Abstract
We have isolated a cDNA, frl (formin-related gene in leukocytes), a novel mammalian member of the formin gene family. The frl cDNA encodes a 160-kDa protein, FRL, that possesses FH1, FH2, and FH3 domains that are well conserved among other Formin-related proteins. An FRL protein is mainly localized in the cytosol and is highly expressed in spleen, lymph node, and bone marrow cells. Formin-related genes and proteins have been reported to play crucial roles in morphogenesis, cell polarity, and cytokinesis through interaction with Rho family small GTPases. FRL binds to Rac at its N-terminal region including the FH3 domain and associates with profilin at the FH1 domain. In a macrophage cell line, P388D1, overexpression of a truncated form of FRL containing only the FH3 domain (FH3-FRL) strongly inhibited cell adhesion to fibronectin and migration upon stimulation with a chemokine. Moreover, expression of the truncated FH3-FRL protein resulted in apoptotic cell death of P388D1 cells, suggesting that the truncated FH3-FRL protein may interfere with signals of FRL. Overexpression in the P388D1 cells of full-length FRL or of the truncated protein containing the FH3 and FH1 domains, with simultaneous expression of the truncated FH3-FRL protein, blocked apoptotic cell death and inhibition of cell adhesion and migration. These results suggest that FRL may play a role in the control of reorganization of the actin cytoskeleton in association with Rac and also in the regulation of the signal for cell survival.
formin-related genes comprise a very large family and have been shown to control morphogenesis, embryonic differentiation, cell polarity, and cytokinesis. Mutation in the mouse formin gene, the limb deformity (ld) locus, causes a reduction and fusion of the distal bones and digits of all limbs, accompanied by variable kidney defects (54). Null mutations of the Drosophila diaphanous gene (dia) result in sterility and lethality due to a failure of cytokinesis in all cells (8). Another Drosophila formin-related gene, cap, is required for localization of molecular determinants within the developing Drosophila oocyte (12). Mutation in the Aspergillus nidulans sepA gene prevents septation and causes defects in the maintenance of cellular polarity (17). The Schizosaccharomyces pombe fus1 gene mutant blocks conjugation at a point after cell contact and agglutination.
The Bni1p in Saccharomyces cerevisiae, identified as a mutant synthetically lethal with the Cdc12 (23, 56), has been shown to be a target of Rho1 and Cdc42. Bni1p interacts with profilin, an actin binding protein, resulting in regulation of the actin cytoskeleton (13, 21, 26). p140mDia, a mammalian homologue of Dia, has also been shown to bind RhoA and profilin (51). DFNA1, a mutant of human Diaphanous 1, is responsible for an autosomal dominant, fully penetrant, sensorineural progressive hearing loss that may result from a defect in the actin organization of hairy cells in the inner ear (30). Bni1p, Dia, and p140mDia are regarded as the members of the Diaphanous protein family.
Formin-related proteins have three characteristic domains: formin homology domain 1 (FH1), FH2, and FH3. FH1 is a proline-rich domain that associates with Src homology 3 (SH3) domains (42), WWP/WW domains (4, 9), and profilin (21, 31, 51). FH2 is the conserved 130-amino-acid region at the C terminus of Formin-related proteins. FH3 is located at the N terminus of Formin-related proteins and differs from the Rho binding site of Bni1p and p140mDia. FH3 is required for the intracellular localization of Fus1 in S. pombe (40). Formin-related proteins have been thought of as multidomain proteins mediating various signals including actin reorganization.
The Rho family GTPases, Rho, Rac, and Cdc42, have been implicated in the regulation of diverse biological processes, including cell motility, cell adhesion, cytokinesis, cell morphology, and cell growth (16). A major function of Rho, Rac, and Cdc42 is to regulate the organization of polymerized actin structure and the assembly of associated integrin complexes. In Swiss 3T3 fibroblasts, activation of Rho by extracellular ligands such as lysophosphatidic acid leads to the formation of actin stress fibers and focal adhesion complexes. Rac could be activated by platelet-derived growth factor (PDGF), epidermal growth factor (EGF), or insulin, which leads to the formation of an actin meshwork at the cell periphery to produce lamellipodia and membrane ruffles (44, 45). Activation of Cdc42 by bradykinin leads to the formation of filopodial protrusions (27, 37). Rac and Cdc42 also induce integrin-based adhesion complexes, which are distinct from Rho-induced focal adhesions (19, 37). These integrin-based adhesion complexes contain vinculin, paxillin, p125FAK, and β-integrin (37). Moreover, Cdc42 activation leads to subsequent activation of Rac, which in turn leads to activation of Rho (27, 37). Rho GTPases regulate the c-Jun NH2-terminal kinase or stress-activated protein kinase (JNK/SAPK) (35) and the p38 mitogen-activated protein (MAP) kinase cascades (50). Rho family GTPases have been also reported to stimulate transcription of serum response transcription factor (SRF) through mechanisms other than MAP kinase cascades (18, 53). Rho family GTPases trigger progression of the G1 phase of the cell cycle in quiescent fibroblasts, and their activities are essential for serum-induced G1 progression (38). Activation of G1 progression by Rac correlates well with its ability to stimulate the formation of lamellipodia but not with its ability to regulate JNK (24, 28).
Recent studies have identified many downstream molecular targets for Rho family GTPases. p140mDia and Bni1p, referred to above, are effectors of Rho family GTPases. As effectors of Rac, p65PAK (p21-activated kinase) (32, 33), NADPH p67phox (43), IQGAP (6), phosphatidylinositol-4-phosphate 5-kinase (41, 47), WAVE (34), and LIM-kinase (3, 55) have been identified. WAVE, a member of the Wiskott-Aldrich syndrome protein (WASP) family, has been shown to regulate the actin cytoskeleton required for membrane ruffling in COS7 cells.
We have cloned the frl (formin-related gene in leukocytes) gene, a novel mammalian gene with characteristics of formin-related genes. In the present study, we clarify the functions of FRL in relation to Rac, which are involved in murine macrophage morphogenesis and motility. Moreover, we demonstrate a possible role for FRL in macrophage survival.
MATERIALS AND METHODS
cDNA screening.
In the course of searching for the molecules involved in signal transduction in immune cells, we isolated a cDNA fragment (TE-4) from a mouse thymus cDNA library. TE-4 contained FH1 and FH2 conserved sequences. Since it was strongly expressed in lymphoid tissues, it was designated frl. Additional clones overlapping the primary frl cDNA fragment were obtained by screening a mouse brain cDNA library (c-600 in λgt-10; Clontech) with 32P-labeled TE-4 as a probe. Three positive clones were obtained. To obtain the 5′ end of the cDNA, a 5′ RACE (rapid amplification of cDNA ends) procedure was used. One full-length cDNA was isolated from a brain cDNA and named frlβ. A spleen cDNA library was also screened with a 32P-labeled 860-bp fragment of frlβ cDNA as a probe, and positive clones were obtained. It was named frlα. Both libraries were screened by filter replica hybridization. The filters were hybridized in hybridization buffer (50% formamide, 0.12 M Na2HPO4, 0.25 M NaCl, 7% sodium dodecyl sulfate [SDS], 1 mM EDTA) with a 32P-labeled probe at 42°C overnight. The filters were finally washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS at room temperature and were then subjected to autoradiography. DNA sequencing was performed on an ABI sequencer.
Northern blot analysis.
The filters of mRNA-loaded adult and embryonic mouse tissues were purchased from Clontech. Total RNAs from various mouse cell lines were extracted using Isogen reagent (Invitrogen). Total RNA was separated on a 1% agarose gel containing 6% formaldehyde and transferred to a nylon membrane (Hybond N; Amersham). The filters were hybridized with the 32P-labeled cDNAs corresponding either to the common part of frlα and frlβ or to β-actin. Each hybridization was performed according to the manufacturer's instructions for QuickHybri (Invitrogen), and products were subjected to autoradiography.
Preparation of antibody.
Anti-FRL antibody was prepared as follows. A peptide against the C terminus of FRL (Cys-Leu-Ile-Tyr-Glu-Ser-Asp-Arg-Asp-Gly-Ile-Glu-Asp-Ile-Ile-Thr) was synthesized and purified by high-performance liquid chromatography (HPLC). Prior to injection, rabbits were bled for preimmune sera. Keyhole limpet hemocyanin (KLH)-conjugated peptide mixed with Freund's adjuvant was injected into two rabbits, and sera were collected after a second booster. Using transiently frl cDNA-transfected 293T cells, antisera were screened by their ability to react with FRL. The immunoglobulin G (IgG) fraction of the antibody was purified using a protein G column (Pharmacia).
Western blotting for FRL.
Various mouse tissues were homogenized in 0.25 M saccharose containing 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM dithiothreitol (DTT). The protein content of each lysate was quantified with a BCA Protein Assay Reagent Kit (Pierce). Each tissue lysate equivalent to 100 μg of proteins was subjected to SDS–7.5% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The filters were blotted with anti-FRL or anti-β-actin (PharMingen). The anti-FRL antibody was used at a 1:1,000 dilution, and human β-actin was at a 1:1,000 dilution. Peroxidase-conjugated goat anti-rabbit IgG (Cappel) was used at a 1:10,000 dilution as a secondary antibody. The blot was then developed using an enhanced chemiluminescence system according to the manufacturer's instructions (Amersham).
P388D1 cells were washed twice with phosphate-buffered saline (PBS), and the cell pellet was resuspended in a hypotonic buffer (20 mM HEPES [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 1 μg of aprotonin/ml, 1 mM PMSF, 1 μg of leupeptin/ml) and allowed to swell on ice for 10 min. Cells were then lysed on ice by vigorous Dounce homogenization (100 strokes) using a tight-fitting Dounce homogenizer. The homogenate was spun down at 1,500 × g for 5 min at 4°C. The pellet was washed twice with a hypotonic buffer containing 0.5% NP-40 and then homogenized by Dounce homogenizer with an additional 10 strokes and spun down at 1,500 × g for 5 min at 4°C. The resulting pellet (nuclear fraction) was resuspended in an SDS-polyacrylamide gel electrophoresis sample buffer and sonicated prior to analysis by Western blotting. The supernatant from the first spin was centrifuged at 100,000 × g for 30 min. The supernatant (S100) was used as a cytosolic fraction, while the pellet (P100) was used as a membrane fraction. Each sample was fractionated by SDS–7.5% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. The Western blot was probed with a rabbit anti-FRL antibody.
Transfection.
The plasmids of full-length frlα cDNA (FULL-pcDNA3.1/His) and its various truncated forms (FH3-pcDNA3.1/HisC, FH1+FH3-pcDNA3.1/HisC, and FH2-pcDNA3.1/HisC) were constructed in the pcDNA3.1/HisC vector (Invitrogen). The plasmids of full-length frlα cDNA (FULL-pEGFP-C1) and truncated forms (FH3-pEGFP-C1, FH1+FH3-pEGFP-C1, FH2-pEGFP-C1, N1-pEGFP-C1, and N2-pEGFP-C1) (see Fig. 3C) were also inserted into the C-terminal protein fusion vector pEGFP-C1 (Clontech). COS7 cells and 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; Cell Culture Technologies), 0.1 mM nonessential amino acids solution, 2 mM l-glutamate, 1 mM sodium pyruvate, penicillin (100 IU/ml), and streptomycin (100 mg/ml) (Gibco BRL). The above plasmid DNAs were transfected into COS7 cells or 293T cells by using Lipofectamine (Gibco BRL) according to the manufacturer's protocol. Cells were lysed 24 h after transfection, and the cell extracts were used for Western blotting.
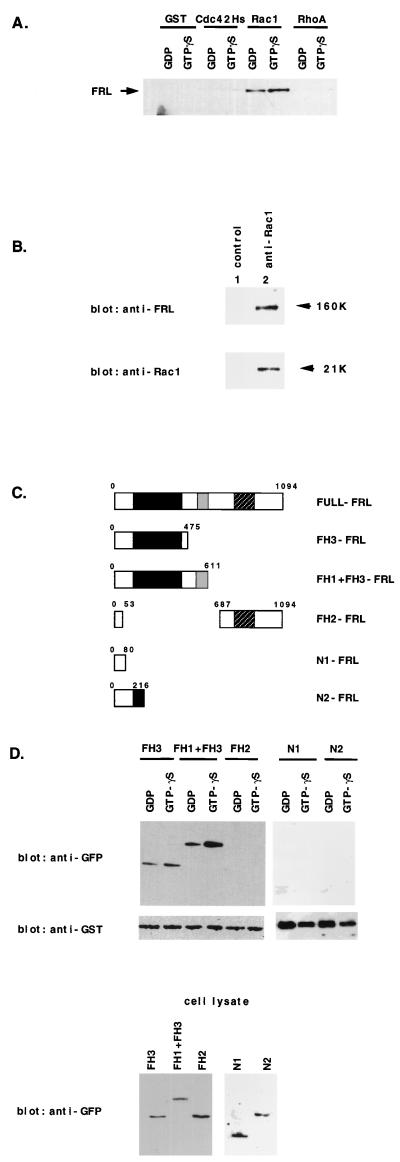
FIG. 3.
Association of FRL and truncated FRL proteins with Rac1 in vitro and in vivo. (A) Precipitation of FRL by the GTPγS- or GDP-bound form of Rac1. P388D1 cell lysates were incubated with GST-RhoA, GST-Rac1, GST-Cdc42Hs, or GST, each of which was preloaded with either GTPγs or GDP. GST fusion proteins were precipitated by glutathione-Sepharose 4B beads, and the pellets were analyzed by immunoblotting with an anti-FRL antibody. (B) In vivo association of FRL with Rac1. P388D1 cell lysates were immobilized on protein A-agarose beads without (lane 1) or with (lane 2) anti-Rac1. The precipitates were analyzed by Western blotting with an anti-FRL antibody and an anti-Rac1 antibody. (C) Structure of truncated FRLs. FH3-FRL contains only the FH3 domain, FH1+FH3-FRL contains the FH1 and FH3 domains, and FH2-FRL contains only the FH2 domain. N1 consists of the N-terminal 80 amino acids, and N2 consists of the N-terminal 216 amino acids. (D) FRL associates with Rac1 at its N-terminal region containing the FH3 domain. COS7 cells transfected with each expression vector (FH3-pEGFP-C1, FH1+FH3-pEGFP-C1, FH2-pEGFP-C1, N1-pEGFP-C1, or N2-pEGFP-C1) were lysed, incubated with GST-Rac1, and precipitated by glutathione-Sepharose 4B. The pellets were analyzed by immunoblotting with an anti-GFP antibody.
The tet-on system was used to obtain stable transformants that can be induced by doxycycline to express full-length or various truncated FRLs. DraI-DraI cDNA fragments of each plasmid (FH3-pcDNA3.1/HisC, FH1+FH3-pcDNA3.1/HisC, FH2-pcDNA3.1/HisC, and FULL-pcDNA3.1/His) that each contained an Xpress tag at the 5′ end were inserted into pUHD 10-3 (15) with a hygromycin resistance gene controlled by the phosphoglycerate kinase (PGK) promoter. These were designated FH3-pUHD 10-3/Hyg, FH1+FH3-pUHD 10-3/Hyg, FH2-pUHD 10-3/Hyg, and FULL-pUHD 10-3/Hyg. P388D1 cells were transfected with pUHD 172-1neo using Lipofectamine (Gibco BRL) according to the manufacturer's protocol and then cultured in RPMI 1640 (Gibco BRL) containing 500 μg of neomycin/ml. The stable transformants (P388D1/pUHD172-1neo) were then transfected by electroporation with each of the pUHD 10-3/Hyg plasmids expressing full-length and truncated forms of FRL proteins. Three to five clones for each type of transformants (FULL/P388D1, FH3/P388D1, FH1+FH3/P388D1, and FH2/P388D1) were obtained by selection with 500 μg of hygromycin/ml. The expression of frlα cDNA and its truncated forms was confirmed by Western blotting with an anti-Xpress antibody (Invitrogen).
For experiments of reintroduction of cDNA encoding full-length or FH1+FH3-FRL in FH3/P388D1 cells, the plasmid DNA of FH1+FH3-pEGFP-C1 or FULL-pEGFP-C1 was transfected into the cells by electroporation. After 24 h, the transfected FH3/P388D1 cells were cultured with doxycycline for another 24 h and then used for assays.
Binding assay.
Glutathione S-transferase (GST) fusion proteins of RhoA, Rac1, and Cdc42Hs were expressed and prepared according to the manufacturer's instructions. The procedure for affinity precipitation has been described previously (22). P388D1 cells (107 cells) were collected and disrupted by sonication in 3.2 ml of buffer A (10 mM morpholineethanesulfonic acid [MES; pH 6.5], 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.5% Triton X-100, 5 mM DTT, 1 mM PMSF, 5 μg of leupeptin/ml). Sonicated homogenates were centrifuged at 10,000 × g for 20 min, and the supernatants were stored. Loading of each nucleotide was carried out by incubating 10 μM GST-Rho GTPases with 1 mM guanosine 5′-O-3-thiotriphosphate (GTPγS) or GDP, or 10 μM GST with 1 mM GTPγS or GDP at 4°C for 1 h with gentle shaking. One-tenth of the supernatants was then incubated with 400 pmol of each nucleotide-loaded GST-Rho GTPase or GST. After incubation at 30°C for 30 min, 5 μl of glutathione-Sepharose 4B beads (Pharmacia) was added to the solution, and the mixture was incubated at 4°C for 1 h. The beads were washed twice with 1 ml of buffer A and then boiled in Laemmli sample buffer. The solubilized extracts were subjected to immunoblotting with an anti-FRL antibody.
To investigate the ability of truncated FRL to bind Rac, COS7 cells were transfected with FH3-pEGFP-C1, FH1+FH3-pEGFP-C1, FH2-pEGFP-C1, N1-pEGFP-C1, or N2-pEGFP-C1. Cell lysates from each COS7 transformant were mixed with GST-Rac at 30°C for 30 min, and then glutathione-Sepharose 4B beads were added (Pharmacia). The mixture was incubated at 4°C for 1 h. The solubilized extracts from the beads were subjected to immunoblotting with an anti-GFP antibody (MBL). GST-profilin I or GST-profilin II was mixed with cell lysates of each of the above transformants, and 20 μl of glutathione-Sepharose 4B beads was added to the mixture. After incubation with rotation for 2 h at 4°C, the beads were washed with PBS plus 0.1% Triton X-100 and suspended in 20 μl of SDS sample buffer. The solubilized extracts from the beads were subjected to immunoblotting with an anti-FRL antibody, an anti-green fluorescent protein (GFP) antibody, and anti-actin.
Immunoprecipitation.
P388D1 cell lysates were mixed with an anti-FRL antibody or anti-Rac1 (Santa Cruz) immobilized on protein A-agarose beads at 4°C for 2 h. The beads were washed with PBS–0.1% Triton X-100 (PBS T) three times. The precipitates were analyzed by Western blotting with an anti-FRL antibody, anti-Rac1, and anti-profilin (Cytoskeleton) antibodies.
Immunofluorescence.
COS7 cells were seeded onto four-chamber slides (Nunc) at a density of 2 × 104 cells and cultured overnight. At 24 h after transfection of cDNAs in pEGFP-C1 vectors, cells were washed in PBS and then fixed in 4% paraformaldehyde for 30 min at room temperature. After three washes with PBS, fluorescence images were photographed with a conventional fluorescence microscope to detect the localization of FRL.
Inducible P388D1 transformants were washed in PBS and then fixed in 4% paraformaldehyde for 30 min at room temperature. After three washes with PBS, cells were incubated in blocking buffer (PBS containing 1% bovine serum albumin [BSA], 0.2% skim milk, and 0.3% Triton X-100) at room temperature for 30 min. Rhodamine-conjugated phalloidin (Molecular Probes) was used for F-actin staining.
Adhesion assay.
Doxycycline-inducible transformants were cultured in RPMI 1640 with 10% FCS containing 1 μg of doxycycline/ml for 24 h. Cells were harvested and resuspended in RPMI 1640 without FCS. Ninety-six-well cluster plates (polystyrene, non-tissue culture treated; Falcon) were coated with 10 mg of fibronectin/ml. Proteins were allowed to bind for 2 h at 37°C before the wells were rinsed and blocked for 1 h with 1% BSA (Sigma) in PBS. Viable cells were distinguished by staining with trypan blue. Viable cells were added to the wells at a concentration of 105/0.1 ml and allowed to attach at 37°C for 20 min. Nonadherent cells were removed by two washes with PBS and were then fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. The fixed cells were then rinsed twice with water, stained with 0.1% crystal violet in water for 30 min at room temperature, and rinsed twice with water. Adhesion was quantitated by adding 10% acetic acid to the crystal violet-stained well and examining the solution in a spectrophotometer at 600 nm. Nonspecific attachment to BSA alone was quantitated and subtracted from values obtained for specific adhesion.
Assay for chemotaxis.
Chemotaxis of inducible P388D1 transformants was examined using stromal cell-derived factor-1 (SDF-1) or macrophage colony-stimulating factor (M-CSF) (Genzyme). SDF-1, a CXC chemokine, has chemotactic activity for CFU–granulocyte-macrophage, burst-forming unit–erythrocyte, and CFU–granulocyte-erythrocyte-macrophage-megakaryocyte (25). The cells were cultured in RPMI 1640 with 10% FCS containing 1 μg of doxycycline/ml for 24 h, switched to RPMI 1640 without FCS, and harvested 24 h later. The in vitro migration of cells was assessed in a Transwell cell culture chamber (Costar) as described previously, with some modification (46). Polycarbonate filters (pore size, 8.0 μm) were precoated with 5 μg of gelatin in a volume of 50 μl on the lower surface and dried overnight at room temperature. The coated filters were washed in PBS and then dried immediately before use. A 100-μl cell suspension (105 cells) was added to the upper compartment of the chamber. SDF-1 (DIACLONE Research) diluted in serum-free culture medium was loaded in the lower compartment at a concentration of 100 ng/ml. After 20 h of incubation, the filters were fixed with methanol and the cells on the upper surface were removed by wiping with cotton swabs. The filters were then cut from the upper compartment and stained with hematoxylin and eosin. The cells that had migrated to the lower surface of the filters were manually counted under a microscope at a magnification of ×400. Data were expressed as the number of migrated cells per field.
Analysis for cell proliferation and viability of P388D1 cells after doxycycline-induced expression of truncated FRLs.
For the cell proliferation assay, the inducible P388D1 transformants (FULL/P388D1, FH3/P388D1, FH1+FH3/P388D1, and FH2/P388D1) were cultured in 96-well-plates without or with 1 μg of doxycycline/ml for 24, 48, or 72 h. The cells were pulsed with [3H]thymidine for the last 8 h of cultures.
For cell cycle analysis, FH3/P388D1- and mock-transformed cells were cultured in media containing 1 μg of doxycycline/ml for 0, 24, 48, or 72 h. Cells were washed in PBS and resuspended in 200 μl of hypotonic buffer (0.1% sodium citrate, 0.1% Triton X-100, and 20 μg of RNaseA/ml) containing 50 μg of propidium iodine (Sigma)/ml. Samples were incubated overnight at 4°C (36). For each sample at least 104 events were collected and analyzed on a FACScan (Becton Dickinson). Further analysis of flow cytometric data was performed using CellQuest software. To examine the plasma membrane phosphatidylserine (PS) transition of inducible transformants, binding buffer (HEPES-buffered saline solution supplemented with 2.5 mM CaCl2 [pH 7.4]) was added to the cell suspension. Cells were incubated with 100 ng of fluorescein isothiocyanate (FITC)-conjugated Annexin-V (Pharmingen)/ml for 15 min at room temperature in the dark. Thereafter, cells were further diluted in 400 μl of binding buffer and analyzed on a FACScan.
Nucleotide sequence accession numbers.
The frlα and frlβ sequences have been assigned GenBank numbers AF215666 and AF006466, respectively.
RESULTS
Isolation of mouse frl cDNA.
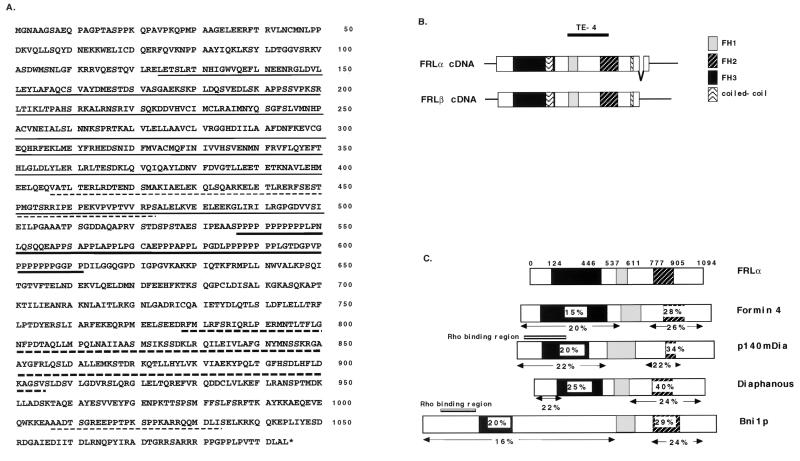
The isolated cDNA fragment (TE-4) had a single open reading frame (ORF), and its deduced amino acid sequences revealed that it contained two domains, FH1 and FH2, that are highly conserved among formin-related proteins. To obtain a full-length coding sequence, a mouse brain cDNA library was screened. The isolated full-length cDNA encoded a protein of 1,064 amino acids. Another type of full-length frl cDNA was obtained by screening a mouse spleen cDNA library, and it encoded a protein of 1,094 amino acids (Fig. 1A). We designated the longer form isolated from spleen cells frlα, and the form from brain cDNA was designated frlβ (Fig. 1B). Two coiled-coil regions are located at the N-terminal side (amino acids [aa] 357 to 422 in FRLα) of FH1 and the C-terminal side (aa 957 to 983) of the FH2 domain. The existence of coiled-coil regions at equivalent positions is also observed in other formin-related proteins.
FIG. 1.
Structure of protein and cDNA of FRL and other formin-related proteins. (A) Deduced amino acid sequence of FRLα. An FH1 domain is indicated by a bold underline, and FH2 by a bold broken line. An FH3 domain is indicated by a thin underline. Coiled-coil regions are indicated by a thin broken line. The accession numbers of the nucleotide sequences in GenBank are AF215666 for frlα and AF006466 for frlβ. (B) Isoforms of frl cDNA. frlα encodes a protein of 1,094 amino acids, and frlβ encodes a protein of 1,064 amino acids. frlα differs from frlβ at its 5′ end and has an in-frame deletion of 99 bp at its 3′ terminus. (C) Schematic presentation of FRL and other formin-related proteins. The homology was calculated by the ratio of the number of identical amino acids to the number of corresponding amino acids of FRL and is expressed as a percentage in each indicated region.
The FH1, FH2, and FH3 domains are commonly found in formin-related proteins. FRL was moderately homologous to formin 4, Dia, p140mDia, and Bni1p (Fig. 1C). FRL shows 24, 25, and 23% identity downstream of the FH1 domain to Formin 4, Dia, and p140mDia, respectively, and shows 40 and 34% identity of the FH2 domain to the respective regions of Dia and p140mDia. FRL shows 22% identity of the N-terminal region, excluding FH3, to the respective region of Dia.
In Western blot analysis using an anti-FRL antibody, FRL proteins were detected strongly in the spleen, thymus and bone marrow (Fig. 2C). Thus, we designated this novel gene frl, for formin-related gene in leukocytes.
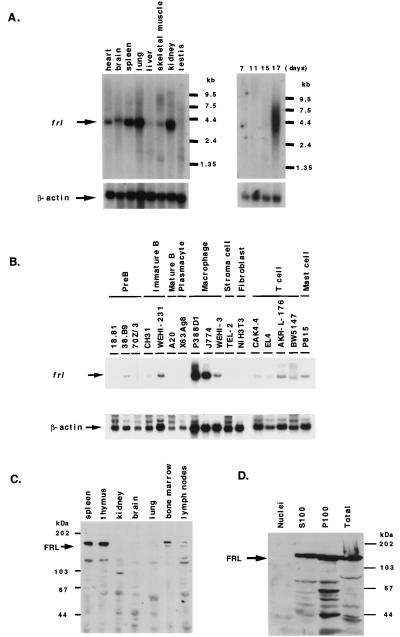
FIG. 2.
Expression of the frl gene and protein in various mouse tissues and cell lines. (A) Northern blot analysis of frl expression in various mouse tissues (left) and in a mouse embryo (right). (B) Northern blot analysis of frl in mouse hematopoietic cell lines. Total RNAs were probed with a common part of frlα and frlβ cDNAs. The amounts of loaded RNA were normalized by the level of β-actin mRNA. (C) Western blot analysis of various mouse tissues. Total tissue extracts (100 μg of protein/lane) from each organ were blotted with a rabbit anti-FRL antibody and a horseradish peroxidase-labeled anti-rabbit IgG antibody. (D) FRL localization in P388D1 cells. P388D1 cells were lysed and separated into three fractions: nuclei, cytosol (S100), and membrane (P100). Each sample was subjected to Western blotting with a rabbit anti-FRL antibody.
Expression of frl gene in mouse tissues and cell lines.
mRNAs from various mouse tissues and hematopoietic cell lines of mice were subjected to Northern blot analysis by using a common part of the frlα and frlβ cDNAs as a probe. Approximately 3.6- to 4.0-kb transcripts for frl were expressed in adult mouse spleen, kidney, lung, heart, brain, and skeletal muscles (Fig. 2A). The expression pattern of transcripts for frl obtained by using a fragment specific for frlα was similar to the above results. The size differences between these transcripts suggested that alternative splicing may have occurred. In analysis of mRNA from a mouse embryo, the expression of frl rapidly increased at day 17 (Fig. 2A). frl transcripts were expressed in several hematopoietic cell lines: 38.B9 (pre-B lymphoid), WEHI-231 (immature B lymphoid), AKR-L-176 (T lymphoid), and P815 (mast cell). The frl mRNA was especially strongly expressed in the P388D1 and J774 macrophage cell lines (Fig. 2B). These results suggested that frl transcripts are widely expressed in hematopoietic cells, especially in macrophages.
Intracellular localization of FRL protein.
Intracellular localization of endogenous FRL was examined by Western blot analysis using subcellular fractions from P388D1 cells. Endogenous FRL was detected in the cytosolic fraction (S100) and the membrane fraction (P100) but was not detected in the nuclear fraction (Fig. 2D). Similar results were obtained using COS7 cells with transient expression of GFP fusion protein of full-length FRL (data not shown).
Specific association of FRL with Rac1 in vitro and in vivo.
p140mDia and Bni1p have been reported to bind Rho family GTPases (13, 26, 51). Therefore we examined whether FRL can bind Rho family GTPases. P388D1 cell lysates were incubated with GST-RhoA, GST-Rac1, GST-Cdc42Hs, or GST, each of which was preloaded with either GTPγs or GDP. GST fusion proteins were precipitated by glutathione-Sepharose 4B beads, and the pellets were analyzed by immunoblotting with an anti-FRL antibody. FRL was precipitated from P388D1 cell lysates by the GTP-bound form of recombinant GST-Rac, and to a lesser extent by the GDP-bound form. FRL was not precipitated by GST-RhoA or GST-Cdc42Hs (Fig. 3A). These results indicated that FRL specifically associates with Rac1 among RhoA, Rac1, and Cdc42Hs and that its binding is stronger in the case of the GTP-bound form of Rac than the GDP-bound form.
In order to examine the interaction between endogenous FRL and Rac, immunoprecipitation analysis was carried out by using an anti-Rac antibody. As shown in Fig. 3B, FRL was coprecipitated with Rac, indicating that FRL associates with Rac in vivo.
Then five types of vectors containing different truncated FRLs were constructed in the form of GFP fusion proteins (Fig. 3C) in order to investigate whether truncated proteins could bind Rac1 in vitro. FH3-FRL consisted of the N-terminal region including the FH3 domain. FH1+FH3-FRL consisted of the FH1 and FH3 domains, and FH2-FRL consisted of the N-terminal 53 amino acids and the FH2 domain. N1-FRL had the N-terminal 80 amino acids, and N2-FRL contained the N-terminal 216 amino acids. Each type of truncated FRL was transiently overexpressed in COS7 cells. FH3-FRL or FH1+FH3-FRL was coprecipitated from COS7 cell lysates by GST-Rac1, but the other truncated FRL proteins were not coprecipitated (Fig. 3D). These results indicated that FRL can bind Rac1 with its N-terminal region, consisting of 216 to 537 amino acids, which contains the FH3 domain.
FRL associates with profilin both in vitro and in vivo.
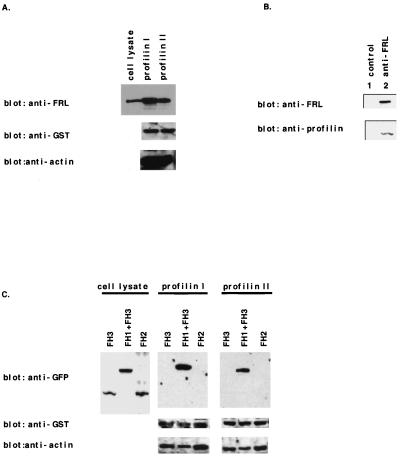
It has been reported that the FH1 domains of p140mDia and Bni1p associate with an actin binding protein, profilin. The association between profilin and FRL was examined in a binding assay using P388D1 cell lysates and the GST-profilin I or GST-profilin II fusion protein. As shown in Fig. 4A, FRL bound to both profilin I and profilin II. Greater amounts of FRL were precipitated with profilin I than with profilin II. Furthermore, actin was coprecipitated with profilins. P388D1 cell lysates were also subjected to immunoprecipitation using an anti-FRL antibody. Profilin was coprecipitated with FRL, indicating that FRL associates with profilin also in vivo (Fig. 4B). To confirm the domain that binds to profilin, the association of several truncated FRL proteins with profilin was investigated by using lysates of COS7 cells expressing GFP fusion proteins with truncated FRLs. Only FH1-FRL could bind to profilin (Fig. 4C), indicating that FRL associates with profilins at the FH1 domain.
FIG. 4.
FRL associates with profilin. (A) Interaction of FRL and profilin in vitro. P388D1 cell lysates were incubated with GST-profilin I or with GST-profilin II and then precipitated by glutathione-Sepharose 4B beads, and the pellets were analyzed by immunoblotting with anti-FRL, anti-GST, and anti-actin antibodies. (B) In vivo association of FRL with profilin. P388D1 cell lysates were immobilized on protein A-agarose beads without (lane 1) or with (lane 2) an anti-FRL antibody. The precipitates were analyzed by Western blotting with anti-FRL and anti-profilin antibodies. (C) The FH1 domain of FRL binds to profilin. COS7 cells transfected with each expression vector (FH3-pEGFP-C1, FH1+FH3-pEGFP-C1, and FH2-pEGFP-C1) were lysed, incubated with GST-profilin I, and precipitated by glutathione-Sepharose 4B beads. The pellets were analyzed by immunoblotting with anti-GFP and anti-actin antibodies.
Expression of the FH3-FRL truncated protein suppressed cell spreading and the formation of lamellipodia.
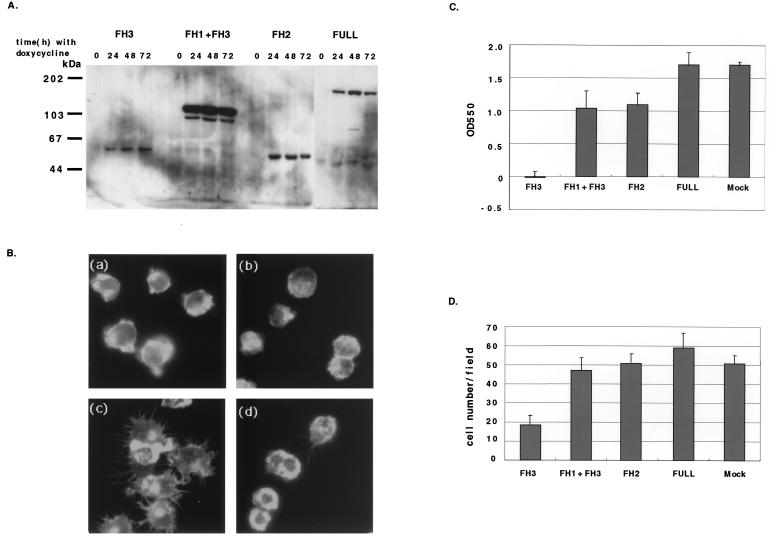
In order to study the functional properties of FRL in macrophages, a doxycycline-regulated inducible vector system (15) was used for expression of the full-length frl cDNA and three types of truncated frl cDNAs (FH3-FRL, FH1+FH3-FRL, and FH2-FRL) in P388D1 cells (Fig. 3C). Addition of doxycycline to these stable transformants for 24 h induced expression of the full length FRL or truncated FRL proteins (Fig. 5A).
FIG. 5.
The inducible P388D1 transformants express various forms of FRL; effects of overexpression of the truncated FH3-FRL protein in P388D1 cells. (A) Cell lysates from doxycycline-inducible transformants of P388D1 (FH3/P388D1, FH1+FH3/P388D1, FH2/P388D1, and FULL/P388D1) were collected after incubation with doxycycline (1.0 μg/ml) at 0, 24, 48, or 72 h and subjected to Western blotting with an anti-Xpress antibody. FRL was strongly expressed from 24 to 72 h after the addition of doxycycline. (B) Inhibition of cell spreading and of formation of lamellipodia in P388D1 cells by doxycycline-inducible overexpression of FH3-FRL. Mock-transformed cells (a and c) and FH3/P388D1 cells (b and d) pretreated with doxycycline were starved for 15 h and then incubated without (a and b) or with (c and d) SDF-1 (100 ng/ml). The cells expressing FH3-FRL protein did not spread well or show the formation of membrane ruffles (d), compared to mock-transformed cells (c). (C) Expression of the truncated FH3-FRL protein strongly inhibited P388D1 cell adherence to fibronectin. After 24 h of incubation with doxycycline, each transformant of P388D1 cells was examined for adhesion to fibronectin-coated wells. Error bars represent standard deviations in triplicate cultures. Similar results were obtained in three separate experiments (P < 0.01 in Student's t test for FH3. (D) Reduction of the chemotaxis of P388D1 cells upon SDF-1 by overexpression of FH3-FRL truncated protein. Inducible P388D1 transformants expressing full-length FRL or each truncated form of FRL and mock-transformed cells were examined for chemotactic activity in response to SDF-1 (100 ng/ml). Random fields of transwell membranes were counted for the number of migrating cells. Similar results were obtained in three separate experiments (P < 0.01 by Student's t test for FH3).
The morphology of transformants that overexpressed various truncated forms of FRL was examined after 15 h of serum starvation. Serum-starved P388D1 cells generally lack lamellipodia, membrane ruffles, and filopodia [Fig. 5B(a) and (b)]. All inducible transformants expressing each form of truncated FRL were stimulated by 100 ng of SDF-1/ml for 20 min. They were then fixed and stained with phalloidin for F-actin. Mock-transformed cells containing only vector showed prominent formation of filopodia and lamellipodia after stimulation with SDF-1 [Fig. 5B(c)]. The cells overexpressing the truncated FH3-FRL protein decreased in size compared to mock-transformed cells and could hardly spread or display membrane ruffling upon SDF-1 stimulation [Fig. 5B(d)]. On the other hand, transformants overexpressing the full-length, FH1+FH3-FRL, or FH2-FRL protein showed responses similar to those of mock-transformed cells upon SDF-1 stimulation. These results suggest that overexpressed FH3-FRL truncated protein inhibited Rac-induced reorganization of the actin cytoskeleton by binding Rac in place of endogenous FRL.
Inhibition of adhesion and chemotaxis of P388D1 cells by the induced expression of the truncated FH3-FRL protein.
As described above, FRL associates with Rac, which suggests that FRL may have a role in cell adhesion and motility. P388D1 cells containing doxycycline-inducible truncated cDNA encoding FH3-FRL (FH3/P388D1 cells) were adhesive to fibronectin-coated plates similarly to the mock cells when cultured without doxycycline. However, 24 h after addition of doxycycline, the number of cells adhesive to fibronectin-coated plates was markedly reduced (Fig. 5C). Transformants overexpressing other truncated FRLs did not show any reduction of adhesion. These results suggested that overexpression of FH3-FRL disturbed cell binding to the extracellular matrix.
Rac has been shown to participate in the migration of macrophages. Migration of the doxycycline-inducible transformants to SDF-1 was examined 24 h after the addition of doxycycline (Fig. 5D). The numbers of migrating P388D1 cells expressing the truncated FH3-FRL protein were decreased to about 35%, compared to those of mock-transformed cells. Similar results were obtained in the case of M-CSF stimulation. The numbers of migrating cells in the absence of these chemokines were fewer than 3 per field in all transformants.
These results indicate that FRL, in cooperation with Rac, is involved in the regulation of adhesion and motility of macrophages, and that the N-terminal region containing the FH3 and FH1 domains is essential for its function.
Inhibition of cell growth by the FH3-FRL truncated protein.
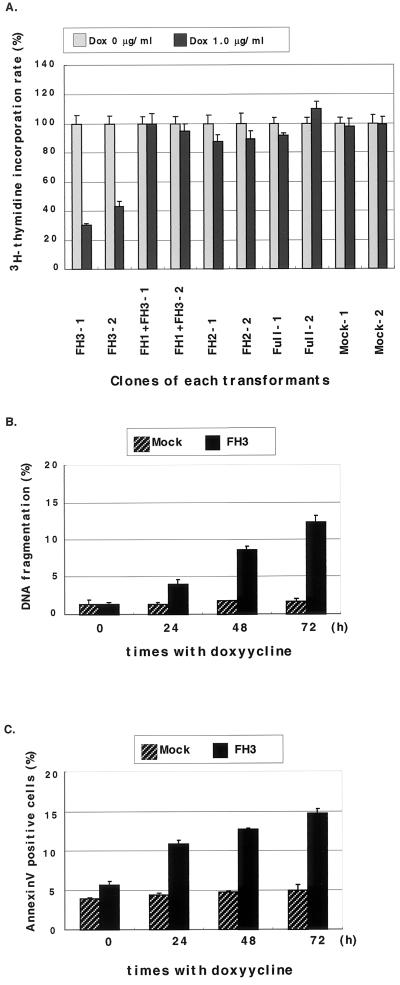
Proliferation of the above transformants in the presence of doxycycline was examined by [3H]thymidine incorporation. In FH3-1 and FH3-2 clones of FH3/P388D1 cells, which overexpressed FH3-FRL upon the addition of doxycycline, [3H]thymidine incorporation was reduced to 30.5 and 43.3%, respectively, at 72 h (Fig. 6A). Expression of full-length and other truncated FRL proteins did not affect [3H]thymidine incorporation by P388D1 cells. Doxycycline alone did not affect the growth of mock-transformed cells. Three other independent experiments showed similar results.
FIG. 6.
Expression of the truncated FH3-FRL protein inhibited cell growth and induced apoptotic cell death in P388D1 cells. (A) [3H]thymidine incorporation of inducible P388D1 transformants at 72 h after the addition of doxycycline (1.0 μg/ml). Overexpression of the truncated FH3-FRL protein strongly inhibited proliferation of P388D1 cells (clone FH3-1 and clone FH3-2). Similar results were obtained in three separate experiments (P < 0.01 by Student's t test for FH3-1 and -2. (B) DNA fragmentation in FH3/P388D1 and mock-transformed cells at 0, 24, 48, and 72 h after the addition of doxycycline (1.0 μg/ml). FH3, FH3/P388D1 cells incubated with doxycycline. (C) FH3/P388D1 and mock-transformed cells were stained with Annexin-V at 0, 24, 48, and 72 h after the addition of doxycycline (1.0 μg/ml). Error bars represent standard deviations in triplicate samples. Similar results were obtained in three separate experiments.
Overexpression of the truncated FH3-FRL protein induced apoptotic cell death in P388D1 cells.
DNA fragmentation and phosphatidylserine (PS) transition in FH3/P388D1 cells were examined at 24, 48, and 72 h after the addition of doxycycline. The percentage of nuclear fragmentation of P388D1 cells overexpressing FH3-FRL was 4.1 ± 0.57% at 24 h, 8.63 ± 0.51% at 48 h, and 12.4 ± 0.87% at 72 h, in contrast to 1.3 to 1.8% in mock-transformed cells (Fig. 6B). The percentages of Annexin-V-positive cells among P388D1 cells overexpressing the FH3-FRL truncated protein were 10.42 ± 0.41% at 24 h, 12.71 ± 0.13% at 48 h, and 14.66 ± 0.62% at 72 h, in contrast to 4.0 to 4.9% in mock-transformed cells at all times (Fig. 6C). Three other independent measurements showed similar results. Overexpression of either FULL-FRL, FH1+FH3-FRL, or FH2-FRL protein did not induce apoptosis. From these results, it appeared that overexpression of FH3-FRL truncated protein induced apoptosis in P388D1 cells, suggesting that a block of FRL binding to Rac by the FH3 truncated protein impaired the cell survival function of FRL.
Rescue of the FH3 truncated protein-induced inhibition of cell adhesion, migration, and proliferation by overexpression of FRL or FH1+FH3-FRL protein.
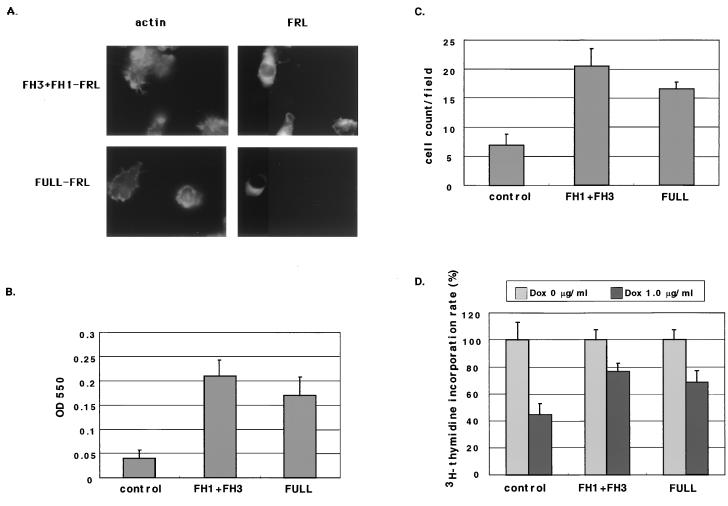
To confirm whether the phenomena observed in P388D1 cells expressing the truncated FH3-FRL protein were caused by its interference with FRL-Rac association, GFP fusion proteins of FULL-FRL or FH1+FH3-FRL were overexpressed in FH3/P388D1 cells. There were no significant differences in the amount of FH3-FRL protein induced by doxycycline among these cells transfected with FULL-pEGFP-C1, FH1+FH3-pEGFP-C1, or pEGFP-C1 vector (data not shown). In spite of the presence of the doxycycline-induced truncated FH3-FRL protein, the cells expressing FULL-FRL or FH1+FH3-FRL protein could spread and form ruffles upon stimulation by SDF-1 (Fig. 7A). In the adhesion assay, overexpression of the FH1+FH3-FRL or FULL-FRL protein recovered the ability of the FH3 protein-producing P388D1 cells to adhere to fibronectin (Fig. 7B). Expression of FH1+FH3-FRL or FULL-FRL in FH3/P388D1 cells could also increase their ability to migrate toward SDF-1 (Fig. 7C). Similar results were obtained in the case of cell migration toward M-CSF (data not shown). Furthermore, these transformants regained their ability to proliferate, as shown in Fig. 7D.
FIG. 7.
Overexpression of FH1+FH3-FRL or FULL-FRL protein restored the cell adhesion, migration, and proliferation abilities of doxycycline-treated FH3/P388D1 cells. FH3/P388D1 cells were transfected with FH1+FH3-pEGFP-C1 (FH1+FH3), FULL-pEGFP-C1 (FULL), and pEGFP-C1 vector only (control), and then cultured with doxycycline for 24 h. (A) The cells expressing FH1+FH3-FRL or FULL-FRL together with FH3-FRL truncated protein spread and formed ruffles upon stimulation with SDF-1. In the panels on the left, cells were stained with rhodamine-conjugated phalloidin for actin staining. Right panels show GFP-positive cells. Note that the cell without expression of transfected FRL-GFP fusion protein does not show spreading or the formation of membrane ruffles. (B) Adhesion assay. Expression of FH1+FH3 or FULL-FRL also rescued the failure of FH3/P388D1 cells to adhere to fibronectin. (C) Migration of FH3/P388D1 against SDF-1 after expression of FH1+FH3-FRL or FULL-FRL protein together with FH3-FRL truncated protein. (D) [3H]thymidine incorporation of FULL-FRL- or FH1+FH3-FRL-transfected FH3/P388D1 cells at 72 h after the addition of doxycycline (1.0 μg/ml). Proliferation of FH3/P388D1 cells was restored by expression of FH1+FH3-FRL or FULL-FRL protein, even in the presence of the truncated FH3-FRL induced by doxycycline.
DISCUSSION
FRL contains three domains, FH1, FH2 and FH3, that characterize Formin-related proteins. Among these proteins, FRL shows moderate homology to Diaphanous proteins p140mDia and Bni1p in the region outside the FH domains. FH1 has been shown to bind the SH3 domain (42), the WWP/WW domain (9), and profilin (13, 21). The Diaphanous proteins p140mDia and Bni1p regulate actin polymerization by binding profilin. In this study, it was shown that the FH1 domain of FRL associates with profilin, suggesting that FRL function may also be mediated by profilin. Since the FH1 domain of FRL contains three PPLP motifs that can bind to both WWP/WW and SH3 domains (4), FRL may express its function by binding several proteins that contain these domains. The function of the FH2 domain has not yet been clarified. Considering that both FH1 and FH2 are well conserved and contained in all Formin-related proteins, the two domains could function together. However, morphology, adhesion, migration, and proliferation were not impaired in P388D1 cells expressing truncated FRL proteins that contained both FH3 and FH1, but not FH2, indicating that the region containing FH3 and FH1 is enough to exhibit the function of FRL. The results of overexpression of FH1+FH3-FRL in FH3/P388D1 cells strongly support this notion (Fig. 7). Thus, the FH2 domain in FRL seems to have little effect on FRL function. The FH3 domain is located at the N terminus in most formin-related proteins. It has been reported that FH3 differs from the Rho binding site of Bni1p and p140mDia. Fus1, one of the Formin-related proteins, is required for conjugation and is localized at the projection tips of mating pairs in S. pombe. The FH3 of Fus1 was shown to localize at the projection tips of mating pairs and has been thought to control localization of Fus1 during conjugation (40). Although the N-terminal region of FRL shows homology (26%) to the Rho binding sites of p140mDia, FRL does not bind Rho, but Rac. The N-terminal region of FRL containing FH3 was shown to be required for associating with Rac. It has been shown that proteins containing the Cdc42/Rac interactive binding (CRIB) motif bind to Cdc42 and/or Rac in a GTP-dependent manner (7). FRL binds preferentially to the GTP-bound form of Rac. However, considering that FRL does not possess the CRIB motif and could also associate weakly with the GDP-bound form of Rac, the interaction of FRL with Rac seems to be independent of the activation of Rac.
Diaphanous proteins have been shown to associate with Rho GTPases and profilin and to contribute to the regulation of actin cytoskeleton (13, 21). Rac induces lamellipodia and regulates integrin adhesion complexes in Swiss 3T3 cells (19, 37, 44, 45). The role of Rho GTPases in regulating cytoskeletal organization has been examined in macrophages, which lack stress fibers. Cells of the monocyte-macrophage lineage are particularly interesting because they are highly locomotive, and their ability to migrate towards a source of a chemoattractant is essential for their recruitment to sites of inflammation (11). In Bac1 cells, a murine macrophage cell line, CSF-1 induces an actin filament-based structure, and the organization of integrin adhesion complexes with the extracellular matrix is regulated by Cdc42 and Rac, but not Rho (1). These adhesion complexes contain several proteins normally associated with focal adhesion in fibroblasts. Rac is also required for the process of macrophage locomotion, whereas Cdc42 is involved in responding to a gradient of the chemokine (2). FRL shows abundant expression in macrophage cell lines, suggesting that FRL might be involved in regulation of actin cytoskeleton organization in macrophages. Expressing the truncated FH3-FRL in P388D1 cells led to reduction in spreading and in the formation of lamellipodia upon stimulation by a chemokine, SDF-1. FH3-FRL also inhibited adhesion and migration in P388D1 cells. Cell migration is a complex process involving dynamic and coordinate changes in cell adhesion and in the cytoskeleton (29). Therefore, the decrease in the migrating activity of P388D1 cells following overexpression of the FH3-FRL truncated protein can be regarded as a result of a combination of a defect in adhesion and a defect in morphological changes in the cytoskeleton. The truncated FH3-FRL protein has an N-terminal region, which can bind Rac but lacks the C-terminal region, including the FH1 domain, which interacts with profilin. Various defects in P388D1 cells resulting from FH3-FRL expression can be explained by the direct binding of the FH3-FRL truncated protein to Rac, which interfered with the Rac binding of endogenous FRL; thus, the normal function of FRL was inhibited. Recently, it was reported that RhoA and the C terminus of p140mDia competitively bind to the N-terminal region of p140mDia, and it was proposed that p140mDia shows intramolecular binding between its N and C termini (52). This finding might be considered one explanation for the mechanism by which FH3-FRL exerts dominant inhibitory effects on endogenous FRL function. Overexpression of FH1+FH3-FRL or FULL-FRL in FH3/P388D1 cells rescued the defects in cell adhesion, migration, and proliferation which were induced by the expression of the truncated FH3 protein. Taken together, it appears that FRL has a crucial role in the actin cytoskeleton, adhesion, and migration by virtue of its association with Rac.
Rac plays a critical role in cell growth. Continuous activation of Rho, Rac, or Cdc42 induces G1 progression in quiescent Swiss 3T3 cells, leading to DNA synthesis (38). Rac and Cdc42 regulate JNK/SAPK and p38 MAP kinase cascades (10, 35, 38, 57). It has been suggested that Rac activates the JNK/SAPK cascade through binding to PAK and induces actin polymerization and proliferation through binding to POR1 and p160ROCK (24, 28, 48). Activation of G1 progression by Rac correlates well with its ability to stimulate lamellipodia. The results of cell proliferation assays and the observation of morphological changes in response to SDF-1 in P388D1 cells expressing the truncated FH3-FRL protein suggest that FRL regulates the cell proliferation of macrophages in a similar fashion. However, overexpression of FH3-FRL also led to apoptosis in P388D1 cells. Therefore, several possibilities can be considered. Integrin-based adhesion complexes that are induced by Rac are also thought to be the source of signals required for cell cycle progression and survival (24, 28). On the other hand, it has been suggested that cell adhesion on fibronectin plays a critical role in cytoskeletal organization, cell cycle progression, and cell survival (14, 20). Detachment of epithelial and endothelial cells from the extracellular matrix leads to programmed cell death (14). It is also suggested that signals from the fibronectin matrix seem to control cell proliferation, whereas cell substrate adhesion provides a survival signal (5). Considering these facts, apoptosis induced by the overexpression of the FH3-FRL truncated protein may occur due to the defects in formation of integrin-based adhesion complexes. Since FRL did not associate with PAK (data not shown), it is unlikely that FRL controls the signal from Rac to PAK, which leads to activation of the JNK/SAPK cascade. Rho GTPases also stimulate transcription from the cyclin D promotor and activate the SRF in the pathway which does not correlate with activation of MAP kinases (ERK, SAPK/JNK, or p38) (18, 53). FRL may be operative in the latter pathway, transmitting a signal essential to survival. Alternatively, FRL may mediate the signal to protect cells from apoptosis through a signaling pathway entirely independent of Rac. Of note, stable transformants of 293 cells or L cells with overexpression of full-length FRL have not yet been obtained. The same phenomenon was also reported with the formin gene (49). It suggests that cells overexpressing FRL may be at a selective disadvantage. FRL might stop the cell cycle or induce cell death in these cell lines. Rho GTPases have been shown to induce various cell responses specific for each cell type, so that expression of FRL in nonhematopoietic cells might lead an the outcome different from that observed in P388D1 cells. In conclusion, from the present studies using P388D1 transformants expressing truncated FRL proteins, the N-terminal region containing FH3 is required for association with Rac, and the FH1 domain is critical for various functions of FRL in P388D1 cells.
Macrophages play an important role in specific immune responses by T and B cells as well as in other host defenses, including phagocytosis and antitumor activities. Most of these macrophage activities are not expressed constitutively but are acquired after exposure to stimuli encountered in the tissue microenvironment. During this activation process, macrophages become usually bigger and more metabolically active, and gain an increased capacity for adherence. In these processes, FRL is expected to display a special function. Furthermore, FRL is expressed in various hematopoietic as well as lymphoid cells. All stages in lymphocyte differentiation and activation are associated with profound changes in cell morphology that mainly depend on functional tubulin and the actin cytoskeleton (39). FRL might be one of the key molecules involved in actin cytoskeleton organization in the immune cells. We are now generating frl gene-targeted mice. They may provide us with further information about the biological function of FRL in vivo.
ACKNOWLEDGMENTS
We thank T. Takenawa (University of Tokyo, Tokyo, Japan) for the gift of pGEX-profilin I and pGEX-profilin II, and N. Watanabe (University of Kyoto, Kyoto, Japan) for pGEX-RhoA, pGEX-Rac1, and pGEX-Cdc42Hs vectors. We are also grateful to Peter D. Burrows for critical reading of the manuscript.
REFERENCES
- 1.Allen W E, Jones G E, Pollard J W, Ridley A J. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J Cell Sci. 1997;110:707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- 2.Allen W E, Zicha D, Ridley A J, Jones G E. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber S, Barbayannis F A, Hanser H, Schneider C, Stanyon C A, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 4.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourdoulous S, Orend G, MacKenna D A, Pasqualini R, Ruoslahti E. Fibronectin matrix regulates activation of Rho and Cdc42 GTPases and cell cycle progression. J Cell Biol. 1998;143:267–276. doi: 10.1083/jcb.143.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brill S, Li S, Lyman C W, Church D M, Wasmuth J J, Weissbach L, Bernards A, Snijders A J. The Ras GTP-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 8.Castrillon D H, Wasserman S A. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 9.Chan D C, Bedford M T, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 10.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 11.Downey G P. Mechanisms of leukocyte motility and chemotaxis. Curr Opin Immunol. 1994;6:113–124. doi: 10.1016/0952-7915(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 12.Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- 13.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 14.Frisch S M, Ruoslahti E. Integrin and anoikis. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- 15.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 16.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 17.Harris S D, Hamer L, Sharpless K E, Hamer J E. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 1997;16:3474–3483. doi: 10.1093/emboj/16.12.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and Cdc42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 19.Hotchin N A, Hall A. The assembly of integrin adhesion complexes requires both extracellular matrix and intracellular rho/rac GTPases. J Cell Biol. 1995;131:1857–1865. doi: 10.1083/jcb.131.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes R O. Fibronectins. New York, N.Y: Springer-Verlag; 1990. [Google Scholar]
- 21.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen R P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 24.Joneson T, McDonough M, Bar-Sagi D, van Aslet L. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 25.Kim C H, Broxmeyer H E. In vitro behavior of hematopoietic progenitor cells under the influence of chemoattractants: SDF-1, steel factor and the bone-marrow environment. Blood. 1998;91:100. [PubMed] [Google Scholar]
- 26.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 27.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 29.Lauffenburger D A, Horwitz A F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 30.Lynch E D, Lee M K, Morrow J E, Welcsh P L, Leon P E, King M-C. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–1318. [PubMed] [Google Scholar]
- 31.Machesky L M, Pollard T D. Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 1993;3:381–385. doi: 10.1016/0962-8924(93)90087-h. [DOI] [PubMed] [Google Scholar]
- 32.Manser E, Leung T, Salihuddin H, Zhao Z S, Lim L. A brain serine/threonine kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 33.Martin G A, Bollag G, McCormick F, Abo A. A novel serine kinase activated by rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;23:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 36.Nicoletti I, Migliorati G, Pagliacci M C, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 37.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 38.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 39.Penninger J M, Crabtree G R. The actin cytoskeleton and lymphocyte activation. Cell. 1999;96:9–12. doi: 10.1016/s0092-8674(00)80954-x. [DOI] [PubMed] [Google Scholar]
- 40.Peterson, J., O. Nielsen, R. Egel, and I. M. Hagen. 1998. FH3, a domain found in formins, targets the fission yeast Fus1 to the projection tip during conjugation. J. Cell Biol. 141. [DOI] [PMC free article] [PubMed]
- 41.Rameh L E, Tolias K F, Duckworth B C, Cantley L C. A new pathway for synthesis of phosphatidylinositol-4,5-biphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 42.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 43.Ridley A J. Intracellular regulation. Rac and Bcr regulate phagocytic phoxes. Curr Biol. 1995;5:710–712. doi: 10.1016/s0960-9822(95)00140-0. [DOI] [PubMed] [Google Scholar]
- 44.Ridley A J, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and stress fibers in response to growth factor. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 45.Ridley A J, Peterson H F, Joneston C L, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 46.Sato K, Nagayama H, Tadokoro K, Juji T, Takahashi T A. Extracellular signal-regulated kinase, stress-activated protein kinase/c-Jun N-terminal kinase, and p38mapk are involved in IL-10-mediated selective repression of TNF-α-induced activation and maturation of human peripheral blood monocyte-derived dendritic cells. J Immunol. 1999;162:3865–3872. [PubMed] [Google Scholar]
- 47.Tolias K F, Cantley L C, Carpenter C L. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 48.van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- 49.Vogt T F, Jackson-Grusby L, Rush J, Leder P. Formins: phosphoprotein isoforms encoded by the mouse limb deformity locus. Proc Natl Acad Sci USA. 1993;90:5554–5558. doi: 10.1073/pnas.90.12.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vojtek A B, Cooper J A. Rho family members: activation of MAP kinase cascades. Cell. 1995;82:527–529. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakano K, Jockusch B M, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 53.Westwick J K, Lambert Q T, Clark G J, Symons M, Van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woychik R P, Maas R L, Zeller R, Vogt T F, Leder P. ‘Formins’: proteins deduced from the alternative transcripts of the limb deformity gene. Nature. 1990;346:850–852. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- 55.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 56.Zahner J E, Harkins H A, Pringle J R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]