Abstract
Multicopper oxidases (MCOs) are a diverse group of enzymes that could catalyze the oxidation of different xenobiotic compounds, with simultaneous reduction in oxygen to water. Aside from laccase, one member of the MCO superfamily has shown great potential in the biodegradation of mycotoxins; however, the mycotoxin degradation ability of other MCOs is uncertain. In this study, a novel MCO-encoding gene, StMCO, from Streptomyces thermocarboxydus, was identified, cloned, and heterologously expressed in Escherichia coli. The purified recombinant StMCO exhibited the characteristic blue color and bivalent copper ion-dependent enzyme activity. It was capable of oxidizing the model substrate ABTS, phenolic compound DMP, and azo dye RB5. Notably, StMCO could directly degrade aflatoxin B1 (AFB1) and zearalenone (ZEN) in the absence of mediators. Meanwhile, the presence of various lignin unit-derived natural mediators or ABTS could significantly accelerate the degradation of AFB1 and ZEN by StMCO. Furthermore, the biological toxicities of their corresponding degradation products, AFQ1 and 13-OH-ZEN-quinone, were remarkably decreased. Our findings suggested that efficient degradation of mycotoxins with mediators might be a common feature of the MCOs superfamily. In summary, the unique properties of MCOs make them good candidates for degrading multiple major mycotoxins in contaminated feed and food.
Keywords: multicopper oxidase, mycotoxin, aflatoxin, zearalenone, degradation, mediator
1. Introduction
Mycotoxins are toxic fungal secondary metabolites that are widely distributed in contaminated feed and food, bringing about many adverse health effects on livestock and humans, as well as huge economic losses in animal husbandry and the food industry [1]. As of now, there are hundreds of types of mycotoxins that have been identified, but the most frequently observed mycotoxins in contaminated feed and food are aflatoxin B1 (AFB1), zearalenone (ZEN), deoxynivalenol, fumonisin B1, and ochratoxin A [2]. AFB1 is mainly produced by Aspergillus flavus and A. parasitica, displaying carcinogenic, teratogenic, and immunosuppressive toxicity [3], and has been recognized as a group I carcinogen by the International Agency for Research on Cancer [4]. ZEN is primarily produced by Fusarium graminearum, F. culmorum, F. cerealis, F. equiseti, and F. verticillioides, exerting reproductive toxicity, hepatotoxicity, immunotoxicity, and genotoxicity [5,6]. Moreover, according to the Food and Agriculture Organization of the United Nations report, about 25% of global food crops are contaminated with these mycotoxins, resulting in an economic loss of billions of dollars each year [7]. Therefore, efficient mycotoxin detoxification strategies are in great demand.
In comparison with traditional physical and chemical detoxification methods, the biological detoxification of mycotoxins using microorganisms and enzymes is one of the most promising methods because of its high efficiency, irreversibility, and environmental friendliness [8]. During the past three decades, a variety of pre- and post-harvest biological control strategies have been developed to reduce mycotoxin contamination in feed and food [9,10,11,12]. On the one hand, bacteria, such as Bacillus and Pseudomonas, and fungi belonging the genus Trichoderma are used as the main biocontrol agents to limit the growth of mycotoxin-producing molds at the pre-harvest stage [9]. On the other hand, different microorganisms, including bacteria, yeast, and fungi, as well as their enzymes, are adopted to transform mycotoxins into less toxic or nontoxic metabolites during the post-harvest period [12].
In recent years, the degradation of mycotoxins with ligninolytic microorganisms and their corresponding ligninolytic enzymes has received more and more attention from researchers [13,14,15,16,17,18]. Interestingly, the broad substrate specificity of ligninolytic enzymes enables them to degrade different structural types of mycotoxins, including AFB1, ZEN, deoxynivalenol, fumonisin B1, and ochratoxin A [16,17]. Meanwhile, ligninolytic enzymes, such as laccase and dye-decolorizing peroxidase, can significantly accelerate the degradation of mycotoxins in the presence of mediators [19,20]. These catalytic properties of ligninolytic enzymes make them promising candidates for mycotoxin degradation. Significantly, laccase belongs to a member of the multicopper oxidase superfamily that contains laccase, laccase-like multicopper oxidase, ferroxidase, and so on [21], and it is not yet clear whether other MCOs could be able to degrade multiple major mycotoxins. In addition, there is a lack of systematic assessments of lignocellulose-derived compounds as the natural mediators of MCOs for mycotoxin degradation.
Streptomyces species are well-known bacteria capable of lignin degradation, and their ligninolytic enzyme system comprises multicopper oxidase, dye-decolorizing peroxidase, and lignin peroxidase, based on the genome-wide annotation analysis [20,22]. In this study, a novel laccase-like multicopper oxidase, StMCO, from Streptomyces thermocarboxydus 41291, was heterogeneously expressed, purified, and biochemically characterized. Moreover, the AFB1 and ZEN degradation properties of purified recombinant StMCO, in the presence of different structural lignin model compounds or ABTS, were analyzed and evaluated. Furthermore, their degradation products were identified by UPLC-MS/MS.
2. Results and Discussion
2.1. Gene Cloning and Sequence Analysis of StMCO from S. thermocarboxydus
It had been reported that the ligninolytic enzyme system of S. thermocarboxydus 41291 consisted of multicopper oxidase and dye-decolorizing peroxidase [20]. In this study, one novel multicopper oxidase-encoding gene, StMCO, was cloned from the genomic DNA of S. thermocarboxydus 41291. It was composed of a 990 bp open reading frame encoding 329 amino acid residues with a calculated molecular weight of approximately 36 kDa. The deduced amino acid sequence contained a putative twin-arginine signal peptide of 31 amino acid residues for secretory expression. Based on the BLAST search in NCBI, StMCO only contained two cupredoxin domains, while most multicopper oxidases consisted of three domains [23,24]. Additionally, each cupredoxin domain of StMCO encompassed one copper binding site. The multiple sequence alignment further revealed that there was one T2/T3 trinuclear copper binding site (two conserved HxH motifs) and one T1 copper binding site (conserved HxxHxH and HCHxxxH motifs) in the first and second cupredoxin domain, respectively (Figure S1). Moreover, according to the number and location of the T1 copper binding sites, the two-domain multicopper oxidase (2dMCO) superfamily was subdivided into the following three subfamilies: A, B, and C [24]. Type A 2dMCO contained two T1 copper binding sites. In contrast, type B and C 2dMCO included one T1 copper binding site in the second and first domain, respectively. Taken together, StMCO belonged to type B 2dMCO.
2.2. Expression and Purification of StMCO
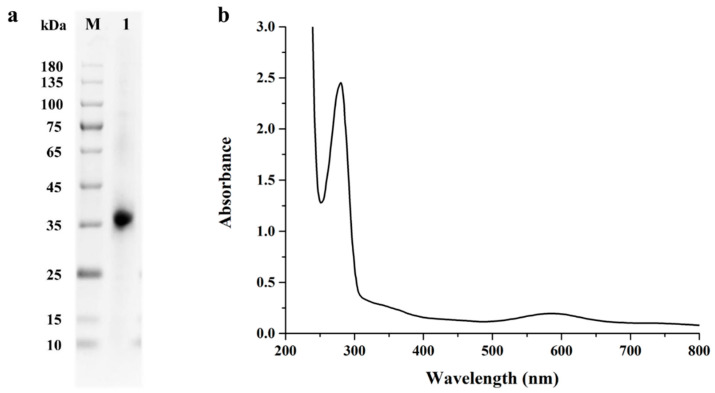
Given that Escherichia coli was the most popular approach for producing recombinant proteins, the recombinant plasmid pCold I-StMCO was transformed into E. coli Transetta (DE3) for heterologous expression. After isopropyl-β-D-thiogalactoside (IPTG) induction, there was obvious multicopper oxidase activity in the supernatant of the cell lysates, indicating that StMCO was successfully heterologously expressed in E. coli Transetta (DE3). After purification via nickel-immobilized metal ion affinity chromatography, a strong blue color was observed in the eluted fraction. Meanwhile, the SDS-PAGE analysis of the eluted fraction corresponding to StMCO exhibited a signal protein band at ~36 kDa (Figure 1a). Significantly, StMCO was found to generate a multimer with a molecular mass >100 kDa in native PAGE (Figure S2), which was consistent with the structural feature of active 2dMCO in multimeric forms (homotrimer or homohexamer) [25,26].
Figure 1.
The SDS-PAGE (a) and UV–visible spectroscopy (b) analysis of purified recombinant StMCO from S. thermocarboxydus. Lane M, the protein molecular mass marker; lane 1, the purified recombinant StMCO.
2.3. Biochemical Characterization of Purified Recombinant StMCO
Multicopper oxidase was reported to include at least two highly conserved copper centers, which are as follows: one T1 copper center for substrate oxidation and one T2/T3 trinuclear copper center for oxygen reduction [27]. Notably, the T1 copper center exhibited an absorption peak around 600 nm, giving rise to the characteristic blue color [28]. As shown in Figure 1b, the UV–visible spectrum of purified recombinant StMCO displayed a maximum absorption peak at ~600 nm, indicating that it was a typical blue multicopper oxidase.
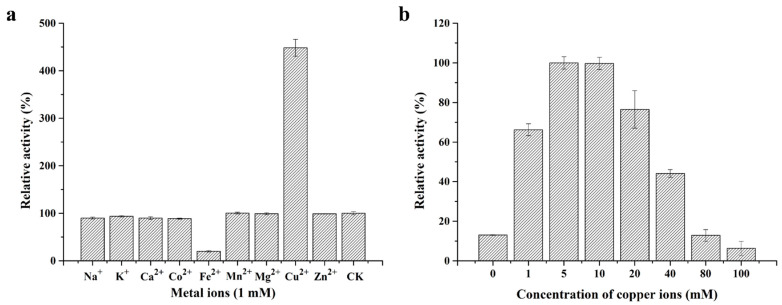
Considering that multicopper oxidases used metal ions as cofactors to oxidize different substrates [29], the effect of metal ions, at a concentration of 1 mM, on the activity of purified recombinant StMCO was evaluated. As shown in Figure 2a, most metal ions, such as Na+, K+, Ca2+, Co2+, Mn2+, Mg2+, and Zn2+, had no effect on multicopper oxidase activity. It was worth noting that Cu2+ could remarkably increase enzymatic activity by 348%, while Fe2+ displayed 80% inhibition of enzymatic activity. Moreover, the enzymatic activity of StMCO was further enhanced with increasing Cu2+ concentrations, and reached its maximum in the range of 5 to 10 mM Cu2+ (Figure 2b). When the concentration of copper ions was higher than 10 mM, the multicopper oxidase activity began to decrease, suggesting that the optimal Cu2+ concentration was 5−10 mM for maximal activity. These results were in agreement with those obtained from previous studies that found that increased copper concentrations led to higher enzymatic activity because Cu2+ was the essential cofactor for multicopper oxidases [29,30,31].
Figure 2.
The effect of metal ion (a) and copper ion concentration (b) on the activity of purified recombinant StMCO.
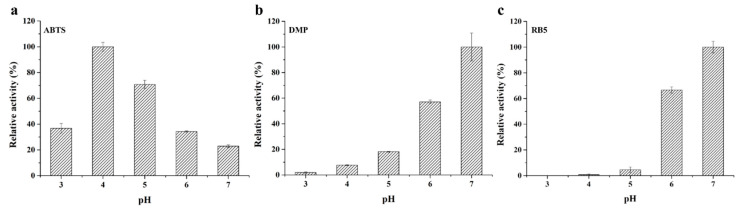
Multicopper oxidases, particularly laccase, were able to catalyze reactions involving a broad range of substrates, such as the model substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), lignin-related aromatic compounds, metal ions, and so on [32]. The purified recombinant StMCO was capable of oxidizing various substrates, including the model substrate ABTS, phenolic compound 2,6-dimethylphenol (DMP), and azo dye reactive black 5 (RB5), but it could not oxidize the phenolic compound guaiacol (GUA), non-phenolic compound veratryl alcohol (VA), and anthraquinone dye reactive blue 19 (RB19), which was similar to the substrate specificity of most other reported bacterial laccases [33,34]. However, the accurate classification of multicopper oxidase, assigned as laccase, still remained unclear [33], thus StMCO was termed a laccase-like multicopper oxidase. In addition, the optimum pH for the oxidation of different substrates by StMCO was 4.0 for ABTS, 7.0 for DMP, and 7.0 for RB5, respectively (Figure 3), exhibiting a substrate-dependent shift of optimum pH. The specific activity of purified recombinant StMCO towards ABTS, DMP, and RB5 at optimum pH was 0.259±0.009, 0.207±0.023, and 0.051±0.002 U/mg, respectively. Surprisingly, the specific activity of StMCO against DMP was one order of magnitude lower than that of ABTS, which might be attributed to the different bisubstrate reaction mechanism. It was reported that the bisubstrate models of ABTS and DMP oxidation by multicopper oxidases were ping-pong and Theorell–Chance, respectively [35].
Figure 3.
The optimum pH of purified recombinant StMCO for the oxidation of the following different substrates: ABTS (a), DMP (b), and RB5 (c).
2.4. Enzymatic Degradation of AFB1 and ZEN by StMCO
Recently, several laccases have been reported to be able to degrade multiple major mycotoxins, such as AFB1 and ZEN, in the presence of various mediators [19,36,37]. However, it was not clear whether mycotoxin degradation is the common feature of the multicopper oxidase superfamily. Besides, lignin-derived compounds as the natural mediators of MCOs for mycotoxin degradation lacked systematic evaluation. Herein, the degradation capacity of AFB1 and ZEN by the laccase-like multicopper oxidase StMCO, in the absence and presence of various structural lignin unit-derived mediators, was further evaluated.
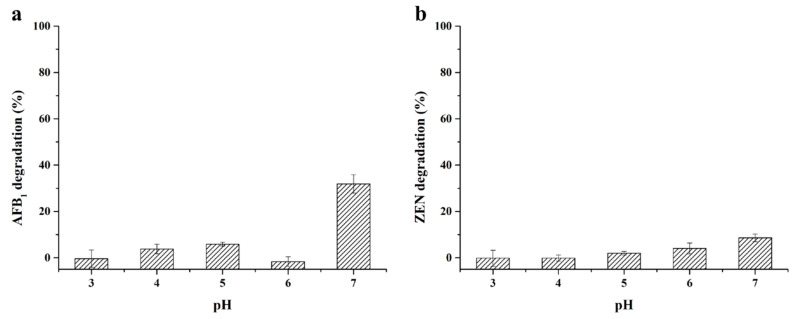
As reported, Lac2 from Pleurotus pulmonarius [36], Ery4 from P. eryngii [37], and BsCotA from Bacillus subtilis [19] were not able to directly degrade mycotoxins. However, as shown in Figure 4, StMCO could directly degrade AFB1 and ZEN in the absence of mediators, with pH 7 being the optimum pH. The degradation percentage of AFB1 and ZEN after the 24 h reaction was 31.87 ± 3.99% and 8.58 ± 1.63%, respectively, suggesting that enzyme–substrate interactions might exist between StMCO and mycotoxins.
Figure 4.
The optimum pH of purified recombinant StMCO for direct degradation of AFB1 (a) and ZEN (b) in 50 mM acetate buffer supplemented with 5 mM CuSO4 for 24 h at 30 °C.
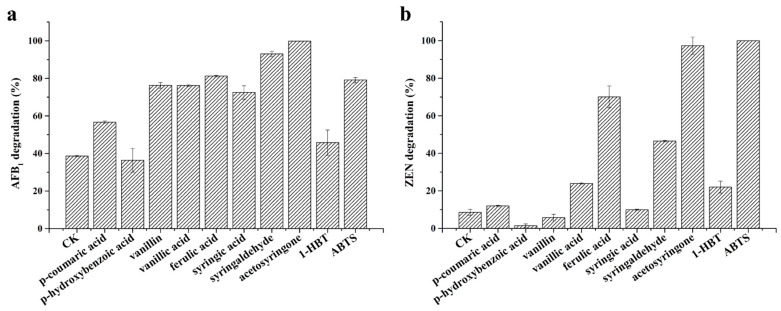
Moreover, different lignin unit-derived natural mediators, including H-type monomers (p-coumaric acid and p-hydroxybenzoic acid), G-type monomers (vanillin, vanillic acid, and ferulic acid), S-type monomers (syringic acid, syringaldehyde, and acetosyringone), 1-hydroxybenzotriazole (1-HBT), and ABTS, were chosen to explore the effect on the degradation of AFB1 and ZEN by StMCO. As shown in Figure 5, most mediators were found to significantly increase the degradation percentage of AFB1 and ZEN. As for AFB1, acetosyringone was the best mediator, with 99.85% degradation, followed by syringaldehyde (93.03%), ferulic acid (81.19%), ABTS (79.11%), vanillin (76.26%), vanillic acid (76.22%), syringic acid (72.48%), and p-coumaric acid (56.66%), while p-hydroxybenzoic acid and 1-HBT were ineffective (Figure 5a). With regards to ZEN, ABTS was the best performing mediator, with a degradation percentage of 100%, followed by 97.35% for acetosyringone, 70.05% for ferulic acid, 46.53% for syringaldehyde, 23.98% for vanillic acid, and 21.96% for 1-HBT, but no improvement in the degradation of ZEN was observed for p-coumaric acid, p-hydroxybenzoic acid, vanillin, and syringic acid (Figure 5b). These results indicated that lignin unit-derived natural mediators might be alternative mediators for mycotoxin degradation by StMCO, in terms of the economic cost and environmental friendliness. Moreover, the great improvement in AFB1 and ZEN degradation in the presence of acetosyringone and ABTS might be attributed to the generation of high potential radicals, aryloxy radicals, and ABTS++, respectively [36]. Generally speaking, these results proved that StMCO might be a promising candidate for the efficient and simultaneous degradation of multiple major mycotoxins, with the use of a single or multiple mediators.
Figure 5.
The effect of various mediators on the degradation of AFB1 (a) and ZEN (b) by 0.2 U/mL StMCO in 50 mM acetate buffer (pH 7.0) containing 1 mg/L AFB1 or ZEN, 5 mM CuSO4, and 1 mM mediator for 24 h at 30 °C.
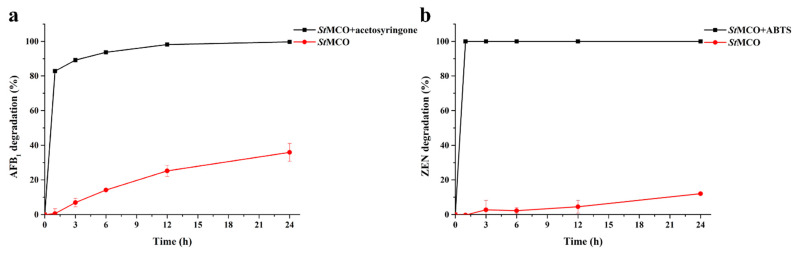
Furthermore, the time courses of AFB1 and ZEN degradation by StMCO, in the presence of their most efficient mediators, acetosyringone and ABTS, were determined. As shown in Figure 6, there was no significant change in the degradation of AFB1 and ZEN by StMCO in the absence of mediators after a 1 h reaction. In contrast, it was notable that AFB1 and ZEN were rapidly removed by StMCO in the presence of acetosyringone and ABTS, respectively.
Figure 6.
The time course analysis of AFB1 (a) and ZEN (b) degradation by 0.2 U/mL StMCO in 50 mM acetate buffer (pH 7.0) containing 1 mg/L AFB1 or ZEN, 5 mM CuSO4, and 1 mM acetosyringone or ABTS at 30 °C.
2.5. Identification of AFB1 and ZEN Degradation Products
Considering that the biological detoxification of mycotoxins was defined as the degradation or enzymatic transformation of mycotoxins into less toxic or nontoxic compounds [38], the degradation products of AFB1 and ZEN by StMCO, in the presence of the most efficient mediator, were identified by UPLC-MS/MS, and their biological toxicities were further elucidated.
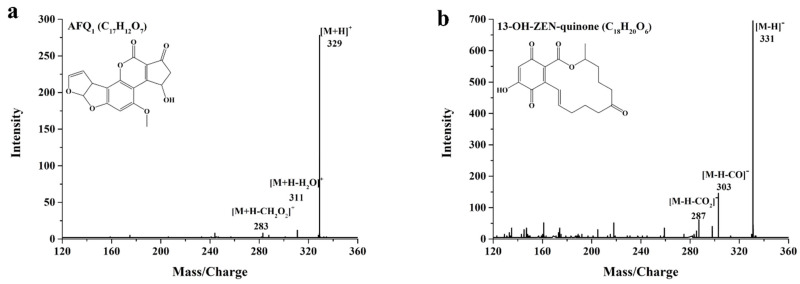
AFQ1 was the main degradation product of AFB1, corresponding to the parent ion at m/z 329 [M+H]+, and daughter ions at m/z 311 [M+H-H2O]+ and m/z 283 [M+H-CH2O2]+ (Figure 7a), though the AFQ1 content decreased slightly over time (Figure S3a). The identified degradation product AFQ1 was in accordance with the product of AFB1 degradation catalyzed by laccases [14,18], indicating that the ability to degrade mycotoxins might be the common feature of the multicopper oxidase superfamily. Notably, the toxic effects of AFQ1 on the chicken embryo were reported to be about 18 times less than AFB1 [39]. Moreover, AFQ1 had a lack of cytotoxicity in the human liver cells L-02 [18]. In addition, 13-OH-ZEN-quinone was found to be the main intermediate degradation product of ZEN, exhibiting the parent ion at m/z 311 [M-H]−, and daughter ions at m/z 303 [M-H-CO]− and m/z 287 [M-H-CO2]− (Figure 7b). With the prolongation of the reaction time, 13-OH-ZEN-quinone was further metabolized (Figure S3b). As reported for the relationship between the chemical structure and biological toxicity, hydroxylation of the aromatic moiety in ZEN exhibited a remarkable decrease in estrogenic activity when compared with ZEN [40]. Therefore, the biological toxicity of 13-OH-ZEN-quinone might be less toxic than ZEN.
Figure 7.
The UPLC-MS/MS analysis of AFB1 and ZEN degradation products, including AFQ1 (a) and 13-OH-ZEN-quinone (b) by 0.2 U/mL StMCO in 50 mM acetate buffer (pH 7.0) containing 1 mg/L AFB1 or ZEN, 5 mM CuSO4, and 1 mM acetosyringone or ABTS at 30 °C.
3. Conclusions
In this study, a novel laccase-like multicopper oxidase, StMCO, from Streptomyces thermocarboxydus, was identified, cloned, and heterologously expressed in E. coli. StMCO was a typical blue multicopper oxidase and showed copper-dependent enzyme activity. Most importantly, StMCO could not only directly degrade multiple major mycotoxins, including AFB1 and ZEN, but also accelerate the degradation of AFB1 and ZEN in the presence of various lignin unit-derived natural mediators and ABTS. Moreover, biological toxicities of their corresponding degradation products, AFQ1 and 13-OH-ZEN-quinone, were significantly removed. These findings indicated that MCOs could serve as a promising biological tool for degrading multiple major mycotoxins in contaminated feed and food.
4. Material and Methods
4.1. Substrates and Chemicals
Substrates ABTS, DMP, GUA, VA, RB5, RB19, AFB1, ZEN, and various structural mediators including p-coumaric acid, p-hydroxybenzoic acid, vanillin, vanillic acid, ferulic acid, syringic acid, syringaldehyde, acetosyringone, and 1-HBT were purchased from Sigma-Aldrich (St. Louis, USA). All other chemicals were of analytical grade and purchased from Sinopharm Chemical Reagent (Beijing, China).
4.2. Strains and Plasmid
S. thermocarboxydus 41291 was obtained from the Agricultural Culture Collection of China (Beijing, China). The E. coli strains Trans1-T1 and Transetta (DE3) were purchased from TransGen (Beijing, China). The expression vector pCold I was purchased from Takara (Beijing, China).
4.3. Cloning, Expression and Purification of StMCO
The StMCO-encoding gene devoid of its signal sequence was amplified from the mycelia of S. thermocarboxydus 41291 with gene-specific primers (StMCO-Nde I-F: 5′ ATCATCATATCGAAGGTAGGCATATGTCCACCACGGCGAGAACCGCG 3′; StMCO-Xba I-R: 5′ TTTTAAGCAGAGATTACCTATCTAGAGTGCGCGTGCCCGG ACTTCTC 3′). The PCR product was then ligated into the expression vector pCold I predigested with Nde I and Xba I to generate the recombinant plasmid pCold I-StMCO, which was transformed into E. coli Trans1-T1 for cloning and sequencing. After confirmation by sequencing, the recombinant plasmid pCold I-StMCO was extracted and subsequently transformed into Transetta (DE3) expression strain.
The E. coli Transetta (DE3) transformant containing pCold I-StMCO was picked and grown in LB broth supplemented with 100 μg/mL ampicillin and 20 μg/mL chloramphenicol at 37 °C to OD600 of 0.8−1.0, followed by adding IPTG and CuSO4 at a final concentration of 0.5 and 1 mM, respectively. After induction at 16 °C for 12 h, the cells were harvested by centrifugation, resuspended in binding buffer (20 mM sodium phosphate, 500 mM NaCl, pH 7.4), and lysed by sonication. The sonicated supernatant containing recombinant StMCO was then purified by nickel-immobilized metal ion affinity chromatography. The purity of recombinant StMCO was verified by 12% SDS-PAGE.
4.4. Biochemical Characterization of StMCO
The multicopper oxidase activity was determined by monitoring the oxidation of ABTS (ε420 = 36,000 M−1·cm−1) at 420 nm in 50 mM acetate buffer containing 1 mM ABTS, 5 mM CuSO4, and the appropriate diluted enzyme solution. One unit (U) of enzyme activity was defined as the amount of enzyme that oxidized 1 μmol of ABTS per min at 25 °C.
The UV–visible spectrum of purified recombinant StMCO was measured in phosphate buffer (20 mM, pH 7.4) in the range from 230 to 800 nm. The effect of metal ions such as Na+, K+, Ca2+, Co2+, Fe2+, Mn2+, Mg2+, Cu2+, and Zn2+ at a concentration of 1 mM on the activity of purified recombinant StMCO was evaluated in 50 mM acetate buffer (pH 4.0). The effect of copper ion concentration ranging from 1 to 100 mM on the activity of purified recombinant StMCO was determined in 50 mM acetate buffer (pH 4.0). The substrate specificity of purified recombinant StMCO was studied for the oxidation of six different substrates ABTS, DMP (ε470 = 12,100 M−1·cm−1), GUA (ε465 = 49,600 M−1·cm−1), VA (ε310 = 9,300 M−1·cm−1), RB5 (ε596 = 30,000 M−1·cm−1), and RB19 (ε595 = 10,000 M−1·cm−1) in 50 mM acetate buffer with pH ranging from 3.0 to 7.0.
4.5. Enzymatic Degradation of AFB1 and ZEN by StMCO
The evaluation of StMCO for AFB1 and ZEN degradation ability was carried out in 50 mM acetate buffer with pH ranging from 3.0 to 7.0 containing 1 mg/L AFB1 or ZEN, 5 mM CuSO4, and 0.2 U/mL StMCO for 24 h at 30 °C.
The effect of mediators, including p-coumaric acid, p-hydroxybenzoic acid, vanillin, vanillic acid, ferulic acid, syringic acid, syringaldehyde, AS, 1-HBT, and ABTS, on AFB1 and ZEN degradation by StMCO was determined in 50 mM acetate buffer (pH 7.0) containing 1 mg/L AFB1 or ZEN, 5 mM CuSO4, 1 mM mediator, and 0.2 U/mL StMCO for 24 h at 30 °C.
The time course of AFB1 and ZEN degradation by StMCO in the presence of the most efficient mediator was determined in 50 mM acetate buffer (pH 7.0) containing 1 mg/L AFB1 or ZEN, 5 mM CuSO4, 1 mM AS or ABTS, and 0.2 U/mL StMCO for 1, 3, 6, 9, 12, and 24 h at 30 °C.
The degradation of AFB1 and ZEN was analyzed by high-performance liquid chromatography coupled to RF-20A fluorescence detector using previous analytical methods [16]. In addition, their degradation products were identified by UPLC-MS/MS methods described in our previous studies [16,20].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13110754/s1, Figure S1. The amino acid sequence alignment of StMCO with a two domain multicopper oxidase SLAC from Streptomyces coelicolor. Blue boxes indicate the copper binding motifs. Figure S2. The native PAGE analysis of StMCO from S. thermocarboxydus. Lane M, the protein molecular mass marker; lane 1, the purified recombinant StMCO. Figure S3. The time-course analysis of AFB1 and ZEN degradation products by UPLC-MS/MS, including AFQ1 (a) and 13-OH-ZEN-quinone (b).
Author Contributions
X.Q.: Conceptualization, Methodology, Investigation, Data Curation, Writing—Original Draft, Visualization. Y.X.: Investigation. J.Z. (Jiahuan Zou): Investigation. X.S.: Conceptualization. X.W.: Methodology. Y.W.: Resources. J.Z. (Jie Zhang): Visualization. T.T.: Funding acquisition. B.Y.: Supervision, Project administration, Funding acquisition. H.L.: Conceptualization, Project administration, Writing—Review & Editing, Funding acquisition. H.H.: Project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2021YFC2100200), the State Key Laboratory of Animal Nutrition Project (2004DA125184G2101), and the China Agriculture Research System of MOF and MARA (CARS-41).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that they have no competing interests.
Key Contribution
The laccase-like multicopper oxidase StMCO could effectively degrade aflatoxin B1 and zearalenone in the presence of mediators, especially various lignin unit-derived natural mediators.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang C., Song G., Lim W. Effects of mycotoxin-contaminated feed on farm animals. J. Hazard. Mater. 2020;389:122087. doi: 10.1016/j.jhazmat.2020.122087. [DOI] [PubMed] [Google Scholar]
- 2.Pleadin J., Frece J., Markov K. Chapter eight—Mycotoxins in food and feed. In: Toldrá F., editor. Advances in Food and Nutrition Research. Academic Press; Cambridge, MA, USA: 2019. pp. 297–345. [DOI] [PubMed] [Google Scholar]
- 3.Pickova D., Ostry V., Toman J., Malir F. Aflatoxins: History, significant milestones, recent data on their toxicity and ways to mitigation. Toxins. 2021;13:399. doi: 10.3390/toxins13060399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q., Yang Q., Wu W. Progress on structured biosensors for monitoring aflatoxin B1 from biofilms: A review. Front. Microbiol. 2020;11:408. doi: 10.3389/fmicb.2020.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ropejko K., Twarużek M. Zearalenone and its metabolites-general overview, occurrence, and toxicity. Toxins. 2021;13:35. doi: 10.3390/toxins13010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai A., Das M., Tripathi A. Occurrence and toxicity of a fusarium mycotoxin, zearalenone. Crit. Rev. Food Sci. Nutr. 2020;60:2710–2729. doi: 10.1080/10408398.2019.1655388. [DOI] [PubMed] [Google Scholar]
- 7.Eskola M., Kos G., Elliott C.T., Hajšlová J., Mayar S., Krska R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25% Crit. Rev. Food Sci. Nutr. 2020;60:2773–2789. doi: 10.1080/10408398.2019.1658570. [DOI] [PubMed] [Google Scholar]
- 8.Ji C., Fan Y., Zhao L. Review on biological degradation of mycotoxins. Anim. Nutr. 2016;2:127–133. doi: 10.1016/j.aninu.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nešić K., Habschied K., Mastanjević K. Possibilities for the biological control of mycotoxins in food and feed. Toxins. 2021;13:198. doi: 10.3390/toxins13030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonçalves A., Gkrillas A., Dorne J.L., Dall’Asta C., Palumbo R., Lima N., Battilani P., Venâncio A., Giorni P. Pre- and postharvest strategies to minimize mycotoxin contamination in the rice food chain. Compr. Rev. Food Sci. Food Saf. 2019;18:441–454. doi: 10.1111/1541-4337.12420. [DOI] [PubMed] [Google Scholar]
- 11.Haque M.A., Wang Y., Shen Z., Li X., Saleemi M.K., He C. Mycotoxin contamination and control strategy in human, domestic animal and poultry: A review. Microb. Pathog. 2020;142:104095. doi: 10.1016/j.micpath.2020.104095. [DOI] [PubMed] [Google Scholar]
- 12.Li P., Su R., Yin R., Lai D., Wang M., Liu Y., Zhou L. Detoxification of mycotoxins through biotransformation. Toxins. 2020;12:121. doi: 10.3390/toxins12020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branà M.T., Sergio L., Haidukowski M., Logrieco A.F., Altomare C. Degradation of aflatoxin B1 by a sustainable enzymatic extract from spent mushroom substrate of Pleurotus eryngii. Toxins. 2020;12:49. doi: 10.3390/toxins12010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z., Li R., Ng T.B., Lai Y., Yang J., Ye X. A new laccase of Lac 2 from the white rot fungus Cerrena unicolor 6884 and Lac 2-mediated degradation of aflatoxin B1. Toxins. 2020;12:476. doi: 10.3390/toxins12080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tso K.-H., Lumsangkul C., Ju J.-C., Fan Y.-K., Chiang H.-I. The potential of peroxidases extracted from the spent mushroom (Flammulina velutipes) substrate significantly degrade mycotoxin deoxynivalenol. Toxins. 2021;13:72. doi: 10.3390/toxins13010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin X., Su X., Tu T., Zhang J., Wang X., Wang Y., Wang Y., Bai Y., Yao B., Luo H., et al. Enzymatic degradation of multiple major mycotoxins by dye-decolorizing peroxidase from Bacillus subtilis. Toxins. 2021;13:429. doi: 10.3390/toxins13060429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Qin X., Hao Z., Luo H., Yao B., Su X. Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins. 2019;11:566. doi: 10.3390/toxins11100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y., Qin X., Tang Y., Ma Q., Zhang J., Zhao L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020;325:126877. doi: 10.1016/j.foodchem.2020.126877. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Bai Y., Huang H., Tu T., Wang Y., Wang Y., Luo H., Yao B., Su X. Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins. 2019;11:609. doi: 10.3390/toxins11100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin X., Xin Y., Su X., Wang X., Wang Y., Zhang J., Tu T., Yao B., Luo H., Huang H. Efficient degradation of zearalenone by dye-decolorizing peroxidase from Streptomyces thermocarboxydus combining catalytic properties of manganese peroxidase and laccase. Toxins. 2021;13:602. doi: 10.3390/toxins13090602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janusz G., Pawlik A., Świderska-Burek U., Polak J., Sulej J., Jarosz-Wilkołazka A., Paszczyński A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020;21:966. doi: 10.3390/ijms21030966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riyadi F.A., Tahir A.A., Yusof N., Sabri N.S.A., Noor M.J.M.M., Akhir F.N.M.D., Othman N.a., Zakaria Z., Hara H. Enzymatic and genetic characterization of lignin depolymerization by Streptomyces sp. S6 isolated from a tropical environment. Sci. Rep. 2020;10:7813. doi: 10.1038/s41598-020-64817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawton T.J., Sayavedra-Soto L.A., Arp D.J., Rosenzweig A.C. Crystal structure of a two-domain multicopper oxidase: Implications for the evolution of multicopper blue proteins. J. Biol. Chem. 2009;284:10174–10180. doi: 10.1074/jbc.M900179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gräff M., Buchholz P.C.F., le Roes-Hill M., Pleiss J. Multicopper oxidases: Modular structure, sequence space, and evolutionary relationships. Proteins Struct. Funct. Bioinform. 2020;88:1329–1339. doi: 10.1002/prot.25952. [DOI] [PubMed] [Google Scholar]
- 25.Trubitsina L.I., Abdullatypov A.V., Larionova A.P., Trubitsin I.V., Alferov S.V., Ponamoreva O.N., Leontievsky A.A. Expression of thermophilic two-domain laccase from Catenuloplanes japonicus in Escherichia coli and its activity against triarylmethane and azo dyes. PeerJ. 2021;9:e11646. doi: 10.7717/peerj.11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisov A.V., Trubitsina L.I., Lisova Z.A., Trubitsin I.V., Zavarzina A.G., Leontievsky A.A. Transformation of humic acids by two-domain laccase from Streptomyces anulatus. Process. Biochem. 2019;76:128–135. doi: 10.1016/j.procbio.2018.11.001. [DOI] [Google Scholar]
- 27.Lang M., Kanost M.R., Gorman M.J. Multicopper oxidase-3 is a laccase associated with the peritrophic matrix of Anopheles gambiae. PLoS ONE. 2012;7:e33985. doi: 10.1371/journal.pone.0033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatzikonstantinou A.V., Gkantzou E., Gournis D., Patila M., Stamatis H. Chapter three—Stabilization of laccase through immobilization on functionalized GO-derivatives. In: Kumar C.V., editor. Methods in Enzymology. Academic Press; Cambridge, MA, USA: 2018. pp. 47–81. [DOI] [PubMed] [Google Scholar]
- 29.Su J., Bao P., Bai T., Deng L., Wu H., Liu F., He J. CotA a multicopper oxidase from Bacillus pumilus WH4, exhibits manganese-oxidase activity. PLoS ONE. 2013;8:e60573. doi: 10.1371/journal.pone.0060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ausec L., Berini F., Casciello C., Cretoiu M.S., van Elsas J.D., Marinelli F., Mandic-Mulec I. The first acidobacterial laccase-like multicopper oxidase revealed by metagenomics shows high salt and thermo-tolerance. Appl. Microbiol. Biotechnol. 2017;101:6261–6276. doi: 10.1007/s00253-017-8345-y. [DOI] [PubMed] [Google Scholar]
- 31.Sherif M., Waung D., Korbeci B., Mavisakalyan V., Flick R., Brown G., Abou-Zaid M., Yakunin A.F., Master E.R. Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive phytochemicals and site-directed mutagenesis. Microb. Biotechnol. 2013;6:588–597. doi: 10.1111/1751-7915.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur K., Sharma A., Capalash N., Sharma P. Multicopper oxidases: Biocatalysts in microbial pathogenesis and stress management. Microbiol. Res. 2019;222:1–13. doi: 10.1016/j.micres.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Reiss R., Ihssen J., Richter M., Eichhorn E., Schilling B., Thöny-Meyer L. Laccase versus laccase-like multi-copper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PLoS ONE. 2013;8:e65633. doi: 10.1371/journal.pone.0065633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zerva A., Pentari C., Termentzi A., America A.H.P., Zouraris D., Bhattacharya S.K., Karantonis A., Zervakis G.I., Topakas E. Discovery of two novel laccase-like multicopper oxidases from Pleurotus citrinopileatus and their application in phenolic oligomer synthesis. Biotechnol. Biofuels. 2021;14:83. doi: 10.1186/s13068-021-01937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto P.A., Fraga I., Bezerra R.M.F., Dias A.A. Phenolic and non-phenolic substrates oxidation by laccase at variable oxygen concentrations: Selection of bisubstrate kinetic models from polarographic data. Biochem. Eng. J. 2020;153:107423. doi: 10.1016/j.bej.2019.107423. [DOI] [Google Scholar]
- 36.Song Y., Wang Y., Guo Y., Qiao Y., Ma Q., Ji C., Zhao L. Degradation of zearalenone and aflatoxin B1 by Lac2 from Pleurotus pulmonarius in the presence of mediators. Toxicon. 2021;201:1–8. doi: 10.1016/j.toxicon.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Loi M., Fanelli F., Cimmarusti M.T., Mirabelli V., Haidukowski M., Logrieco A.F., Caliandro R., Mule G. In vitro single and combined mycotoxins degradation by Ery4 laccase from Pleurotus eryngii and redox mediators. Food Control. 2018;90:401–406. doi: 10.1016/j.foodcont.2018.02.032. [DOI] [Google Scholar]
- 38.Hathout A.S., Aly S.E. Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014;64:905–919. doi: 10.1007/s13213-014-0899-7. [DOI] [Google Scholar]
- 39.Hsieh D.P.H., Salhab A.S., Wong J.J., Yang S.L. Toxicity of aflatoxin Q1 as evaluated with the chicken embryo and bacterial auxotrophs. Toxicol. Appl. Pharmacol. 1974;30:237–242. doi: 10.1016/0041-008X(74)90095-7. [DOI] [Google Scholar]
- 40.Drzymala S.S., Binder J., Brodehl A., Penkert M., Rosowski M., Garbe L.-A., Koch M. Estrogenicity of novel phase I and phase II metabolites of zearalenone and cis-zearalenone. Toxicon. 2015;105:10–12. doi: 10.1016/j.toxicon.2015.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.