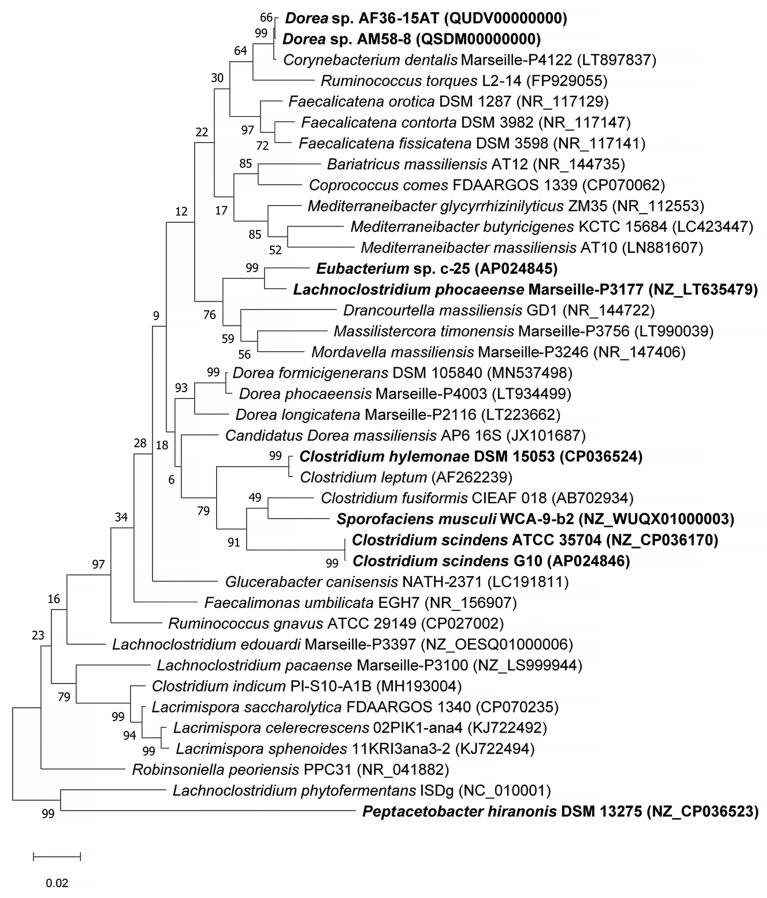
Figure 5.
Maximum Likelihood phylogenetic tree of Eubacterium sp. c-25 and related species of the intestinal bacteria including known DCA producers. Strains included in the tree construction consisted of the 29 closest 16S rDNA sequence BLASTn matches to Eubacterium sp. c-25, excluding uncultured and unidentified strains, and 10 other manually added strains: L. phocaeense Marseille-P3177, C. scindens G10, C. scindens ATCC 35704T, P. hiranonis DSM 13275, S. musculi WCA-9-b2, Dorea sp. AF36-15AT, Dorea sp. AM 58-8, Lachnoclostridium edouardi Marseille-P3397 (NZ_OESQ01000006), Lachnoclostridium pacaense Marseille-P3100 (NZ_LS999944), and Lachnoclostridium phytofermentans ISDg344 (NC_010001). The strains used in bioinformatics analyses in this study are bolded. The tree was constructed in MEGA X [38] using the Tamura–Nei model [39] with 1000 bootstraps and was rooted at the midpoint. Bootstrap support values are listed next to the branches.