Abstract
Background and purpose:
The preponderance of microbial infections remains a global challenge. In the present study, synthesis of novel cysteine-based antimicrobial agents and their biological evaluation is reported.
Experimental approach:
The reaction of p-toluenesulphonyl chloride with cysteine afforded 2-{[(4-methylphenyl)sulphonyl]amino}-3-sulphanylpropanoic acid (3) which was acetylated based on Lumiere-Barbier method using acetic anhydride. The ammonolysis of the acetylated compound (4) gave the carboxamide derivative (5) which reacted with aniline, aminopyridine and diaminopyrimidine via nickel catalyzed Buchwald-Hartwig amidation reaction to afford compounds 6a, 6b, and 6c, respectively. The compounds were characterized using FTIR, 1H-NMR, 13C-NMR, and elemental analysis. The in vitro antimicrobial activities were determined. Their physicochemical properties were generated in silico and the molecular docking studied bacterial and fungal infections.
Findings/Results:
Compounds 4, 6b, and 6c exhibited excellent in vitro antibacterial activities while compound 4 had the best antifungal activities. From the in silico antimicrobial results, compound 3 had a better binding affinity (-10.95 kcal/mol) than penicillin (-10.89 kcal/mol) while compounds 3 and 4 had binding affinities (-10.07 and -10.62kcal/mol) comparable to ketoconazole (-10.85 kcal/mol).
Conclusion and implication:
All the synthesized compounds exhibited significant antibacterial and antifungal activities and were confirmed to be potential antimicrobial agents.
Keywords: Antimicrobial activity, Carboxamide, Cysteine, Molecular docking, Sulphonamide
INTRODUCTION
The fact that virtually all pathogenic microorganisms have the ability to develop antimicrobial resistance calls for the consistent development of novel antimicrobial agents (1,2). Amino acids have been found to exhibit excellent antimicrobial activities (3,4). Cysteine being an amino acid has immense biological functions (5) and plays a central role in protein synthesis, detoxification, metabolism of co-enzyme A and biotin (6), provision of sulfide in human metabolism (7), and consequently serves as a precursor in pharmaceutical industries (8). Previously, some α-amino acids such as methionine (9) and serine (10) in combination with sulphonamide moiety had been employed as potential antimicrobials in various capacities.
Similarly, sulphonamide derivatives are notable antimicrobial (11,12,13,14) and anticancer (15) agents while carboxamides are used in HIV (16) and hypertension (17) management. Moreover, aniline, pyridine, and pyrimidine as coupling partners are of great pharmacological importance (18). Aniline is used in paracetamol, acetaminophen, and Tylenol production (19) while pyridine and Pyrimidine serve as central nervous system (CNS) stimulants, anesthetics (20), and antifolates (21), respectively. The fact that multi-therapy drug usage occasioned by the prevalence of drug-resistant microorganisms has not yet eliminated the problem of antimicrobial resistance is quite worrisome. Sadly, about 70% of the 2 million patients who acquired bacterial infection annually in US hospitals displayed resistance to at least a drug (22). Therefore, the solution to this antimicrobial resistance menace is to develop drug molecules that can work on new drug target sites that are yet unexploited (23).
The objective of the current study was to design, synthesize cysteine-based p-toluenesulphonamide derivatives, and evaluate their antimicrobial activities for enhanced drug potency. The need for novel antimicrobial agents with improved drug potency necessitated this research work in which numerous pharmacophores were combined in one drug molecule.
MATERIALS AND METHOD
Materials
Sigma-Aldrich Corporation, St. Louis, Missouri, United States of America was the source of the chemicals used. The melting points of compounds were carried out with electrothermal melting point apparatus IA9200 model of Cole-Parmer Ltd, Staffordshire, UK. Fourier-transform infrared spectroscopy (FTIR) spectroscopy of title compounds was recorded on 8400s FTIR. Nuclear magnetic resonance (1H-NMR and 13C-NMR) were run in dimethyl sulfoxide (DMSO) on 400 MHz using NMR spectrophotometer in Prof. Sandeep Verma Laboratory, Department of Chemistry, Indian Institute of Technology, Kanpur, India. Chemical shifts were recorded in part per million (ppm). The reaction processes were monitored using thin-layer chromatography (TLC, methanol/dichloromethane, 1:8). Nitrogen was used for the inert atmosphere. Compounds were prepared in analytical grade. The microorganisms used were clinical isolates gotten from the Department of Pharmaceutical Microbiology, University of Nigeria, Nsukka.
Synthesis
Synthesis of 2-{[(4-methylphenyl)sulphonyl] amino}-3-sulphanylpropanoic acid
Cysteine (25 mmol) was dissolved in 30 mL of water and Na2CO3 (52.50 mmol) was added and stirred. The solution was cooled to 0 °C followed by the addition of 4-methylsulphonyl chloride (1) (30 mmol) in five portions for 1 h intervals and was stirred for 4 h at room temperature and the reaction mixture was acidified to pH 2 with 2 M hydrochloric acid to enable recrystallization. The content was kept for 12 h and the products were obtained by suction filtration. The crude product was washed with tartaric acid and dried in a desiccator to obtain compound 3.
Acylation of 2-{[(4-methylphenyl)sulphonyl] amino}-3-sulphanylpropanoic acid (3)
Two g of compound 3 was taken to a 100 ml beaker; 9 mL of concentrated HCl and 25 mL of distilled water were added to the beaker and vigorously stirred. 16 g of sodium acetate was dissolved in 50 mL distilled water in another beaker. Then, 13 mL of acetic anhydride was added in three portions over an interval of 1 h after which it was poured into the sodium acetate solution. The reaction mixture was stirred with a glass rod and immersed in an ice bath for 1 h and filtered to obtain compound 4.
Chlorination and amonnolysis of 2-{acetyl [(4-methylphenyl)sulphonyl]amino}-3-sulphanyl-propanoic acid (4) Chlorination
Two hundred and fifty mL three-necked flask equipped with a magnetic stirring bar was charged with compound 4 (1 mmol) and acetone (10 mL). The flask was stoppered and the content cooled to 0 °C. Then the mixture was stirred at 80 °C under reflux for 3 h, after which it was taken to a water bath at 80 °C to evaporate excess thionyl chloride. 20 mL acetone was added and evaporated twice and the acid chloride was obtained.
Ammonolysis
The resulting acid chloride obtained above was immediately dissolved in acetone (20 mL) and cooled to 0-5 °C, crystallized with ammonia (2 mL) and allowed to stay overnight after which it was filtered and washed with acetone to afford compound 5.
Nickel-catalysed synthesis of cysteine-based sulphanamoylcarboxamides derivatives. Preparation of bis(triphenylphosphine)nickel (ii)chloride
Using L.M Venanzi procedure (24), nickel (II) chloridehexahydrate catalyst (2.37 g, 10 mmol) in distilled water (2 mL) followed by dilution with 50 mL glacial acetic acid and addition of a mixture of triphenylphosphine ligand (5.25 g, 20 mmol) in 25 mL glacial acetic acid. A green precipitate was formed and allowed to be in contact with the glacial acetic acid solution for 24 h. A dark blue crystal (the complex compound) was afforded on filtration, washed with glacial acetic acid and dried in desiccators.
Cysteine-based sulphamoyl carboxamide derivatives
Bis (triphenylphosphine) nickel (II) chloride (6.54 g, 10 mmol) and triphenylphosphine (5.25 g, 30 mmol) were both introduced into a 250 mL three-necked flask. The solvent t-butanol (4 mL) and distilled water (2 mL) were added using a syringe and the mixture was stirred for 10 min at room temperature under inert nitrogen atmosphere. The mixture was heated at 80 °C for 1.5 min. Then, compound 5 (10 mmol), potassium carbonate, K2CO3 (1.38 g, 10 mmol), substituted aryl and heteroaryl halides (4-chloroaniline, 4-amino-3-chloropyridine and 5-chloro-4,6-diaminopyrimidine) added with t-butanol and H2O in the ratio of 2:1 under inert atmosphere. Then, it was refluxed and stirred for 1 h at 100-110 °C, cooled to room temperature, crystallized with ethyl acetate and washed with water to afford compounds 6a-6c.
Antimicrobial studies
The Wiegand et al. method was employed (25). There was a further incubation of plates for another 24 h at 37 °C and 48 h at 25 °C in order to determine whether the activity was bacteriostatic or bactericidal. The procedure was repeated for ofloxacin (a standard antibacterial agent) and fluconazole (a standard antifungal agent).
In silico methodology
Physicochemical properties
The physicochemical properties of the title compounds were generated in silico and important parameters were recorded. The descriptors calculator in Swiss dock online servers was used in the computation of these parameters and the drug-likeness of compounds was investigated using Lipinski’s rule of five.
Molecular docking
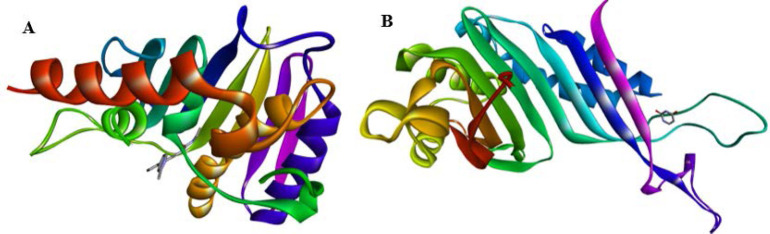
The molecular docking studied bacterial and fungal infections. The drug targets for the antibacterial assay were E. coli DNA gyrase in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3yl] urea (PDB code: 5MMN) and antifungal was urate oxidase from Aspergillus flavus complexed with uracil (PDB code: 1WS3) (Fig. 1). The 3-dimensional structures of the drug targets were downloaded from the Protein Data Bank (PDB; http://www.pdb.org) database. Through molecular docking using PyRx, the prepared compounds interacted with each of the receptors. The protocol enables flexible compound docking for various compound conformers within the rigid receptor. Best conformation for each title compound was chosen and interaction visualized using the Discovery studio.
Fig. 1.
(A) Escherichia coli DNA gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea (26). (B) Crystal structure of urate oxidase from Aspergillus flavus complexed with uracil (27).
Urate oxidase (Uox) catalyzes the oxidation of urate to allantoin and is used to reduce toxic urate accumulation during chemotherapy (28). This enzyme has been found to be of therapeutic interest, and so useful as a drug target. On the other hand, DNA gyrase is a type II topoisomerase found in all bacteria consisting of two subunits: GyrA and GryB of E. coli gyrase. These enzymes are responsible for catalyzing topological changes in DNA and have proved to be drug targets for therapeutic agents.
RESULTS
Chemistry
The synthesis of cysteine-based sulphamoyl carboxylic acids (3) was achieved by the reaction of L-cysteine (1) with p-toluenesulphonyl chloride (2), this nucleophilic substitution reaction afforded the cysteine-based sulphamoyl carboxylic acid (3). The synthesis of N-acetylated sulphamoyl carboxylic acid (4) was carried out via Lumiere-Barbiere method of acetylation with the purpose of protecting the amino group of the sulphonamide moiety from side reactions during subsequent chlorination and ammonolysis. The chlorination of the N-acetylated sulphamoyl carboxylic acid was an activation process which enabled its conversion to a reactive acid chloride intermediate (29) that reacted directly with aqueous ammonia in a basic medium (ammonolysis) to afford sulphamoyl carboxamide (5) via Schotten-Baumann reaction (30,31). Through Buchwald-Hartwig cross-coupling reaction (32), sulphamoyl carboxamides were coupled with aryl and heteroaryl halides such as 4-chloroaniline, 4-amino-3-chloropyridine, 5-chloro-4,6-diaminopyrimidine to afford their sulphamoyl carboxamide derivatives (6a-c) as represented in Scheme 1.
Scheme 1.
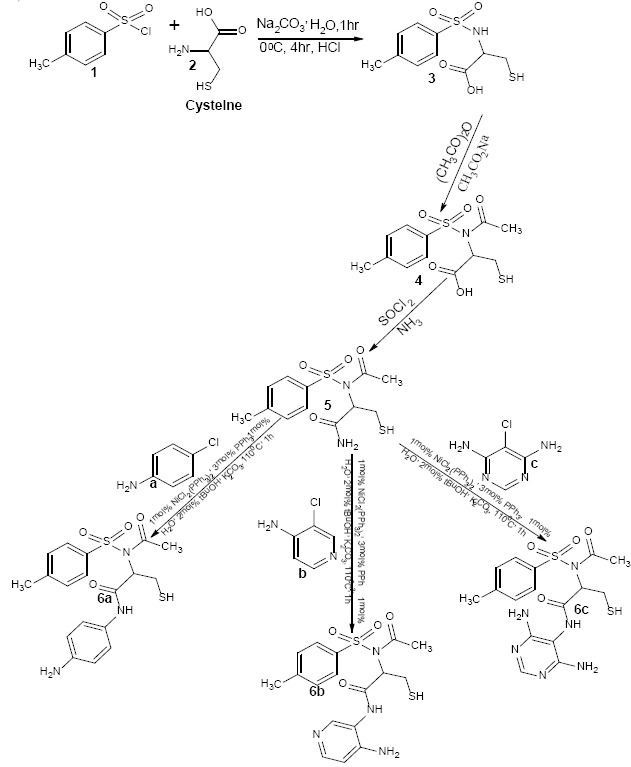
Synthetic pathway for the synthesis of cysteine-based sulphonamide derivatives.
2-{[(4-methylphenyl)sulphonyl]amino}-3-sulphanylpropanoic acid (3)
Dark tan solids, yield: 3.01g (87.8%), mp. 134-135 °C. IR (KBr) cm-1: 3462(N-H), 2997 (C-H aromatic), 2898 (C-H aliphatic), 2651 (OH of COOH), 2573 (S-H), 1715 (C=O of COOH), 1658, 1651 (C=C), 1336, 1164 (S=O two bands), 772 (Ar-H). 1H-NMR (DMSO, 400 MHz) δ: 8.63-8.58 (s, 1H, SH), 8.33-8.31 (d, J = 8.3 Hz, 1H, NH-CH), 7.75-7.71(m, 1H, OH), 7.69-7.60 (d, J = 8 Hz, 2H, Ar-H), 7.50-7.48 (d, J = 7.7 Hz, 2H, Ar-H), 7.16-7.14 (d, J = 7.5 Hz, IH, CH of CH2), 2.41 (s, 3H, CH3-Ar). 13C-NMR (DMSO, 400 MHz) δ (ppm): 170.124 (C=O), 142.721, 135.213, 131.421, 129.511, 126.921, 123.513 (aromatic carbons), 78.445, 76.109, 75.885 (aliphatic carbons). Anal.calcd. for C10H13NO4S2 (275.104): C, 43.64, H, 4.78, N, 5.11, S, 23.26. Found: C, 43.60, H, 4.82, N, 5.15, S, 23.21.
2-{Acetyl[(4-methylphenyl)sulphonyl]amino}-3-sulfanylpropanoic acid (4)
Yield: 1.86 g (91.7%), mp. 120-121 °C, IR (KBr) cm-1: 3311 (O-H), 2929 (C-H alphatic), 2914 (S-H), 2083 (C-H aromatic), 1725, 1690 (C=O), 1659, 1651 (C=C), 1281, 1188 (2S=O), 1175, 1162 (SO2NH), 1136 (C-N), 779 (Ar-H). 1HNMR (DMSO, 400 MHz) δ: 7.933 (d, J = 7.4 Hz, 2H, ArH), 7.713 (d, J = 8.1 Hz, 2H, ArH) 7.645-7.619 (t, J = 7.8 Hz, IH, ArH), 6.577 (s, IH, OH), 3.355 (s, 1H, SH), 2.996 (m, 2H, CH2), 2.493 (s, 3H, Ar-CH3), 2.488 (s, 3H, CH3-C=O). 13CNMR (DMSO, 400 MHz) δ: 171.223, 170.212 (C=O), 138.243, 137.971, 134.811, 133.764. 132.601, 130.723 (aromatic carbons), 41.632, 40.431, 40.211, 39.901 (aliphatic carbons). Anal.calcd. for C12H15NO5S2 (317.38): C, 45.39, H, 4.75, N, 4.43, S, 20.19. Found: C, 45.42, H, 4.77, N, 4.41, S, 20.23.
2-{acetyl[(4-methylphenyl)sulphonyl]amino}-3- sulphanylpropanamide (5)
Yield: 3.34 g (98.2%), mp. 204-205 °C, IR (KBr) cm-1:3391 (N-H), 2079 (C-H aliphatic), 1982 (C-H aromatic), 1871,1673 (C=O), 1582, 1405 (C=C), 1372, 1267 (2S=O), 1136, 1134 (SO2-NH), 1088 (C-N), 738 (Ar-H). 1HNMR (DMSO, 400 MHz) δ: 9.453 (d, J = 7.3 Hz, 2H, ArH), 7.196 (t, J = 7.5 Hz, 2H, ArH), 5.112 (s, IH, SH), 3.310 (s, 2H, NH2), 2.485-2.407 (d, J = 31.2 Hz, 3H, CH3-C=O), 1.54-1.012 (d, J = 56.8 Hz, 3H, CH3-Ar). 13CNMR (CD3CN, 400 MHz) δ: 172.112, 169.883 (C=O), 146.927, 142.121, 131.604, 128.465, 125.346, 118.347 (aromatic carbons), 79.335, 67.329, 61.779, 39.908 (aliphatic carbons). Anal.calcd. for C12H16N2 O4S2 (316.40): C, 45.51, H, 5.10, N, 8.85, S, 20.23. Found: C, 45.46, H, 5.05, N, 8.78, S, 20.18.
2-{acetyl[(4-methylphenyl)sulphonyl]amino}-N-(4-aminophenyl)-3-sulphanylpropanamide (6a)
Yield: 3.04 g (91.5%), mp. 93-94 °C, IR (KBr) cm-1: 3400, 3300 (N-H), 2877 (C-H aliphatic), 2553 (S-H), 1927 (C-H aromatic), 1718, 1670 (2C=O), 1620, 1619 (C=C), 1330, 1196 (2S=O), 1151, 1150 (SO2-NH), 1088 (C-N), 771 (Ar-H). IHNMR (CDCl3, 400 MHz) δ: 8.197 (d, J = 8.4 Hz, 2H, ArH), 7.608-7.522 (m, 2H, ArH), δ 6.772 (d, J = 7.6 Hz, 2H, ArH), δ 6.044 (t, J = 8.1 Hz, 2H, ArH), 4.835 (s, br, IH, SH), 2.767 (s, IH, NH), 2.7693 (s, 2H, NH2), 2.662 (s, 3H, CH3-C=O), 2.259-2.253 (m, 3H, CH3-Ar), 2.241 (t, J = 2.6 Hz, 1H, CH). 13C-NMR (CDCl3, 400 MHz) δ: 172.444, 169.333 (C=O), 137.189, 133. 737, 133.554, 129.062, 128.827, 128.410, 126.445, 124.889, 123.675, 122.321, 121.114, 120.456 (aromatic carbon), 79.472, 79.146, 78.820, 40.385 (aliphatic carbons). Anal.calcd. for C18H21N3O4S2 (407.51): C, 53.00, H, 5.15, N, 10.31, S, 15.70. Found: C, 52.97, H, 5.11, N, 10.28, S, 15.67.
2-{acetyl[(4-methylphenyl)sulphonyl]amino}-N-(4-aminopyridine-3-yl)-3-sulphanylpropanamide (6b)
Yield: 2.90 g (88.9%), mp. 83-84 °C, IR (KBr) cm-1: 3409, 3347 (N-H), 3064 (C-H aliphatic), 2,600 (S-H), 1893 (C-H aromatic), 1707,1690 (C=O), 1650 (C=N), 1397, 1181 (2S=O), 1153, 1151 (SO2-NH), 1118, 1084 (C-N), 995 (C=C), 719 (Ar-H). 1HNMR (CDCl3, 400 MHz) δ: 8.465 (d, J = 7.0 Hz, 2H, ArH), 7.960 (t, J = 7.4 Hz, 2H, ArH), 2.218 (d, J = 7.2 Hz, 2H, ArH), 6.994 (m, 2H, ArH). 5.104 (s, IH, NH), 5.954 (s, 2H, NH2), 2.4 (d, J = 7.0 Hz, 3H, CH3-Ar), 2.2 (s, 3H, CH3-C=O). 13(NMR (CDCl3/DMSO, 400MH2) δ: 170.441, 169.889 (C=O), 159.591 (C=N), 138.343, 133.638, 133.426, 131.446, 130.643, 128.669, 126.433, 125.678, 123.441, 122.678 (aromatic carbons), 85.949, 83.282, 75. 628. 72.991 (aliphatic carbons). Anal.calcd. for C17H20N4O4S2 (408.50): C, 49.94, H, 4.90, N, 13.71, S, 15.67. Found: C, 49.88, H, 4.87, N, 13.74, S, 15.71.
2-{acetyl[(4-methylphenyl)sulphonyl]amino}-N-(4,6-diaminopyrimidine-5-yl)-3-sulphanyl-propanamide (6c)
Yield: 2.90 g (89.8%), mp. 109-110 °C, IR (KBr) cm-1: 3451, 3328, 3318(N-H), 3145 (C-H aliphatic), 1190 (C-H aromatic), 1690, 1685(C=O), 1676,1650 (C=N), 1367, 1274 (2S=O), 1118, 1115 (SO2N), 1025 (C-N), 745 (Ar-H). 1HNMR (DMSO, 400 MHz) δ: 7.248 (d, J = 8.0 Hz, 2H, ArH), 6.396 (t, J = 8.1 Hz, 2H, ArH), 6.011 (d, J = 7.8 Hz, IH, Ar), 5.000 (d, J = 7.5 Hz, 1H, SH-CH2) 5.735 (d, J = 7.3 Hz, 2H, NH2), 3.462 (s, 1H, NH), 2.470 (s, 3H, CH3-Ar), 2.043 (s, 3H, CH3-C=O). 13CNMR (DMSO, 400 MHz) δ: 170.843, 169.445 (C=O) 159.211, 159.012 (C=N), 137.136, 133.706, 133.516, 132.280, 131.946, 129.009, 128.781, 126.442 (aromatic carbons), 93.093, 76.252, 78.918, 78.584 (aliphatic carbons). Anal.calcd. for C16H20N6O4S2 (424.50): C, 45.23, H, 4.71, N, 19.79, S, 15.08. Found: C, 45.27, H, 4.75, N, 19.81, S, 15.12.
Antimicrobial studies
Table 1 shows that all the compounds displayed considerable inhibitory effects against all the bacteria used. Obviously, only compounds 4, 5, 6b, and 6c exhibited antifungal activities against the two fungi Aspergillus niger and Candida Albicans.
Table 1.
Antimicrobial activities of synthesized compounds. Ofloxacin and fluconazole were used as antibacterial and antifungal standard drugs, respectively.
| Minimum inhibitory concentration (mg/mL) | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Compounds | Escherichia coli | Salmonella typhi | Staphylococcus aureus | Bacillus subtilis | Pseudomonas aeruginosa | Candida albicans | Aspergillus niger |
| 3 | 0.9 | - | 0.9 | 0.8 | - | - | - |
| 4 | 0.6 | 0.9 | 0.7 | 0.4 | 0.9 | 0.6 | 0.9 |
| 5 | 0.9 | 0.9 | 0.7 | 0.8 | 0.9 | - | 0.8 |
| 6a | 0.9 | 0.8 | 0.9 | 0.6 | - | - | - |
| 6b | 0.8 | 0.9 | 0.7 | 0.5 | 0.9 | 0.8 | - |
| 6c | 0.8 | 0.9 | 0.7 | 0.5 | 0.9 | 0.8 | - |
| Ofloxacin | 0.005 | 0.005 | 0.010 | 0.020 | 0.025 | - | - |
| Fluconazole | - | - | - | - | - | 0.020 | 0.005 |
(-) Implies no activity.
Molecular docking studies
From Table 2, the hydrogen bond acceptor (HBA) ≤ 6, hydrogen bond donor (HBD) ≤ 3, number of rotatable bonds (NRB) ≤ 7, octanol/water partition coefficient logP (o/w) ≤ 2.68, aqueous solubility (SlogP) ≤ 1.93, topological polar surface area (TPSA) ≤ 200.17 and molecular weight (MW) ≤ 424.51. In comparison with Lipinski’s rule of 5, LogP ≤ 5, MW ≤ 500, HBA ≤ 10, and HBD ≤ 5 is the benchmark for a drug candidate.
Table 2.
Physicochemical properties of the synthesized compounds.
| Compounds | HBA | HBD | NRB | logP(o/w) | SlogP | TPSA | MW | lip_violation |
|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 3 | 5 | 1.38 | 0.66 | 122.27 | 275.35 | 0 |
| 4 | 5 | 2 | 5 | 1.40 | 0.87 | 130.55 | 303.36 | 0 |
| 5 | 4 | 1 | 6 | 0.59 | 0.32 | 136.34 | 316.40 | 0 |
| 6a | 4 | 2 | 7 | 1.93 | 2.05 | 148.37 | 407.51 | 0 |
| 6b | 5 | 2 | 7 | 0.70 | 1.45 | 161.26 | 408.50 | 0 |
| 6c | 6 | 3 | 7 | 0.31 | 0.42 | 200.17 | 424.51 | 0 |
HBA, Hydrogen bond acceptor; HBD, hydrogen bond donor; NRB, number of rotatable bonds; logP(o/w), octanol/water partition coefficient; SlogP, aqueous solubility, TPSA, topological polar surface area; MW, molecular
In silico antibacterial and antifungal activities results
Table 3 shows that compounds 3 and 4 had the highest antibacterial and antifungal binding energies of -10.95 and -10.62 kcal/mol, respectively. Table 4 shows the binding interactions of compound 3 with 5MMN and 1WS3. Validation of docking protocols using 1WS3 is represented in Fig. 2. The crystal structure of 3 bound to the binding sites of 5MMN (A) and 1WS3 (B) is shown in Fig. 3. Compound 3 (green) and its co-crystallized ligands overlaid on each other in the binding site of 5MMN and 1WS3 are indicated in Figs. 4 and 5 respectively.
Table 3.
In silico antibacterial and antifungal activities results. Penicillin and ketoconazole were used as standard drugs for 5MMN and 1WS3, respectively.
| Compounds | Antibacterial | Antifungal |
|---|---|---|
|
| ||
| 5MMN | 1WS3 | |
| 3 | -10.95 | -10.07 |
| 4 | -10.17 | -10.62 |
| 5 | -9.18 | -8.60 |
| 6a | -10.41 | -9.22 |
| 6b | -9.65 | -9.56 |
| 6c | -9.90 | -9.58 |
| Standard drug | -10.89 | -10.85 |
Table 4.
Binding interactions of compound 3 with 5MMN and 1WS3.
| Complex | Ligand | Receptor | Interaction | Distance (Å) |
|---|---|---|---|---|
| 3_5MMN | S 14 | VAL 43 | H-bonding | 3.95 |
| S 14 | ASP 73 | H-donor | 3.31 | |
| S 14 | ALA 47 | H-Acceptor | 4.09 | |
| 6-ring | ILE 78 | pi-H | 3.89 | |
| 6-ring | PRO 79 | pi-H | 4.67 | |
|
| ||||
| 3_1WS3 | S 14 | GLN 228 | H-donor | 3.76 |
| O 16 | ARG 176 | H-Acceptor | 3.27 | |
| O 16 | ARG 176 | H-Acceptor | 2.96 | |
| N 11 | PHE 156 | pi-H | 3.96 | |
Fig. 2.
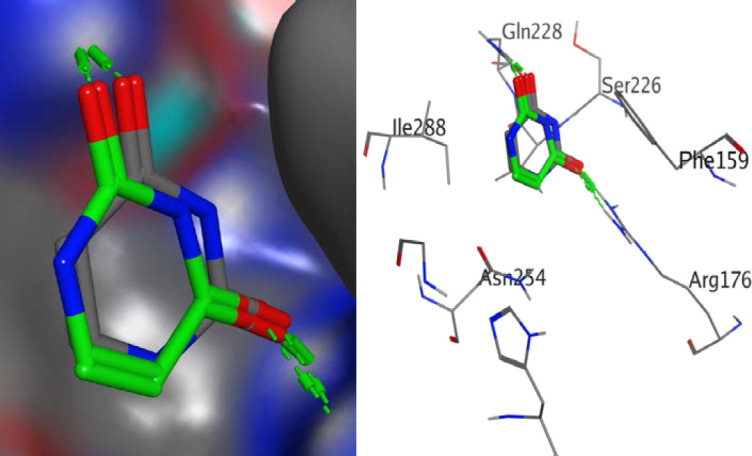
Validation of docking protocols using 1WS3. Uracil (green) was docked into the binding cavity of 1WS3 containing the co-crystallized uracil (grey). The root means square deviation (RMSD) was found to be 1.9 Å. This shows that docked ligands are within the binding site of 1WS3.
Fig. 3.
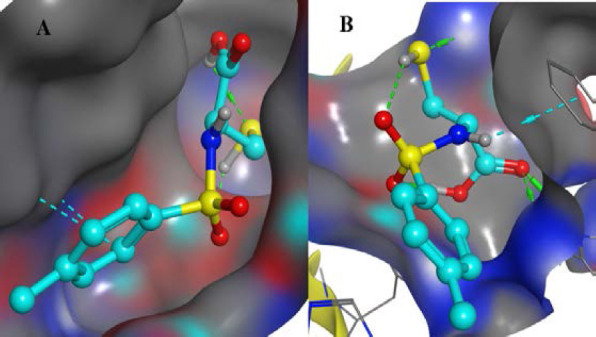
Crystal structure of 3 bound to the binding sites of (A) Escherichia coli GyrB (PDB code: 5MMN) and (B) Aspergillus flavus urate oxidase (PDB code: 1WS3)
Fig. 4.
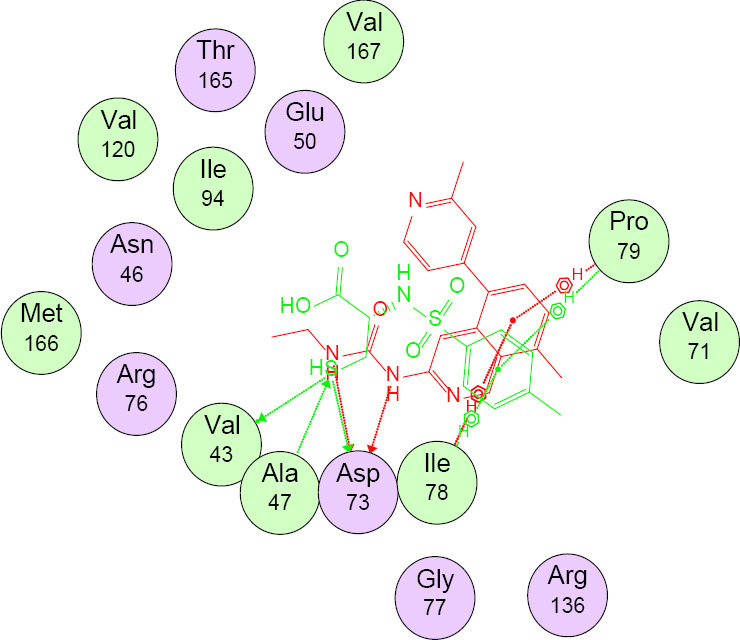
Compound 3 (green) and co-crystallized ligand (1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea) overlaid on each other in the binding site of 5MMN.
Fig. 5.
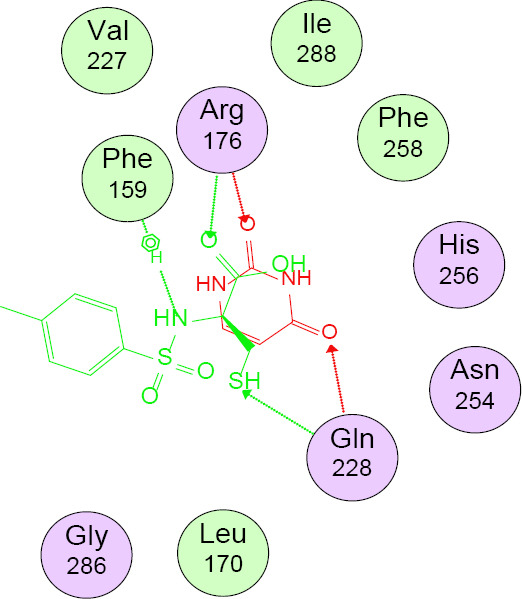
Compound 3 (green) and co-crystallized ligand (uracil) overlaid on each other in the binding site of 1WS3.
DISCUSSION
The presence of the diagnostic peaks at 2651 cm-1 due to OH of COOH, 2573 cm-1 due to S-H, 1715 cm-1 due to C=O of COOH, 1658 cm-1 and 1651 cm-1 due to C=C, 1336 cm-1 and 1164 cm-1 due to two bands of S=O frequencies in FTIR spectrum, the peculiar peaks of 8.63-8.58 (s, 1H, SH), 7.75-7.71 (m,1H, OH), 7.69-7.60 (d, J = 8 Hz, 2H, Ar-H) and 7.50-7.48 (d, J = 7.7 Hz, 2H, Ar-H) in 1H-NMR spectra, the diagnostic peak of 170.124 (C=O) in 13C-NMR spectra confirmed the successful synthesis of sulfamoyl carboxylic acid (compound 3) from cysteine and paratoluene sulphonyl chloride. The presence of double peaks of 1725 cm-1 and 1690 cm-1 due to C=O of the acetyl group in FTIR spectrum, 2.488 δ (s, 3H, CH3-C=O) in 1H-NMR and 171.223 δ and 170.212 δ due to C=O in 13C-NMR spectra were indicative of the successful acetylation to obtain compound 4. In compound 5, the appearance of the peak at 3391 cm-1(N-H of amide) in FTIR spectrum and 3.310 δ (s, 2H, NH2), in 1H-NMR spectrum indicated a successful amonnolysis (addition of ammonia).
The diagnostic peaks in compound 6a, were 3400 cm-1 and 3300 cm-1 due to two N-H bands, 1620 cm-1, 1619 cm-1 due to C=C of aromatic in FTIR, 2.767 (s, IH, NH), 2.7693 (s, 2H, NH2), in 1H-NMR spectrum confirmed the successful coupling of 4-chloroaniline. In compound 6b, the appearance of diagnostic peaks such as 1650 cm-1 due C=N heterocyclic in the FTIR spectrum and 159.591 (C=N) in the 13C-NMR spectrum confirmed the successful coupling of 4-amino-3-chloropyridine. Similarly, in compound 6c, the double peaks at 1676 cm-1 and 1650 cm-1 due to C=N in FTIR spectrum and 159.211 cm-1 and 159.012 cm-1 due to C=N in the 13C-NMR spectrum confirmed the successful coupling of 5-chloro-4,6-diaminopyrimidine derivative of sulphamoyl carboxamide.
The results in Table 1 showed that all the target compounds exhibited significant antimicrobial activities. Generally, compounds 4 exhibited the best antimicrobial activities being the compound with significant inhibitory effect against all the tested bacteria, namely Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Bacillus subtilis, and Salmonella typhi and all the tested fungi namely Candida albicans and Aspergillus niger. Specifically, compound 4 was the most potent antibacterial agent against E.coli and Bacillus subtilis with MIC 0.60 mg/mL and 0.4 mg/mL, respectively. Compound 6a (MIC 0.8 mg/mL) was the most active antibacterial agent against S. typhi, while compounds 4, 5, 6b, and 6c (MIC: 0.7 mg/mL) were the most potent agents against S. ureus. Although it was previously reported that sulphonamides do not inhibit the growth of Pseudomonas aeruginosa, a recalcitrant bacterium (33-35), compounds 4, 5, 6b, and 6c (MIC 0.9 mg/mL) exhibited significant inhibitory activities against this bacterium and this could be attributed to the synergistic antimicrobial antagonism arising from the combination of sulphonamide, carboxamide, and cysteine moieties in a single molecule.
The antifungal studies revealed that Aspergillus niger resisted many of the target compounds because sulphonamides scarcely inhibit the growth of Aspergillus niger (36,37), nevertheless, compounds 5 (MIC 0.8 mg/mL) displayed the best antifungal activities against the fungus. Compound 4 (MIC 0.6 mg/mL) was the most potent antifungal agent against Candida albican. The high resistance exhibited by Candida albican against several cysteine-based sulfonamide derivatives could be as a result of the fact that this fungus has high cysteine tolerance due to its ability to excrete sulfite when confronted with a high amount of cysteine (38). Summarily, the antimicrobial activities of some of the sulfonamide derivatives were possibly potentiated by the incorporated carboxamide and cysteine moieties.
With reference to Table 2, Lipinski’s rule of 5 states that a drug candidate is said to be drug-like if it has lipophilicity (log P) ≤ 5, number of HBA ≤ 10, MW ≤ 500, and number of HBD ≤ 5. Compounds with TPSA ≤ 140 Å2 can penetrate the cell and have a good oral bioavailability in rats while NRB ≤ 10 is generally required for good oral bioavailability (39). Compounds with TPSA ≤ 90 Å2 can penetrate the blood-brain barrier and the central nervous system (40). It implies that compounds 3, 4, and 5 can permeate the cell but none can permeate the blood-brain-barriers. Based on the above principles, all our compounds were confirmed drug candidates with good oral bioavailability.
From Table 3, all compounds had strong binding affinities with the receptors used. For the antibacterial, compound 3 had a better binding affinity (-10.95 kcal/mol) than penicillin (-10.89 kcal/mol) while antifungal study revealed that compounds 3 and 4 had binding affinities (-10.07 and -10.62 kcal/mol, respectively) comparable to ketoconazole (-10.85 kcal/mol). The high binding affinity of compound 3 is due to the fact that sulphamoyl carboxylic acids have excellent in silico antibacterial activities (41). It implies that compounds 3 and 4 had the best in silico antimicrobial activities. The root means square deviation (RMSD) was found to be 1.9 Å in the validation of docking protocols using 1WS3 as shown in Fig. 2. This confirms that docked ligands are within the binding site of 1WS3. Compound 3 is well fitted into the binding cavity of 5MMN and 1WS3 (Fig. 3). Strong conventional H-bonding interaction was observed between compound 3 and VAL 43 amino acid via an H-bond distance of 3.95Å as seen in table 4. Figs. 4 and 5 show that compound 3 and its ligands, (1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea) and uracil are well overlaid on each other in the binding sites of the drug targets 5MMN and 1WS3 respectively. This further confirms the antibacterial potentials of compound 3.
CONCLUSION
In conclusion, facile synthesis of cysteine-based sulphonamide derivatives having carboxamide pyridine, aniline, and pyrimidine pharmacophores was achieved. The assigned structures agreed with the spectral data. Compounds 4 was the most potent antibacterial agents against Escherichia coli and Bacillus subtilis (MIC: 0.6mg/mL and 0.4 mg/mL, respectively). Compound 6a (MIC: 0.8 mg/mL) had the best antibacterial activity against Salmonella typhi while compounds 4, 5, 6b, and 6c (MIC: 0.7 mg/mL) were the most potent antibacterial agents against Staphylococcus aureus. Compounds 4, 5, 6b, and 6c (MIC: 0.9 mg/mL) were equally active against Pseudomonas aeruginosa. Compound 4 (MIC: 0.6 mg/mL) and compound 5 (MIC: 0.8 mg/mL) showed the best antifungal activities against Candida albicans, and Aspergillus niger, respectively. Compounds 3 and 4 showed the best in silico antimicrobial activities, compound 4 (-10.17 kcal/mol and -10.62 kcal/mol) had comparable binding energy with penicillin (-10.89 kcal/mol) and ketoconazole (-10.85 kcal/mol), respectively. All the synthesized compounds were found to be potential antimicrobial agents with good drug-likeness.
Conflict of interest statement
The authors declared no conflicts of interest in this study.
Authors’ contribution
M.C. Egbujor and U.C. Okoro contributed to the concept and idea development. The experimental studies were performed by M.C. Egbujor, and supervised by U.C. Okoro. Molecular docking was done by S.N. Okafor. Manuscript editing and review were done by O.R. Umeh, S.A. Egu, I.S. Amasiatu, P.I. Egwuatu, and E.M. Ibo.
Acknowledgements
The authors appreciate Renaissance University, Ugbawka Enugu State, Nigeria and the University of Nigeria Nsukka, Nigeria for providing the laboratories for the research.
REFERENCES
- 1.Goossens H, Ferech M, Vander Stichele R, Elseviers M. Outpatient antibiotic use in Europe and association with resistance: a crossnational database study. Lancet. 2005;365((9459)):579–587. doi: 10.1016/S0140-6736(05)17907-0. DOI: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 2.Cassir N, Rolain JM, Brouqui P. A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front Microb. 2014;5:551–565. doi: 10.3389/fmicb.2014.00551. DOI: 10.3389/fmicb.2014.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michael G, Schomburg D. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology. 2nd ed. Oxford: Wiley-Blackwell; 2012. p. 366. [Google Scholar]
- 4.Siebert A, Wysocka M, Krawczyk B, Cholewinski G, Rachon J. Synthesis and antimicrobial activity of amino acid and peptide derivatives of mycophenolic acid. Eur J Med Chem. 2018;143:646–655. doi: 10.1016/j.ejmech.2017.11.094. DOI: 10.1016/j.ejmech.2017.11.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulaj G, Kortemme T, Goldenberg DP. Ionization-reactivity relationships for cysteine thiol in polypeptides. Biochemistry. 1998;37((25)):8965–8972. doi: 10.1021/bi973101r. DOI: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- 6.Hell R, Wirtz M. Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. Arabidopsis Book. 2011:e0154. doi: 10.1199/tab.0154. 1-14. DOI: 10.1199/tab.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lill R, Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. DOI: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 8.Hui YH, Nip W, Rogers RW, Young OA. Meat science and application. 1th ed. New York: CRC Press; 2001. pp. 207–220. DOI: 10.1201/9780203908082. [Google Scholar]
- 9.Egbujor MC, Okoro UC. New methionine-based p-toluenesulphonamoylcarboxamide derivatives as antimicrobial and antioxidant agents: design, synthesis, and molecular docking. J Pharm Res Int. 2019;28((1)):1–12. DOI: 10.9734/jpri/2019/v28i130192. [Google Scholar]
- 10.Egbujor MC, Okoro UC, Okafor S, Nwankwo NE. Design, synthesis and molecular docking of novel serine-based sulphonamide bioactive compounds as potential antioxidant and antimicrobial agents. Indo Am J P Sci. 2019;06((06)):12232–12240. DOI: 10.5281/zenodo.3250306. [Google Scholar]
- 11.Henry RJ. The mode of action of sulfonsmides. Bacteriol Rev. 1943;7((4)):175–262. doi: 10.1128/br.7.4.175-262.1943. PMID: 16350088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egbujor MC, Okoro UC, Okafor S, Nwankwo NE. Synthesis, characterization and in silico studies of novel alkanoylated 4-methylphenyl sulphonamoyl carboxylic acids as potential antimicrobial and antioxidantagents. Int J Pharm Phytopharmacological Res. 2019;9((3)):89–97. [Google Scholar]
- 13.Egbujor MC, Okoro UC, Okafor SN, Amasiatu IS, Amadi UB, Egwuatu PI. Synthesis, molecular docking and pharmacological evaluation of new 4-methylphenylsulphamoyl carboxylic acids analogs. Int J Res Pharm Sci. 2020;11((4)):5357–5366. DOI: 10.26452/ijrps.v11i4.3157. [Google Scholar]
- 14.Egbujor MC, Okoro UC, Egu SA, Egwuatu PI, Eze FU, Amasiatu IS. Synthesis and biological evaluation of alanine derived bioactive p-toluenesulphonamide analogs. Int J Res Pharm Sci. 2020;11((4)):6449–6458. DOI: 10.26452/ijrps.v11i4.3440. [Google Scholar]
- 15.Casini A, Scozzafava A, Mastrolorenzo A, Supuran LT. Sulfonamides and sulfonylated derivatives as anticancer agents. Curr Cancer Drug Targets. 2002;2((1)):55–75. doi: 10.2174/1568009023334060. DOI: 10.2174/1568009023334060. [DOI] [PubMed] [Google Scholar]
- 16.Roskoski RJ. STI-571: Anticancer protein-tyrosine kinase inhibitor. Biochem Biophys Res Commun. 2003;309((4)):709–717. doi: 10.1016/j.bbrc.2003.08.055. DOI: 10.1016/j.bbrc.2003.08.055. [DOI] [PubMed] [Google Scholar]
- 17.Ananthanarayanan VS, Tetreault S, Saint-Jean A. Interaction of calcium channel antagonists with calcium: spectroscopic and modeling studies on diltiazem and its Ca2+ complex. J Med Chem. 1993;36((10)):1324–1332. doi: 10.1021/jm00062a004. DOI: 10.1021/jm00062a004. [DOI] [PubMed] [Google Scholar]
- 18.Ju Y, Verma RS. Aqeous N-hererocylization of primary anuries and hydrazines with dihalids microwave-assistant synthesis of N-azacycloalkanes, isoindole, pyrazole, pyrazolidine and phthalazine derivatives. J Org Chem. 2006;71((1)):135–141. doi: 10.1021/jo051878h. DOI: 10.1021/jo051878h. [DOI] [PubMed] [Google Scholar]
- 19.Parke DV. The Biochemistry of Foreign Compounds. 1th ed. Oxford: Pergamon press; 1968. p. 224. [Google Scholar]
- 20.Ju Y, Kumar D, Verma RS. Revisiting nucleophilic substifation reactions: microwave-assisted synthesis of azides, thiocyanats and sulfones in an aqeous medium. J Org Chem. 2006;71((17)):6697–6700. doi: 10.1021/jo061114h. DOI: 1021/jo061114h. [DOI] [PubMed] [Google Scholar]
- 21.Jain KS, Chitre TS, Miniyar PB, Muthu K, Bendre VS, Veer VS, et al. Biological and medical significance of pyrimidine. Curr Sci. 2006;90((6)):793–803. [Google Scholar]
- 22.Infectious Society of America, Statement of the IDSA Concerning. Bioshield II: Responding to an Ever-Changing Threat. Alexandria, Va, USA: IDSA; 2004. pp. 2–4. [Google Scholar]
- 23.Coates ARM, Hu Y. Novel approaches to developing new antibiotics for bacterial infections. Br J Pharmacol. 2007;152((8)):1147–1154. doi: 10.1038/sj.bjp.0707432. DOI: 10.1038/sj.bjp.0707432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venanzi LM. Tetrahedral nickel (II) complexes and the factors determining their formation. Part I. Bistriphenylphosphine nickel(II) compounds. J Chem Soc. 1958;3((4)):719–724. DOI: 10.1039/JR9580000719. [Google Scholar]
- 25.Wiegand I, Hilpert K, Hancock REW. Agar and Broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3:163–175. doi: 10.1038/nprot.2007.521. DOI: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 26.Panchaud P, Bruyère T, Blumstein AC, Bur D, Chambovey A, Ertel EA, et al. Discovery and optimization of isoquinoline ethyl ureas as antibacterial agents. J Med Chem. 2017;60((9)):3755–3775. doi: 10.1021/acs.jmedchem.6b01834. DOI: 10.1021/acs.jmedchem.6b01834. [DOI] [PubMed] [Google Scholar]
- 27.Retailleau P, Colloc’h N, Vivarès D, Bonneté F, Castro B, El Hajji M, et al. Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site. Acta Crystallogr D Biol Crystallogr. 2005;61((Pt 3)):218–229. doi: 10.1107/S0907444904031531. DOI: 10.1107/S0907444904031531. [DOI] [PubMed] [Google Scholar]
- 28.Oksanen E, Blakeley MP, Bonneté F, Dauvergne MT, Dauvergne F, Budayova-Spano M. Large crystal growth by thermal control allows combined X-ray and neutron crystallographic studies to elucidate the protonation states in Aspergillus flavus urate oxidase. J R Soc Interface. 2009;6(Suppl 5(Suppl 5)):S599–S610. doi: 10.1098/rsif.2009.0162.focus. DOI: 10.1098/rsif.2009.0162.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clayden J, Greves N, Warren S, Wothers P. Organic Chemistry. 1th ed. Oxford: Oxford University Press; 2001. p. 188. [Google Scholar]
- 30.Schotten C. Ueber die oxidation des piperidins. Chem Ber. 1884;17((2)):2544–2547. DOI: 10.1002/cber.188401702178. [Google Scholar]
- 31.Baumann E. Ueber eine einfache method der darstellung von benzoesaureathern. Chem Ber. 1886;19((2)):3218–3222. DOI: 10.1002/cber.188601902348. [Google Scholar]
- 32.Fors BP, Krattiger P, Strieter E, Buchwald SL. Water-mediated catalyst preactivation: an efficient protocol for C-N cross-coupling reactions. Org Lett. 2008;10((16)):3505–3508. doi: 10.1021/ol801285g. DOI: 10.1021/ol801285g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig CR, Stitzel RE. Modern Pharmacology with Clinical Applications. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 515–525. [Google Scholar]
- 34.Zessel K, Mohring S, Hamscher G, Kietzmann M, Stahl J. Biocompatibility and antibacterial activity of photolytic products of sulfonamides. Chemosphere. 2014;100:167–174. doi: 10.1016/j.chemosphere.2013.11.038. DOI: 10.1016/j.chemosphere.2013.11.038. [DOI] [PubMed] [Google Scholar]
- 35.Egbujor MC, Nwobodo DC, Egwuatu PI, Abu IP, Ezeagu CU. Sulphonamide drugs and pseudomonas aeruginosa resistance: a review. Int J Modern Pharm Res. 2020;4((1)):78–83. [Google Scholar]
- 36.Muhammad AQ, Mahmood A, Hina A, Sadia W, Muhammad IS. Amide sulfonamides and benzene sulfonamides: synthesis and their biological evaluation. J Chem. 2015;2015:524056,1–8. DOI: 101155/2015/524056. [Google Scholar]
- 37.Egbujor MC, Okoro UC, Okafor S. Novel alanine-based antimicrobial and antioxidant agents: synthesis and molecular docking. Indian J Sci Technol. 2020;13((09)):1003–1014. DOI: 10.17485/ijst/2020/v013i09/146687. [Google Scholar]
- 38.Hennicke F, Grumbt M, Lermann U, Ueberschaar N, Palige K, Bottcher B, et al. Factors supporting cysteine tolerance and sulfite production in Candida albicans. Eukaryot Cell. 2013;12((4)):604–613. doi: 10.1128/EC.00336-12. DOI: 10.1128/EC.00336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45((12)):2615–2623. doi: 10.1021/jm020017n. DOI: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 40.Van de waterbeemd H, Carter RE, Grassy G, Kubinyi H, Martins YC, Tute MS, et al. Grossory of terms used in computational drug design. Pure Appl Chem. 1997;69((5)):1137–1152. [Google Scholar]
- 41.Egbujor MC, Okoro UC, Okafor S. Design, synthesis, molecular docking, antimicrobial and antioxidant activities of new phenylsulfamoyl carboxylic acids of pharmacological interest. Med Chem Res. 2019;28:2118–2127. DOI: 10.1007/s00044-019-02440-3. [Google Scholar]