Abstract
COVID-19 patients reveal various clinical manifestations; however, the specific mechanisms and factors contributing to rapid recovery remain unclear. We performed serum cytokine profiling using a bead-based immunoassay in six COVID-19 patients with mild symptoms who experienced rapid recovery. All patients had fever that resolved within 4 days. During the study, the interferon gamma-related protein 10 (IP-10) level rapidly increased initially, and then rapidly decreased in all six patients. Similarly, the interferon (IFN)-λ2/3 levels rapidly increased initially, and then decreased in five of the six patients. IP-10 and IFN-λ2/3 may play a key role in the rapid recovery of mild COVID-19.
Keywords: SARS-CoV-2, COVID-19, IP-10, IFN-λ2/3
INTRODUCTION
Coronaviruses are positive-stranded RNA viruses that cause illnesses of varying severity, ranging from the common cold to fatal pneumonia. Two coronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), are known to cause severe pneumonia (7). COVID-19, caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), was identified as a novel coronavirus in December 2019. Since early 2020, COVID-19 has been a pandemic worldwide.
The clinical manifestations of COVID-19 range from asymptomatic to critical symptoms. Wu et al. (22) reported that 81% of symptomatic patients with COVID-19 had a mild disease, 14% had a severe disease, and 5% had a critical disease. The defense mechanisms against acute viral infection consist of innate immune and adaptive immune responses, including the production of pathogen-specific antibodies. SARS-CoV-2-specific antibodies develop 10–15 days after the onset of symptoms (2,13). As some patients with a mild disease recover within a few days, innate immunity, including the production of inflammatory cytokines, seems to play a key role in the rapid recovery from COVID-19.
Innate immunity against a virus involves blocking of infection by protecting cells from infection, rapidly recognizing and destroying any cells that are infected, and regulating the antiviral inflammatory response (15). The cytokine profile has been suggested to differentiate the clinical manifestations of patients with Hantavirus infection (10). Therefore, to investigate the rapid recovery mechanisms, we performed cytokine profiling in 6 patients with COVID-19, whose fever resolved within 4 days as case series.
PATIENTS AND METHODS
Peripheral blood samples were collected from patients who rapidly recovered from COVID-19 in Kobe University Hospital between August 2020 and November 2020. The diagnosis of COVID-19 was confirmed using a quantitative reverse transcription-polymerase chain reaction assay for SARS-CoV-2. Fever was defined as an axillary temperature higher than or equal to 37.5 °C. Since the Japanese Ministry of Health, Labor and Welfare recommended to receive testing of COVID-19 who have cold symptoms or a fever of 37.5 °C or over for 4 days or more in April 2020, patients with fever resolution within 4 days were defined as rapid recovery (14). To determine disease severity, the National Institutes of Health COVID-19 Treatment Guidelines were used (6). The study was approved by the Kobe University Hospital Ethics Committee (No. B2056704) and was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent for the use of their blood samples in this study.
Sample collection and processing
Serum samples were obtained by centrifuging the blood samples for 10 min at 1000 × g at room temperature and were immediately transferred to a −80 °C freezer. Before analysis, SARS-CoV-2 inactivation in all serum samples was performed by ultraviolet light-C irradiation.
Cytokine profiling
The levels of the following human cytokines and enzymes associated with the antiviral response in serum samples were measured using LEGENDplex (BioLegend), a bead-based immunoassay with the same basic principles as a sandwich immunoassay: interleukin (IL)-1β, IL-6, IL-8, IL-10, IL-12p70, interferon (IFN)-α2, IFN-β, IFN-λ1, IFN-λ2/3, IFN-γ, tumor necrosis factor (TNF)-α, interferon gamma-induced protein 10 (IP-10), and granulocyte macrophage colony-stimulating factor (GM-CSF). The capture bead with the antibody for a particular molecule conjugated on its surface was mixed with serum samples, and the mixture was incubated for 2 h. After washing, biotinylated detection antibodies were added, and samples were incubated for 1 h to allow the formation of bead–molecule–detection antibody sandwiches. Streptavidin–phycoerythrin was subsequently added, and the samples were incubated for 30 min. For each bead population, the phycoerythrin signal fluorescence intensity was quantified using a BD FACSCanto™ II cell analyzer. The concentration of a particular molecule was determined using a standard curve of known concentration with LEGENDplex data analysis software.
RESULTS
Patient characteristics
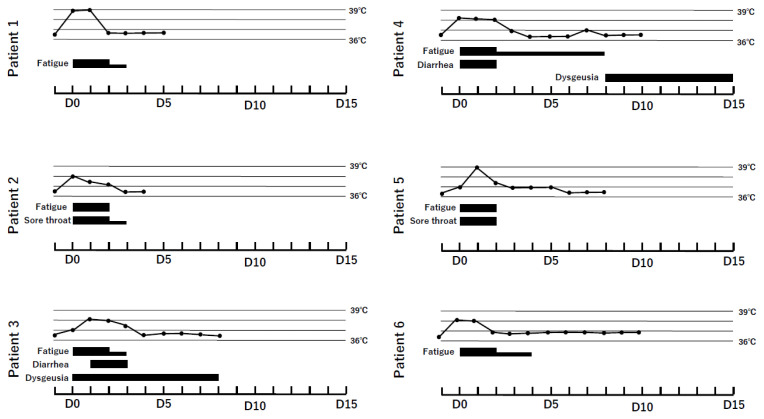
Six patients were included in this study (Table). Clinical course of six patients is shown in Figure 1. All six patients had fever and fatigue for a few days, and were classified to have mild illness. The median age of the patients was 30.5 (range, 22–46) years, and four patients were women. Patient 2 had diffuse large B-cell lymphoma that was under observation after chemotherapy. All patients were of Asian ethnicity.
Table.
Patient characteristics
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
|---|---|---|---|---|---|---|
| Age | 30 | 29 | 31 | 41 | 46 | 22 |
| Sex | Female | Male | Male | Male | Female | Male |
| Comorbidity | None | DLBCL | PDA | None | Hashimoto disease | None |
| Occupation | Office worker | Bus driver | Bar manager | Dance teacher | Care giver | Student |
| Symptoms other than fever | Fatigue | Fatigue Sore throat |
Fatigue Diarrhea Dysgeusia |
Fatigue Diarrhea Dysgeusia |
Fatigue Sore throat |
Fatigue |
| Laboratory values within 48 h of hospital admission | ||||||
| WBC | 2200 /μL | 3300 /μL | 3500 /μL | 3600 /μL | 4800 /μL | 2900 /μL |
| Neutrophil | 1146 /μL | 1155 /μL | 1582 /μL | 2361 /μL | 2976 /μL | 1311 /μL |
| Lymphocyte | 761 /μL | 1683 /μL | 1554 /μL | 986 /μL | 1478 /μL | 1012/μL |
| CRP | N/A | 0.84 mg/dL | N/A | N/A | 0.78 mg/dL | N/A |
| LDH | 135 U/L | 188 U/L | 170 U/L | 150 U/L | 179 U/L | 131 U/L |
| Ferritin | N/A | N/A | 100 ng/mL | N/A | N/A | N/A |
| D-dimer | <0.5 μg/mL | N/A | <0.5 μg/mL | <0.5 μg/mL | <0.5 μg/mL | N/A |
DLBCL; diffuse large B-cell lymphoma, PDA; patent ductus arteriosus,
N/A; not available
Figure 1.
Clinical course of the six COVID-19 patients. The black line represents body temperature. The black box shows the symptoms of COVID-19. Days indicate the day after symptom onset.
Fluctuations in cytokine production
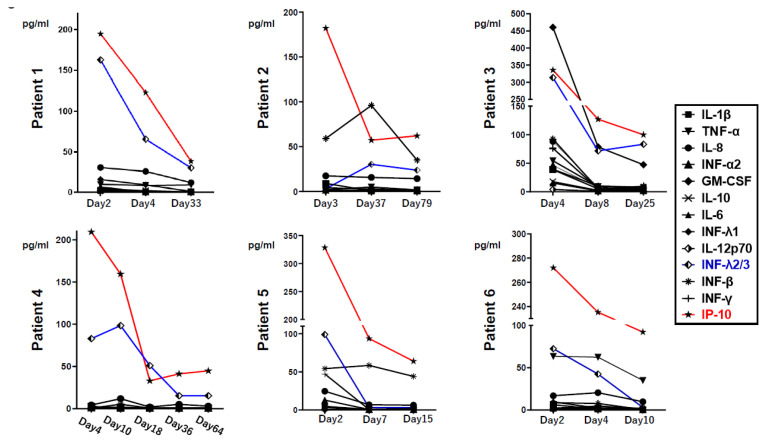
As shown in Figure 2, the IP-10 level was apparently high in all six patients, and the IFN-λ2/3 levels were also high in five out of the six patients at fever resolution. Thereafter, the levels of these cytokines rapidly decreased. The GM-CSF level increased in one patient (Patient 3). There were no notable trends in the levels of other proinflammatory cytokines (IL-1β, IL-6, IL-8, IL-10, IL-12p70, IFN-α2, IFN-β, IFN-λ1, IFN-γ, and TNF-α) during illness and recovery in these six patients.
Figure 2.
Alterations of serum levels of interleukin (IL)-1β, IL-6, IL-8, IL-10, IL-12p70, interferon (IFN)-α2, IFN-β, IFN-λ1, IFN-λ2/3 (blue line), IFN-γ, tumor necrosis factor (TNF)-α, interferon gamma-induced protein 10 (IP-10) (red line), and granulocyte macrophage colony-stimulating factor (GM-CSF) in six COVID-19 patients. Days indicate the day after symptoms onset.
DISCUSSION
In our study, all six patients with COVID-19 showed a pattern of an initial rapid increase followed by a rapid decrease in the levels of IP-10 and IFN-λ2/3, whereas no apparent trend was observed in the production of proinflammatory cytokines of the IL and TNF families.
As all six patients recovered within 4 days after the onset of symptoms and neutralizing antibodies usually develop 10–15 days after symptom onset, our results suggest that pathogen-specific immune responses do not seem to play a key role in rapid recovery.
IFN-λ2/3 are members of the IFN-λ family of human cells (5,12), which stimulate the expression of IFN-stimulated genes and activate the antiviral defense response. IFN-λ could be induced in epithelial cells in mouse models and human primary nasal epithelial cells by several viruses, including respiratory viruses such as influenza virus, whereas type 1 IFNs were not strongly induced in these models (9,17). Furthermore, IFN-λ could prevent the spread of influenza virus from the upper airways to the lungs in a mouse model (11). Based on these reports and our findings, we suggest that IFN-λ2/3 may play a key role in rapid recovery. Whereas, because IFN-λ has been shown to cause excessive inflammation during bacterial infection (16), the rapid decrease in the IFN-λ2/3 levels may be important for the mild clinical manifestations of patients with COVID-19. Rapidly increase and decrease of IFN-λ2/3 may be a marker for rapid recovery as seen in our cohort. On the other hand, it had been reported that IFN-λ3 and IP-10 rapidly increase and decrease in critical forms of COVID-19 (20). In terms of the relationship between cytokine level and severity of COVID-19, further study is required.
Although the significance of cytokine levels in COVID-19 are still unclear, previous studies have shown that IL-1β, IL-8 and TNF-α significantly elevated in ICU patients compared to non-ICU patients (1,3,18). Th1 cytokines, including IL-1β and TNF-α can be associated with the cytokine storm and organ damage. We observed non notable increase trends in these Th1 cytokines in our rapid recovery cases. Our findings may also indicate that elevated Th1 cytokine levels cause severe form of COVID-19.
IP-10, also known as C-X-C motif chemokine 10 (CXCL10), is a pro-inflammatory cytokine secreted by immune cells. IP-10 has been associated with the defense mechanism against several infections (21) and has been highlighted as a potential biomarker of several viral infections (4,19). Yang et al. (23) reported an increase in the serum IP-10 and MCP-3 levels in patients with COVID-19. This is in line with our finding of a rapid increase, followed by a rapid decrease in the serum IP-10 level in patients with mild COVID-19 symptoms. These findings suggest that IP-10 is not only a key player but also a potential biomarker for patients with mild COVID-19.
GM-CSF is an immune modulator as well as a hematopoietic growth factor. It has been reported that the induction of GM-CSF could reduce the mortality of influenza virus infection in an animal model (8). GM-CSF may also be one of key players for rapid recovery from COVID-19. However, an increase in the GM-CSF level was observed in only one patient in our cohort. Further research is needed to determine the role of GM-CSF.
Our study had some limitations. First, only six patients were analyzed during the study period. To confirm the significance of changes in the IFN-λ2/3 and IP-10 levels during recovery, further investigation with a larger number of patients is needed. Furthermore, only a few points (3–5 points) per case were analyzed. To clarify rapid fluctuation of cytokines, more detailed analysis must be evaluated. Second, we did not measure the cytokine profiles in non-rapid recovery patients or critical COVID-19 patients. Analysis of these patients is necessary to endorse our hypothesis. Therefore, we plan to measure the cytokine profiles in these patients with COVID-19.
CONCLUSION
In conclusion, IFN-λ2/3 and IP-10 may play a key role in rapid recovery from mild SARS-CoV-2 infection. Additionally, the rapid decrease in the serum levels of IFN-λ2/3 and IP-10 may be potential biomarkers of mild infection. Our data is not enough to confirm the precise mechanism of rapid recovery. Further study is necessary to assess the importance of these cytokines.
Footnotes
FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Alosaimi B, Mubarak A, Hamed ME, Almutairi AZ, Alrashed AA, AlJuryyan A, et al. Complement Anaphylatoxins and Inflammatory Cytokines as Prognostic Markers for COVID-19 Severity and In-Hospital Mortality. Front Immunol. 2021;12:668725. doi: 10.3389/fimmu.2021.668725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eden E, Srugo I, Gottlieb T, Navon R, Boico O, Cohen A, et al. Diagnostic accuracy of a TRAIL, IP-10 and CRP combination for discriminating bacterial and viral etiologies at the Emergency Department. J Infect. 2016;73(2):177–180. doi: 10.1016/j.jinf.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Hamming OJ, Terczynska-Dyla E, Vieyres G, Dijkman R, Jorgensen SE, Akhtar H, et al. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 2013;32(23):3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National of Instutute of Health. Clinical Spectrum of SARS-CoV-2 Infection. Dec 17, 2020. Retrieved from https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/
- 7.Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2020;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang FF, Barnes PF, Feng Y, Donis R, Chroneos ZC, Idell S, et al. GM-CSF in the lung protects against lethal influenza infection. Am J Respir Crit Care Med. 2011;184(2):259–268. doi: 10.1164/rccm.201012-2036OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, Schindler C, et al. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol. 2010;84(21):11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaiboullina SF, Levis S, Morzunov SP, Martynova EV, Anokhin VA, Gusev OA, et al. Serum Cytokine Profiles Differentiating Hemorrhagic Fever with Renal Syndrome and Hantavirus Pulmonary Syndrome. Front Immunol. 2017;8:567. doi: 10.3389/fimmu.2017.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, et al. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018;7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 13.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 14.Ministry of Health, Labour and Welfare. Q & A on Coronavirus Disease 2019 (COVID-19) Apr 1, 2020. Retrieved from: https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/kenkou_iryou/dengue_fever_qa_00014.html.
- 15.Mueller SN, Rouse BT. Immune responses to viruses. Clinical Immunology. 2008:422–431. [Google Scholar]
- 16.Odendall C, Voak AA, Kagan JC. Type III IFNs Are Commonly Induced by Bacteria-Sensing TLRs and Reinforce Epithelial Barriers during Infection. J Immunol. 2017;199(9):3270–3279. doi: 10.4049/jimmunol.1700250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okabayashi T, Kojima T, Masaki T, Yokota S, Imaizumi T, Tsutsumi H, et al. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011;160(1–2):360–366. doi: 10.1016/j.virusres.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quint JK, Donaldson GC, Goldring JJ, Baghai-Ravary R, Hurst JR, Wedzicha JA. Serum IP-10 as a biomarker of human rhinovirus infection at exacerbation of COPD. Chest. 2010;137(4):812–822. doi: 10.1378/chest.09-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiyama M, Kinoshita N, Ide S, Nomoto H, Nakamoto T, Saito S, et al. Serum CCL17 level becomes a predictive marker to distinguish between mild/moderate and severe/critical disease in patients with COVID-19. Gene. 2021;766:145145. doi: 10.1016/j.gene.2020.145145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sumino KC, Walter MJ, Mikols CL, Thompson SA, Gaudreault-Keener M, Arens MQ, et al. Detection of respiratory viruses and the associated chemokine responses in serious acute respiratory illness. Thorax. 2010;65(7):639–644. doi: 10.1136/thx.2009.132480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Shen C, Li J, Yuan J, Wei J, Huang F, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol. 2020;146(1):119–127e114. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]