Abstract
Background and purpose:
Glioblastoma multiforme (GBM) is the most invasive type of cancer which starts inside the brain. GBM cells were found to have similar properties to glioblastoma cancer stem cells. CD44 can be used as a marker of the cancer stem cells in a subset of glioblastoma tumor cells. Recent studies showed that CD44 is involved in developing cancer cells via the protein kinase B (PKB or AKT) signaling pathway. Therefore, this study aimed to investigate the CD44 mRNA silencing effects on the glioblastoma cell cycle via AKT signaling pathway.
Experimental approach:
To determine CD44 expression in the samples of the patients with GBM, we used the analysis of data extracted from TCGA database. qRT-PCR and western blotting were used to evaluate the expression level of genes and proteins. Different cell cycles were evaluated by DAPI staining and flow cytometry.
Findings/Results:
Bioinformatics results showed that CD44 expression level in GBM tumor samples is higher than in normal samples. Effects of poly (ethylene imine)-polyethylene glycol (PEI-PEG)-loaded CD44 siRNA in cell cycle showed that CD44 silencing could inhibit cell cycle in G0-G1 phase by more than 20% compared to the negative control (P < 0.05). Furthermore, PEI-PEG-loaded CD44 siRNA reduces the expression of cyclin D1 and CKD-4. According to our findings, this structure also prevented AKT phosphorylation at Thr-308 and Ser-473.
Conclusion and implications:
Our results suggest that PEI-PEG-loaded CD44 siRNA may attenuate the cell cycle by suppressing AKT signaling pathway.
Keywords: AKT, CD44, Cell cycle, Glioblastoma
INTRODUCTION
Glioblastoma multiforme (GBM) is an aggressive and rapidly growing type of central nervous system tumor which accounts for 57% of gliomas and 48% of central nervous system tumors (1). This tumor has a poor prognosis, and the median survival is often less than 2 years for these patients (2). This tumor often invades surrounding tissues in the brain and rarely metastases to distant organs (3). Studies in recent years show that the main responsibility for progress, recurrence, metastasis, and resistance to treatment in GBM tumor cells is called glioblastoma stem cells (GSCs) (4,5). As a result, targeting GSCs might be employed as a potential therapeutic technique to enhance the treatment of GBM patients.
Cancer stem cells are a small subpopulation within the tumor which gives cancer cells the ability for proliferation, self-renewal, drug resistance development, and metastasis (6).
Current evidence indicates that the existence of cancer stem cells in the GBM is a key source of phenotypic, genetic, and epigenetic heterogeneity, as well as the primary cause of non-response to radiotherapy and chemotherapy by regulating DNA damage response (7). Cancer stem cells are often identified using cell membrane surface markers. Cancer stem cells isolated from GBM express high levels of markers cluster of differentiation (CD)133, CD15/SSEA, CD44, or A2B5, of which CD44 is the most important.
CD44 is one of the most prevalent markers of cancer stem cells which is correlated to many cancer cell hallmarks, including growth and proliferation, survival, metastasis, cell invasion, and so on (8). A growing body of evidence indicated that the high expression level of CD44 in GSCs is often associated with a poor prognosis of the disease. Mechanistic studies showed that the protein kinase B (PKB or AKT) signaling pathway is one of the most well-known signaling pathways which link CD44 with tumor progression. CD44 was implicated in the proliferation and metastasis of tumors through the AKT signaling pathway in several types of tumors. In contrast, the knockdown of CD44 by downregulating phosphorylated-AKT (p-AKT) expression level leads to attenuate cell proliferation and metastasis (9,10). AKT signaling is one of the most important signaling pathways in GBM, which plays a key role in the proliferation, inhibition of apoptosis, and induction of metastasis (11).
Supportive care for GBM patients focuses on symptom reduction and neurological function improvement (12). Surgery is the first step in the treatment of GBM. However, when a tumor is located near sensitive areas of the brain, the risk of tumor resection increases. Following surgery, radiotherapy and chemotherapy (typically with temozolomide) are used to suppress and delay the recurrence of the tumor (13,14). Given the above, as well as the fact that cancer is one of the leading causes of death worldwide, over the past decade, there were many research studies on finding new treatments to reduce side effects of conventional therapies, and fundamental advances are required for the development of new therapeutic strategies (or models) for brain cancer (15).
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a group of non-coding RNA molecules that are 20-24 bp in length. These molecules disrupt the expression of specific genes with complementary nucleotide sequences by inhibiting the translation process of mRNA (16,17). In our previous study, we showed that targeting CD44 by siRNA inhibited the proliferation, invasion, and migration of glioma cells via suppression of aldehyde dehydrogenase 1 (ALDH1) and NOTCH1, as well as RANKL1 expression levels. We evaluated, for the first time, the effect of poly(ethylene imine)-polyethylene glycol (PEI-PEG)-loaded CD44 siRNA on the cell cycle via the CD44-AKT signaling pathway in GBM (U87MG) cells.
MATERIALS AND METHODS
Materials
siRNAs were purchased from Microsynth company (Switzerland). RPMI 1640, fetal bovine serum (FBS), and penicillin/streptomycin were provided from Gibco, (Invitrogen, Grand Island, NY, USA). RiboEx solution was provided from GeneAll biotechnology, Republic of Korea. BIO FACT synthesis kit and Syber Green qPCR Master Mix were purchased from South Korea and primers were also provided from Pishgam Biotech, Tehran, Iran. Primary antibody against p-AKT, cyclin-dependent kinase (CDK) 4, cyclin D1, and β-actin, and the secondary antibody, was obtained from Santa Cruz Biotechnology (CA, USA). Primary antibody against AKT was provided from Elabscience Biotechnology (Wuhan, China).
Bioinformatic analysis
The expression level of CD44 gene in TCGA data was collected for tumor samples as well as healthy samples from USCS Xena server databases, Cancer Immunome Atlas, GTEx GBM datasets, as well as TIMER 2.0 database according to the following details: TCIA dataset, CD44 RNAseq dataset available in TCGA database including 156 tumor tissue expression and five matched normal tissue expressions; USCS Xena dataset, CD44 RNAseq dataset available in TCGA database including 155 tumor tissue expression and five matched normal tissue expression; GEPIA 2.0 dataset, the dataset is composed of TCGA GBM tumor datasets (n = 163) in comparison to GTEx GBM normal tissue expression datasets (n = 207); and TIMER 2.0 dataset, CD44 RNAseq dataset available in TCGA database including 153 tumor tissue expression and five matched normal tissue expression.
Selection of cell line
To select the appropriate cell line from U87MG LN229, SF126, M059K, SNB75, SNU-1105, SNU-201, SNU-466, SNU-489, SNU-626, CAS-1, GMS-10, 42-MG, 8-MG-BA, and AM38 cell lines, finally, based on transcripts per million (TPM, a normalized measure of gene expression), U87MG cell line (Fig. 1) was selected as a model to investigate the inhibitory effect of PEI-PEG-loaded CD44 siRNA on CD44 expression. PEI-PEG-loaded CD44 siRNA was prepared as per the method developed previously (18). Besides, U87MG cell line selection, PEI-PEG synthesis, and loading process were performed in our previous study. In brief, PEG was functionalized using bromoacetyl chloride and then attached to the PEI surface. Then, siRNA against CD44 and non-targeting siRNA, as negative control were used. PEI-PEG solution was added to the siRNA-containing vial and mixed and incubated for 30 min. The concentration of siRNA was 60 pmol. The size of generated PEI-PEG-siRNA nanoparticles was 174.3 nm (18).
Fig. 1.
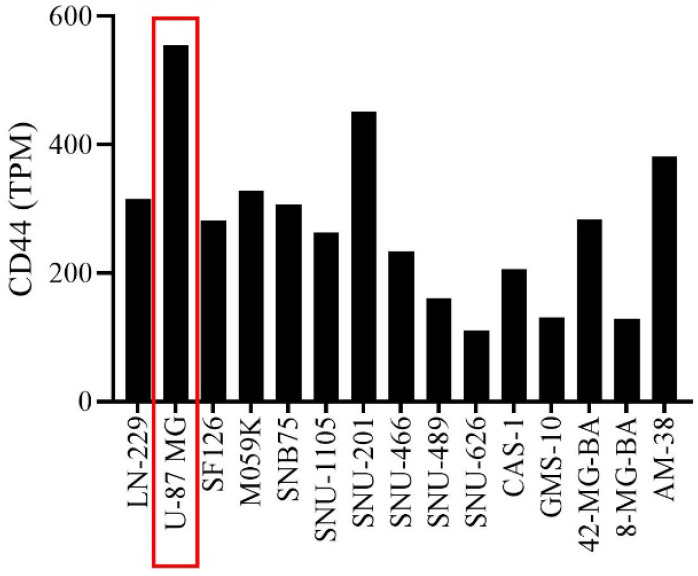
TPM of the U87MG cell line related to CD44 among the glioblastoma multiforme cell lines. CD44-related U87MG cell line TPM was higher among the target cell lines than the others. TPM, Transcripts per million
Cell culture
U87MG glioblastoma cancer human cell line was obtained from Pasteur Institute of Iran Cell Bank (Tehran, Iran). Cells were cultured in RPMI-1640 medium containing 10% FBS and 100 U/mL penicillin/100 U/mL streptomycin and incubated at 37 °C and 5% CO2. After cells were approximately 80-90% confluent, first washed with PBS and detached using trypsin-EDTA for subculture or treatment. The morphology of cells was performed using an inverted microscope and the cell culture medium was changed every 2 days.
RNA extraction and quantitative real-time polymerase chain reaction analysis
Total RNA extraction was performed using RiboEx solution (Gene All biotechnology, Republic of Korea). Approximately 5 × 105 cells/well were cultured in 6-well plates. After 24 h, the cells were treated with PEI-PEG-loaded CD44 siRNA for 48 h (18). After 48 h, RiboEx was added to the wells and RNA extraction was performed according to the manufacturer’s instructions. Finally, NanoDrop 2000 (Thermo, USA) and agarose gel electrophoresis were used to evaluate the quantity and quality of the extracted RNA, respectively. According to the manufacturer’s instructions, the extracted RNA was stored at -80 °C. Complementary DNA (cDNA) was synthesized by the BIO FACT (South Korea) synthesis kit. Quantitative real-time polymerase chain reaction (qRT-PCR) performed using BIO FACT (South Korea) 2X Real-Time PCR Master Mix and assessed using One-step Real-Time PCR System (Applied Biosystems) based on the following procedure: initial denaturation step, 10 min at 95 °C; followed by 35 cycles of 10 sec denaturation at 95 °C; annealing step, 59 °C for 35 s; and extension step, 72 °C for 20 s. The data were analyzed based on the 2-ΔΔCT(Livak) method. The sequence of primers was listed in Table 1.
Table 1.
The sequences of the used primers.
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| CD44 | 5’-CAAGCCACTCCAGGACAAGG-3’ | 5’-ATCCAAGTGAGGGACTACAACAG-3’ |
| β-actin | 5’-CACTCTTCCAGCCTTCCTTCCT-3’ | 5’-GTGATCTCCTTCTGCATCCTGTCG -3’ |
Western blot analysis
Approximately 5 × 105 cells/well were cultured in 6-well plates. After 24 h, the cells were treated with PEI-PEG-loaded CD44 siRNA for 48 h. The cells were lysed using radioimmunoprecipitation assay (RIPA) buffer. The cell lysates were centrifuged at 12,000 g, and the Bradford protein assay was used to evaluate the amount of protein. Total protein was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis -polyacrylamide gel electrophoresis and separated proteins transferred to polyvinylidene fluoride (PVDF) membrane. The membrane was then blocked with 5% skimmed milk and incubated overnight at 4 °C with primary antibodies against p-AKT, AKT, CDK4, cyclin D1, and β-actin. The membrane was then washed and incubated with secondary antibody conjugated with horseradish peroxidase for 2 h at room temperature. Visualization of protein bands was performed using enhanced chemiluminescence reagents and the results were analyzed using ImageJ software.
Cell cycle analysis
Approximately 5 × 105 cells/well were seeded in 6-well plates. After 24 h, the cells were treated with PEI-PEG-loaded CD44 siRNA for 48 h. The cells were washed twice in PBS solution. Then 75% cold ethanol was added dropwise with vortex on the cell precipitate and incubated at -20 °C for 24 h. The next day, after centrifugation, the supernatant was removed. After washing, 200 μL of the solution containing 1 μL of RNAse A and 0.001% 4’,6-diamidino-2-phenylindole (DAPI) dye was added to the cells and after 10 min incubation at room temperature. The cell cycle was analyzed using MACSquant10 (Milteni Biotech, Germany). The results were analyzed using FlowJo software (Treestar, Ashland, OR).
Statistical analysis
Data analysis was performed using GraphPad Prism (GraphPad Software, La Jolla, CA). The difference between the control group and the treatment group was evaluated using a two-tailed Student’s t-test. Results expressed as the mean ± standard deviation (SD) and P-values less than 0.05 were considered statistically significant.
RESULTS
CD44 expression in GBM patients
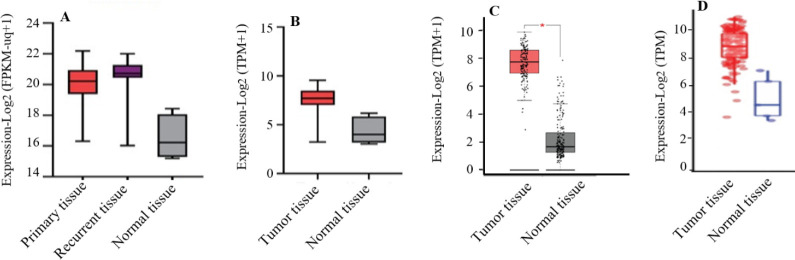
The expression of the CD44 gene in the samples of GBM patients extracted from the TCGA dataset showed that the CD44 expression level is higher in the samples of the patients with GBM than in the control group (normal samples) (Fig. 2).
Fig. 2.
CD44 gene expression in GBM cases. Gene expression in normal vs tumoral tissue is shown by box plots. Gene expression values are demonstrated in Log scale to make the comparison more obvious. (A) CD44 gene expression RNASeq from USCS Xena server based on TCGA GBM cohort data (solid normal tissue: n = 5; primary tumor: n = 155; recurrent tumor: n = 13); (B) CD44 gene expression RNASeq in The Cancer Immunome Atlas (TCIA) server based on TCGA datasets (tumor tissue: n = 156; normal: n = 5); (C) CD44 gene expression in GEPIA 2.0 database based on TCGA and GTEx GBM datasets (tumor samples: n = 163; normal samples: n = 207); (D) gene expression of CD44 in TIMER 2.0 database based on TCGA datasets (tumor tissue: n = 153; normal tissue: n = 5). GMB, Glioblastoma multiforme; FPKM, fragment per kilobase of transcript per million mapped reads, a normalized measure of gene expression; TPM, transcripts per million, a normalized measure of gene expression.
Effect of PEI-PEG-loaded anti-CD44 siRNA on CD44 expression in GBM cells
U87MG glioblastoma cells were treated with PEI-PEG-loaded anti-CD44 siRNA and CD44 expression level by qRT-PCR method showed that the group treated with PEI-PEG-loaded anti-CD44 siRNA significantly reduced CD44 gene expression by less than 50% (P < 0.001, Fig. 3).
Fig. 3.
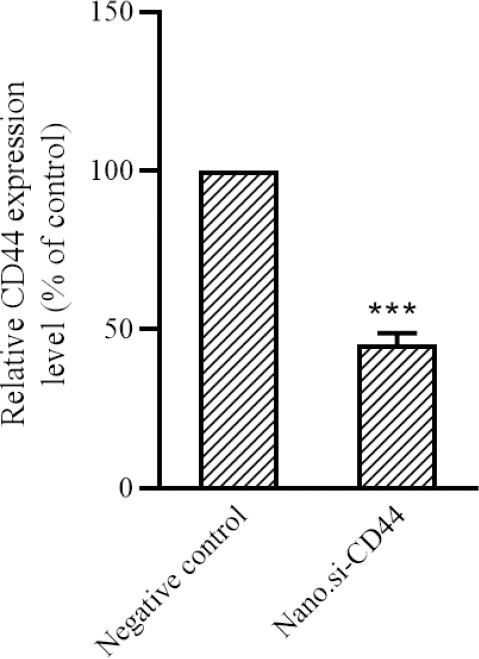
PEI-PEG-loaded anti-CD44 siRNA reduces the expression of CD44 gene in glioblastoma multiforme cells compared to the negative P< 0.001. PEI-PEG, Poly (ethylene imine)-polyethylene glycol.
Effect of PEI-PEG-loaded anti-CD44 siRNA on GBM cell cycle
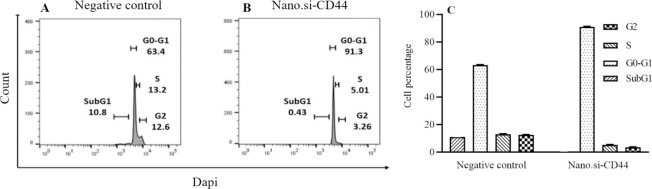
The effects of PEI-PEG-loaded anti-CD44 siRNA on different phases of the cell cycle showed that CD44 silencing could inhibit the cell cycle in the G0-G1 phase up to 20% compared to the negative control (P < 0.05, (Fig. 4). It has been concluded that the decrease of phase S and phase G2 is probably due to the inhibition of phase G0-G1. The flow cytometry results were verified by western blot, and PEI-PEG-loaded anti-CD44 siRNA decreased the expression of cyclin D1 and CDK-4 proteins (Fig. 5).
Fig. 4.
(A-B) PEI-PEG-loaded anti CD44 siRNA inhibits the G0-G1 phase of the cell cycle in glioblastoma multiforme cells. (C) The results of cell cycle experiments showed that siRNA against CD44 has the ability to inhibit the cell cycle in G0-G1 phase compared to the negative control group. *P< 0.05. PEI-PEG, Poly (ethylene imine)-polyethylene glycol.
Fig. 5.
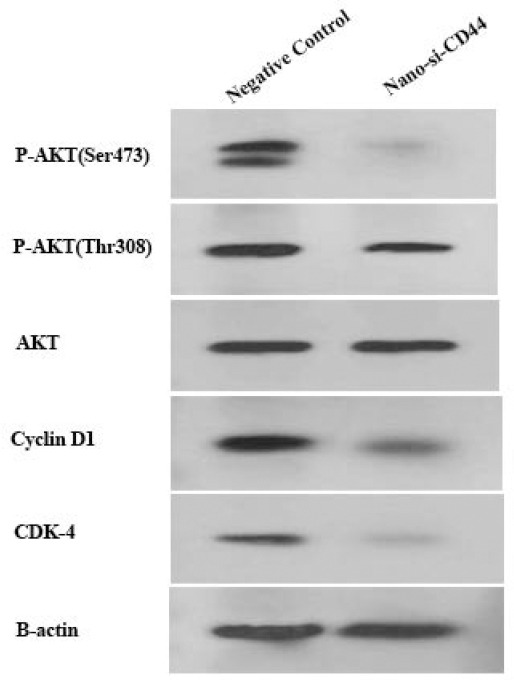
PEI-PEG-loaded anti CD44 siRNA inhibits P-AKT expression levels in Ser473, Thr308, CD1, and CDK-4 in glioblastoma multiforme cells. The expression level of phosphorylated AKT in Ser473 and Thr308. and CD1, and CDK-4 decreased in the PEI-PEG-loaded anti-CD44 siRNA treatment group compared to the control group. Protein expression levels of all targets were measured by western blotting. PEI-PEG, Poly (ethylene imine)-polyethylene glycol, p-AKT, phosphorylated protein kinase B.
Effect of EI-PEG-loaded anti-CD44 siRNA on p-AKT expression
Previous studies on CD44 showed that this cellular surface marker is involved in tumor progression through the AKT signaling pathway. For this reason, we examined the expression levels of p-AKT in Ser-473 and Thr-308. Our results showed that PEI-PEG-loaded anti-CD44 siRNA downregulated the expression level of p-AKT in Ser-473 and Thr-308 compared to the control group (Fig. 5). These findings suggest that a decrease in CD44 expression by PEI-PEG-loaded anti-CD44 siRNA may inhibit AKT phosphorylation and AKT signaling pathway, resulting in inhibition of cell cycle (Fig. 6).
Fig. 6.
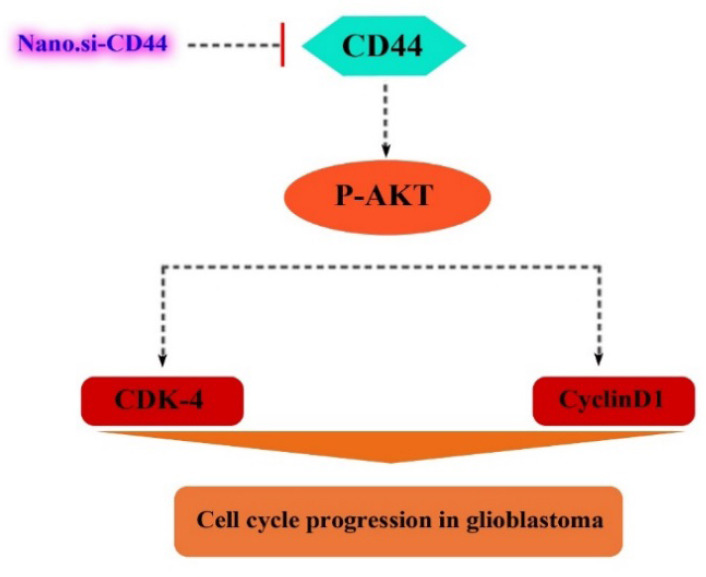
A summarizing illustration of the possible molecular mechanism of PEI-PEG-loaded anti-CD44 siRNA on cell cycle arrest in glioblastoma multiforme cells. CDK, Cyclin-dependent kinase. PEI-PEG, Poly (ethylene imine)-polyethylene glycol.
DISCUSSION
Supportive care for GBM patients focuses on symptom reduction and neurological function improvement. In the present study, according to treatment strategies based on targeting GBM-related stem cells, CD44, one of the common surface markers in cancer stem cells and GSC, was selected as a target. We hypothesized that inhibiting CD44 expression, which is essential for the maintenance of GSCs, could inhibit the growth and proliferation of cancer cells. In our previous study, the potential for efficient siRNA delivery using PEI-PEG nanoparticles was proposed to achieve multiple therapeutic roles by directly reducing GBM tumor growth, reducing invasion and migration, and increasing apoptosis. Our findings suggest that PEI-PEG-loaded anti-CD44 siRNA, by inhibiting CD44 expression, may suppress AKT signaling pathway and ultimately caused cell cycle arrest in G0-G1 phase in the GBM cell line (U87MG).
CD44 molecule is a cell surface glycoprotein and an essential factor in the interaction of the cell and the cell-matrix, and the most common cell surface markers of the GSCs. It was shown that the highest expression of the CD44 gene in gliomas is related to GBM. Fast-growing tumors such as GBM with more CD44 expression and other markers have more cancer stem cell markers than slow-growing tumors such as astrocytoma (19).
Mihić et al. found that brain tumors show the high expression of the CD44 gene (20). The results of Xu et al. research showed that selecting appropriate anti-CD44 small hairpin RNA (shRNA) and transferring them to U87MG, and U251 cell lines with lentivirus effectively reduced CD44 gene expression at mRNA and protein levels in GBM cells (21). However, Wang et al. reported different results. They showed that there was no significant relationship between CD44 gene expression and GSC. Decreasing CD44 expression while promoting the expression of GSC markers such as nestin, CD133, and OCT-4 causes enhanced GSC properties and long-term proliferation of GSC cells (22). There has been no investigation of CD44 gene suppression based on PEI-PEG/CD44 siRNA-based siRNA delivery for GBM. In similar to results from the study by Xu et al. we found that inhibiting the expression of CD44 gene by PEI-PEG-loaded anti-CD44 siRNA significantly reduced the expression level of CD44 in U87MG cells.
As mentioned, CD44 plays a key role in tumor progression and cell cycle in GBM. For example, Yeh et al. suggested that miR-138 via binding to the 3’ untranslated regions of CD44 suppresses its expression level which caused cell cycle arrest in GBM cells (23). According to studies by Park et al. it was demonstrated thatCD44 knockdown by shRNA induced cell cycle arrest in the G0-G1 phase and a decrease in the S phase, hence inhibited cell proliferation and induced cell apoptosis in colon cancer cells (24). In addition, Papadopoulos et al. showed that alteration in the expression of CD24 and CD44 genes using haloperidol in GBM U87MG, U251, and T98 cell lines induces M/G2 cell cycle arrest and increases the population of cells in the G0/G1 phase (25).
In agreement with these studies, our results from the cell cycle assay showed that in the group treated with PEI-PEG-loaded anti-CD44 siRNA, cell cycle inhibition was seen in phase G0-G1. Moreover, the reason for the decrease in entry into other phases of the cell cycle, including phase S and phase G2 in the siRNA-treated group against CD44, is probably due to the inhibition of phase G0-G1. This finding suggests that the inhibition of CD44 may arrest the cell cycle by suppressing the expression of proteins effective in cell cycle regulation in GBM. Although studies have been performed on the cell cycle related to the role of CD44, the effect of CD44 inhibition on the arresting of different phases of the cell cycle in GBM cells requires further studies.
Studies to clarify the association between CD44 and tumor progression showed that the AKT signaling pathway is one of the most important signaling pathways in this regard. The activation of the AKT signaling pathway by CD44 can be crucial in maintaining stemness characteristics in cancer cells. According to reports from various malignancies, CD44 knockdown exhibits antitumor activity via reducing p-AKT and blocking the AKT signaling pathway (10,26). Moreover, Park et al. reported that CD44 inhibition by shRNA led to decreased phosphorylated Akt in colon cancer cells. They concluded that CD44 suppression mediated anticancer effects on colon cancer cells through inhibiting AKT signaling (24). AKT signaling is one of the most important signaling pathways in GMB, which plays a key role in proliferation, inhibition of apoptosis, and induction of metastasis (11). Our results indicated that PEM-PEG-loaded anti-CD44 siRNA-treated GBM cells reduced p-AKT expression levels in Ser473 and Thr308 compared to control cells. This structure does not affect total AKT. Our results were also confirmed by Yeh et al. who demonstrated miR-138 as a CD44 inhibitor by reducing the expression of CD44 and suppressing the CD44/AKT signaling pathway to cell cycle arrest in GBM (23).
CONCLUSION
In summary, our findings reveal that PEI-PEG-loaded anti-CD44 siRNA by inhibiting CD44 in GBM could inhibit AKT signaling pathway and ultimately arrest the cell cycle. These findings suggest that PEI-PEG-loaded anti-CD44 siRNA might be a potential therapeutic strategy to treat patients who suffer from GBM by targeting GSCs.
Conflict of interest statement
All authors declared no conflict of interest in this study.
Authors’ contribution
P. Mahinfar implemented in vitro techniques concerning cell culture, bioinformatic analysis, RT PCR, western blot analysis, and cell cycle analysis,; E. Siasi Torbati contributed to the study consultation, conceptualization, and editing of the manuscript; and B. Baradaran and A. Mokhtarzadeh guided the present scientific team, data analysis, statistical analysis, wrote and revised the article. All the authors studied and approved the final version of the manuscript.
Acknowledgments
The authors would like to thank all the technical staff of the Immunology Research Center of Tabriz University of Medical Science, Tabriz, Iran for their assistance.
REFERENCES
- 1.Abbasi A, Hajialyani M, Hosseinzadeh L, Jalilian F, Yaghmaei P, Navid SJ, et al. Evaluation of the cytotoxic and apoptogenic effects of cinnamaldehyde on U87MG cells alone and in combination with doxorubicin. Res Pharm Sci. 2020;15((1)):26–35. doi: 10.4103/1735-5362.278712. DOI: 10.4103/1735-5362.278712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin. 2020;70((4)):299–312. doi: 10.3322/caac.21613. DOI: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 3.Mahinfar P, Baradaran B, Davoudian S, Vahidian F, Cho WCS, Mansoori B. Long non-coding RNAs in multidrug resistance of glioblastoma. Genes. 2021;12((3)):455–473. doi: 10.3390/genes12030455. DOI: 10.3390/genes12030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gersey ZC, Rodriguez GA, Barbarite E, Sanchez A, Walters WM, Ohaeto KC, et al. Curcumin decreases malignant characteristics of glioblastoma stem cells via induction of reactive oxygen species. BMC Cancer. 2017;17((1)):99–109. doi: 10.1186/s12885-017-3058-2. DOI: 10.1186/s12885-017-3058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volovetz J, Berezovsky AD, Alban T, Chen Y, Lauko A, Aranjuez GF, et al. Identifying conserved molecular targets required for cell migration of glioblastoma cancer stem cells. Cell Death Dis. 2020;11((2)):152–163. doi: 10.1038/s41419-020-2342-2. DOI: 10.1038/s41419-020-2342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babaei G, Ansari MHK, Aziz SGG, Bazl MR. Alantolactone inhibits stem-like cell phenotype, chemoresistance and metastasis in PC3 cells through STAT3 signaling pathway. Res Pharm Sci. 2020;15((6)):551–562. doi: 10.4103/1735-5362.301340. DOI: 10.4103/1735-5362.301340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K. Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis. 2015;2((2)):152–163. doi: 10.1016/j.gendis.2015.02.001. DOI: 10.1016/j.gendis.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu G, Song X, Liu J, Li S, Gao W, Qiu M, et al. Expression of CD44 and the survival in glioma: a meta-analysis. Biosci Rep. 2020;40((4)):BSR20200520. doi: 10.1042/BSR20200520. 1-10. DOI: 10.1042/BSR20200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XP, Zhang XW, Zheng LZ, Guo WJ. Expression of CD44 in pancreatic cancer and its significance. Int J Clin Exp Pathol. 2015;8((6)):6724–6731. PMID: 26261555. [PMC free article] [PubMed] [Google Scholar]
- 10.Xiaoping L, Xiaowei Z, Leizhen Z, Weijian G. Expression and significance of CD44 and p-AKT in pancreatic head cancer. World J Surg Oncol. 2015;13:334–340. doi: 10.1186/s12957-015-0746-8. DOI: 10.1186/s12957-015-0746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YJ, Xu ZG, Li SQ, He LJ, Tang Y, Chen ZZ, et al. Benzimidazoisoquinoline derivatives inhibit glioblastoma cell proliferation through down-regulating Raf/MEK/ERK and PI3K/AKT pathways. Cancer Cell Int. 2018;18((1)):90–101. doi: 10.1186/s12935-018-0588-x. DOI: 10.1186/s12935-018-0588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlsson SK, Brothers SP, Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med. 2014;6((11)):1359–1370. doi: 10.15252/emmm.201302627. DOI: 10.15252/emmm.201302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens GHJ. Antiepileptic therapy in patients with central nervous system malignancies. Curr Neurol Neurosci Rep. 2006;6((4)):311–318. doi: 10.1007/s11910-006-0024-9. DOI: 10.1007/s11910-006-0024-9. [DOI] [PubMed] [Google Scholar]
- 14.Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95((2)):190–198. doi: 10.3171/jns.2001.95.2.0190. DOI: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 15.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68((6)):394–424. doi: 10.3322/caac.21492. DOI: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Nair A, Wang S, Wang L. Quality control of RNA-seq experiments. Methods Mol Biol. 2015;1269:137–146. doi: 10.1007/978-1-4939-2291-8_8. DOI: 10.1007/978-1-4939-2291-8_8. [DOI] [PubMed] [Google Scholar]
- 17.Safari F, Tamadon A, Zarghami N, Abolmali S, Najafi H. Effect of degree of polyethyleneimine PEGylation on biological and cellular activity of hTERT siRNA. Res Pharm Sci. 2012;7((5)):S1033. [Google Scholar]
- 18.Mahinfar P, Mokhtarzadeh A, Baradaran B, Torbati ES. Effects of PEI-PEG nanoparticles loaded with CD44 siRNA on inhibition of growth, invasion, and migration of glioblastoma cells. Crescent J Med Biol Sci. 2021;8((3)):215–222. [Google Scholar]
- 19.Brown DV, Filiz G, Daniel PM, Hollande F, Dworkin S, Amiridis S, et al. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS One. 2017;12((2)):e0172791. doi: 10.1371/journal.pone.0172791. DOI: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mihić J, Rotim K, Vučić M, Hude Dragičević I, Borić M, Lugović-Mihić L. Prognostic role of CD44 expression and neovascularization determined by endoglin (CD105) in glioblastoma patients. Acta Clin Croat. 2019;58((3)):455–462. doi: 10.20471/acc.2019.58.03.08. DOI: 10.20471/acc.2019.58.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70((6)):2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. DOI: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang HH, Liao CC, Chow NH, Huang LL, Chuang JI, Wei KC, et al. Whether CD44 is an applicable marker for glioma stem cells. Am J Transl Res. 2017;9((11)):4785–4806. PMID: 29218080. [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh M, Wang YY, Yoo JY, Oh C, Otani Y, Kang JM, et al. MicroRNA-138 suppresses glioblastoma proliferation through downregulation of CD44. Sci Rep. 2021;11((1)):9219–9229. doi: 10.1038/s41598-021-88615-8. DOI: 10.1038/s41598-021-88615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park YS, Huh JW, Lee JH, Kim HR. shRNA against CD44 inhibits cell proliferation, invasion and migration, and promotes apoptosis of colon carcinoma cells. Oncol Rep. 2012;27((2)):339–346. doi: 10.3892/or.2011.1532. DOI: 10.3892/or.2011.1532. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulos F, Isihou R, Alexiou GA, Tsalios T, Vartholomatos E, Markopoulos GS, et al. Haloperidol induced cell cycle arrest and apoptosis in glioblastoma cells. Biomedicines. 2020;8((12)):595–606. doi: 10.3390/biomedicines8120595. DOI: 10.3390/biomedicines8120595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai CJ, Lin CY, Liao WY, Hour TC, Wang HD, Chuu CP. CD44 promotes migration and invasion of docetaxel-resistant prostate cancer cells likely via induction of hippo-yap signaling. Cells. 2019;8((4)):295–307. doi: 10.3390/cells8040295. DOI: 10.3390/cells8040295. [DOI] [PMC free article] [PubMed] [Google Scholar]