Abstract
While previous studies suggest that both genetic and environmental factors play an important role in the development of autism-related traits, little is known about potential biological mechanisms underlying these associations. Using data from the Avon Longitudinal Study of Parents and Children (ALSPAC), we examined prospective associations between DNA methylation (DNAm: nbirth = 804, nage 7 = 877) and trajectories of social communication deficits at age 8–17 years. Methylomic variation at three loci across the genome (false discovery rate = 0.048) differentiated children following high (n = 80) versus low (n = 724) trajectories of social communication deficits. This differential DNAm was specific to the neonatal period and not observed at 7 years of age. Associations between DNAm and trajectory membership remained robust after controlling for co-occurring mental health problems (i.e., hyperactivity/inattention, conduct problems). The three loci identified at birth were not replicated in the Generation R Study. However, to the best of our knowledge, ALSPAC is the only study to date that is prospective enough to examine DNAm in relation to longitudinal trajectories of social communication deficits from childhood to adolescence. Although the present findings might point to potentially novel sites that differentiate between a high versus low trajectory of social communication deficits, the results should be considered tentative until further replicated.
Keywords: ALSPAC, autistic traits, DNA methylation, longitudinal, methylome-wide
Introduction
Autism is associated with high health care utilization because of its persistence, long-term impairments, and high comorbidity with other psychiatric disorders (Ganz, 2007; Joshi et al., 2010; Simonoff et al., 2008). Like most complex phenotypes, autism is thought to have a multifactorial etiology that includes a dynamic interplay of genetic and environmental factors. It has been shown that autism has a strong heritable basis (64%–91%) (Tick, Bolton, Happe, Rutter, & Rijsdijk, 2016) and shares a common genetic vulnerability with other neuropsychiatric disorders such as attention-deficit/hyperactivity disorder (ADHD) (Lichtenstein, Carlstrom, Rastam, Gillberg, & Anckarsater, 2010). Environmental factors such as prenatal exposure to maternal smoking or stress also play a role in the etiology of various neuropsychiatric disorders, including autism (Ronald, Pennell, & Whitehouse, 2010; St Pourcain et al., 2011). However, little is known about the biological mechanisms through which these genetic and environmental effects on autism manifest.
Epigenetic mechanisms that regulate gene expression, such as DNA methylation (DNAm), have been shown to (a) respond to both genetic and environmental factors (Meaney, 2010) and (b) associate with various psychiatric conditions, including autism (Vogel Ciernia & LaSalle, 2016). As such, DNAm may represent a potential biological mechanism that could explain autism susceptibility across the life span. However, findings to date are mixed (Dall’Aglio et al., 2018) and may reflect the primarily clinical focus of methylome-wide studies on autism. For example, the common etiology of neuropsychiatric disorders suggests caution in relying solely on the diagnostic criteria of autism, without considering subthreshold manifestations. Core characteristics of autism, including social communication deficits, exist in all individuals to varying degrees, resulting in a dimensional distribution in the general population (Bolte, Westerwald, Holtmann, Freitag, & Poustka, 2011; Skuse, Mandy, & Scourfield, 2005).
The vast majority of methylome-wide studies have utilized clinical samples, relying primarily on clinical diagnosis of autism in case-control designs (e.g., Andrews et al., 2018; Hannon et al., 2018b). One notable exception is the recent population-based study by Massrali et al. (2019), examining the association between DNAm at birth and childhood social communication deficits – a core component of autism-related traits in childhood. Although the authors did not identify a significant association between DNAm and social communication deficits, they did observe a significant correlation with the results from a methylome-wide study of a similar childhood phenotype (i.e., pragmatic communication) measured in the same cohort. However, as this study involved measures at a single time point, longitudinal evidence of the developmental trajectories of social communication deficits is lacking. The use of developmental trajectories to describe the longitudinal course of autism-related traits may help unravel biological mechanisms for subgroups of social communication deficits that differentiate over time (Lord, Bishop, & Anderson, 2015).
The present study sought to address this issue by conducting the first epigenome-wide association study (EWAS) of developmental trajectories of social communication deficits in a large, prospective, population-based sample featuring repeated measures of both DNAm and social communication deficits spanning birth to early adolescence. These data enabled us to uniquely investigate the longitudinal course of autism-related traits in relation to genome-wide DNAm at birth and childhood. Our aim was to address the following key questions.
Are DNAm patterns at birth and early childhood prospectively associated with trajectories of social communication deficits (age 8–17 years)?
Do the identified DNAm markers associate with genetic and environmental risk exposures?
Are associations with DNAm unique to social communication deficits trajectories or shared with – or even entirely confounded by – other mental health symptom domains?
Method
Participants
Participants were drawn from the Accessible Resource for Integrated Epigenomics Studies (ARIES; Relton et al., 2015) (www.ariesepigenomics.org.uk), containing DNAm data for a subset of 1,018 mother–offspring pairs and nested within the Avon Longitudinal Study of Parents and Children (ALSPAC). ALSPAC is an ongoing epidemiological study of children born from 14,541 pregnant women residing in Avon, United Kingdom, with an expected delivery date between April 1991 and December 1992 (85% of the eligible population; Fraser et al., 2013). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee as well as local research ethics committees. Informed consent was obtained from all ALSPAC participants. The original ALSPAC sample is representative of the general population (Boyd et al., 2013). The study website contains details of all the data available through a fully searchable data dictionary and variable search tool (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/). For this study, we included children from ARIES who had available data on social communication ratings (at ages 8–17 years) as well as DNAm data at birth (n = 804, 49% male) or at 7 years of age (n = 877, 50% male; 88% overlapping with the data at birth). A total of 769 children with social communication ratings had DNAm data at both time points, while a minority had DNAm data only at birth (n = 35) or only at age 7 (n = 108).
Measures
Social communication deficits
Social communication skills were assessed using the 12-item Social Communication Disorder Checklist (SCDC; Skuse et al., 2005), which is a validated screening instrument of social reciprocity and verbal/nonverbal communication with high sensitivity and specificity for autism. Mother-reported SCDC scores for children and adolescents were computed at ages 8, 11, 14, and 17 years, with higher scores reflecting more social communication deficits (score range 0–24).
DNAm data
Array-based methylation quantification was conducted by ARIES (Relton et al., 2015). DNA was measured from cord blood drawn from the umbilical cord upon delivery and from whole blood at 7 years of age. Following extraction, 500 ng of genomic DNA was bisulfite-converted using the EZ-DNA methylation kit (Zymo Research, Orange, CA). DNAm was quantified using the Illumina HumanMethylation450 BeadChip (Illumina, San Diego, CA) according to the standard protocol. The arrays were scanned using an Illumina iScan (software version 3.3.28). Initial data quality control was conducted using GenomeStudio (version 2011.1, Illumina). For further details on the DNAm preprocessing pipeline see the Supplementary Material, section 1.
Samples (nbirth = 25; nage 7 = 8) or probes (nbirth = 7873; nage 7 = 4861) that failed quality control employed by the ARIES team (>1% probes/samples with background detection p value ≥ .05) were excluded from further analysis. Furthermore, multiple checks for sample mismatch were carried out. Specifically, samples were checked by calculating concordance with (a) genome-wide association data from the same participants, (b) single nucleotide polymorphism (SNP) probes on the Illumina 450K array across mother and child, and (c) sex. Data were quantile normalized using the dasen function as part of the wateRmelon package (wateRmelon_1.0.3; Pidsley et al., 2013) within the R statistical analysis environment.
We removed probes previously reported to be cross-reactive or polymorphic (Chen et al., 2013; Price et al., 2013), in addition to SNP (i.e., “rs”) probes (total probes removed = 72,068). We further removed participants with non-Caucasian or missing ethnicity (based on self-reports; n = 61). This left a total of 397,879 probes and 828 samples (cord blood at birth) and 397,791 probes and 903 samples (blood at age 7 years) after quality control.
For each probe, DNAm levels were indexed by beta values (i.e., the ratio of methylated signal divided by the sum of the methylated and unmethylated signal [M/(M + U + 100)], ranging from 0 (no methylation) to 1 (complete methylation). Probes were annotated using the Illumina HumanMethylation450 BeadChip annotation file.
Risk exposures
As risk exposures, we included prenatal maternal smoking and exposure to stress. Maternal smoking was measured during the first trimester of pregnancy via maternal ratings, using a dichotomous (12.2% yes) variable. With regards to prenatal stress exposure, we included a cumulative risk score of prenatal (18–32 weeks) adversity. This score was based on 56 dichotomous items from the Life Events Inventory (adapted in ALSPAC based on the work of Barnett, Hanna, & Parker (1983) and Brown, Sklair, Harris, & Birley (1973)) and the Family Adversity Index (Bowen, Heron, Waylen, & Wolke, 2005; Wolke, Steer, & Bowen, 2004), which are widely used (e.g., Barker, Oliver, Viding, Salekin, & Maughan, 2011; Mesirow, Cecil, Maughan, & Barker, 2017). Briefly, the items were organized into four conceptually distinct but related risk domains, and summed to create the following cumulative scores covering the following four domains: (a) life events (e.g., death in family, accident), (b) contextual risks (e.g., poor housing, financial problems), (c) parental risks (e.g., psychopathology, criminal behavior), and (d) interpersonal risks (e.g., partner abuse, family conflict). The overall cumulative risk score was estimated using confirmatory factor analysis, as described elsewhere (Cecil et al., 2014; Rijlaarsdam et al., 2016). For the full list of items contained in these cumulative scores, see Cecil et al. (2014).
Child characteristics
We assessed autism-related phenotypes using Pupil Level Annual School Census (PLASC) records and the Development and Well-Being Assessment (DAWBA; Goodman, Ford, Richards, Gatward, & Meltzer, 2000). The UK Department for Children, Schools and Families has a database referred to as PLASC, to which ALSPAC data are routinely linked. It encompasses all pupils who pass through the state-maintained educational system in the UK, providing data on children with special educational needs (www.dcsf.gov.uk). The DAWBA is a validated semi-structured interview. Parents complete open and closed questions about a range of symptoms relevant to youth psychiatric disorders. This study used the DAWBA Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV) diagnosis of pervasive developmental disorder at 7 years of age.
Child mental health symptoms were assessed using the Strengths and Difficulties Questionnaire (SDQ; Goodman, 2001), which is a widely used screening questionnaire with high reliability and validity. We included mothers’ reports on their children’s conduct problems (e.g., “often fights with other children or bullies them”), hyperactivity/inattention (e.g., “restless, overactive, cannot stay still for long”), emotional difficulties (e.g., “often unhappy, downhearted or tearful”), and peer problems (e.g., “rather solitary, tends to play alone”). For each subscale, scores were combined across three time points – at age 8 (n = 734), 10 (n = 755), and 13 (n = 719) – using confirmatory factor analysis (see Table S1 in the Supplementary Material for descriptive statistics and factor loadings). Child IQ was assessed at 8 years of age using the Wechsler Intelligence Scale for Children (WISC-III; Wechsler, 1991).
Statistical analysis
Social communication deficits trajectories
Developmental trajectories of social communication deficits were modeled through longitudinal latent profile analysis as implemented within Mplus (version 7.11; Muthén & Muthén, 2011). This type of analysis aims to identify a latent class variable that corresponds to different developmental patterns of social communication deficits across multiple time points (e.g., high vs. low levels at ages 8–17 years). A series of models was fitted, beginning with a one-class model and moving to a four-class model. In line with previous research documenting both latent profile and methylome-wide analyses (Barker et al., 2018), model selection was captured through (a) entropy, with values closer to 1 suggesting a higher model classification accuracy, and (b) the overall percentage of children estimated in the different trajectories. Currently, to our knowledge, there is no software program that can both estimate mixture models and perform bias-adjusted methods within a methylome-wide association analysis. Hence, as we estimated the trajectories in Mplus and then ran the methylome-wide association analyses in R, we paid close attention to potential bias that could affect confidence in the results. According to a simulation study using ALSPAC data (Heron, Croudace, Barker, & Tilling, 2015), there is little detriment to assessing covariates (i.e., low bias) when using a classification approach with high levels of entropy and high class-separation (i.e., low overlap in class membership).
We further assessed group contrasts for child characteristics including autism-related phenotypes, mental health symptoms, and IQ. These group contrasts indicated the descriptive statistics of given variables with the likelihood of trajectory membership relative to the contrast group.
Methylome-wide analysis of social communication trajectories
Methylome-wide analysis (DNAm beta values at birth and at 7 years of age) of social communication trajectories was performed in R version 3.4.3 (R Core Team, 2017) using the CpGassoc package (Barfield, Kilaru, Smith, & Conneely, 2012). Analyses were conducted using linear regression adjusting for sex, sample plate, and estimates of cell type proportions (CD8T lymphocytes, CD4T lymphocytes, natural killer cells, B lymphocytes, and monocytes), as detailed by Houseman et al. (2012). Differentially methylated probes (DMPs) were considered statistically significant if they passed a false discovery rate (FDR) correction of q < 0.05. For these DMPs, we additionally examined the extent to which DNAm levels at birth correlated with those at age 7 (i.e., autocorrelations or temporal stability). The probes were winsorized prior to analyses to reduce the influence of potential outliers (>3 SD).
Genetic and environmental influences underlying the top hits
We then investigated to what extent identified associations were explained by genetic factors or known prenatal environmental risks, including maternal smoking and stress exposure. Smoking is particularly important to account for, being one of the most consistently associated exposures to DNAm alterations (e.g., see Hannon et al., 2019).
As our sample was underpowered to directly examine genetic effects, we used the ALSPAC-derived methylation quantitative trait loci database (mQTLdb) resource (http://www.mqtldb.org/; Gaunt et al., 2016) to search for known mQTLs associated with our DMPs. This mQTLdb characterizes genome-wide significant cis effects (i.e., SNP within ±1,000 base pairs of the DNAm site) and trans effects (i.e., ±1 million base pairs) on DNAm levels across Illumina 450K probes at five different life stages, including cord blood DNAm at birth (Gaunt et al., 2016). We used mQTLdb results from the conditional genome-wide complex trait analysis (GCTA) set to identify mQTLs with the most representative, independent effect on each DNAm site in order to account for linkage disequilibrium (Gaunt et al., 2016). We then examined the associations between prenatal environmental risk exposures and DMPs, using linear regression correcting for sex, sample plate, and cell type.
Univariate and multivariate models: mental health symptoms
To assesses whether potentially co-occurring mental health problems (conduct problems, hyperactivity/inattention, emotional difficulties, peer problems) confound links between social communication deficits and DNAm, we examined their associations with the DMPs. To this end, we used both univariate and multivariate linear regression, correcting for sex, sample plate, and cell type. The multivariate linear regression model was designed to examine whether epigenetic associations are unique to social communication deficits trajectories or shared with – or even entirely confounded by – other mental health symptom domains.
Replication
We used data from the Generation R Study (Kooijman et al., 2016) to examine the replicability of our initial observations in ALSPAC. The Generation R Study is a population-based prospective cohort study from fetal life onwards. Pregnant women residing in Rotterdam, the Netherlands, with expected delivery dates between April 2002 and January 2006 were invited to participate. The study was approved by the Medical Ethical Committee of the Erasmus MC University Medical Center Rotterdam, and written consent was obtained for all the participants. The Generation R Study Biobank has DNAm results from 1,396 cord blood samples measured using the Illumina 450 Infinium BeadChip (Kruithof et al., 2014).
In the Generation R Study, children’s social responsiveness was assessed via parental ratings using an 18-item short form of the Social Responsiveness Scale (SRS; Constantino et al., 2003) when children were 6 years of age. In this study, the subscale indexing social communication deficits (eight items, e.g., “slow or awkward in interactions with peers”) was used. Items were rated on a 4-point scale, ranging from 0 (= never true) to 3 (= almost always true). In the absence of the repeated measures that are required for latent profile analysis, we used the social communication score at age 6 years both continuously and categorically (mean + SD). To our knowledge, ALSPAC is the only cohort that is prospective enough to examine the association between neonatal DNAm patterns and trajectories of social communication difficulties from childhood to adolescence.
DMPs were extracted from the Generation R epigenome-wide data set and regressed on the social communication score, using linear regression correcting for sex, sample plate, and cell type (Houseman et al., 2012).
Results
Social communication deficits trajectories
Latent profile analyses yielded a two-trajectory solution (Figure 1) for social communication problems, showing clearly discernible “high” (10%, n = 80) and “low” (90%, n = 724) groups between ages 8 and 17 years (entropy = 0.955). Table S2 of the Supplementary Material shows that the separation of the two-trajectory model was better than that of the three-trajectory (entropy = 0.916) and above models, where further trajectories were often additional low trajectories, which can result in high overlap of trajectory contrasts. Furthermore, the three-trajectory model (and above) resulted in a much smaller percentage of children in the high trajectory (3.5%), challenging methylome-wide analyses.
Figure 1.
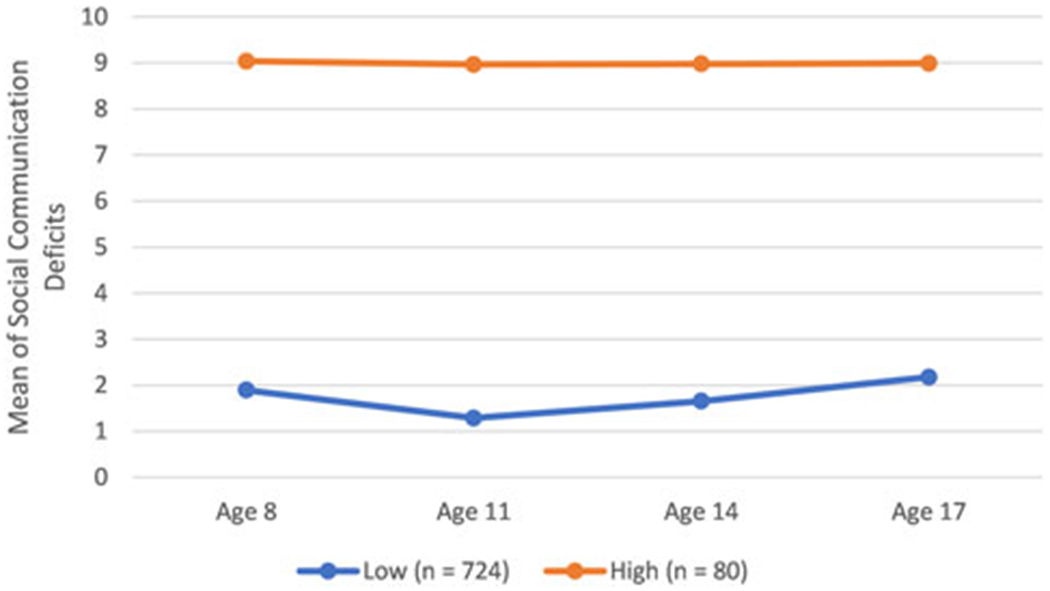
Trajectory of social communication deficits.
Social communication deficits group contrasts for this two-trajectory solution are displayed in Table 1. There were no sex differences in the likelihood of high (58.8% boys) versus low (47.7% boys) trajectory membership, p = .059. Regarding the autism-related phenotypes, records from the UK Department for Children, Schools and Families (PLASC, n = 668) did not indicate the presence of autism disorder in our study sample. Only one child (total n = 764) met criteria for a DAWBA-based DSM-IV diagnosis of pervasive developmental disorder, again hampering group comparisons. Group contrasts for conduct problems, hyperactivity/inattention, emotional difficulties, and peer problems were all significant and not specific to any individual domain (all p ≤ 0.001, see Table 1). IQ did not differ between the groups (p = .215).
Table 1.
Characteristics of the study sample by trajectories of social communication deficits
| Variable | Total sample (n = 804) | Social communication deficits trajectories | ||
|---|---|---|---|---|
| High (n = 80) | Low (n = 724) | p value for difference | ||
| Sex (n = 804): % boys | 48.9 | 58.8 | 47.7 | .059 |
| Risk exposures | ||||
| Cumulative risk (n = 804): score | −0.31 (0.72) | −0.30 (0.91) | −0.31 (0.72) | .847 |
| Maternal smoking (n = 798): % yes | 12.2 | 13.9 | 11.8 | .586 |
| Child characteristics | ||||
| Conduct problems (n = 793): score | −0.30 (1.26) | 1.34 (1.64) | −0.43 (1.04) | <.001 |
| Hyperactivity/inattention (n = 792): score | −0.48 (2.53) | 1.80 (3.11) | −0.56 (2.32) | <.001 |
| Emotional difficulties (n = 792): score | −0.44 (1.26) | 0.06 (2.05) | −0.47 (1.20) | .001 |
| Peer problems (n = 793): score | −0.38 (1.02) | 0.13 (2.02) | −0.45 (0.94) | <.001 |
| IQ (n = 768) | 107.32 ± 15.61 | 103.95 ± 19.68 | 107.70 ± 15.04 | .215 |
Note: Values represent median (interquartile range) or mean ± SD unless otherwise specified; p values are derived from chi-square tests (categorical variables) or Mann–Whitney–Wilcoxon tests (continuous variables).
Methylome-wide analysis of the social communication trajectories
At birth, three probes were associated with low versus high social communication deficits trajectories after genome-wide correction (q < 0.05; Table 2). The full EWAS results of DNAm collected at birth are available in the Zenodo repository (https://doi.org/10.5281/zenodo.4031357) and the EWAS Catalog (http://www.ewascatalog.org/). The mean percent methylation difference between the high-and low-trajectory groups for the three DMPs identified at birth ranged from 3% to 4% (Table 2), with small effect sizes consistent with what has been reported in the wider psychiatric epigenetic literature (Barker et al., 2018; Walton et al., 2017). Of the DMPs, both cg23234820 and cg04112471 were hypomethylated in the high social communication deficits trajectory, whereas cg06825512 was hypermethylated. Cg23234820 is annotated to A4GALT (Alpha 1,4-Galactosyltransferase (P Blood Group)), a gene involved in the biosynthesis of the P(k) blood antigen (Iwamura et al., 2003). Cg06825512 is annotated to APCDD1 (APC Down-Regulated 1), an inhibitor of the Wnt signaling pathway (Shimomura et al., 2010) that has previously been linked to autism (Cusco et al., 2009). Cg04112471 is annotated to SPSB4 (SplA/Ryanodine Receptor Domain And SOCS Box Containing 4), a gene regulating EphB2-dependent cell repulsive responses (Okumura et al., 2017) that has also been associated with autism (Wang et al., 2014).
Table 2.
DNA methylation loci at birth that prospectively associate with social communication deficits (SCD) trajectories
| Probe | Gene | Chr | Genomic location | Position | F | p | FDR (q value) | Mean ± SD | % Difference | mQTL | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| High SCD trajectory (n = 80) | Low SCD trajectory (n = 724) | ||||||||||
| cg23234820 | A4GALT | 22 | Body | 43089041 | 27.11 | 2.54E−07 | 0.048 | 0.79 ± 0.06 | 0.82 ± 0.05 | 3 | – |
| cg06825512 | APCDD1 | 18 | TSS1500 | 10453969 | 26.64 | 3.21E−07 | 0.048 | 0.51 ± 0.08 | 0.47 ± 0.07 | 4 | – |
| cg04112471 | SPSB4 | 3 | 5’UTR | 140783430 | 26.42 | 3.58E−07 | 0.048 | 0.76 ± 0.07 | 0.80 ± 0.05 | 4 | – |
Note: Genomic location refers to the gene body, transcription start sites (TSS1500), and untranslated regions (5′UTR). Chr = chromosome; F = F-statistic for ANOVA; p = uncorrected p value; q = FDR-corrected value; mQTL = methylation quantitative trait loci; FDR = false discovery rate.
With regards to temporal stability, we first tested, for each DMP, the extent to which DNAm levels at birth correlated with those at age 7 (i.e., autocorrelations). Only cg06825512 showed a significant (p < .05) autocorrelation, which was in the positive direction (r = .27). However, none of the DMPs identified at birth continued to prospectively associate with social communication deficits trajectories by age 7 (at FDR corrected [q < 0.05] and even nominal threshold [p < .05]). More generally at age 7, we identified no genome-wide corrected DMPs between low versus high social communication deficits trajectories. The full EWAS results of DNAm collected at age 7 are available in the Zenodo repository (https://doi.org/10.5281/zenodo.4031387) and the EWAS Catalog (http://www.ewascatalog.org/).
Genetic and environmental influences underlying the top hits
The three DMPs identified at birth were carried forward to explore associations with genetic and environmental risk. Based on a mQTLdb search, we found that none of the DMPs were associated with genetic variations known to have an effect on DNAm levels. This lack of mQTLs seems to be supported by a heritability tool characterizing additive genetic influences as opposed to shared and nonshared environmental influences on DNAm, based on data from monozygotic and dizygotic twins (Hannon et al., 2018a). Both cg23234820 and cg04112471 showed low additive genetic influences (r = 4.08E−12 and r = 9.58E−15, respectively), whereas cg06825512 showed moderate genetic influences (r = 0.64). However, as shown in Table 3, the associations between the two environmental risk exposures (i.e., prenatal stress exposure and smoking) and the three DMPs were generally weak and nonsignificant after Bonferroni correction for multiple testing (corrected p value = .05/three DMPs × two risk factors = 0.0083).
Table 3.
Associations of differentially methylated probes with risk exposures and child characteristics
| Differentially methylated probes | ||||||
|---|---|---|---|---|---|---|
| cg23234820 | cg06825512 | cg04112471 | ||||
| β | p value | β | p value | β | p value | |
| Risk exposures | ||||||
| Cumulative risk: score | −0.04 | .2977 | 0.06 | .0962 | −0.06 | .0777 |
| Maternal smoking: yes vs. no | −0.01 | .7693 | 0.02 | .5632 | −0.05 | .1461 |
| Child characteristics | ||||||
| IQ | 0.00 | .9775 | 0.00 | .9943 | 0.03 | .4446 |
| Mental health symptoms: scores | ||||||
| Conduct problems | −0.09 | .0127 | 0.12 | .0004 | −0.09 | .0063 |
| Hyperactivity/inattention | −0.07 | .0675 | 0.00 | .3369 | −0.12 | .0006 |
| Emotional difficulties | −0.04 | .2566 | 0.02 | .6021 | 0.00 | .8929 |
| Peer problems | −0.10 | .0053 | 0.01 | .8632 | −0.03 | .3574 |
| SCD trajectories: low vs. high | −0.18 | 2.54E−07 | 0.17 | 3.21E−07 | −0.17 | 3.58E−07 |
Note: Standardized regression coefficients and p values are presented. SCD = social communication deficits.
Univariate and multivariate models: mental health symptoms
As can be seen in Table 3, the univariate models showed that peer problems and emotional difficulties were unrelated to all three DMPs after Bonferroni correction for multiple testing (corrected p value = .05/three DMPs × four mental health scores = 0.0042). However, conduct problem scores associated with cg06825512 (β = 0.12, p < .001), while hyperactivity/inattention scores associated with cg04112471 (β = −0.12, p < .001). The multivariate models showed that social communication deficits trajectories remained significantly (corrected p value = .05/three DMPs = 0.0167) associated with cg06825512 (β = 0.14, p < .001) after potentially co-occurring conduct problems were accounted for. Similarly, controlling for hyperactivity/inattention left the association between social communication deficits trajectories and cg04112471 (β = −0.15, p < .001) essentially unchanged.
Replication
The results from the EWAS in ALSPAC did not replicate in the Generation R sample (see Table 4). Of the three DMPs that were associated with social communication deficits trajectories (8–17 years) in ALSPAC, two were associated with the continuous score of social deficits (6 years) in Generation R with the same direction of effect. One of the three DMPs identified in ALSPAC was associated with the categorical (mean + SD) score of social communication deficits in Generation R with the same direction of effect. None of them reached statistical significance.
Table 4.
Replication in the Generation R Study of the differentially methylated probes identified in ALSPAC
| Probe | Social communication deficits | |||||
|---|---|---|---|---|---|---|
| ALSPAC (age 8–17 years) | Generation R Study (age 6 years) | |||||
| Trajectories | Continuous | Categorical | ||||
| β | p value | β | p value | β | p value | |
| cg23234820 | −0.18 | 2.54E-07 | 0.001 | .9821 | 0.004 | .8655 |
| cg06825512 | 0.17 | 3.21E-07 | 0.017 | .4750 | 0.014 | .5589 |
| cg04112471 | −0.17 | 3.58E-07 | −0.030 | .3026 | 0.001 | .9766 |
Note: Standardized regression coefficients and p values are presented.
Discussion
Using longitudinal data spanning gestation to early adolescence, this study examined the extent to which genome-wide DNAm patterns (at birth and at 7 years of age) prospectively associate with trajectories of social communication deficits (at 8–17 years). We highlight here three key findings: (a) three loci at birth were differently methylated between high and low trajectories of social communication deficits in a temporally specific way; (b) none of the loci identified at birth associated with known mQTLs or prenatal maternal risk exposure; (c) the observed links between DNAm and social communication deficits trajectories were not accounted for by co-occurring mental health problems
Our genome-wide analysis showed that epigenetic variation across three loci at birth differed between the high versus low trajectories of social communication deficits. These loci were annotated to A4GALT, APCDD1, and SPSB4 – genes involved in the biosynthesis of the P(k) blood antigen, the Wnt signaling pathway, and EphB2-dependent cell repulsive responses, respectively. Interestingly, both APCDD1 and SPSB4 have previously been linked to autism spectrum disorder in clinical settings (Cusco et al., 2009; Wang et al., 2014). For example, SPSB4 was found to be similarly hypomethylated in a cross-sectional study of 131 children (age 3–12 years) diagnosed with autism spectrum disorders versus controls (Wang et al., 2014). Besides this study, the two that are most comparable to ours in terms of sample size, time point (i.e., DNAm at birth), analytical methods (i.e., methylome-wide analysis), and design (i.e., prospective study) found no significant associations between DNAm and autism (Hannon et al., 2018b; Massrali et al., 2019). Specifically, Hannon et al. (2018b) found no significant DNAm differences between patients diagnosed with autism spectrum disorder (n = 629) and healthy controls (n = 634). Similarly, Massrali et al. (2019) did not identify significant CpGs associated with social communication deficits (age 8 years). However, using DNAm data for autism from postmortem brain tissues (vs. peripheral data), the authors identified a significant concordance in effect direction of top hits in the social communication deficits EWAS. The current study is the first to investigate DNAm underlying longitudinal trajectories of social communication deficits (age 8–17 years). These trajectories are useful in that they provide us with a longitudinal case-control comparison that is based on dimensional, repeated measures of core characteristics of autism. The current results emphasize the advantage of using trajectories as autism-related phenotypes, especially in population-based samples where the prevalence of autism might be too small for meaningful methylome-wide analyses. By identifying two trajectories (high vs. low) with high entropy and high class-separation (i.e., low overlap in class membership), our study should provide results reasonably close to the best possible scenario in terms of latent profile analysis (Heron et al., 2015).
The EWAS associating DNAm at age 7 with social communication deficits trajectories revealed no genome-wide significant associations. The finding that none of the three DMPs identified at birth continued to associate with social communication deficits trajectories by age 7 is consistent with previous longitudinal epigenetic studies investigating other psychiatric phenotypes (ADHD, conduct problems, substance-use risk) in the ALSPAC cohort (Cecil et al., 2016; Rijlaarsdam et al., 2017; Walton et al., 2017) and the larger Pregnancy and Childhood Epigenetics (PACE) consortium (Neumann et al., in press). The low and mostly nonsignificant autocorrelations (birth, age 7) among the three CpGs support another ALSPAC-based study reporting low genome-wide continuity in DNAm patterns across development (Gaunt et al., 2016). Given that sample sizes were comparable across the two time points, with 88% participants represented at both time points, the temporally specific associations observed in our study might be explained by several reasons other than differences in statistical power. A possible explanation that cannot be ruled out is that temporal differences reflect tissue-specific DNAm patterns, as data were extracted from cord blood at birth and whole blood at age 7 (Bakulski et al., 2016). An alternative explanation is that the neonatal period represents a particular window of vulnerability for DNAm patterns and social communication deficits. That is, in light of the dynamic nature of both environmental and epigenetic factors, differences in DNAm patterns at birth and age 7 may reflect (a) the specific timing of environmental influences (Richmond et al., 2015) or (b) DNAm patterns at birth that induce long-lasting developmental changes but are not necessarily maintained over time (Numata et al., 2012). It is possible that, while epigenetic effects early in life may be stronger during early development (as observed in our study), weaker effects may still be observed later in childhood, particularly within high-risk clinical samples. However, given that this is the first longitudinal epigenome-wide study of trajectories of autism-related traits, we still know little about what triggers continuity versus discontinuity in epigenetic patterns (Dall’Aglio et al., 2018). Hence, the above explanations remain inevitably speculative and necessitate further investigation.
None of the DMPs identified at birth associated with genetic variations known to have an effect on DNAm levels (mQTLs; Gaunt et al., 2016). This lack of mQTLs seems to be supported by twin heritability data (Hannon et al., 2018a) showing that methylomic variation of the DMPs was primarily explained by environmental as opposed to genetic factors. However, the three DMPs were unrelated to prenatal cumulative stress exposure and smoking after multiple testing correction. Environmental effects on these DMPs may be explained by exposures other than those assessed in the current study. Furthermore, associations with environmental factors may be dependent on genetic factors. Recent evidence suggests that interactions between genotype and environmental exposures might explain more variation in DNAm than environmental exposures alone (Czamara et al., 2019; Teh et al., 2014). These Gene × Environment interactions, potentially accounting for variation in DNAm underlying social communication deficits trajectories, warrant investigation in larger studies with more statistical power.
Children following the high versus low trajectory of social communication deficits presented with more conduct problems, hyperactivity/inattention, emotional difficulties, or peer problems. Both conduct problems and hyperactivity/inattention associated with one of the three DMPs identified at birth. These results are of interest, given that early social–cognitive abilities have been implicated in the emergence and maintenance of conduct problems (Gilmour, Hill, Place, & Skuse, 2004; Oliver, Barker, Mandy, Skuse, & Maughan, 2011), with deficits for early-onset persistent conduct problems especially marked (Oliver et al., 2011). Despite the strong overlaps of social communication deficits trajectories with conduct problems and hyperactivity/inattention, controls for these mental health problems had little impact on the associations between the trajectories and DMPs. This result suggests that the observed links between DNAm and social communication deficits trajectories are not simply a consequence of co-occurrence with conduct problems or hyperactivity/inattention.
Although the three DMPs identified in ALSPAC were unrelated to social communication deficits in the Generation R Study, this lack of replication might partially reflect differences in how the phenotype was operationalized between cohorts. ALSPAC was unique in that repeated assessments of social communication deficits were collected from age 8 to 17 years, enabling the estimation of longitudinal developmental trajectories (i.e., a person-centered approach). In contrast, in the Generation R Study, social communication deficits were measured only at one time point (age 6 years) and thus examined continuously and dichotomously (i.e., a variable-centered approach). According to a recent genome-wide analysis on major depressive disorder, a more strictly defined phenotype (i.e., standardized lifetime diagnosis) captures the underlying biological mechanisms better than minimal phenotyping (i.e., phenotype definitions based on self-reports) (Cai et al., 2020). As such, trajectory-based approaches may provide us with a longitudinal case-control comparison that better captures epigenetic signal underlying general psychopathology. However, this will need to be investigated more systematically as more cohorts become available with longitudinal data spanning childhood to adolescence.
It is important to bear in mind a number of limitations while interpreting the results of this study. First, the Illumina HumanMethylation450 BeadChip covers less than 2% of all CpG sites in the genome. Thus, differentially methylated loci significantly associated with social communication deficits trajectories may be located in areas outside of the array. Second, because DNAm is tissue-specific, observations in the peripheral blood may not reflect other tissues of interest (e.g., the brain, assuming that we are looking at epigenetic changes associated to autism-related traits) that are unavailable for population-based studies of living individuals. Using the Blood–Brain Epigenetic Concordance tool (Edgar, Jones, Meaney, Turecki, & Kobor, 2017) and the Iowa Methylation Array Graphing for Experimental Comparison of Peripheral Tissue & Gray Matter (IMAGE-CpG) tool (Braun et al., 2019), we found no evidence that DNAm levels of our three DMPs correlate between blood and brain samples. However, these correlations tag specific brain areas in individuals (mostly adults) with unclear social communication deficits histories. Because DNAm patterns vary across brain regions and we do not have access to the most relevant regions to our study (e.g., amygdala), it will be important in future to establish the extent to which our findings reflect associations in the brain. The integration of transcriptomic data will also be important for assessing the functional relevance of DNAm changes to gene expression. Third, the study focused exclusively on DNAm. Other epigenetic processes (e.g., histone modifications) are likely to be important in the etiology of autism-related traits. Fourth, the exclusion of individuals with non-European ethnicity based on self-report (n = 61) does not imply that additional sources of unwanted variation linked to genetic ancestry are accounted for. Fifth, identifying subgroups of children with distinct trajectories using latent profile analysis is only one way of looking at longitudinal data of social communication deficits. For example, if interested in the longitudinal change in social communication deficits, one could use a mixed model approach to examine changes of the direction of the effect associated with age (see, e.g., Simpkin et al., 2015). Finally, the analyses were correlational in nature and, hence, causality cannot be inferred. The present results should be considered hypothesis-generating and in need of replication.
In summary, the present findings lend potentially novel insights into epigenetic patterns that might differentiate between high versus low trajectories of social communication deficits from childhood to adolescence, pinpointing temporally-specific markers for further interrogation. These associations were not replicated in an independent cohort. However, given that our sample was unique in that person-centered developmental trajectories could be modeled, this lack of replication might partially reflect biological heterogeneity underlying different phenotype operationalizations. The current findings should be viewed as preliminary until they have been further replicated.
Supplementary Material
Acknowledgments.
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. With regard to the ALSPAC DNAm data, we thank all involved, particularly the laboratory scientists and bioinformaticians who contributed considerable time and expertise to the data generation and quality control.
The Generation R Study is conducted by the Erasmus MC in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam, and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives, and pharmacies in Rotterdam. The generation and management of the Illumina 450K methylation array data (EWAS data) for the Generation R Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. The authors thank all colleagues involved in the generation and management of methylation data and genotyping.
Financial Statement. The UK Medical Research Council and the Wellcome Trust (grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. ARIES was funded by the BBSRC (BBI025751/1 and BB/I025263/1). ARIES is maintained under the auspices of the MRC Integrative Epidemiology Unit at the University of Bristol (MC UU 12013/2 and MC UU 12013/8). A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf).
The general design of the Generation R Study was made possible by financial support from Erasmus MC, Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development, and the Ministry of Health, Welfare and Sport. The EWAS data were funded by a grant from the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA; project no. 050-060-810), by funds from the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by a grant from the National Institute of Child and Human Development (R01HD068437).
This publication is the work of the authors who will serve as guarantors for the contents of this article. This study was funded by research awards from the National Institute of Child and Human Development (R01HD068437) and the Economic and Social Research Council (ES/R005516/1) to EDB. The work of JR was supported by the Netherlands Organization for Scientific Research (NWO ZonMw VENI, grant no. 91618147). The work of CAMC received funding from the European Union’s Horizon 2020 research and innovation programme under Marie Sklodowska-Curie grant agreement no. 707404.
Footnotes
Supplementary Material. The supplementary material for this article can be found at https://doi.org/10.1017/S0954579420001662
Conflicts of Interest. None
References
- Andrews SV, Sheppard B, Windham GC, Schieve LA, Schendel DE, Croen LA, … Ladd-Acosta C (2018). Case-control meta-analysis of blood DNA methylation and autism spectrum disorder. Molecular Autism, 9, 40. doi: 10.1186/s13229-018-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, McKenney SL, … Fallin MD (2016). DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics, 11, 354–362. doi: 10.1080/15592294.2016.1161875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield RT, Kilaru V, Smith AK, & Conneely KN (2012). CpGassoc: An R function for analysis of DNA methylation microarray data. Bioinformatics, 28, 1280–1281. doi:bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker ED, Oliver BR, Viding E, Salekin RT, & Maughan B (2011). The impact of prenatal maternal risk, fearless temperament and early parenting on adolescent callous-unemotional traits: A 14-year longitudinal investigation. Journal of Child Psychology and Psychiatry and Allied Disciplines, 52, 878–888. doi: 10.1111/j.1469-7610.2011.02397.x [DOI] [PubMed] [Google Scholar]
- Barker ED, Walton E, Cecil CAM, Rowe R, Jaffee SR, Maughan B, … Gaunt TR (2018). A methylome-wide association study of trajectories of oppositional defiant behaviors and biological overlap with attention deficit hyperactivity disorder. Child Development, 89(5), 1839–1855. doi: 10.1111/cdev.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett BE, Hanna B, & Parker G (1983). Life event scales for obstetric groups. Journal of Psychosomatic Research, 27, 313–320. [DOI] [PubMed] [Google Scholar]
- Bolte S, Westerwald E, Holtmann M, Freitag C, & Poustka F (2011). Autistic traits and autism spectrum disorders: The clinical validity of two measures presuming a continuum of social communication skills. Journal of Autism and Developmental Disorders, 41, 66–72. doi: 10.1007/S10803-010-1024-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen E, Heron J, Waylen A, & Wolke D (2005). Domestic violence risk during and after pregnancy: Findings from a British longitudinal study. BJOG: An International Journal of Obstetrics and Gynaecology, 112, 1083–1089. doi: 10.1111/j.1471-0528.2005.00653.X [DOI] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, … Davey Smith G (2013). Cohort Profile: The ‘children of the 90s’ – the index offspring of the Avon Longitudinal Study of Parents and Children. International Journal of Epidemiology, 42, 111–127. doi: 10.1093/ije/dys064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT, … Shinozaki G (2019). Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Translational Psychiatry, 9, 47. doi: 10.1038/s41398-019-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Sklair F, Harris TO, & Birley JL (1973). Life-events and psychiatric disorders. 1. Some methodological issues. Psychological Medicine, 3, 74–87. [DOI] [PubMed] [Google Scholar]
- Cai N, Revez JA, Adams MJ, Andlauer TFM, Breen G, Byrne EM, … Flint J (2020). Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nature Genetics, 52, 437–447. doi: 10.1038/s41588-020-0594-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil CA, Lysenko LJ, Jaffee SR, Pingault JB, Smith RG, Relton CL, … Barker ED (2014). Environmental risk, oxytocin receptor gene (OXTR) methylation and youth callous-unemotional traits: A 13-year longitudinal study. Molecular Psychiatry, 19, 1071–1077. doi: 10.1038/mp.2014.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil CA, Walton E, Smith RG, Viding E, McCrory EJ, Relton CL, … Barker ED (2016). DNA methylation and substance-use risk: A prospective, genome-wide study spanning gestation to adolescence. Translational Psychiatry, 6, e976. doi: 10.1038/tp.2016.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, … Weksberg R (2013). Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics, 8, 203–209. doi: 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, … Reich W (2003). Validation of a brief quantitative measure of autistic traits: Comparison of the Social Responsiveness Scale with the Autism Diagnostic Interview-revised. Journal of Autism and Developmental Disorders, 33, 427–433. doi: 10.1023/a:1025014929212 [DOI] [PubMed] [Google Scholar]
- Cusco I, Medrano A, Gener B, Vilardell M, Gallastegui F, Villa O, … Perez-Jurado LA (2009). Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Human Molecular Genetics, 18, 1795–1804. doi: 10.1093/hmg/ddp092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czamara D, Eraslan G, Page CM, Lahti J, Lahti-Pulkkinen M, Hamalainen E, … Binder EB (2019). Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns. Nature Communications, 10, 2548. doi: 10.1038/s41467-019-10461-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Aglio L, Muka T, Cecil CAM, Bramer WM, Verbiest M, Nano J, … Tiemeier H (2018). The role of epigenetic modifications in neurodevelopmental disorders: A systematic review. Neuroscience and Biobehavioral Reviews, 94, 17–30. doi: 10.1016/j.neubiorev.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Edgar RD, Jones MJ, Meaney MJ, Turecki G, & Kobor MS (2017). BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Translational Psychiatry, 7, e1187. doi: 10.1038/tp.2017.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, … Lawlor DA (2013). Cohort profile: The Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. International Journal of Epidemiology, 42, 97–110. doi: 10.1093/ije/dys066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz ML (2007). The lifetime distribution of the incremental societal costs of autism. Archives of Pediatrics and Adolescent Medicine, 161, 343–349. doi: 10.1001/archpedi.161.4.343 [DOI] [PubMed] [Google Scholar]
- Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, … Relton CL (2016). Systematic identification of genetic influences on methylation across the human life course. Genome Biology, 17, 61. doi: 10.1186/s13059-016-0926-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour J, Hill B, Place M, & Skuse DH (2004). Social communication deficits in conduct disorder: A clinical and community survey. Journal of Child Psychology and Psychiatry and Allied Disciplines, 45, 967–978. doi: 10.1111/j.1469-7610.2004.t01-1-00289.x [DOI] [PubMed] [Google Scholar]
- Goodman R (2001). Psychometric properties of the Strengths and Difficulties Questionnaire. Journal of the American Academy of Child and Adolescent Psychiatry, 40, 1337–1345. doi:S0890-8567(09)60543-8. [DOI] [PubMed] [Google Scholar]
- Goodman R, Ford T, Richards H, Gatward R, & Meltzer H (2000). The Development and Well-Being Assessment: Description and initial validation of an integrated assessment of child and adolescent psychopathology. Journal of Child Psychology and Psychiatry and Allied Disciplines, 41, 645–655. [PubMed] [Google Scholar]
- Hannon E, Knox O, Sugden K, Burrage J, Wong CCY, Belsky DW, … Mill J (2018a). Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. Plos Genetics, 14, e1007544. doi: 10.1371/journal.pgen.1007544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Schendel D, Ladd-Acosta C, Grove J, Hansen CS, Hougaard DM, … Mill J (2019). Variable DNA methylation in neonates mediates the association between prenatal smoking and birth weight. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 374, 20180120. doi: 10.1098/rstb.2018.0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Schendel D, Ladd-Acosta C, Grove J, iPSYCH-Broad ASD Group, Hansen CS, & Mill J (2018b). Elevated polygenic burden for autism is associated with differential DNA methylation at birth. Genome Medicine, 10, 19. doi: 10.1186/s13073-018-0527-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron J, Croudace TJ, Barker ED, & Tilling K (2015). A comparison of approaches for assessing covariate effects in latent class analysis. Longitudinal and Life Course Studies, 6, 420–434. doi: 10.14301/llcs.v6i4.322 [DOI] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, … Kelsey KT (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamura K, Furukawa K, Uchikawa M, Sojka BN, Kojima Y, Wiels J, … Furukawa K (2003). The blood group P1 synthase gene is identical to the Gb3/CD77 synthase gene. A clue to the solution of the P1/P2/p puzzle. Journal of Biological Chemistry, 278, 44429–44438. doi: 10.1074/jbc.M301609200 [DOI] [PubMed] [Google Scholar]
- Joshi G, Petty C, Wozniak J, Henin A, Fried R, Galdo M, … Biederman J (2010). The heavy burden of psychiatric comorbidity in youth with autism spectrum disorders: A large comparative study of a psychiatrically referred population. Journal of Autism and Developmental Disorders, 40, 1361–1370. doi: 10.1007/s10803-010-0996-9 [DOI] [PubMed] [Google Scholar]
- Kooijman MN, Kruithof CJ, van Duijn CM, Duijts L, Franco ΟH, van IMH, … Jaddoe VW (2016). The Generation R Study: Design and cohort update 2017. European Journal of Epidemiology, 31, 1243–1264. doi: 10.1007/s10654-016-0224-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruithof CJ, Kooijman MN, van Duijn CM, Franco OH, de Jongste JC, Klaver CC, … Jaddoe VW (2014). The Generation R Study: Biobank update 2015. European Journal of Epidemiology, 29, 911–927. doi: 10.1007/s10654-014-9980-6 [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, & Anckarsater H (2010). The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. American Journal of Psychiatry, 167, 1357–1363. doi: 10.1176/appi.ajp.2010.10020223 [DOI] [PubMed] [Google Scholar]
- Lord C, Bishop S, & Anderson D (2015). Developmental trajectories as autism phenotypes. American Journal of Medical Genetics. Part C: Seminars in Medical Genetics, 169, 198–208. doi: 10.1002/ajmg.c.31440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massrali A, Brunel H, Hannon E, Wong C, iPSYCH-MINERvA Epigenetics Group, Baron-Cohen S, & Warrier V (2019). Integrated genetic and methylomic analyses identify shared biology between autism and autistic traits. Molecular Autism, 10, 31. doi: 10.1186/s13229-019-0279-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ (2010). Epigenetics and the biological definition of gene x environment interactions. Child Development, 81, 41–79. doi: 10.1111/j.1467-8624.2009.01381.x [DOI] [PubMed] [Google Scholar]
- Mesirow MS, Cecil C, Maughan B, & Barker ED (2017). Associations between prenatal and early childhood fish and processed food intake, conduct problems, and co-occurring difficulties. Journal of Abnormal Child Psychology, 45, 1039–1049. doi: 10.1007/s10802-016-0224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén LK, & Muthén BO (2011). Mplus User’s Guide, 1998–2010 (6th ed.). Los Angeles, CA: Author. [Google Scholar]
- Neumann A, Walton E, Alemany S, Cecil C, Gonzalez JR, Jima DD, … Tiemeier H (in press). Association between DNA methylation and ADHD symptoms from birth to school age: A prospective meta-analysis. Translational Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, … Lipska BK (2012). DNA methylation signatures in development and aging of the human prefrontal cortex. American Journal of Human Genetics, 90, 260–272. doi:S0002-9297(11)00555-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Joo-Okumura A, Obara K, Petersen A, Nishikimi A, Fukui Y, … Kamura T (2017). Ubiquitin ligase SPSB4 diminishes cell repulsive responses mediated by EphB2. Molecular Biology of the Cell, 28, 3532–3541. doi: 10.1091/mbc.E17-07-0450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver BR, Barker ED, Mandy WP, Skuse DH, & Maughan B (2011). Social cognition and conduct problems: A developmental approach. Journal of the American Academy of Child and Adolescent Psychiatry, 50, 385–394. doi: 10.1016/j.jaac.2011.01.006 [DOI] [PubMed] [Google Scholar]
- Pidsley R, Wong CCY, Volta M, Lunnon K, Mill J, & Schalkwyk LC (2013). A data-driven approach to preprocessing Illumina 450 K methylation array data. BMC Genomics, 14, 293. doi: 10.1186/1471-2164-14-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ME, Cotton AM, Lam LL, Farre P, Emberly E, Brown CJ, … Kobor MS (2013). Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin, 6, 4. doi: 10.1186/1756-8935-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A language and environment for statistical computing. Retrieved from https://www.R-project.org/.
- Relton CL, Gaunt T, McArdle W, Ho K, Duggirala A, Shihab H, … Davey Smith G (2015). Data resource profile: Accessible Resource for Integrated Epigenomic Studies (ARIES). International Journal of Epidemiology, 44, 1181–1190. doi: 10.1093/ije/dyv072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, … Relton CL (2015). Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: Findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Human Molecular Genetics, 24, 2201–2217. doi: 10.1093/hmg/ddu739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam J, Cecil CA, Walton E, Mesirow MS, Relton CL, Gaunt TR, … Barker ED (2017). Prenatal unhealthy diet, insulin-like growth factor 2 gene (IGF2) methylation, and attention deficit hyperactivity disorder symptoms in youth with early-onset conduct problems. Journal of Child Psychology and Psychiatry and Allied Disciplines, 58, 19–27. doi: 10.1111/jcpp.12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam J, Pappa I, Walton E, Bakermans-Kranenburg MJ, Mileva-Seitz VR, Rippe RC, … van IMH (2016). An epigenome-wide association meta-analysis of prenatal maternal stress in neonates: A model approach for replication. Epigenetics, 11, 140–149. doi: 10.1080/15592294.2016.1145329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Pennell CE, & Whitehouse AJ (2010). Prenatal maternal stress associated with ADHD and autistic traits in early childhood. Frontiers in Psychology, 1, 223. doi: 10.3389/fpsyg.2010.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, … Christiano AM (2010). APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature, 464, 1043–1047. doi: 10.1038/nature08875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonoff E, Pickles A, Charman T, Chandler S, Loucas T, & Baird G (2008). Psychiatric disorders in children with autism spectrum disorders: Prevalence, comorbidity, and associated factors in a population-derived sample. Journal of the American Academy of Child and Adolescent Psychiatry, 47, 921–929. doi: 10.1097/CHI.0b013e318179964f [DOI] [PubMed] [Google Scholar]
- Simpkin AJ, Suderman M, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, … Relton CL (2015). Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Human Molecular Genetics, 24, 3752–3763. doi: 10.1093/hmg/ddv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, Mandy WP, & Scourfield J (2005). Measuring autistic traits: Heritability, reliability and validity of the Social and Communication Disorders Checklist. British Journal of Psychiatry, 187, 568–572. doi: 10.1192/bjp.187.6.568 [DOI] [PubMed] [Google Scholar]
- St Pourcain B, Mandy WP, Heron J, Golding J, Davey Smith G, & Skuse DH (2011). Links between co-occurring social-communication and hyperactive-inattentive trait trajectories. Journal of the American Academy of Child and Adolescent Psychiatry, 50, 892–902 e895. doi: 10.1016/j.jaac.2011.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh AL, Pan H, Chen L, Ong ML, Dogra S, Wong J, … Holbrook JD (2014). The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Research, 24, 1064–1074. doi: 10.1101/gr.171439.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tick B, Bolton P, Happe F, Rutter M, & Rijsdijk F (2016). Heritability of autism spectrum disorders: A meta-analysis of twin studies. Journal of Child Psychology and Psychiatry and Allied Disciplines, 57, 585–595. doi: 10.1111/jcpp.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel Ciernia A, & LaSalle J (2016). The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nature Reviews: Neuroscience, 17, 411–423. doi: 10.1038/nrn.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E, Pingault JB, Cecil CA, Gaunt TR, Relton CL, Mill J, & Barker ED (2017). Epigenetic profiling of ADHD symptoms trajectories: A prospective, methylome-wide study. Molecular Psychiatry, 22, 250–256. doi: 10.1038/mp.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fang Y, Zhang F, Xu M, Zhang J, Yan J, … Zhong N (2014). Hypermethylation of the enolase gene (ENO2) in autism. European Journal of Pediatrics, 173, 1233–1244. doi: 10.1007/s00431-014-2311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D (1991). Manual for the Wechsler Intelligence Scale for Children (3rd ed.). San Antonio, TX: Psychological Corporation. [Google Scholar]
- Wolke D, Steer C, & Bowen E (2004). The ALSPAC Family Adversity Index (FAI): Background and development. Unpublished manuscript, University of Bristol. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


