Abstract
Of 179 methicillin-resistant Staphylococcus aureus strains isolated from 1997 to 1998, two strains (1.1%) gave subclones for which the vancomycin MICs were 8 mg/liter. Pulsed-field gel electrophoresis showed identical restriction patterns for both isolates, suggesting transfer of a single clone between two different patients.
Since the first report describing a Japanese clinical isolate of vancomycin-resistant Staphylococcus aureus (VRSA) (4), several papers have documented the emergence of these microorganisms elsewhere, including Michigan (2), New Jersey (2), France (12), and New York (14), causing growing concern. All VRSA strains described were also methicillin resistant (MRSA). Subsequent to the description of VRSA, Hiramatsu and colleagues (5) observed in Japan the presence of S. aureus strains among MRSA that were heterogeneously resistant to vancomycin (hetero-VRSA). Recently, heterogeneous resistance to vancomycin has also been reported in Spain (1). These findings might explain some therapeutic failures experienced with vancomycin in the treatment of MRSA infections (1, 5).
Since the development and spread of this phenomenon are perceived as a fearsome threat to the already challenging therapy of nosocomial infections, we have investigated the prevalence of heterogeneous vancomycin resistance in our hospital, where the incidence of MRSA exceeds 35%.
Nonrepetitive MRSA strains (179 strains) isolated at the Institute of Microbiology of the University Hospital of Genoa from August 1997 to December 1998 were studied. All 179 strains were isolated from 179 different hospitalized patients with infections attributed by the clinicians to MRSA. The clinical strains were isolated from the respiratory tract (96 strains), blood (20 strains), urine (12 strains), liquor (1 strain), drainage fluid (6 strains), feces (6 strains), pus (11 strains), and other sites (27 strains). Microorganisms were identified with the API Staph system (Biomerièux Italia S.p.A., Rome, Italy). Screening for detection of possible hetero-VRSA on brain heart infusion (BHI) agar plates containing 4 mg of vancomycin per liter and analysis of resistant subpopulations of bacteria (population analysis) were carried out as described by Hiramatsu et al. (5).
In detail, this test was carried out by spreading 100 μl of the starting cell suspension (prepared by diluting overnight cultures to an optical density of 0.3 at 540 nm) and its serial dilutions over BHI agar plates containing vancomycin at concentrations ranging from 1 to 10 mg/liter. The plates were incubated at 37°C for 48 h, and the number of colonies was counted. The number of cells contained in 100 μl of the starting cell suspensions growing on the different concentrations of vancomycin was calculated and plotted on a semilogarithmic graph.
Microdilution MIC determinations were performed with strains grown in cation-adjusted Mueller-Hinton broth according to National Committee for Clinical Laboratory Standards guidelines (10). Microplates were incubated for a full 24 h as recommended by Tenover et al. (18). Vancomycin, rifampin, chloramphenicol, and cotrimoxazole were purchased from commercial sources (Sigma-Aldrich, Milan, Italy). Erythromycin was supplied by Abbott S.p.a., Campoverde, Italy; linezolid was provided by Pharmacia & Upjohn, Milan, Italy; and quinupristin/dalfopristin was provided by Rhone Poulenc Rorer, Milan, Italy.
PCR was performed with specific primers for vanA, vanB, and vanC1–3 genes as described by Tenover et al. (17).
Analysis of colony size was carried out by plating strains on both Mueller-Hinton agar and BHI agar. Size was visually inspected after 24 and 48 h of incubation.
Rate of growth was evaluated as described by Sieradzky and Tomasz (13).
Morphologic changes in vancomycin-resistant isolates were assessed by thin-section electron microscopy as described by Tacchetti et al. (16), with minor modifications.
Chromosomal DNA was prepared and digested with the SmaI endonuclease (New England Biolabs, Italian distributor Celbio S.r.l., Milan, Italy), and the DNA fragments were separated by pulsed-field gel electrophoresis (PFGE) with a CHEF-DRII apparatus (Bio-Rad Laboratories S.r.l., Milan, Italy) as described by de Lencastre and Tomasz (3).
The banding patterns were interpreted visually by published guidelines (18).
Of the 179 MRSA clinical strains screened, 38 grew on BHI agar plates containing vancomycin (4 mg/liter) after 48 h of incubation. Vancomycin MICs for all 38 strains ranged from 1 to 2 mg/liter. Of 38 strains, 5 (G1, G64, G65, G260, and G375) produced subclones when grown on BHI agar plates in the presence of 8 mg of vancomycin per liter, after 48 h of incubation, with frequencies ranging from 1.6 × 10−6 to 8.2 × 10−6. Only two strains (G1 and G65) produced subclones for which the vancomycin MIC was 8 mg/liter, with frequencies of 1.4 × 10−7 and 1 × 10−6, respectively.
PFGE of the five strains producing colonies when grown on BHI agar containing vancomycin (8 mg/liter) revealed that G1 was identical to G65 while G64 was identical to G260 and G375 (Fig. 1).
FIG. 1.
PFGE patterns of some MRSA strains studied. Lanes 1, 19, 20, and 26, molecular weight standards; lanes 2 and 18, S. aureus NCTC 8325; lane 3, MRSA G1; lane 4, MRSA G65; lanes 21, 22, and 23, MRSA G64, G260, and G375, respectively.
The strain possessing the higher frequency of VR subclones (G65) was further characterized.
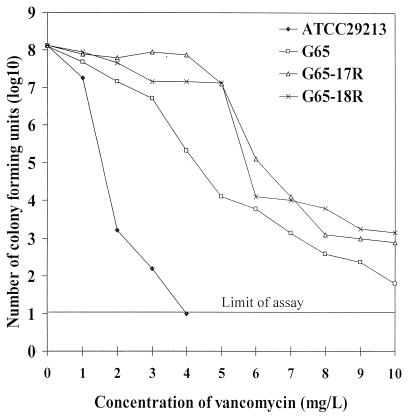
Figure 2 shows a comparison of the population analysis of G65, G65-derived substrains (G65-17R and G65-18R), and S. aureus ATCC 29213. Approximately 90% of G65-17R and G65-18R cells grew in 4 mg of vancomycin per liter, and 0.00001% of the population grew in the presence of 10 mg vancomycin per liter. G65 was more susceptible to vancomycin than its subclone, as evidenced by the 99.9% inhibition of the population by vancomycin at a concentration of 4 mg/liter. S. aureus ATCC 29213 was completely inhibited by vancomycin at 4 mg/liter. In addition to being resistant to vancomycin, G65-17R and 18R showed were more highly resistant to teicoplanin (MIC, 8 mg/liter) than to G65 (MIC, 4 mg/liter). G65 and its subclones were also resistant to erythromycin, ciprofloxacin, rifampin, and gentamicin but were susceptible to chloramphenicol and cotrimoxazole. The MICs of quinupristin/dalfopristin and linezolid were 0.5 and 2 mg/liter.
FIG. 2.
Analysis of resistant subpopulations of G65 and G65-derived substrains. Strains G65-17R and 18R were G65-derived substrains obtained by selecting G65 with 8 mg of vancomycin per liter.
G65 and its subclones were negative when tested for the vanA, vanB, and vanC1–3 genes. This pattern of resistance and the absence of van genes were observed in previously described VRSA strains (17).
The resistant derivatives of G65 produced smaller colonies on solid medium after 24 h of incubation and a reduced rate of growth in Trypticase soy broth (doubling time, about 1 h) than did the parental culture (doubling time, about 30 min). These characteristics have also been described for in vitro mutants (9, 13).
Sections of G65-17R examined by electron microscopy showed a thicker cell wall than did vancomycin-susceptible strains (Fig. 3).
FIG. 3.
Morphologic changes assessed by thin-section electron microscopy. (Top) Vancomycin-susceptible S. aureus; (bottom) S. aureus G65-17R (vancomycin MIC, 8 mg/liter). Bars, 0.5 μm.
Strain G1 was isolated on 8 August 1997 from a 62-year-old patient admitted to the rehabilitation unit after pancreas transplantation. In July, an MRSA strain was isolated from drainage fluid from the patient. Strain G1 was recovered after 15 days of treatment with vancomycin (1 g every 48 h) and ceftazidime (2 g twice a day). The patient was also treated with cotrimoxazole (1 g twice a day) during the last 10 days before isolation of G1. Therapy was effective, and on day 16 the drainage fluid became sterile. By the end of August, the patient was discharged from the hospital.
Strain G65 was isolated on 12 October 1997 from sputum produced by a 58-year-old patient who had undergone primary lung cancer surgery. In August, the patient was hospitalized in a rehabilitation unit, and on September 25, the patient became infected with an MRSA strain. He was treated with piperacillin-tazobactam (4 g) for 2 days, teicoplanin (400 mg) for 1 day, and pefloxacin (800 mg) for 1 day. The treatment did not eradicate the strain. The general condition of the patient was compromised, antibiotic therapy was discontinued, and on October 16 the patient died because of his serious underlying illness.
All microorganisms isolated in Japan (4) and in the United States (2, 14) that showed decreased susceptibility to vancomycin after prolonged exposure to that drug were MRSA. The French isolate originated from a patient treated with vancomycin (10 days) and teicoplanin (2 days) (11). In our experience, we isolated S. aureus G1 from a patient treated with vancomycin for 15 days that produced subclones for which the vancomycin MIC of 8 mg/liter occurred at a lower frequency (1.4 × 10−7) than for strain G65 (1 × 10−6) that derived from a patient treated with piperacillin-tazobactam for 2 days and teicoplanin for 1 day. Since the two patients were hospitalized during the same time period, it seems plausible that G1, selected by vancomycin in the first patient, was transmitted to the second and that, in addition, although short, teicoplanin therapy increased the frequency of subclones.
Does a connection exist between methicillin and vancomycin resistance? According to the data available, VRSA or hetero-VRSA strains have been described in areas where the prevalence of MRSA is high (Japan, 60%; Spain, >30%; Italy, 44%; United States, 35%) (6, 8, 12). However, it also evident that the ensuing incidence of heterogeneous drug resistance, which is extremely variable (Japan, 5 to 26% in university hospitals and 1.3% in non-university Hospitals; Spain, 65%; Italy, 1.1%), cannot be predicted on the basis of the incidence of MRSA alone. In addition, it is far from clear whether methicillin-susceptible S. aureus (MSSA) strains are able, under vancomycin-selective pressure, to lose their susceptibility to this drug. A single MSSA strain that developed resistance to teicoplanin during therapy has been described previously (6). If this also holds true for vancomycin, additional factors, other than methicillin resistance, should be taken into consideration. Data reported in Japan (5), New York (14), and Spain (1) indicate that VRSA and hetero-VRSA strains emerge from clonotypes largely diffused in those areas. Whether strains identical to G1 and G65 are widely circulating in Italy remains to be established. PFGE of 25 of the 179 MRSA strains studied, randomly collected from patients in different units, revealed that 15 (60%) strains were identical or related to G1 and G65. This fact raises obvious concerns, even if the prevalence of hetero-VRSA in our hospital is lower than that observed in Japan and Spain. At present, since the exact mechanism of vancomycin resistance has not been determined and very little epidemiological data is available, the differences observed may generally be ascribed to the patterns of antibiotic usage or to the spread of different clones.
What is the clinical impact of glycopeptide resistance in S. aureus? Ariza et al. (1) support the view that the finding of MRSA isolates in clinical samples displaying heterogeneous vancomycin resistance may be a more common problem than has generally been recognized and that this event may be clinically relevant only under certain conditions. Our results show that hetero-VRSA strains circulate in Italy and that in the two cases observed no therapeutic failures have been caused by these organisms. On the other hand, it seems plausible that VRSA strains represent a more-threatening aspect of the problem than hetero-VRSA strains, but it is also apparent that in the world, the number of cases in which VRSA strains have been isolated is very limited. Glycopeptide resistance in S. aureus is not a new phenomenon, given that the first clinical isolate of S. aureus with reduced susceptibility to teicoplanin was described in 1990 (6). Ten years later, these microorganisms still represent an uncommon finding and, with this in mind, a slow development of VRSA cannot be anticipated on the basis of the available experience with teicoplanin. Data on vancomycin and teicoplanin mutants indicate that these microorganisms have common properties, including an increase in penicillin-binding protein 2 production (9). However, teicoplanin-resistant clinical isolates may not be characterized by a significant change in vancomycin susceptibility (6, 7), while VRSA strains show borderline susceptibility or resistance to teicoplanin. These observations suggest the existence of differences that may play a role in the evolution of resistance to the glycopeptides.
Acknowledgments
We are grateful to R. Comai and C. Tacchetti (Department of Experimental Medicine, University of Genoa, Genoa, Italy) for preparation of electron microscopy samples and expert assistance.
REFERENCES
- 1.Ariza J, Pujol M, Cabo J, Pena C, Fernandez N, Linares J, Ayats J, Gudiol F. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet. 1999;353:1587–1588. doi: 10.1016/s0140-6736(99)01017-x. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin-United States. Morbid Mortal Weekly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 3.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguzi T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 5.Hiramatsu K, Aritaka N, Hanki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 6.Kaatz G W, Seo S M, Dorman N J, Lerner S A. Emergence of teicoplanin resistance during therapy of Staphylococcus aureus endocarditis. J Infect Dis. 1990;162:103–108. doi: 10.1093/infdis/162.1.103. [DOI] [PubMed] [Google Scholar]
- 7.Mainardi J, Shales D M, Goering R V, Shales J H, Acar J F, Goldstein F W. Decreased teicoplanin susceptibility of methicillin-resistant strains of Staphylococcus aureus. J Infect Dis. 1995;171:1646–1650. doi: 10.1093/infdis/171.6.1646. [DOI] [PubMed] [Google Scholar]
- 8.Marchese A, Debbia E A, Bacca D, Balistreri G, Musolino B, Schito G C. Multidrug-resistant gram-positive pathogens. Drugs. 1997;54(Suppl. 6):11–20. doi: 10.2165/00003495-199700546-00005. [DOI] [PubMed] [Google Scholar]
- 9.Moreira B, Boyle-Vara S, deJonge B L M, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Vol. 17. 1997. , no. 2. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 11.Ploy M C, Greland C, Martin C, de Lumley L, Denis F. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet. 1998;351:1212. doi: 10.1016/s0140-6736(05)79166-2. [DOI] [PubMed] [Google Scholar]
- 12.Roberts R B, de Lencastre A, Eisner W, Severina E P, Shopsin B, Kreiswirth B N, Tomasz A the MRSA Collaborative Study Group. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- 13.Sieradzky K, Tomasz A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siradzki K, Roberts R B, Harber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 15.Spencer R C, Bauernfeind A, Garcia-Rodriguez J, Jarlier V, Pfaller M, Turnidge J, Voss A. Surveillance of the current resistance of nosocomial pathogens to antimicrobials. Clin Microbiol Infect. 1997;3(Suppl. 1):S21–S35. [Google Scholar]
- 16.Tacchetti C, Quarto R, Nitsch L, Hartmann D J, Cancedda R. In vitro morphogenesis of chick embryo hypertrophic cartilage. J Cell Biol. 1987;105:999–1006. doi: 10.1083/jcb.105.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]