Abstract
We describe the design and attributes of a linear pentapeptide-like derivative (C14(ω5)OOc10O) screened for its ability to elicit bactericidal competences of plasma constituents against Gram-negative bacteria (GNB). In simpler culture media, the lipopeptide revealed high aptitudes to sensitize resilient GNB to hydrophobic and/or efflux-substrate antibiotics, whereas in their absence, C14(ω5)OOc10O only briefly delayed bacterial proliferation. Instead, at low micromolar concentrations, the lipopeptide has rapidly lowered bacterial proton and ATP levels, although significantly less than upon treatment with its bactericidal analog. Mechanistic studies support a two-step scenario providing a plausible explanation for the lipopeptide’s biological outcomes against GNB: initially, C14(ω5)OOc10O permeabilizes the outer membrane similarly to polymyxin B, albeit in a manner not necessitating as much LPS-binding affinity. Subsequently, C14(ω5)OOc10O would interact with the inner membrane gently yet intensively enough to restrain membrane-protein functions such as drug efflux and/or ATP generation, while averting the harsher inner membrane perturbations that mediate the fatal outcome associated with bactericidal peers. Preliminary in vivo studies where skin wound infections were introduced in mice, revealed a significant efficacy in affecting bacterial viability upon topical treatment with creams containing C14(ω5)OOc10O, whereas synergistic combination therapies were able to secure the pathogen’s eradication. Further, capitalizing on the finding that C14(ω5)OOc10O plasma-potentiating concentrations were attainable in mice blood at sub-maximal tolerated doses, we used a urinary tract infection model to acquire evidence for the lipopeptide’s systemic capacity to reduce the kidney’s bacterial loads. Collectively, the data establish the role of C14(ω5)OOc10O as a compelling antibacterial potentiator and suggest its drug-like potential.
Keywords: antibiotic resistance, innate immunity, synergism of action, peptidomimetics, mechanism of action
1. Introduction
Towards tackling the ongoing antibiotic resistance crisis, the search for antibiotics potentiators is gaining increasing interest [1,2,3] as a backup alternative for development of brand new substitutes. Namely, broadening the activity spectrum of established antibiotics counts as a tempting approach for minimizing the emergence and impact of resistance, particularly when the antibiotics inefficacy against Gram-negative bacteria (GNB) emanates from low permeability across the outer membrane (OM) [4,5]. In this sense, antimicrobial peptides (AMPs) represent appealing potential substitutes [6,7,8] as their antibacterial properties largely depend on molecular hydrophobicity which, in turn, can be synthetically fine-tuned with relative ease. Indeed, unlike outright hydrophobic AMPs that tend to disrupt both membranes of GNB abruptly [9,10,11], borderline hydrophobic analogs were proposed to maintain the OM permeabilization capacity but may additionally instigate little more than transient superficial damages to the inner membrane (IM) [12,13,14,15]. While not fully understood, the latter activity was linked to a variety of processes (such as restricted efflux [16], inhibited expression of antibiotic resistance factors [17,18,19] and pathogens sensitization to diverse antimicrobials [20,21,22,23,24]) some of which, may open a window of opportunity for therapeutic exploitation. Thus, potentiated agents would encompass exogenous antibiotics as well as endogenous bactericidal capabilities of innate plasma complements [25].
From this perspective, the AMP mimetic approach based on oligomeric acylated cations (OAC) [26] appears particularly suitable for engineering membrane active selective compounds [10,27,28], as it provides a simple, sensitive, and systematic tool for dissecting the relative importance of two most critical AMP attributes, charge and hydrophobicity, as will be illustrated herein. Recent OAC designs [29,30,31,32,33] have concentrated on the pentameric formula A1C1C2A2C3, where As and Cs represent acyl derivatives and cationic amino acids, respectively. Among the sequences investigated so far, C14KKc12K (Figure 1a) revealed broad-spectrum bactericidal properties [32]. However, at least from therapeutic perspectives, this compound also exhibited caveat properties (such as aggregation in aqueous media) [32] that seem interconnected to its high hydrophobicity. In addition, as will be revealed in the present study, C14KKc12K holds potential for high hemolytic activity and for inactivity in plasma. Therefore, we set out to address these flaws by investigating a series of lower hydrophobicity analogs working under the hypothesis that combining several fine-tuning strategies for gently reducing molecular hydrophobicity might succeed in converting the bactericidal pentapeptide to a borderline hydrophobic, more useful analog.
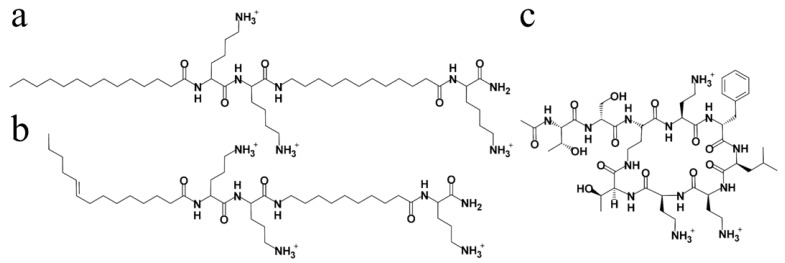
Figure 1.
Molecular structures of two main tested OACs and a comparator. (a) C14KKc12K (MW: 809); (b) C14(ω5)OOc10O (MW: 737); (c) PMB derivative SPR741 (MW: 992). In (a,b) the C-terminus is amidated, the letters C and c, respectively, denote an acyl and aminoacyl whose length (number of carbon atoms) is defined by the subscript; the parenthesis (ω5) in (b) denotes the position of a double bond; K and O represent the amino acids lysine and ornithine, respectively.
2. Materials and Methods
Bacteria:Escherichia coli strains: 25922 and 35218 (ATCC, Manassas, VA, USA), 14182 (clinical isolate), Ag100 and Ag100A [34] (ΔacrAB) are two K-12 isogenic mutantsand the engineered mutant ML-35p [35]. Additional ESKAPE species [36] tested: Klebsiella pneumoniae strains 1287 and 224 (clinical isolates), Acinetobacter baumannii ATCC strain 19606, Pseudomonas aeruginosa ATCC strains 9027 and 27853. Bacteria were grown in Luria–Bertani (LB) broth (0.5% NaCl, 0.5% yeast extract, 1% tryptone, pH = 7), except for E. coli ML-35p that was grown in tryptic-soy broth. Note that LB was used for comparison purposes with previous OAC publications, and that replacing LB with cation adjusted Mueller Hinton broth resulted in essentially identical outcomes [32].
Peptides: Unless otherwise stated, all peptides were produced in house by the solid phase method using 9-fluorenylmethoxycarbonyl active ester chemistry on rink amide 4-Methylbenzhydrylamine resin (100–200 mesh, Iris Biotech, Germany), as described [37].
Peptide self-assembly was determined by static light scattering measurements as described [32]. Hemolysis was determined by measuring hemoglobin leakage from washed human RBCs, as described [31]. Minimal inhibitory concentration (MIC) was determined using the microdilution assay, as described [32]. Bactericidal kinetics were determined by mixing bacteria with rifampin, OAC, or their combinations, as described [38]. Data were obtained from three independent assays performed in duplicate.
Bacterial sensitization: Sensitization to antibiotics was determined using the checkerboard method in presence of sub-MIC OAC (2.5, 5, and 10 μM), as described [31]. The synergistic effect of the combinations was expressed in terms of the Sensitization Factor (SF), where SF = (MIC antibiotic alone)/(MIC antibiotic upon combination). Data were obtained from three independent assays performed in duplicate.
Sensitization to plasma components was assessed by mixing bacteria with serial two-fold OAC dilutions in 80% human plasma (Israel Blood Bank) or plasma from the specified species (Technion preclinical research authority or VetSource), as described [29]. Data were obtained from three independent experiments.
Outer Membrane damages: OM permeabilization was investigated using the OM impermeable hydrophobic fluorescent dye 1-N-phenylnapthylamine (NPN), as described [39]. Data were obtained from three independent experiments performed in triplicate. For maximal fluorescence, 10 μM PMB [40,41] were used.
Dansyl-polymyxin displacement assay: Commercial PMB sulfate (Sigma P4119) was covalently attached to dansyl chloride and assessed as described [42]. Mono-dansyl Polymyxin B (DPMB) was purified by RP-HPLC. Next, 180 μL of 5 mM HEPES containing 3 μg/mL LPS (from E. coli or P. aeruginosa) and 2 μM mono-DPMB were incubated in a 96-well plate with 20 μL of the tested compound for 1.5 h at room temperature and fluorescence (excitation: 340 nm, emission: 485 nm) was measured immediately (Synergy HT, BioTek Instruments, Winooski, VT, USA).
Inner Membrane Damages: Damage inflicted to the cytoplasmic membrane was assessed using 3,3-dipropylthiadicarbocyanine iodide (DiSC3(5)), a lipophilic potentiometric dye that changes its fluorescence intensity in response to changes in transmembrane potential. Bacteria were grown overnight, diluted and at mid-log were adjusted to O.D = 0.1 (600 nm), centrifuged (10,000 RCF, 5 min), and re-suspended in the assay buffer (5 mM HEPES containing 20 mM glucose, 0.2 mM EDTA, and 50 mM KCl). Then, DiSC3(5) dye was added (to final concentration 4 μM) and incubated at 37 °C for 60 min in the dark to allow dye uptake. An aliquot (180 μL) of the bacterial suspension was placed in a 96-well plate and fluorescence was monitored until baseline stabilization (excitation, 622 nm; emission, 670 nm, monitored using Synergy HT, BioTek Instruments, Winooski, VT, USA). A solution (20 μL) containing OAC was added to obtain the desired final concentration. Fluorescence was immediately monitored continuously for 30 min. Reported results are from three independent experiments.
Intracellular ATP levels of E. coli 25922 (1.5 × 108 CFU/mL) were determined 1 h after incubation with or without OACs using commercial Luciferase-based bioluminescence Assay Kit HSII (Roche diagnostics GmbH, Mannheim, Germany), according to the manufacturer’s instructions.
Animals: All animal studies were performed using male ICR mice (25 ± 2 gr). The Technion Animal Care and Use committee approved all procedures, care, and handling of animals. Ethics approval codes: IL0800519, IL0640421, IL0550618, IL1811217.
The maximum tolerated dose (MTD) was determined after a single-dose subcutaneous (S.C) administration of C14(ω5)OOc10O using three mice per dose. Animals were monitored for adverse effects for 7 days after injection.
For efficacy assessments, three infection models were used including one with topical treatment and two with systemic treatment.
Excisional skin wound infection model: mice were anesthetized by intraperitoneal administration of a mixture of ketamine 100 mg/kg and xylazine 5 mg/kg in PBS and their backs shaved with electric clippers. The following day mice were similarly anesthetized and were administrated (S.C) 0.05 mg/kg buprenorphine (for pain relief). A 5 mm diameter piece of skin was removed from the middle of the shaved back, with sterile biopsy punch resulting in a full-thickness injury. A total of 20 µL of a mid-logarithmic culture, containing 5 × 106 CFU P. aeruginosa 27853 were applied on the wound. Then, 15 min after inoculation, about 50 µL of hypromellose gel (prepared as described [43]) containing OAC, antibiotic, or their combination were applied on the skin and covered with a piece of medical tape. As a control, the vehicle (drug-free gel) was similarly applied on the skin. After a treatment period of 4 h, about 8 mm diameter of skin surrounding the infected area was removed, suspended in PBS, homogenized, serially diluted 10-fold, and plated for CFU enumeration. The number of viable bacteria was determined after overnight incubation at 37 °C. The lower limit of detection was 50 CFU/wound.
Thigh infection model (TI): mice were inoculated intramuscularly with 1 × 106 CFU/mouse of mid-logarithmic E. coli 25922 and treated subcutaneously 1 and 3 h after inoculation. Mice were sacrificed 24 h after infection, their thighs excised, homogenized, serially diluted 10-fold, and plated for CFU enumeration. The number of viable bacteria was determined after overnight incubation at 37 °C. The lower limit of detection was 50 CFU/thigh.
-
Urinary tract infection model (UTI): mice were anesthetized via intraperitoneal injection of 100 mg/kg ketamine and 5 mg/kg xylazine. Mice penises were lubricated with an analgesic 2% lidocaine gel. Then, mice were infected with 50 µL of 1 × 108 CFU/mouse of E. coli UPEC CFT073, administrated by an intra-urethral injection using a catheter (24 GA, 0.156 IN, 0.7 × 14 mm). Mice were subcutaneously treated with OAC at 1 and 6 h post infection.
Mice were sacrificed 24 h post inoculation, the bladder and kidneys were excised, homogenized, serially diluted 10-fold, and plated for CFU enumeration. The number of viable bacteria was determined after overnight incubation at 37 °C. The lower limit of detection was 50 CFU/organ.
Blood Circulating Concentrations and Organ Bio-Distribution of C14(ω5)OOc10O
C14(ω5)OOc10O was subjected to preliminary pharmacokinetics (PK) analysis to determine its plasma concentrations or organ bio-distribution following S.C administration to non-neutropenic pathogen-free mice. OAC quantification was performed by LC-MS as follows: blood was drawn at various time intervals and plasma was separated by centrifugation (5000 RCF, 10 min). Organs were excised and homogenized. Then, OAC was extracted from both plasma and organ homogenates by incubation with 50% acetonitrile: 50% methanol at room temperature with shaking for 30 min, and subsequent centrifugation (5000 RCF, 10 min). Supernatants were diluted two-fold in distilled water and analyzed by LC-MS (ULC ultimate 3000 DIONEX, MS BRUKER Maxis impact). Quantification was based on standard calibration curves, prepared by spiking fresh mice plasma (for PK) or water (for organ bio-distribution) with various amounts of the tested OAC (final concentrations from 2.5 to 50 µg/mL) and subjected to an identical procedure as described above.
Statistics:p-values were calculated using a 1-tailed t test (assuming unequal variance). A p-value of <0.05 is considered statistically significant.
3. Results and Discussion
3.1. Derivatives Design and In Vitro Assessment
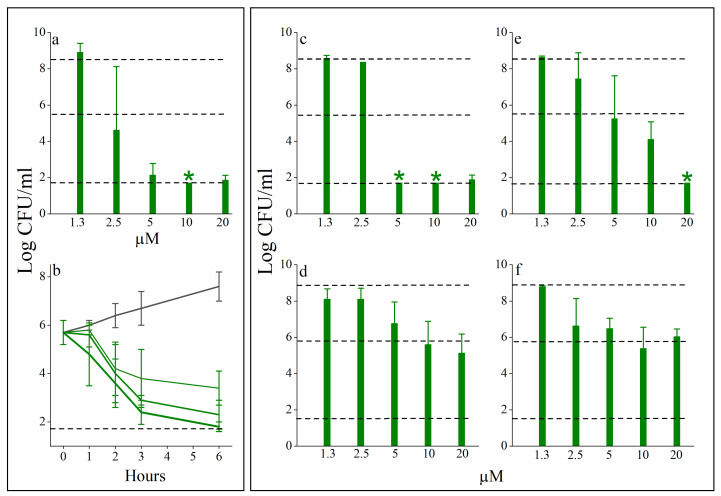
Table 1 outlines relevant biophysical properties of the reference sequence C14KKc12K [32] and those of six new derivatives. The first new derivative (C14OOc12O) represents a sequence alteration involving three lysine-to-ornithine substitutions, presumed to lead to some reduction in molecular hydrophobicity due to ornithine’s shorter side chain [29]. However, these substitutions appear not to lead to biophysical changes substantial enough to be detected by the assays used, including HPLC elution time and bactericidal activity (Figure 2). In contrast, the next alteration (C14OOc10O) which involved an additional dodecanoyl-to-decanoyl substitution of the A2 position, has affected each one of the tested properties. Namely, molecular hydrophobicity was reduced whereas self assembly and hemolysis were deferred to higher concentrations. Moreover, the growth inhibitory concentration in LB has risen (average MIC increased from 3–6 to 12–25 µM, as determined against 12 GNB strains selected from four different species). This was considered a pivotal step towards the desired conversion, as corroborated by the fact that C14OOc10O also induced a weak but significant antibacterial activity in plasma, namely an activity that was absent when C14KKc12K or C14OOc12O were similarly assessed. These effects have further intensified upon additionally substituting the A1 position with its unsaturated acyl derivative, yielding C14(ω5)OOc10O (Figure 1b). In fact, this alteration succeeded in reducing both aggregation and hemolysis to more significant extents (CAC and HC50 values became superior to the highest tested concentration, i.e., >100 µM each). Concomitantly, the antibacterial activity in LB was reduced (MIC became >50 µM), yet C14(ω5)OOc10O has induced a concentration-dependent bacterial killing effect in human plasma (Figure 3a) where the CFU counts were reduced to below the limit of detection at 5 or 10 µM (depending on the plasma donor). This effect revealed to correspond to a bactericidal activity (defined as the ability to reduce the CFU count by three orders of magnitude within 3 h) as evident from time-kill experiments (Figure 3b). Antibacterial activity was not specific to human plasma, as evidenced after a brief survey of diverse animal species (Figure 3 panels c to f). Thus, plasma from human and pig origins appear to be similarly affected by C14(ω5)OOc10O (Figure 3a,c, respectively) displaying higher antibacterial activity. Note, however, that even though cat and dog plasma samples exhibited high MIC values (≥20 µM) they have nonetheless significantly inhibited proliferation of bacterial inoculums at sub-minimal inhibitory concentrations (Figure 3d,f).
Table 1.
Biophysical attributes of relevant lipopeptide analogs.
| Lipopeptide Sequence | H (%) |
CAC (µM) |
HC50 (µM) |
MIC (µM) | |
|---|---|---|---|---|---|
| LB a Medium |
Human b Plasma |
||||
| * C14KKc12K | 55 | 20 ± 5 | 12 ± 1 | 3–6 | >20 |
| C14OOc12O | 55 | 15 ± 1 | 14 ± 4 | 3–6 | >20 |
| C14OOc10O | 53 | 45 ± 14 | 28 ± 2 | 12.5–25 | 10–20 |
| C14(ω5)OOc10O | 50 | >100 | >100 | >50 | 2.5–5 |
| C14(ω5)OOc8O | 48 | >100 | >100 | >50 | 2.5–5 |
| C14(ω5)OOc6O | 47 | >100 | >100 | >50 | 5 |
| OOc12O | 24 | >100 | >100 | >50 | >20 |
*, Reference peptide [32], shown for comparison purposes. Grey background specifies published data; H, hydrophobicity, defined as % acetonitrile required for elution in reversed phase HPLC using a C18 column. Values were rounded to nearest whole number; CAC, critical aggregation concentration, determined by light scattering in PBS; HC50, lipopeptide concentration that caused 50% hemolysis compared to water (determined by measuring hemoglobin leakage after 3 h incubation in PBS at 37 °C, using 1% washed human erythrocytes); MIC, minimal inhibitory concentration, determined in LB medium and in plasma, using OD measurements and CFU counts, respectively; a, mean of 12 GNB strains, specified in Section 2; b, mean of three donors, assessed on E. coli 25922.
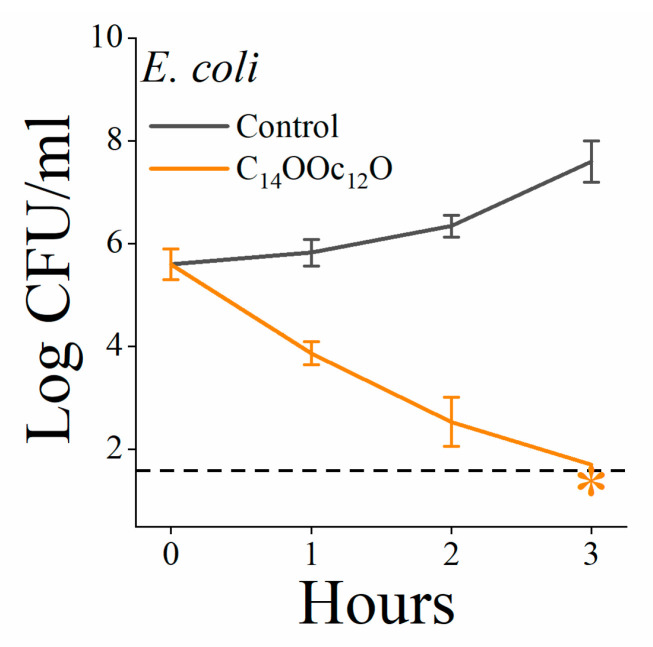
Figure 2.
Time-kill kinetics. The bactericidal activity of 10 µM C14OOc12O (orange trace) is demonstrated in comparison to the untreated control (black trace) in LB. The dashed horizontal line represents the limit of detection (log10 50 CFU/mL = 1.69). Orange asterisk (*) denotes lack of detectable CFU.
Figure 3.
Animal plasma antibacterial activities against E. coli 25922. (a) Dose-dependent reduction of colony forming units (CFU) as determined 24 h after exposure to C14(ω5)OOc10O in human plasma (80% final plasma concentration in PBS). Values represent the mean ± SD obtained from three individual plasma donors (mean inoculum was 1.6 ± 0.8 × 106 CFU/mL). (b) Time-kill kinetics determined upon E. coli exposure to 0, 2.5, 5, and 10 µM C14(ω5)OOc10O (from top to bottom, respectively). Values represent the mean ± SD obtained from two individual plasma samples, each subjected to two independent assays. Panels (c–f), respectively, show the OAC’s concentration-dependent activities in plasma samples obtained from pig, cat, sheep, and dog. The three dashed horizontal lines respectively represent the average CFU count of the vehicle control (upper line), the inoculum (middle line), and the limit of detection (lower line), i.e., log10 50 CFU/mL = 1.69. Green stars (*) denote lack of detectable CFU.
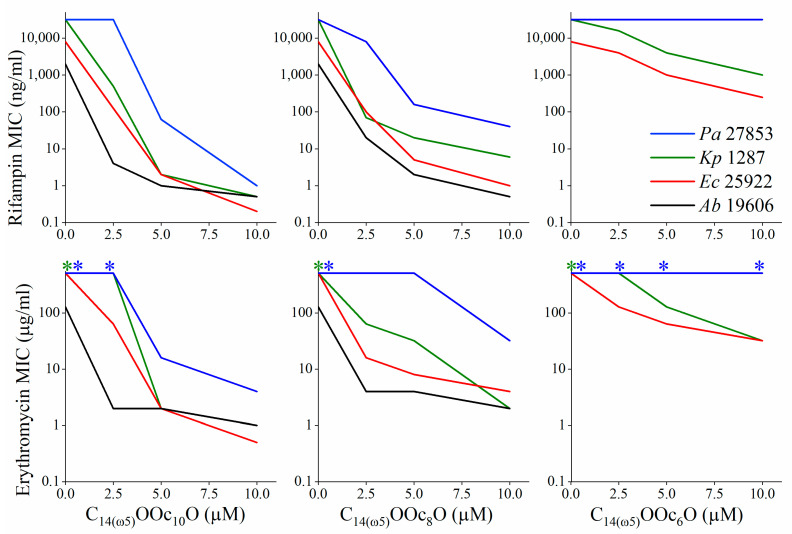
Additional efforts to reduce hydrophobicity by substituting the A2 position with less hydrophobic acyls led to no apparent improvement. Rather, C14(ω5)OOc8O and C14(ω5)OOc6O displayed a gradually weaker capacity to potentiate plasma (and antibiotics, as shown in Figure 4), suggesting that acyl length bridging between the cationic residues represents a critical parameter in defining the lipopeptide’s interaction with target bacteria and that it might optimally correspond to a c8–c10 aminoacyl. Finally, deleting the A1 position yielded a significantly more hydrophilic and virtually inactive analog (OOc12O) supporting the notion that the interactions leading to potentiation require an optimal hydrophobicity.
Figure 4.
Hydrophobicity dependence of antibiotics potentiation by lipopeptide analogs. Sensitization of GNB to rifampin and erythromycin (upper and lower panel, respectively) was assessed by MIC experiments performed in LB medium, in presence or absence of an OAC analog whose A2 position was gradually shortened (from c10 to c6). Pseudomonas aeruginosa 27853 (Pa, blue), Klebsiella pneumoniae 1287 (Kp, green), Escherichia coli 25922 (Ec, red), and Acinetobacter baumannii 19606 (Ab, black). Asterisks (*) denote MIC values greater than the highest tested concentration (i.e., >512 µg/mL).
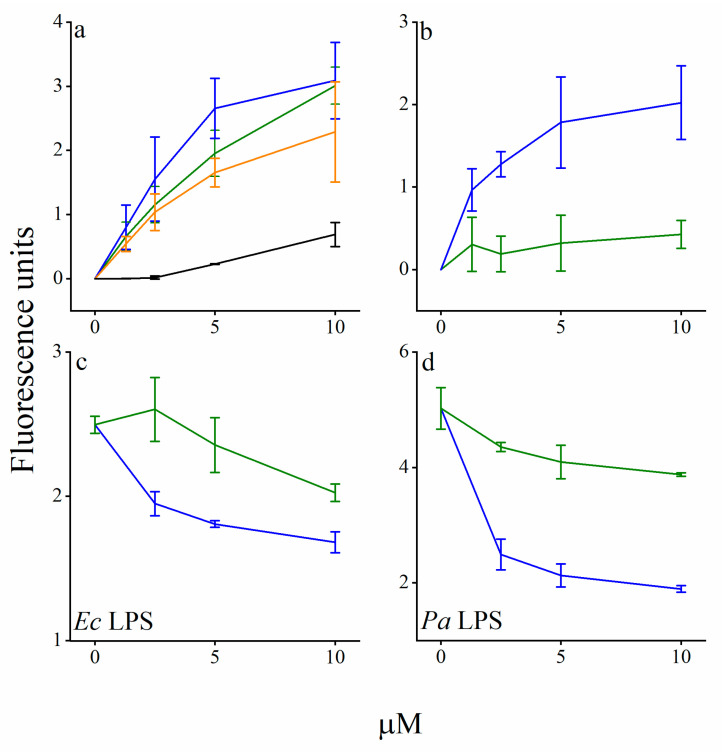
Whereas peptide hydrophobicity plays a critical role in the interactions with membranes [44,45,46], our finding that reducing hydrophobicity has increased the plasma antibacterial activity may be rather counter intuitive. Yet, it is reminiscent of reports concerning PMB [47] and related cyclic lipopeptides whose direct antibacterial potency was de-facto substituted for potentiation activity [48] merely by reducing molecular hydrophobicity (e.g., by deleting the fatty acid tail). Furthermore, the overall properties itemized in Table 1 were also reminiscent of previous findings using an OAC analog C10OOc12O [29,30] proposed to elicit improved activity of innate antibacterial proteins, allegedly through increasing OM permeability. Thus, to corroborate the possibility that similar principles might prevail for C14(ω5)OOc10O, this analog was similarly tested. Based on the NPN uptake assay (Figure 5a) both C14OOc12O and C14(ω5)OOc10O were able to permeabilize the E. coli OM but the unsaturated analog appeared more efficient and practically as efficient as PMB (p > 0.05) often considered as the “gold standard” for LPS-sequestering agents [49]. Figure 5b provides evidence for the notion that the NPN permeative capacities in presence of C14(ω5)OOc10O and PMB were significantly hampered by Mg2+ thereby suggesting a common (or similar) mode of interaction with the OM. Yet, the fact that PMB was less affected (i.e., the curves in presence and in absence of Mg2+ are quite similar for PMB as opposed to the greater change observed for the OAC), could argue for its higher affinity for the cation’s binding sites. This view is supported by additional experiments comparing their abilities to displace binding of dansylated PMB to LPS from E. coli (Figure 5c) or from P. aeruginosa (Figure 5d), thereby advocating that C14(ω5)OOc10O is nearly as efficient as PMB in OM permeabilization despite its lower LPS binding affinity. Note that the binding issue will be further elaborated below, in the proposed mechanism of action.
Figure 5.
Lipopeptide capacities to affect E. coli outer membrane permeability. (a) Outer membrane (OM) permeabilization to the hydrophobic dye NPN was determined 10 min after bacteria (E. coli 25922, 2 × 108 CFU/mL) were exposed to each peptide (5 µM) in NPN-containing HEPES at 37 °C. p << 0.05 for comparing C14OOc12O to C14(ω5)OOc10O or to PMB, and p > 0.05 for comparing C14(ω5)OOc10O to PMB. Color code (panels (a–d)): green, C14(ω5)OOc10O; orange, C14OOc12O; black, OOc12O; blue, polymyxin B (PMB). (b) OM permeabilization (as in panel a) in presence of 10 mM MgCl2; (c,d), Dansyl-PMB displacement assay using LPS from Escherichia coli and Pseudomonas aeruginosa, respectively, as measured 1.5 h after incubation in HEPES with C14(ω5)OOc10O (green) or PMB (blue).
3.2. C14(ω5)OOc10O Is a Remarkable Antibiotics Potentiator against GNB
Figure 4 shows the antibiotic’s MICs evolution in absence versus in presence of an adjuvant (C14(ω5)OOc10O and analogs) at a specified sub-MIC concentration as assessed for rifampin and erythromycin against four GNB species. Figure 4 (left-most upper panel) indicates that while the concentration-dependent trends exhibited some interspecies differences, C14(ω5)OOc10O was nonetheless able to potentiate rifampin’s action against all four bacterial species, reducing the MIC against E. coli and P. aeruginosa, from 8 and 32 μg/mL to 0.25 and 1 ng/mL, respectively (i.e., at 10 μM C14(ω5)OOc10O, rifampin’s MIC were reduced by 32,000 fold for both species). Similarly, rifampin’s MIC against K. pneumoniae and A. baumannii were both reduced from 32 and 2 μg/mL, respectively, to 0.5 ng/mL. Remarkably, C14(ω5)OOc10O has reduced rifampin’s MIC values against all four GNB species to values well below the susceptibility breakpoint of staphylococcus species (i.e., 1 µg/mL, according to the Clinical Standards Institute) [50]. Noteworthy also is the fact that the lipopeptide’s capacity to reduce the antibiotic’s MIC was hydrophobicity dependent (compare left panel to middle and right panels), thereby correlating rifampin potentiation against GNB and their OM permeabilization (evidenced for E. coli in Figure 5). These data again strengthen the notion of a possible role (yet to be determined) for acyl bridge length in rifampin’s permeation and, moreover, highlight a possible causative parallelism between potentiation of antibiotics and potentiation of plasma antimicrobial constituents.
Table 2 shows the sensitization factor (SF) values of two additional published rifampin potentiators, as compared at a single concentration (8 µg/mL each, i.e., 10 µM for C14(ω5)OOc10O). C14(ω5)OOc10O was often more potent than the most effective potentiator OAC published so far, i.e., C10OOc12O [29]. C14(ω5)OOc10O was also more potent than the PMB derivative SPR741 [48,51] (Figure 1c). Combined, these data suggest that flexible smaller compounds may be more advantageous for efficient antimicrobials potentiation against GNB. Possibly, the OAC’s relatively lower LPS binding affinity (Figure 5) could play a facilitating role as such compounds would be less restrained from engaging in additional interactions, for example.
Table 2.
Sensitization of Gram-negative bacteria to rifampin.
| Sensitization Factor at 8 µg/mL | |||
|---|---|---|---|
| Bacteria | C14(ω5)OOc10O | C10OOc12O | SPR741 |
| Kp | 64,000 | 8000 [30] | 32 [51] |
| Ec | 32,000 | 16,000 [30] | 8192 [51] |
| Pa | 32,000 | 1000 | 5 [48] |
| Ab | 4000 | 4000 | 256 [48] |
Comparing C14(ω5)OOc10O sensitization extents with those of two published adjuvants; sensitization factor is the ratio (rifampin MIC alone)/(rifampin MIC in combination) at the specified adjuvant concentration; Kp, Klebsiella pneumoniae; Ec, Escherichia coli 25922; Pa, Pseudomonas aeruginosa 27853; Ab, Acinetobacter baumannii 19606; highlighted in bold fonts are values determined in the present study. Note: SF values of the PMB analog SPR741 were obtained using the same Ab, Pa, and Ec (but not Kp) strains.
3.3. Mechanistic Studies
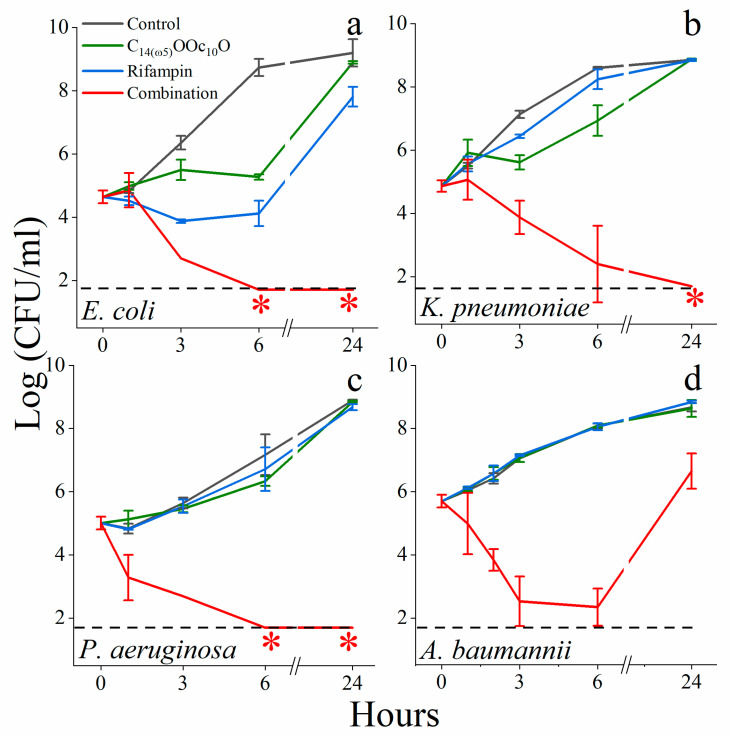
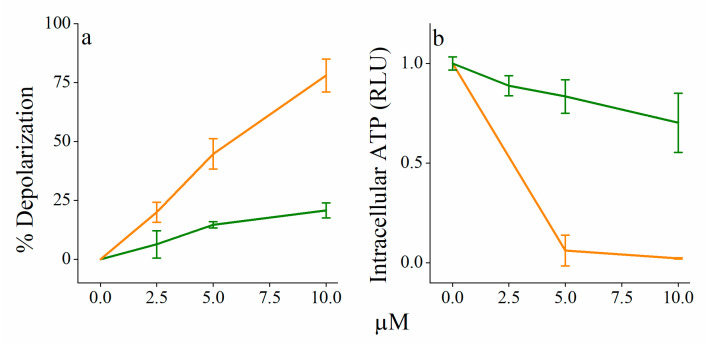
To gain insight into the specific role of each protagonist in the synergistic pair, we determined the survival kinetics under synergistic conditions (i.e., bacteria were exposed to solutions composed of 10 μM C14(ω5)OOc10O or/and 4 ng/mL rifampin) as summarized in Figure 6. The data suggest some interspecies fluctuations in terms of relative effect(s) exerted by each compound on each bacterial species. However, C14(ω5)OOc10O and rifampin were individually only capable of delaying proliferation (at most), whereas their combination was bactericidal against each of the tested species. Such an outcome sits well with the notion that C14(ω5)OOc10O merely facilitates rifampin’s inherent bactericidal mode of action by increasing its cytoplasmic accumulation. A similar view was proposed for C10OOc12O [29] and polymyxin analogs [20,47,48]. The individual time-kill curves obtained with different GNB species may well illustrate this general idea, where Figure 6a, in particular, was key to our interpretation of the survival kinetics, as follows: upon exposure to rifampin alone, E. coli bacteria exhibited a transient static phase that lasted at least 6 h before eventually fading out, reaching normal growth levels after 24 h. Figure 6a also indicates that in absence of rifampin, C14(ω5)OOc10O too has weakly inhibited bacterial proliferation, unlike its saturated analog that was responsible for rapid bacterial death at this concentration range (Figure 2) allegedly due to abrupt IM disruption (Figure 7). Conceivably, therefore, this lack of drastic IM damage in itself raises the possibility that C14(ω5)OOc10O exerts a similar but weaker damage as reported for equivalent lipophilic compounds that mildly affect IM functions (such as delocalization of membrane proteins [14,52], partial respiration inhibition [53], and/or dissipation of the transmembrane potential [15,54]). Such damages were proposed for various borderline hydrophobic membrane-active compounds found to have temporarily halted proliferation [12] and thus prompted us to monitor the lipopeptide’s effect on the transmembrane potential. For lack of available direct methods, we used the transmembrane potential sensitive dye, DiSC3(5) considering the fluorescent signal released in presence of the bactericidal OAC C12K-7α8 (used as positive control) to reflect lethal depolarization [26]. Indeed, depolarization by the bactericidal analog, C14OOc12O, displayed a significant dose-response (Figure 7a), whereas the concentration-dependent depolarization obtained at sub-MIC values of C14(ω5)OOc10O supports the notion that even at the high concentration of 10 μM, only partial depolarization was produced, thereby reinforcing its borderline hydrophobic status.
Figure 6.
Time-kill of selected ESKAPE bacteria.Bacteria were cultured in LB medium in absence of a drug (black traces) or in presence of 10 μM C14(ω5)OOc10O (green traces), 4 ng/mL rifampin (blue traces), or their combination (red traces). Error bars represent standard deviations. The dashed horizontal line represents the limit of detection (log10 50 CFU/mL = 1.69). Red asterisks denote lack of detectable CFU.
Figure 7.
Evidence for proton and ATP leakage across the inner membrane. (a) Dissipation of the transmembrane potential in E. coli 25922 (8.8 ± 1.8 × 107 CFU/mL) pre-incubated with DiSC3(5) as determined 15 min after exposure to C14OOc12O (orange) or to C14(ω5)OOc10O (green). Data represent percent depolarization as compared to the positive control, 50 µM C12K-7α8 [10]. (b) Intracellular ATP concentrations were determined after 1 h incubation with C14(ω5)OOc10O (green) or C14OOc12O (orange) using E. coli 25922 at 5 × 107 CFU/mL. RLU, relative luminescence units.
Furthermore, bacterial ATP concentrations provided additional evidence that support the occurrence of such mild IM damages (Figure 7b) as both the bactericidal and adjuvant analogs (C14OOc12O and C14(ω5)OOc10O, respectively) have reduced the intracellular ATP content but the unsaturated analog was less potent, consistently exhibiting significantly lower ATP levels. We submit that lower ATP content could represent a direct consequence of depolarization and perhaps even reflect its extent, for instance if the periplasmic protons required for ATP production [55] leak back into the cytoplasm through cracks allegedly produced by lipopeptide–IM interaction, as proposed for respiration decoupling agents [56].
Another line of supporting evidence emerged by inquiring expected consequences of these alleged damages. Since both protons and ATP are required to fuel efflux pumps [57] actions, their reduced concentrations might decrease drug expulsion, which could translate into increased potency of efflux substrates. This hypothesis is supported by the fact that LL-37 (a host defense peptide) [58] or erythromycin [59] (a macrolide antibiotic), are considered as established efflux substrates of the resistance-nodulation-division (RND) superfamily of pump proteins. Indeed, both are inefficient against GNB (Table 3), yet, both have exhibited higher potency against the efflux-deficient mutant strain Ag100A [60]. Under these conditions, C14(ω5)OOc10O (but not C14OOc12O) also behaved as an efflux substrate. Surprisingly (despite its efflux) C14(ω5)OOc10O managed to overcome resilience to erythromycin in all four GNB species, including in extremely resistant strains whose MIC was >512 µg/mL (lower panels of Figure 4). The fact that Pseudomonas bacteria were least sensitized may be precisely linked to their propensity to counteract the action of toxic drugs via overexpression of efflux pumps [57]. In addition, it is worth noting the striking resemblance between erythromycin’s potentiation trend and that of rifampin (compare upper and lower panels of Figure 4) as it argues that OM permeabilization could be implicated in potentiation of both drugs.
Table 3.
Effect of RND pumps on MIC values of diverse antimicrobials.
| Tested Compound | MIC (µM) | |
|---|---|---|
| Ag100 | Ag100A | |
| LL-37 | 22.2 | 1.1 |
| Erythromycin | 174.4 | 10.9 |
| C14(ω5)OOc10O | 25 | 6.2 |
| C14OOc12O | 3.1 | 3.1 |
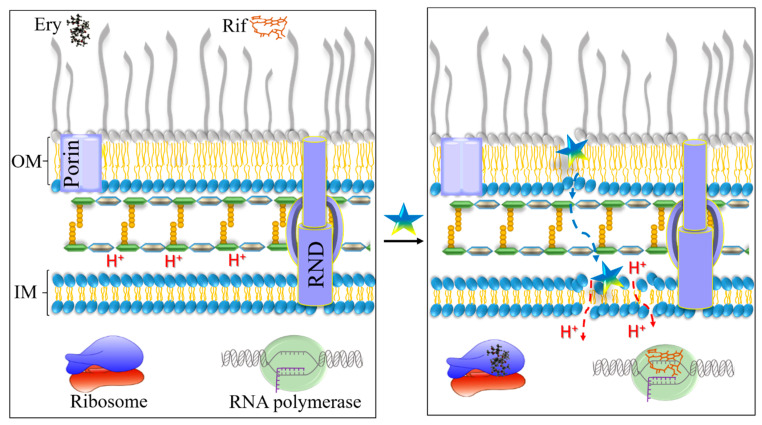
The overall findings can be assembled to draw a plausible two-stage scenario attempting to explain how C14(ω5)OOc10O might potentiate the action of such diverse antibacterial substances against GNB (Figure 8). The first stage follows the path described for many cationic host defense peptides, e.g., PMB [61]. Accordingly, bactericidal and potentiator analogs would readily cross the OM, namely as portrayed by the self-promoted uptake hypothesis [62] where the peptide’s bulkier molecular volume causes the OM permeabilization by forcing local reorganization of peptide–lipid A complexes into unstable mixtures that facilitate OM crossing and periplasm invasion. Once in the periplasm, however, these lipopeptide analogs display drastically divergent behaviors: C14OOc12O may imbed deeply within the IM thereby inducing its disruption and rapid death, as observed experimentally (Figure 2). Such an outcome is less likely with C14(ω5)OOc10O for two tightly linked reasons: binding affinity and efflux pumps. Indeed, less hydrophobic analogs typically display a lower membrane-binding affinity [33,63]. As a result, they are more likely to be expelled due to their lingering in the aqueous phase, instead of building up high membrane-bound concentrations leading to lethal membrane perturbations. Data shown in Table 3 argue that C14(ω5)OOc10O is an efflux substrate, unlike C14OOc12O. Thus, if part of C14(ω5)OOc10O molecules manage only a superficial integration of the IM [33,36,60] they could generate milder membrane perturbations (e.g., proton leaks?), eventually leading to partial dissipation of the transmembrane potential. The ensuing dwindled level of periplasmic protons is likely to affect a broad range of membrane functions including efflux which, in GNB, is often carried out by RND [64] and/or ABC [57] pumps. Collectively, therefore, the data support the view that GNB sensitization to erythromycin could be a consequence of lower bacterial respiration and ATP chemiosmosis (Figure 7). We propose that bacterial sensitization to animal plasma could be explained by these or similar considerations, i.e., if plasma resistance of the tested bacteria is related to low permeability of host defense proteins and peptides and/or to their efflux [16], both problems would be addressed by C14(ω5)OOc10O, as reported herein. Alternatively, plasma resistance may be linked to bacterial virulence factors (e.g., pseudomonal alkaline protease which cleaves C2 complements, thereby blocking both classical and lectin pathways) [65]. In this case too, C14(ω5)OOc10O might overcome the problem, as suggested by investigations of analogous borderline hydrophobic OACs linking partial depolarization of staphylococci to inhibition of virulence and resistance factors [18,19].
Figure 8.
Proposed mechanism for GNB sensitization by facilitating drug interactions with their cytoplasmic targets. The left panel depicts the general organization of the two-membrane system prevailing in GNB cell wall. The outer membrane (OM) and inner membrane (IM) are separated by a peptidoglycan-containing periplasmic space where protons (H+) normally accumulate to support the trans-membrane potential. Porin and RND, respectively, represent a passive and an energy-dependent metabolic protein gate. Hydrophobic antibiotics such as rifampin (Rif) and erythromycin (Ery) are depicted floating above the OM layer, to reflect their low permeability, impeding interaction with their cytoplasmic targets (the RNA polymerase and ribosome, respectively). The right panel highlights the reported effects of C14(ω5)OOc10O (represented by a pentameric star): initially, the lipopeptide destabilizes the OM thereby facilitating OM translocation of itself and that of low permeability antibiotics. Its subsequent superficial interaction with the IM would partially perturb various IM-linked functions such as active transport, hence the observed potentiation of efflux substrates, allegedly resulting from cytoplasmic accumulation, as exemplified by Ery.
The wild-type E. coli strain AG100 and its isogenic ΔacrAB mutant AG100A were used to determine the MIC of OACs and of two known acrAB-TolC substrates: the AMP LL-37 and the macrolide antibiotic erythromycin that are normally inefficient against GNB.
3.4. In Vivo Studies
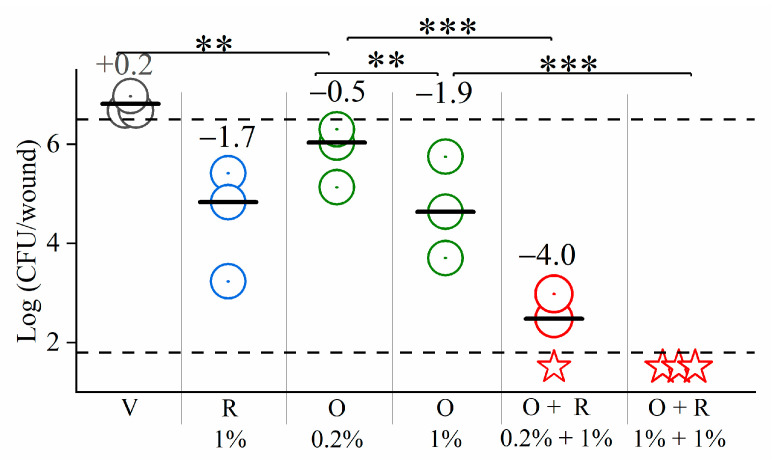
To evaluate the potential for therapeutic applications we performed preliminary toxicity, biodistribution, and efficacy experiments using a variety of mouse models. First, we tested the excisional skin wound infection model to assess the effect of topical treatment of P. aeruginosa, which was selected for its clinical importance and staggering ability to colonize skin wounds. As shown in Figure 9, the vehicle-treated control experiment enabled some increase in CFU count (i.e., displayed an average difference with initial inoculum of +0.2 log10 CFU) whereas application of high concentrations of rifampin or lipopeptide revealed a biocidal capacity. Thus, the rifampin containing gel (1%, i.e., 10 mg/mL, representing standard drug concentration for therapeutic gels and corresponding to MIC × 312) led to a reduction by 1.7 log10 units and the C14(ω5)OOc10O-containing gels (applied at two concentrations: 0.2 and 1%, respectively, corresponding to MIC × 20 and 200, assuming a MIC of 100 µM) succeeded in reducing the CFU counts significantly (i.e., by 0.5 and 1.9 log10 units, respectively). Yet, upon combination (i.e., when the wounds were treated with gels containing mixtures of both rifampin and C14(ω5)OOc10O) the treatments exhibited synergy of action at both concentrations (i.e., CFU counts were reduced by ~4 and >4 orders of magnitude, respectively). The ability to eradicate infecting organisms can be highly desirable, particularly for P. aeruginosa, known to render infection resolution very difficult, namely due to weak penetration of antibiotics and host clearance mediators such as antibodies and phagocytes [66,67].
Figure 9.
Excisional skin wound infection model. Plotted are colony forming units (CFU) counts of P. aeruginosa 27853 in each infected mouse (n = 3 per group) upon topical treatment by a drug-containing hypromellose gel. V, vehicle (drug-free hypromellose gel); R, rifampin; O, C14(ω5)OOc10O. Percentages denote the drugs concentration (w/v) in the gel. The numbers above each column indicate the change from the initial inoculum (represented by the upper dashed line). Horizontal bars specify the median. The limit of detection is represented by the lower dashed line (log10 50 CFU/mL = 1.69). Statistically significant differences are denoted by double (**) and triple (***) asterisks for p-value < 0.05 and p-value < 0.005, respectively. Red stars denote lack of detectable CFU.
Towards assessing C14(ω5)OOc10O ability to affect infections after systemic treatment, we initially determined the maximal tolerated dose (MTD) following subcutaneous injections to ICR uninfected mice at increasing doses (i.e., 0, 10, 20, 30, and 40 mg/kg/mouse). All mice survived the administered doses after monitoring for 7 days and no signs of toxicity-related stress were visible, arguing for an MTD value higher than 40 mg/kg, which is considerably higher than the highest MTD so far observed with published, systemically active OACs (i.e., typically 20–30 mg/kg) [29].
Guided by these findings, we next assessed the effect of systemic sub-MTD treatments using two E. coli mouse infection models: the thigh muscle infection and the urinary tract infection.
In the thigh model, mice were inoculated intramuscularly, treated subcutaneously, and their CFU/thigh enumerated 24 h after infection. In the UTI model, mice were infected by an intra-urethral injection, treated subcutaneously, and their CFU enumerated in bladder and kidneys, 24 h post inoculation. It should be noted that the treatment here involved only C14(ω5)OOc10O, in order to verify its capability to sensitize GNB to plasma bactericidal components, which could then result in curbing bacterial infections as observed in combination with the bactericidal antibiotic, rifampin (Figure 9).
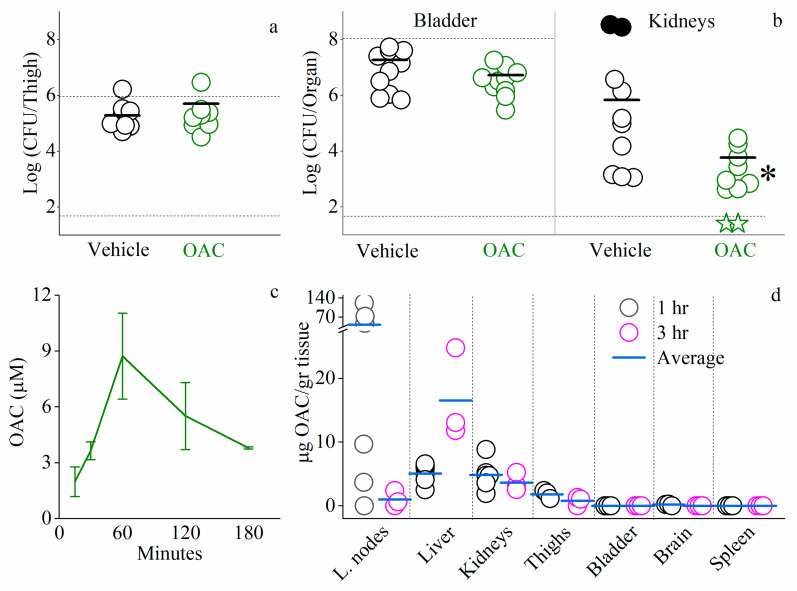
Figure 10a shows that C14(ω5)OOc10O was unable to reduce bacterial loads in the thigh model. The same treatment, however, was efficient in the urinary tract infection model (Figure 10b), having reduced the kidneys’ CFU counts (by up to >2 orders of magnitude) but not those of the bladder. Note that in the PBS-treated control experiment two infected mice died at ~14 h post-infection and hence, their CFU counts were not taken into consideration (if they were, then the gap with the treated group would be of four orders of magnitude).
Figure 10.
Systemic efficacy studies using mouse infection models. (a,b) Data points represent colony forming unit (CFU) counts harvested from infected mice (8 and 10 mice per group, respectively). Panel (a) depicts results of the thigh infection model where mice were inoculated intramuscularly with E. coli 25922 (upper dashed line represents the inoculum) and treated subcutaneously with C14(ω5)OOc10O (12.5 mg/kg) at 1 and 3 h post-infection. Panel (b) depicts results of the urinary tract infection model where mice were infected with E. coli UPEC CFT073 by intra-urethral injection and treated subcutaneously with C14(ω5)OOc10O (12.5 mg/kg) at 1 and 6 h post-infection. Solid circles denote mice that died before the experimental endpoint. Green stars denote lack of detectable CFU. Horizontal bars specify the average. The juxtaposed asterisk (*) denotes statistically significant difference (p-value = 0.003 or 0.008, if the dead mice are not included). The limit of detection is represented by the lower dashed line (log10 50 CFU/mL = 1.69). Panels (c,d) show the evolution of C14(ω5)OOc10O concentrations in mouse plasma (c) or in organs (d) after single subcutaneous administration (12.5 mg/kg) to ICR mice (data in c are from two mice/time points except for 60 and 180 that were 8 and 3, respectively) as determined by liquid chromatography–mass spectrometry. Error bars represent standard deviations. The limit of detection was 0.5 ppm (0.68 µM).
To shed some light into these seemingly conflicting in vivo findings, we attempted to evaluate the lipopeptide’s biodistribution. As shown in Figure 10c, its plasma concentrations (as determined by quantitative liquid chromatography–mass spectrometry following subcutaneous administration) argue that intact C14(ω5)OOc10O has rapidly achieved circulating levels of magnitudes comparable to those of classical antibiotics [68] and C10OOc12O [29]. Thus, the lipopeptide’s maximal plasma concentration (8.7 ± 2.3 µM) was reached approximately after 1 h from administration but potentiating concentrations (i.e., >2.5 µM) were maintained for at least 3 h thereafter. Based on these findings, we next estimated the lipopeptide’s organ biodistribution, targeting seven additional tissues of interest. Values presented in Figure 10d suggest that 3 h post administration, the lipopeptide preferentially accumulated in liver and kidneys at concentrations averaging at least 3 times higher than in the lymph nodes, thighs, bladder, brain, or spleen. To some extent, therefore, these findings tend to align well with those observed in Figure 10a,b and might explain them in the sense that the treatments efficacies correlate well with the adequate lipopeptide’s presence. Beyond findings evidently pertaining to the efficacy outcomes, the lipopeptide’s rapid passage in the lymph nodes and conversely, its accumulation in the liver (Figure 10d), are noteworthy for suggesting merits of future follow-on studies investigating implication on the antibacterial efficacy reported herein, as well as the potential toxicity that might emerge from excessive C14(ω5)OOc10O accumulation and/or its potential efficacy in affecting hepatic infections and tumors [69].
4. Conclusions
The presented data suggest that C14(ω5)OOc10O affects permeability of GNB similarly to reported OACs and polymyxins, albeit exhibiting lesser LPS-binding affinities but significantly higher potentiation capacities. Therefore, C14(ω5)OOc10O represents a new member of a growing family of linear miniature lipopeptides practically devoid of direct antibiotic activity at circulating concentrations but having the ability to facilitate the tasks of diverse and perhaps more appealing antibacterial substances as postulated in our previous study [29] of the analog, C10OOc12O where we showed that the OAC induced antibacterial activity in human plasma, which was antagonized by heat treatment, suggesting the proteinaceous nature of the antibacterial factors. This was supported by the synergistic effect observed between C10OOc12O and exogenous lysozyme in broth and serum media. Moreover, this activity was suppressible by anti-complement antibodies, pointing to the possibility that the lipo-peptides permeabilize GNB to plasma complements as observed with PMBN that sensitized E. coli and S. typhimurium to the bactericidal complements [20,23]. In fact, beyond antibiotics potentiation, sensitization of pathogenic GNB to antibacterial innate immune mechanisms could endow infected animals with the possibility to benefit from more holistic strategies for resolving infections while minimizing the risk of pro-inflammatory complications that might be associated with biocidal drugs.
Author Contributions
F.Z., synthesized reagents, performed research, analyzed data, and wrote the manuscript; O.M., performed research (results for C14KKc12K shown in Table 1 and in vivo experiments), analyzed data, and wrote the manuscript; A.M., designed experiments, analyzed data, and wrote the manuscript. All authors discussed the results and commented on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Israel Science Foundation (grant 1233/18).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Technion Animal Care and Use committee that approved all procedures, care, and handling of animals. Ethics approval codes: IL0800519, IL0640421, IL0550618, IL1811217.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Theuretzbacher U., Outterson K. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020;18:275–285. doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zabawa T.P., Pucci M.J., Parr T.R., Lister T. Treatment of Gram-negative bacterial infections by potentiation of antibiotics. Curr. Opin. Microbiol. 2016;33:7–12. doi: 10.1016/j.mib.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Wright G.D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Masi M., Réfregiers M., Pos K.M., Pagès J.M. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2017;2:17001. doi: 10.1038/nmicrobiol.2017.1. [DOI] [PubMed] [Google Scholar]
- 5.Silver L.L. A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem. 2016;24:6379–6389. doi: 10.1016/j.bmc.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Lazzaro B.P., Zasloff M., Rolff J. Antimicrobial peptides: Application informed by evolution. Science. 2020;368:eaau5480. doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020;19:311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 8.Mishra B., Reiling S., Zarena D., Wang G. Host defense antimicrobial peptides as antibiotics: Design and application strategies. Curr. Opin. Chem. Biol. 2017;38:87–96. doi: 10.1016/j.cbpa.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epand R.F., Maloy W.L., Ramamoorthy A., Epand R.M. Probing the “ Charge Cluster Mechanism ” in Amphipathic Helical Cationic. Biochemistry. 2010;49:4076–4084. doi: 10.1021/bi100378m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epand R.M., Rotem S., Mor A., Berno B., Epand R.F. Bacterial membranes as predictors of antimicrobial potency. J. Am. Chem. Soc. 2008;130:14346–14352. doi: 10.1021/ja8062327. [DOI] [PubMed] [Google Scholar]
- 11.Westerhoff H.V., Juretid D., Hendler R.W., Zasloff M. Magainins and the disruption of membrane-linked free-energy transduction. Proc. Natl. Acad. Sci. USA. 1989;86:6597–6601. doi: 10.1073/pnas.86.17.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneti G., Meir O., Mor A. Controlling bacterial infections by inhibiting proton-dependent processes. BBA Biomembr. 2015;1858:995–1003. doi: 10.1016/j.bbamem.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Hicks D.B., Cohen D.M., Krulwich T.A. Reconstitution of Energy-Linked Activities of the Solubilized F1F0 ATP Synthase from Bacillus subtilis. J. Bacteriol. 1994;176:4192–4195. doi: 10.1128/jb.176.13.4192-4195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel M., Chiriac A.I., Otto A., Zweytick D., May C., Schumacher C., Gust R., Albada H.B., Penkova M., Krämer U., et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc. Natl. Acad. Sci. USA. 2014;111:1409–1418. doi: 10.1073/pnas.1319900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epand R.F., Pollard J.E., Wright J.O., Savage P.B., Epand R.M. Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins. Antimicrob. Agents Chemother. 2010;54:3708–3713. doi: 10.1128/AAC.00380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reens A.L., Crooks A.L., Su C.C., Nagy T.A., Reens D.L., Podoll J.D., Edwards M.E., Yu E.W., Detweiler C.S. A cell-based infection assay identifies efflux pump modulators that reduce bacterial intracellular load. PLoS Pathog. 2018;14:e1007115. doi: 10.1371/journal.ppat.1007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandenburg K., Heinbockel L., Correa W., Lohner K. Peptides with dual mode of action: Killing bacteria and preventing endotoxin-induced sepsis. BBA Biomembr. 2016;1858:971–979. doi: 10.1016/j.bbamem.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Kaneti G., Sarig H., Marjieh I., Fadia Z., Mor A. Simultaneous breakdown of multiple antibiotic resistance mechanisms in S. aureus. FASEB J. 2013;27:4834–4843. doi: 10.1096/fj.13-237610. [DOI] [PubMed] [Google Scholar]
- 19.Hershkovits A.S., Pozdnyakov I., Meir O., Mor A. Sub-inhibitory membrane damage undermines Staphylococcus aureus virulence. BBA Biomembr. 2019;1861:1172–1179. doi: 10.1016/j.bbamem.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Vaara M. Agents That Increase the Permeability of the Outer Membrane. Microbiol. Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker K.R., Jana B., Hansen A.M., Nielsen H.M., Franzyk H., Guardabassi L. Repurposing azithromycin and rifampicin against gram-negative pathogens by combination with peptidomimetics. Front. Cell. Infect. Microbiol. 2019;9:236. doi: 10.3389/fcimb.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L., Nonejuie P., Munguia J., Hollands A., Olson J., Dam Q., Kumaraswamy M., Rivera H., Corriden R., Rohde M., et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine. 2015;2:690–698. doi: 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaara M., Vaara T. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature. 1983;303:526–528. doi: 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- 24.Ruden S., Rieder A., Chis Ster I., Schwartz T., Mikut R., Hilpert K. Synergy Pattern of Short Cationic Antimicrobial Peptides Against Multidrug-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2019;10:2740. doi: 10.3389/fmicb.2019.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor P.W. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol. Rev. 1983;47:46–83. doi: 10.1128/mr.47.1.46-83.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radzishevsky I.S., Rotem S., Bourdetsky D., Navon-Venezia S., Carmeli Y., Mor A. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat. Biotechnol. 2007;25:657–659. doi: 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- 27.Livne L., Kovachi T., Sarig H., Epand R.F., Zaknoon F., Epand R.M., Mor A. Design and Characterization of a Broad -Spectrum Bactericidal Acyl-lysyl Oligomer. Chem. Biol. 2009;16:1250–1258. doi: 10.1016/j.chembiol.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Radzishevsky I.S., Kovachi T., Porat Y., Ziserman L., Zaknoon F., Danino D., Mor A. Structure-Activity Relationships of Antibacterial Acyl-Lysine Oligomers. Chem. Biol. 2008;15:354–362. doi: 10.1016/j.chembiol.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Jammal J., Zaknoon F., Kaneti G., Hershkovits A.S., Mor A. Sensitization of Gram-Negative Bacilli to Host Antibacterial Proteins. J. Infect. Dis. 2017;215:1599–1607. doi: 10.1093/infdis/jix119. [DOI] [PubMed] [Google Scholar]
- 30.Jammal J., Zaknoon F., Mor A. Eliciting improved antibacterial efficacy of host proteins in the presence of antibiotics. FASEB J. 2018;32:369–376. doi: 10.1096/fj.201700652r. [DOI] [PubMed] [Google Scholar]
- 31.Jammal J., Zaknoon F., Kaneti G., Goldberg K., Mor A. Sensitization of Gram-negative bacteria to rifampin and OAK combinations. Sci. Rep. 2015;5:srep09216. doi: 10.1038/srep09216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meir O., Zaknoon F., Cogan U., Mor A. A broad-spectrum bactericidal lipopeptide with anti-biofilm properties. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-02373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarig H., Livne L., Held-Kuznetsov V., Zaknoon F., Ivankin A., Gidalevitz D., Mor A. A miniature mimic of host defense peptides with systemic antibacterial efficacy. FASEB J. 2010;24:1904–1913. doi: 10.1096/fj.09-149427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okusu H., Ma D., Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redfield R. Antibiotic Resistance Threats in the United States. CDC; Atlanta, GA, USA: 2019. [Google Scholar]
- 36.Zaknoon F., Sarig H., Rotem S., Livne L., Ivankin A., Gidalevitz D., Mor A. Antibacterial properties and mode of action of a short acyl-lysyl oligomer. Antimicrob. Agents Chemother. 2009;53:3422–3429. doi: 10.1128/AAC.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaknoon F., Goldberg K., Sarig H., Epand R.F., Epand R.M., Mor A. Antibacterial properties of an oligo-acyl-lysyl hexamer targeting gram-negative species. Antimicrob. Agents Chemother. 2012;56:4827–4832. doi: 10.1128/AAC.00511-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hancock R.E.W., Farmer S.W., Li Z., Poole K. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob. Agents Chemother. 1991;35:1309–1314. doi: 10.1128/AAC.35.7.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daugelavicius R., Bakiene E., Bamford D.H. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 2000;44:2969–2978. doi: 10.1128/AAC.44.11.2969-2978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown P., Dawson M.J. Development of new polymyxin derivatives for multi-drug resistant Gram-negative infections. J. Antibiot. 2017;70:386–394. doi: 10.1038/ja.2016.146. [DOI] [PubMed] [Google Scholar]
- 41.Lehrer R.I., Barton A., Ganz T. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J. Immunol. Methods. 1988;108:153–158. doi: 10.1016/0022-1759(88)90414-0. [DOI] [PubMed] [Google Scholar]
- 42.Moore R., Bates N.C., Hancock R.E.W. Interaction of polycationic antibiotics with Pseudomonas aeruginosa Lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 1986;29:496–500. doi: 10.1128/AAC.29.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haisma E.M., Göblyös A., Ravensbergen B., Adriaans A.E., Cordfunke R.A., Schrumpf J., Limpens R.W.A.L., Schimmel K.J.M., Den Hartigh J., Hiemstra P.S., et al. Antimicrobial peptide P60.4Ac-containing creams and gel for eradication of methicillin-resistant Staphylococcus aureus from cultured skin and airway epithelial surfaces. Antimicrob. Agents Chemother. 2016;60:4063–4072. doi: 10.1128/AAC.03001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Dou X., Song J., Lyu Y., Zhu X., Xu L., Li W., Shan A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019;39:831–859. doi: 10.1002/med.21542. [DOI] [PubMed] [Google Scholar]
- 45.Wiradharma N., Sng M.Y.S., Khan M., Ong Z. Rationally Designed α -Helical Broad-Spectrum Antimicrobial Peptides with Idealized Facial Amphiphilicity. Macromol. Rapid Commun. 2013;34:74–80. doi: 10.1002/marc.201200534. [DOI] [PubMed] [Google Scholar]
- 46.Hollmann A., Martínez M., Noguera M.E., Augusto M.T., Disalvo A., Santos N.C., Semorile L., Maffía P.C. Role of amphipathicity and hydrophobicity in the balance between hemolysis and peptide-membrane interactions of three related antimicrobial peptides. Colloids Surf. B Biointerfaces. 2016;141:528–536. doi: 10.1016/j.colsurfb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Vaara M., Fox J., Loidl G., Siikanen O., Apajalahti J., Hansen F., Frimodt-Møller N., Nagai J., Takano M., Vaara T. Novel polymyxin derivatives carrying only three positive charges are effective antibacterial agents. Antimicrob. Agents Chemother. 2008;52:3229–3236. doi: 10.1128/AAC.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaara M., Siikanen O., Apajalahti J., Fox J., Frimodt-Møller N., He H., Poudyal A., Li J., Nation R.L., Vaara T. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob. Agents Chemother. 2010;54:3341–3346. doi: 10.1128/AAC.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mares J., Kumaran S., Gobbo M., Zerbe O. Interactions of lipopolysaccharide and polymyxin studied by NMR spectroscopy. J. Biol. Chem. 2009;284:11498–11506. doi: 10.1074/jbc.M806587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clinical and Laboratory Standards Institute M100-S26 . Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI; Wayne, PA, USA: 2015. Approved Standard. [Google Scholar]
- 51.Corbett D., Wise A., Langley T., Skinner K., Trimby E., Birchall S., Dorali A., Sandiford S., Williams J., Warn P., et al. Potentiation of antibiotic activity by a novel cationic peptide: Potency and spectrum of activity of SPR741. Antimicrob. Agents Chemother. 2017;61:1–10. doi: 10.1128/AAC.00200-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scocchi M., Mardirossian M., Runti G., Benincasa M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2015;16:76–88. doi: 10.2174/1568026615666150703121009. [DOI] [PubMed] [Google Scholar]
- 53.Lobritz M.A., Belenky P., Porter C.B.M., Gutierrez A., Yang J.H. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. USA. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu M., Maier E., Benz R., Hancock R.E.W. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 55.Boyer P.D. The ATP synthase—A splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 56.Chouchani E.T., Kazak L., Spiegelman B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019;29:27–37. doi: 10.1016/j.cmet.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Du D., Wang-Kan X., Neuberger A., van Veen H.W., Pos K.M., Piddock L.J.V., Luisi B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 58.Shafer W.M., Qu X.D., Waring A.J., Lehrer R.I. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zgurskaya H.I., Nikaido H. Bypassing the periplasm: Reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldberg K., Sarig H., Zaknoon F., Epand R.F., Epand R.M., Mor A. Sensitization of gram-negative bacteria by targeting the membrane potential. FASEB J. 2013;27:3818–3826. doi: 10.1096/fj.13-227942. [DOI] [PubMed] [Google Scholar]
- 61.Trimble M.J., Mlynarcik P., Kolar M., Hancock R.E.W. Polymyxin: Alternative Mechanisms of Action. Cold Spring Harb Perspect. Med. 2016;6:1–22. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hancock R.E.W. Alterations in structure of the cell envelope. Ann. Rev. Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 63.Gaidukov L., Fish A., Mor A. Analysis of Membrane-Binding Properties of Dermaseptin Analogues: Relationships between Binding and Cytotoxicity. Biochemistry. 2003;42:12866–12874. doi: 10.1021/bi034514x. [DOI] [PubMed] [Google Scholar]
- 64.Blair J.M.A., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. V Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 65.Laarman A.J., Bardoel B.W., Ruyken M., Fernie J., Milder F.J., Strijp J.A.G., van Suzan H., Rooijakkers M., Strijp J.A.G., van Rooijakkers S.H.M. Pseudomonas aeruginosa Alkaline Protease Blocks Complement Activation via the Classical and Lectin Pathways. J. Immunol. 2012;188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 66.Mangoni M.L., Mcdermott A.M., Zasloff M. Antimicrobial peptides and wound healing: Biological and therapeutic considerations. Exp. Dermatol. 2016;25:167–173. doi: 10.1111/exd.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfalzgraff A., Brandenburg K., Weindl G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 2018;9:1–23. doi: 10.3389/fphar.2018.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandes P.B., Hardy D.J., McDaniel D., Hanson C.W., Swanson R.N. In vitro and in vivo activities of clarithromycin against Mycobacterium avium. Antimicrob. Agents Chemother. 1989;33:1531–1534. doi: 10.1128/AAC.33.9.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiang Y., Da Y., Zhao Y., Qi S., Liu L., Lu L., Luo Q., Zhang Z.H. Melittin-lipid nanoparticles target to lymph nodesand elicit a systemic anti-tumor immune response. Nat. Commun. 2020;11:1110. doi: 10.1038/s41467-020-14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.