene, hsk1, which specifies a small conductance potassium channel (35). The inhibitory sequence lies just upstream (−91 to −10) of the small (9 nt) hsk1 exon 9. Interestingly, this small exon is absent in some isoforms of the homologous rat mRNA (35).
TABLE 1.
Homologies in inhibitory insert sequences
| Insert name | Size (bp) | Homology (positions in insert) | GenBank accession no. |
|---|---|---|---|
| 7Arev | 105 | PTR5 repeat (1–46) | |
| Human chromosome 20 DNA (41–100) | AL049540.11 | ||
| 3E | 56 | Human X chromosome DNA | AL034384.1 |
| B11 | 64 | Reverse complement of an Alu-Sx repeat | |
| P16 | 81 | Exact homology to the 3′ end of hSK1 gene intron 8 ending in the PPT | AF131946 |
| P6 | 75 | Human class III satellite DNA | |
| B5 | 231 | Human cosmid clone (141–231) | Z68326 |
| B9 | 201 | Human chromosome 6 DNA (2–103); reverse complement of an Alu-Sq repeat (105–200) | Z78663; AB015355 |
| A2 | 158 | None found | |
| B2 | 135 | None found | |
| B36 | 134 | None found |
DISCUSSION
Isolation of inhibitory sequences.
We report here the isolation of sequences capable of inhibiting splicing when placed within an exon. The target exon, exon 2 of the hamster dhfr transcript, is flanked by 3′ and 5′ splice sites that have somewhat above-average consensus scores (52) of 84 and 86, respectively. There is no detectable skipping of this exon in endogenous dhfr transcripts and <5% skipping in cells transfected with the minigene used here. We first used a genetic selection to isolate transfectants that had incorporated minigenes containing inhibitory human DNA inserts. Inhibition of splicing to the extra exon 2 in this construct leads to exon skipping and to the production of functional DHFR and growth in selective medium. Given the low frequency of DHFR+ transfectants, we had to consider the prospect that the skipping phenotype was caused by heritable variation of a trans-acting factor rather than by cis inhibition caused by the insert. We ruled out the former possibility by rescuing the insert from exon-skipping clones, reinserting it into a fresh minigene and retesting the new construct. In all of the eight cases tested, the skipping phenotype followed the insert, indicating that a cis-acting element was responsible for the exon-skipping phenotype. In all of these selected cases, the inserts caused >50% exon 2 skipping. It is interesting to note that although selected for their ability to skip, two of the inserts also exhibited low levels of cryptic splicing to an unmapped position within the insert (A1 and A2 in Fig. 4).
Inhibitory sequences are frequent in the human genome and rare in a bacterial genome.
Perhaps the most striking result of this work was the prevalence of inhibitory sequences found in human genomic DNA. More than one-third (7 of 19) of randomly cloned human genomic restriction fragments proved to be inhibitory in this in vivo splicing assay. In order to analyze as many distinct sequences as possible, we excluded inserts that were highly related; these amounted to about half of the original set (14 of 33). If we assume that these discarded sequences produce the same splicing phenotype as their tested homologues, the statistical results would be much the same, with 15 of 33 sequences being inhibitory. In contrast, DNA from two other sources did not generate inhibitory inserts. The first source was random 20-mers. These synthetic sequences yielded no splicing mutants in genetic selections that screened an estimated 9,000 different sequences. Since the short size of these inserts may have limited their effectiveness, we turned to E. coli as a second source of control DNA. Of 27 constructs containing E. coli restriction fragments with an average size of 120 nt, none skipped the test exon more than 15% of the time (Fig. 6B).
The finding that 7 of 19 human inserts with an average size of 100 nt proved to be inhibitory implies a frequency in the human genome of about one inhibitory sequence per 270 nt. This pervasiveness suggests that one role of enhancers may be to counteract negative effects, a mechanism that has been demonstrated in several systems (3, 10, 33, 68, 69).
Sequence analysis of the inhibitory inserts.
Compared to splicing enhancer sequences, there have been fewer splicing silencer sequences described. Nevertheless, it is interesting to note that the inhibitory inserts isolated here contain sequence elements that have been implicated previously in splicing inhibition.
Eight of the 10 inhibitory inserts contained PPTs, and in six cases an AG dinucleotide can be found just downstream, such that these sequences resemble 3′ splice sites. Thus, it is possible that normal splicing components are involved in the inhibition. Moreover, there is evidence that the PPTs in some of these inserts can function in splicing, given a favorable context. First, when B36 was placed within an aprt exon, we observed splicing to a cryptic site within B36 itself (along with inhibition of splicing at the normal site). Second, P16 contains part of a natural 3′ splice site at the 3′ end of intron 8 in the hsk1 gene. Third, B11 corresponds to the Sx family of Alu repeats; it is present in the reverse complement orientation. This orientation of the Alu repeat contains sequences resembling 3′ splice sites, and these sites are occasionally used as functional 3′ splice sites, resulting in the incorporation of Alu sequences into the mRNA (2, 6, 8; reviewed in reference 38).
Another feature of the set of inserts is an enrichment for G triplets and quartets. GGG motifs were found associated with the 5′ and 3′ prime boundaries of primate introns (23, 46) and have been shown to play a role in small intron definition (41). In a construct containing the G triplets flanked by duplicated 5′ splice sites, the G triplets promoted the use of the upstream splice site. The function of these G triplets to promote intron definition when present within small introns may act to subvert exon definition when they are placed within exons, as has been done here. Later work has shown the G triplets to bind to U1 snRNP (A. J. McCullough, personal communication), so here again we have a possible role of normal splicing components in an inhibitory action.
Alternatively, insert sequences may be binding nonspliceosomal proteins that are known to antagonize splicing. Thus, the splicing inhibitor PTB acts by binding to PPTs. This abundant protein has been shown to antagonize splicing in several well-studied systems (4, 37, 50, 58). PTB can act from sites distinct from the 3′ splice site (26), yet its inhibitory action can be antagonized by U2AF65, suggesting a competition between these two factors for the same site (37). However, insertion of one of the winning PTB-binding sequences isolated by iterative selection did not inhibit splicing of our test exon.
Other hnRNPs are also candidates for splicing inhibitors. Although hnRNP proteins were originally described as being necessary for splicing in vitro (16, 17, 57), more recent work in mammalian cells has implicated hnRNP A1 in the inhibition of splicing in the transcripts of the FGF receptor, hnRNP A1, and HIV tat genes (7, 11, 21). In addition, hrp48, a Drosophila hnRNP belonging to the hnRNPA/B family, plays a role in the inhibition of P-element exon 3 splicing (27). In the regulation of its own message, hnRNP A1 interacts with two intronic binding sites flanking exon 7b (7). However, in HIV tat exon 2 and tat exon 3 (11) and in the FGF receptor K-SAM exon (21) hnRNP A/B or A1 binds the exon that is being repressed, a situation that could apply here. The binding requirements of hnRNP A1 are poorly understood (1). However, a consensus sequence that emerged from a SELEX selection binds this protein with high affinity (9). The core UAGGGU of this sequence is similar to that present in the G-rich consensus sequence GGCAGGGUGG derived from 6 of the 10 inserts isolated here (Fig. 7), raising the possibility that hnRNP A1 may be playing a role in many of the cases seen here. However, a sequence selected for tight binding to hnRNP A1 acted as only a modest splicing inhibitor (23% inhibition) when inserted as a short (19-nt) oligomer. An hnRNP H binding sequence (13) did not inhibit splicing at all.
The hnRNP A1, hnRNP H, and PTB binding sequences had at best a weak effect on this constitutively spliced exon. These inhibitory sequences have been identified as such in alternatively spliced transcripts, and it is possible that they are too weak to interfere with strong splicing signals. Many alternative splicing elements bind factors (e.g., PTB, hnRNP A1, and ASF/SF2) whose level varies in different tissues (28) and so they may be ill suited for a role in determining constitutive splicing. In contrast, insertion of a binding site for the universal splicing factor U2AF65, which presumably does not vary in different tissues, strongly inhibited splicing. Perhaps the U2AF65 binding site is functioning as an extra splice signal here and can interfere with the bridging that underlies exon definition by competing for the interaction with the downstream 5′ splice site or the upstream 3′ site. The end result in either case would be a new (false) exon that is defined in terms of partial or even full spliceosome assembly but that is incapable of proceeding through the catalytic steps of splicing. It should be noted that the U2AF65 preferred binding sequence [U6(U/C)CC(C/U)U8] includes long runs of U's (58). Such runs of U's are rare in exon sequences, with U6 being the least frequent among 4,096 possible hexamers (67). This near absence of U runs could exclude the ectopic binding of U2AF65 in exons.
A negative role for splice-like sites.
Sequences that show good agreement to the consensus splice site sequences (pseudosites) far outnumber real splice sites within large introns (47, 54). A negative role for pseudo-splice sites would help to explain why these sites in vertebrate introns do not function as splice sites. It is an intriguing possibility that they are initially recognized as splice sites, nucleate partial spliceosome formation, and only fail at the later catalytic steps where they are discarded in favor of the sites that border real exons. Evidence from several well-studied systems of negative regulation of splicing supports the idea that splice-like sites can act as negative elements, recruiting components of the spliceosome to inappropriate places where they presumably compete with their legitimate counterpart for protein-protein interactions that are necessary for either recognition of an exon or a catalytic step in the splicing reaction. The exonic splicing silencer in the Drosophila P-element transcript functions by recruiting U1 snRNP into a nonproductive complex at pseudo-5′ splice sites adjacent to the real 5′ splice site (56). In the HIV tat or rev transcript, catalytically inactive complexes containing U1 snRNP and U2 snRNP repress exon 2 splicing (22). Rous sarcoma virus inhibits the splicing of most genomic copies of RNA via an element termed a negative regulator of splicing (NRS). The intronic NRS has two regions, a 5′ purine-rich region that recruits ASF/SF2 and a 3′ region that is capable of interacting with both U1 snRNP and U11 snRNP. The binding of U1 snRNP is predominantly responsible for the inhibition (18). The splicing inhibitor in influenza virus NS1 RNA forms a large complex containing U1, U2, U4, and U6 snRNPs that never proceeds to a functional spliceosome (45). U2 snRNP is also present in a complex associated with the immunoglobulin exon M2 inhibitor. U2AF65 binds to the real 3′ splice site, but U2 snRNP is part of the complex that binds to the inhibitor (33). Mutations in endogenous genes provide additional support for the idea that pseudo-splice sites interact with the splicing machinery. In the hamster dhfr gene, mutations in a pseudo-5′ splice site located 90 nt downstream of the real 5′ splice site allow splicing of an otherwise-inactive mutant form of the site (12).
Several of the inserts isolated here on the basis of promoting exon skipping also exhibit low levels of internal cryptic splicing. Furthermore, B36 caused exon skipping when present in dhfr exon 2 and cryptic splicing when ligated into aprt exon 2. The splicing machinery is obviously interacting with the cryptic sites in the inserts; perhaps this interaction underlies the skipping phenotype as well. Consistent with this idea, a U2AF binding site strongly inhibited splicing, while a PTB site had little effect. Site-directed mutagenesis experiments and cell-free splicing experiments on a selected subset of these inhibitory sequences should clarify the putative roles of the sequence elements described here. The ultimate test for the role of splicing inhibitors in splice site selection will be to examine the consequences of deleting these sequences in their normal context.
ACKNOWLEDGMENTS
This work was supported by grant GM-22629 from the National Institutes of Health.
We thank Hanzhen Sun for useful discussions and a critical reading of the manuscript.
REFERENCES
- 1.Abdul-Manan N, Williams K R. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamkiewicz T V, McSherry C, Bach F H, Houchins J P. Natural killer lectin-like receptors have divergent carboxy-termini, distinct from C-type lectins. Immunogenetics. 1994;39:218. doi: 10.1007/BF00241264. . (Erratum, 40:318.) [DOI] [PubMed] [Google Scholar]
- 3.Amendt B A, Hesslein D, Chang L J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey T L, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Ismb. 1994;2:28–36. [PubMed] [Google Scholar]
- 6.Barnett T R, Drake L, Pickle W D. Human biliary glycoprotein gene: characterization of a family of novel alternatively spliced RNAs and their expressed proteins. Mol Cell Biol. 1993;13:1273–1282. doi: 10.1128/mcb.13.2.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownell E, Mittereder N, Rice N R. A human rel proto-oncogene cDNA containing an Alu fragment as a potential coding exon. Oncogene. 1989;4:935–942. [PubMed] [Google Scholar]
- 9.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi M, Mayeda A, Krainer A R, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carothers A M, Urlaub G, Grunberger D, Chasin L A. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1993;13:5085–5098. doi: 10.1128/mcb.13.8.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C D, Kobayashi R, Helfman D M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen I T, Chasin L A. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 1993;13:289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen I T, Chasin L A. Large exon size does not limit splicing in vivo. Mol Cell Biol. 1994;14:2140–2146. doi: 10.1128/mcb.14.3.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi Y D, Grabowski P J, Sharp P A, Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986;231:1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- 17.Cobianchi F, Biamonti G, Maconi M, Riva S. Human hnRNP protein A1: a model polypeptide for a structural and genetic investigation of a broad family of RNA binding proteins. Genetica. 1994;94:101–114. doi: 10.1007/BF01443425. [DOI] [PubMed] [Google Scholar]
- 18.Cook C R, McNally M T. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J Virol. 1999;73:2394–2400. doi: 10.1128/jvi.73.3.2394-2400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coolidge C J, Seely R J, Patton J G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cote J, Beaudoin J, Tacke R, Chabot B. The U1 small nuclear ribonucleoprotein/5′ splice site interaction affects U2AF65 binding to the downstream 3′ splice site. J Biol Chem. 1995;270:4031–4036. doi: 10.1074/jbc.270.8.4031. [DOI] [PubMed] [Google Scholar]
- 21.Del Gatto-Konczak F, Olive M, Gesnel M C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyhr-Mikkelsen H, Kjems J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J Biol Chem. 1995;270:24060–24066. doi: 10.1074/jbc.270.41.24060. [DOI] [PubMed] [Google Scholar]
- 23.Engelbrecht J, Knudsen S, Brunak S. G+C-rich tract in 5′ end of human introns. J Mol Biol. 1992;227:108–113. doi: 10.1016/0022-2836(92)90685-d. [DOI] [PubMed] [Google Scholar]
- 24.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 25.Furdon P J, Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988;8:860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gooding C, Roberts G C, Smith C W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond L E, Rudner D Z, Kanaar R, Rio D C. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 29.Haut D D, Pintel D J. Intron definition is required for excision of the minute virus of mice small intron and definition of the upstream exon. J Virol. 1998;72:1834–1843. doi: 10.1128/jvi.72.3.1834-1843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M N, High K A. Efficient subcloning of DNA fragments amplified by crude oligonucleotides. BioTechniques. 1990;9:710–711. [PubMed] [Google Scholar]
- 32.Hwang D Y, Cohen J B. U1 small nuclear RNA-promoted exon selection requires a minimal distance between the position of U1 binding and the 3′ splice site across the exon. Mol Cell Biol. 1997;17:7099–7107. doi: 10.1128/mcb.17.12.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan J L, Green M R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler O, Jiang Y, Chasin L A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohler M, Hirschberg B, Bond C T, Kinzie J M, Marrion N V, Maylie J, Adelman J P. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 36.Krawczak M, Reiss J, Cooper D N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 37.Lin C H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 38.Makalowski W, Mitchell G A, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 39.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 40.Mayeda A, Screaton G R, Chandler S D, Fu X D, Krainer A R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell P J, Urlaub G, Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986;6:1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 45.Nemeroff M E, Utans U, Kramer A, Krug R M. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol Cell Biol. 1992;12:962–970. doi: 10.1128/mcb.12.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nussinov R. Conserved quartets near 5′ intron junctions in primate nuclear pre-mRNA. J Theor Biol. 1988;133:73–84. doi: 10.1016/s0022-5193(88)80025-0. [DOI] [PubMed] [Google Scholar]
- 47.Ohshima Y, Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987;195:247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- 48.O'Neill J P, Rogan P K, Cariello N, Nicklas J A. Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat Res. 1998;411:179–214. doi: 10.1016/s1383-5742(98)00013-1. [DOI] [PubMed] [Google Scholar]
- 49.Orkin S H, Kazazian H H., Jr The mutation and polymorphism of the human beta-globin gene and its surrounding DNA. Annu Rev Genet. 1984;18:131–171. doi: 10.1146/annurev.ge.18.120184.001023. [DOI] [PubMed] [Google Scholar]
- 50.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 51.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 52.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaal T D, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senapathy P, Shapiro M B, Harris N L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 55.Siebel C W, Admon A, Rio D C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 56.Siebel C W, Fresco L D, Rio D C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- 57.Sierakowska H, Szer W, Furdon P J, Kole R. Antibodies to hnRNP core proteins inhibit in vitro splicing of human beta-globin pre-mRNA. Nucleic Acids Res. 1986;14:5241–5254. doi: 10.1093/nar/14.13.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 59.Sterner D A, Berget S M. In vivo recognition of a vertebrate mini-exon as an exon-intron-exon unit. Mol Cell Biol. 1993;13:2677–2687. doi: 10.1128/mcb.13.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 61.Talerico M, Berget S M. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Treisman R, Orkin S H, Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 63.Valcarcel J, Singh R, Zamore P D, Green M R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Hoffmann H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 65.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 66.Wu S, Romfo C M, Nilsen T W, Green M R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 67.Zhang M Q. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 68.Zheng Z M, He P, Baker C C. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J Virol. 1996;70:4691–4699. doi: 10.1128/jvi.70.7.4691-4699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Z M, Huynen M, Baker C C. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc Natl Acad Sci USA. 1998;95:14088–14093. doi: 10.1073/pnas.95.24.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]