Abstract
Liver cancer cell lines are frequently used in vitro tools to test candidate anti-cancer agents as well as to elucidate mechanisms of liver carcinogenesis. Among such mechanisms is cellular communication mediated by connexin-based gap junctions. The present study investigated changes in connexin expression and gap junction functionality in liver cancer in vitro. For this purpose, seven human liver cancer cell lines, as well as primary human hepatocytes, were subjected to connexin and gap junction analysis at the transcriptional, translational and activity level. Real-time quantitative reverse transcription polymerase chain reaction analysis showed enhanced expression of connexin43 in the majority of liver cancer cell lines at the expense of connexin32 and connexin26. Some of these changes were paralleled at the protein level, as evidenced by immunoblot analysis and in situ immunocytochemistry. Gap junctional intercellular communication, assessed by the scrape loading/dye transfer assay, was generally low in all liver cancer cell lines. Collectively, these results provide a full scenario of modifications in hepatocyte connexin production and gap junction activity in cultured liver cancer cell lines. The findings may be valuable for the selection of neoplastic hepatocytes for future mechanistic investigation and testing of anti-cancer drugs that target connexins and their channels.
Keywords: connexin, gap junction, liver cancer, cell line, in vitro
1. Introduction
Liver cancer, mainly represented by hepatocellular carcinoma (HCC), is the second and sixth most common cause of cancer death among men and women, respectively [1]. Primary liver cancer is associated with a wide variety of causes, including chemical, viral and dietary factors [2,3]. Many laboratories worldwide have devoted their work to the study of the mechanisms underlying liver cancer, as well as to the characterization of potential targets for the development of new liver cancer therapeutics [4]. A multitude of experimental systems and models has been used throughout this research [5,6,7]. While some animal models of liver cancer can provide a good reflection of the corresponding human disease and thus have high translational value, they come with a number of issues [7,8,9,10]. In this respect, besides the obvious ethical constraints, animal models are not always well-fit for mechanistic investigation because of their complex dynamic nature [11]. In vitro models seem better suited for this purpose, as they provide a more controllable experimental setting that allows the dissection of specific signaling pathways [5,6,8].
Direct intercellular signaling is mainly mediated by gap junctions [12]. Gap junctions establish a pathway for the exchange of small and hydrophilic molecules and ions between neighboring cells, a flux denoted gap junctional intercellular communication (GJIC). Gap junctions consist of two hemichannels of adjacent cells, which in turn are built by six connexin (Cx) proteins [12]. Connexins are expressed in a cell-type-specific fashion [13]. In healthy liver, the most abundantly expressed connexin species by hepatocytes is Cx32 next to small quantities of Cx26 [14]. By contrast, non-parenchymal cell types in the liver, including Kupffer cells [15] and stellate cells [16], mainly produce Cx43 [17]. During chronic liver disease, however, which may burgeon into the onset of liver cancer, a switch takes place in the connexin expression pattern [17]. Specifically, Cx43 becomes increasingly detectable in the HCC tissue of human patients suffering from liver cancer and experimental animal models not only in non-parenchymal liver cells, but equally in neoplastic hepatocytes [18,19,20,21,22]. This is accompanied by a reduction in the expression of Cx26 [21,22,23] and in particular that of Cx32 [21,23,24], as well as an overall deterioration of GJIC [17].
Liver cancer cell lines are frequently used in vitro tools to test candidate anti-cancer agents as well as to elucidate mechanisms of liver carcinogenesis [25], such as cellular communication mediated by connexin-based gap junctions [13]. The present study is set up to investigate connexin expression and gap junction functionality in liver cancer in vitro compared to healthy primary human hepatocytes (PHH). To this end, seven human cancer liver cell lines, including six from HCC and one of adenocarcinoma origin, are subjected to analysis of connexin mRNA and protein abundance, as well as of subcellular localization. This is aligned by measurement of the activity of gap junctions. This will provide a full scenario of modifications in connexin expression and gap junction activity in cultured liver cancer cell lines, which may be valuable for the selection of neoplastic hepatocytes for future mechanistic investigation and testing of anti-cancer drugs targeting connexin proteins.
2. Results
2.1. Selection of Liver Cancer Cell Lines
In order to assess connexin expression in liver cancer in vitro, a panel of seven cancer cell lines was selected (Table 1). The panel contained six HCC cell lines (C3A, SNU-449, SNU-423, SNU-387, SNU-475 and PLC/PRF/5) and one adenocarcinoma cell line (SK-HEP-1), primarily derived from Caucasian or Asian male patients.
Table 1.
Overview of the liver cancer cell lines and primary human hepatocytes (PHH). Specifications for all cell types used in this study, including morphology, disease, age, gender, ethnicity, hepatitis B virus (HBV) infection of the donors and used cell culture medium (HCC, hepatocellular carcinoma; EMEM, Eagle’s minimum essential medium; FBS, fetal bovine serum; RPMI-1640, Roswell Park Memorial Institute 1640 Medium; CHRM, cryopreserved hepatocyte recovery medium; CHPM, cryopreserved hepatocyte plating medium).
| Cell Line | Morphology | Disease | Age | Gender | Ethnicity | HBV | Medium |
|---|---|---|---|---|---|---|---|
| SK-HEP-1 | Epithelial | Adenocarcinoma | 52 | Male | Caucasian | Not detected | EMEM + FBS |
| C3A | Epithelial | HCC | 15 | Male | Caucasian | Not detected | EMEM + FBS |
| SNU-449 | Epithelial | Grade II-III/IV, HCC | 52 | Male | Asian | DNA detected | RPMI-1640 + heat-inactivated FBS |
| SNU-423 | Epithelial | Grade III/IV, pleomorphic HCC | 40 | Male | Asian | DNA detected | RPMI-1640 + heat-inactivated FBS |
| SNU-387 | Epithelial | Grade IV/V, pleomorphic HCC | 41 | Female | Asian | DNA detected | RPMI-1640 + FBS |
| SNU-475 | Epithelial | Grade II-IV/V, HCC | 43 | Male | Asian | DNA detected | RPMI-1640 + heat-inactivated FBS |
| PLC/PRF/5 | Epithelial | HCC | 24 | Male | African | Secreted HbsAg |
EMEM + FBS |
| PHH | Epithelial | Liver metastasis of colorectal adenocarcinoma | 74 | Female | Caucasian | Not detected | CHRM + CHPM |
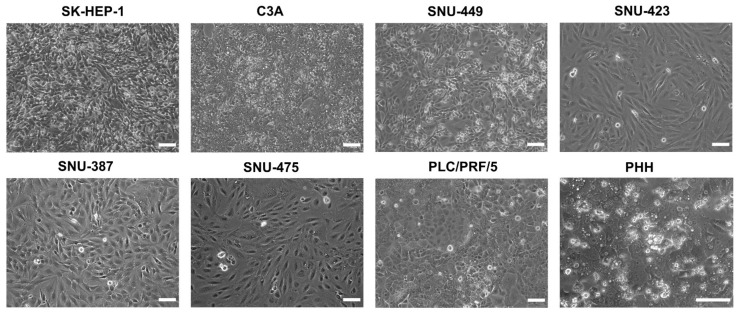
The cell lines all varied in differentiation status and metastatic potential. In this respect, SK-HEP-1 cells are highly metastatic and display mesenchymal stem cell characteristics [26], while C3A cells, which are derived from the commonly used HepG2 cell line, are more differentiated and have retained some metabolic activity [27]. In contrast to C3A cells, the SNU cell lines are less differentiated, more invasive and mesenchymal-like [28]. In order to fully characterize the expression of connexins in HCC, the expression levels were compared to PHH, which represented the normal homeostatic state. Hepatocytes are typically polygonal and can have 1–3 nuclei [29,30]. During carcinogenesis, their morphology changes to a more fibroblast-like phenotype [30]. This was clearly seen in this study (Figure 1).
Figure 1.
Phase contrast images of liver cancer cell lines and primary human hepatocytes (PHH) at 100% confluence. All assays and extractions were performed when liver cancer cell lines reached 100% confluence. Phase contrast images were taken on a Nikon Eclipse Ti-S microscope (Nikon, Tokyo, Japan) with a 10× (liver cancer cell lines) or 20× objective (PHH). Scale bar = 100 µm.
Morphology pictures were taken during the exponential growth phase (Figure S1) of the cancer cell lines or at 100% confluence (Figure 1) and at the last day of the PHH sandwich cultivation period (Figure 1 and Figure S1). Most cancer cell lines displayed a spindle form and appeared to be larger and/or flatter compared to PHH (Figure 1 and Figure S1). The C3A cell line, which is known to be more differentiated [27], exhibited a morphology that is distinct from the other cancer cell lines. The C3A cells were less elongated and had partly retained the polygonal shape and thickness of PHH, which was also the case for PLC/PRF/5 (Figure S1). The retained polygonal shape of PLC/PRF/5 cells was clearer at 100% confluence compared to their appearance during the exponential growth phase ((Figure 1 and Figure S1). In addition, C3A and PLC/PRF/5 cells tended to grow in clusters compared to the other more fibroblast-like cell lines that grew in a looser pattern (Figure S1).
2.2. Connexin Gene Expression in Liver Cancer Cell Lines
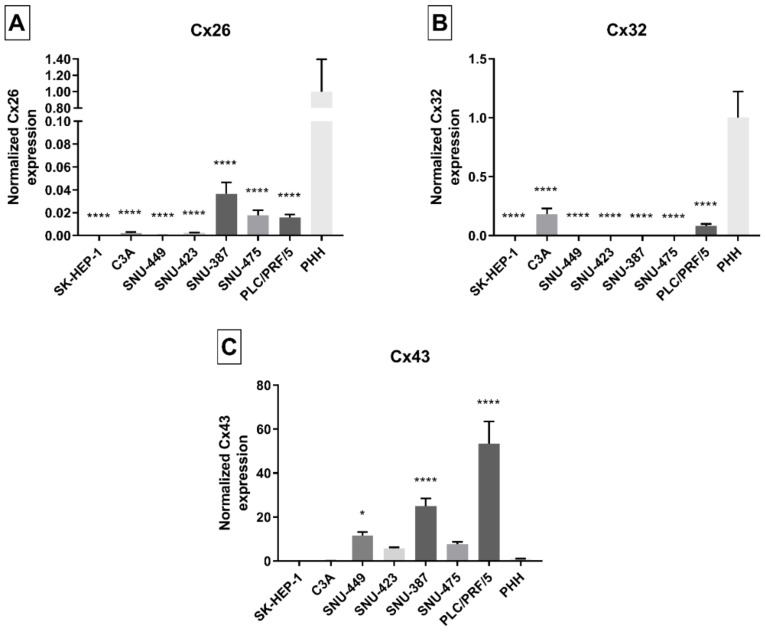
In vivo, human hepatocytes mainly produce Cx32, and to a lesser extent Cx26, which account for 90% and 5% of connexin protein expression, respectively [31]. While Cx32 is ubiquitously expressed [32], Cx26 mRNA is more restricted to the periportal areas [33]. Cx43, on the other hand, is expressed by non-parenchymal cells, such as stellate cells and Kupffer cells [16,34,35,36]. During liver disease, in particular upon acute inflammation, Cx32 mRNA expression is decreased because of increased degradation [37]. In HCC, Cx26 [38] and Cx32 [19,38] gene expression is downregulated, while Cx43 mRNA production becomes promoted [19,20]. However, other studies have shown opposite changes for Cx43 mRNA expression [38] or no changes for Cx32 gene expression [20,21]. Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis in this study detected all connexin species in PHH. Data collected from 100% confluent cancer cell line cultures and PHH confirmed that Cx26 (Figure 2A) and Cx32 (Figure 2B) mRNA quantities were strongly decreased, and even undetectable (Cx26 in SK-HEP-1 cells), in the vast majority of the liver cancer cell lines when compared to PHH. These reductions seemed mildest in C3A and PLC/PRF/5 cells for Cx32, and in SNU-387, SNU-475 and PLC/PRF/5 cells for Cx26 (Figure 2B). The exact same trends could be seen when performing RT-qPCR on cancer cell line cultures during their exponential growth phase (Figure S2A,B).
Figure 2.
Cx26 (A), Cx32 (B) and Cx43 (C) gene expression in liver cancer cell lines and primary human hepatocytes (PHH). Cancer cell lines were grown to 100% confluence, while PHH were used in suspension when total RNA was extracted (n = 1, N = 3). Subsequently, real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis was performed. Relative alterations compared to PHH were calculated according to the Pfaffl method in qbase+ (Biogazelle, Gent, Belgium). Data are expressed as mean ± standard deviation with * p ≤ 0.05 and **** p ≤ 0.0001 compared to the PHH control.
By contrast, Cx43 mRNA abundance was significantly increased in three cell lines, namely SNU-449, SNU-387 and PLC/PRF/5 cells, compared to PHH (Figure 2C). This specifically held true for the PLC/PRF/5 cell line, which showed a 50-fold upregulation in Cx43 production compared to PHH. Cx43 gene expression in SNU-423 cells and SNU-475 cells was higher compared to PHH but was rather negatively affected in C3A and SK-HEP-1 cells. Again, these results were very similar to the results seen during the exponential growth phase of the liver cancer cell lines (Figure S2C).
Additionally, mRNA levels were expressed as a percentage of the corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels (Figure S3) to appreciate the relative levels of Cx26, Cx32 and Cx43 in the liver cancer cell lines or PHH. It was clear that Cx26 (Figure S3A) and Cx32 (Figure S3B) were the main connexin species in PHH and C3A, with Cx32 levels being approximately 13 times higher than Cx26 in PHH. For all cancer cell lines, except C3A cells, the Cx43 levels (Figure S3C) were higher than the Cx32 and Cx26 expression when expressed as a percentage to GAPDH, indicating a clear switch in connexin expression patterns.
2.3. Connexin Protein Expression in Liver Cancer Cell Lines
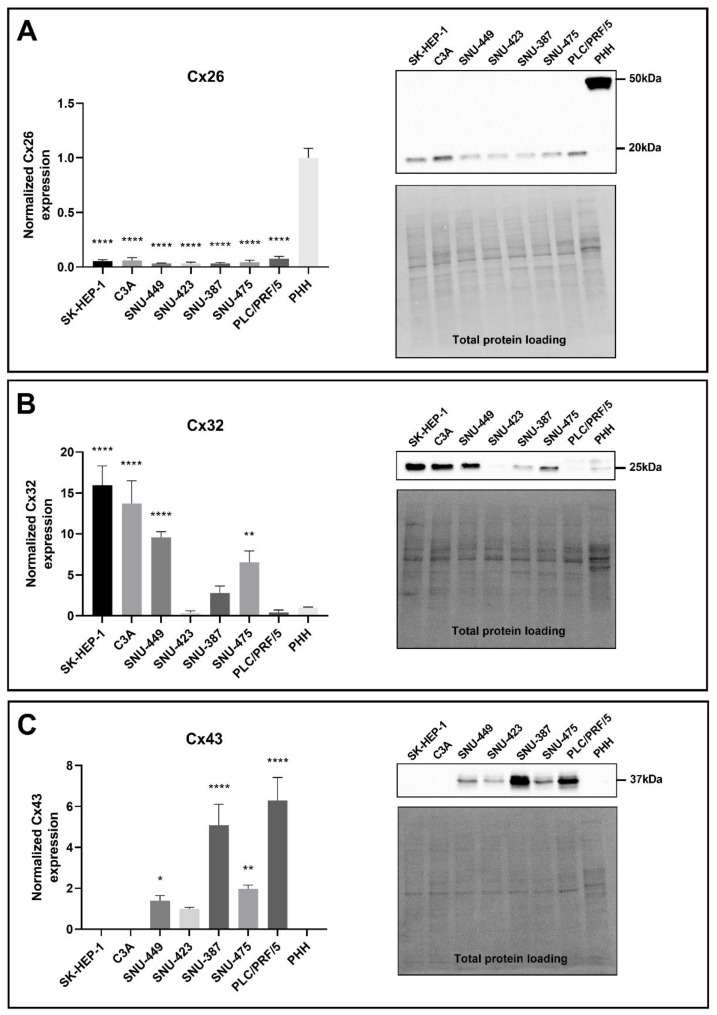
A number of reports have shown differentially affected connexin protein expression in human and rat HCC tumor samples [20,21,22,38] as well as in human and rat liver cancer cell lines [38,39,40]. In this study, both Cx32 and Cx26, but not Cx43, were detected in PHH at the protein level (Figure 3A–C). Cx26 was detected at two different molecular weights, namely at 17 kDa and 50 kDa. Both molecular weights were analyzed, as the band at 50 kDa was presumed to be a dimer [41]. In accordance with the RT-qPCR data (Figure 2A), Cx26 protein moieties were decreased in all liver cancer cell lines compared to PHH (Figure 3A) when immunoblot analysis was performed on 100% confluent cell cultures. The same result was seen during the exponential growth phase of the cancer cell lines (Figure S4A).
Figure 3.
Cx26 (A), Cx32 (B) and Cx43 (C) protein expression in liver cancer cell lines and primary human hepatocytes (PHH). Cancer cell lines (n = 1, N = 3) were grown to 100% confluence, while PHH were used in suspension for protein extraction. Immunoblotting and visualization were done with the Pierce™ ECL Western Blotting Substrate kit (Thermo Fisher Scientific, Waltham, MA, USA) on a ChemiDocTM MP imaging system. All signals were divided by their respective total protein loading signal and normalized by the sum of all data points in a replicate [42]. Unlike Cx43, which was not expressed by PHH, Cx26 and Cx32 are expressed relative to their expression in PHH. Data are expressed as mean ± standard deviation with * p ≤ 0.05, ** p ≤ 0.01 and **** p ≤ 0.0001 compared to the PHH control.
While Cx32 gene expression was suppressed in all liver cancer cell lines compared to PHH (Figure 2B), a downregulation at the translational level could only be observed for SNU-423 and PLC/PRF/5 cells, although not significant (Figure 3B). All other liver cancer cell lines, except for the SNU-387 cells, showed significantly increased Cx32 protein levels (Figure 3B). The same trends were seen during the exponential growth phase of the liver cancer cell lines (Figure S4B). Cx43 was detected as three different signals around 37 kDa in PLC/PRF/5 (Figure 3C). These represent the non-phosphorylated isoform, the phosphorylated isoform and the double-phosphorylated isoform from the lowest to the highest molecular weight, respectively. All bands were included in the quantification of Cx43 in all cell lines. In compliance with the mRNA profile (Figure 2C) and the protein expression during the exponential growth phase (Figure S4C), Cx43 protein production was enhanced in SNU-449, SNU-387, PLC/PRF/5, SNU-475 and SNU-423 cells compared to PHH (Figure 3C), although the latter was not significant. Cx43 was not found in C3A and SK-HEP-1 cells or PHH (Figure 3C), despite being detected during RT-qPCR analysis (Figure 2C), albeit at a very low level (not visible on graph).
2.4. Connexin Protein Subcellular Localization in Liver Cancer Cell Lines
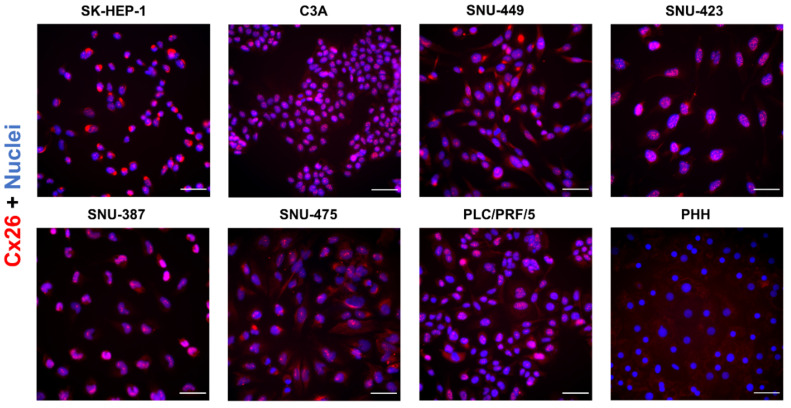
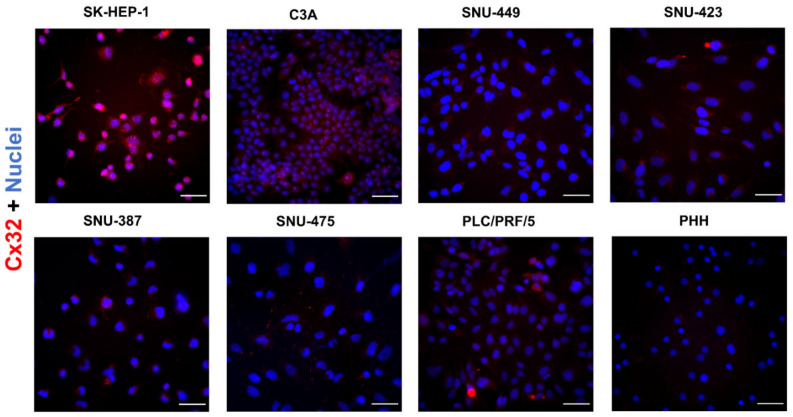
In normal liver, approximately 3% of the hepatocellular membrane surface is occupied by Cx32-based gap junctions [43]. In general, the majority of the connexin proteins are located at the cell plasma membrane [44]. Nevertheless, a substantial amount of them are found in the cytosol, probably due to their rapid turn-over [44,45,46]. Indeed, connexin half-lives are 2–3 h [45,47], while this ranges from 17 to 100 h for most plasma membrane proteins in primary human hepatocytes [46]. In HCC, both in vitro [48,49] and in vivo [19,38,49], a shift in connexin location has been noted from the membrane to the cytoplasm. In addition, the extent of intracellular Cx43 localization has been related to the malignant potential of rat liver epithelial cell lines [39]. The immunocytochemistry data of the present study confirmed Cx26 occurrence in all liver cancer cell lines, appearing predominantly in the perinuclear areas and as a dotted signal coinciding with the signal of the nuclei (Figure 4). This was in contrast to PHH, which did not display this punctuation at the nuclei and only showed a signal in the cytoplasm.
Figure 4.
Cx26 protein localization in liver cancer cell lines and primary human hepatocytes (PHH). Cancer cell lines (n = 1, N = 2) were fixed during the exponential growth phase, while PHH were fixed at the final day of the sandwich cultivation period. All cells were immunostained for Cx26 (red) with nuclear counterstaining using 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar = 50 µm. Images were taken with a 40× objective. A representative image is shown.
In situ immunostaining indicated a merely low presence of Cx32 in all liver cancer cell lines and PHH (Figure 5). SK-HEP-1, C3A, SNU-423 and PLC/PRF/5 cells displayed a dotted Cx32 pattern in the nuclei and a diffuse signal in the cytoplasm.
Figure 5.
Cx32 protein localization in liver cancer cell lines and primary human hepatocytes (PHH). Cancer cell lines (n = 1, N = 2) were fixed during the exponential growth phase, while PHH were fixed at the final day of the sandwich cultivation period. All cells were immunostained for Cx32 (red) with nuclear counterstaining using 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar = 50 µm. Images were taken with a 40× objective. A representative image is shown.
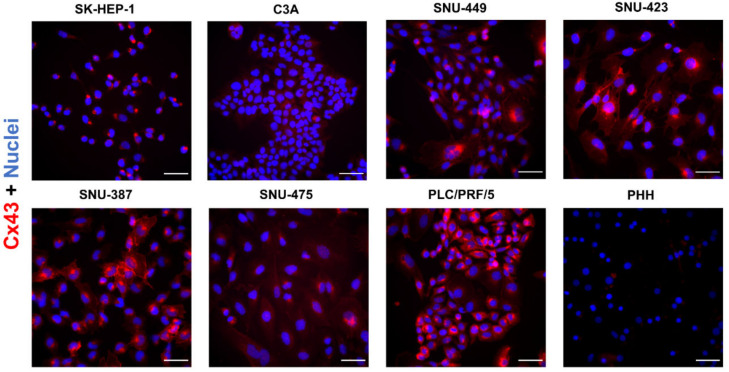
In support of the immunoblot analysis results (Figure 3), Cx43 was most prominently represented in PLC/PRF/5 cells and the SNU cell lines but showed faint staining in C3A and SK-HEP-1 cells and PHH (Figure 6).
Figure 6.
Cx43 protein localization in liver cancer cell lines and primary human hepatocytes (PHH). Cancer cell lines (n = 1, N = 2) were fixed during the exponential growth phase, while PHH were fixed at the final day of the sandwich cultivation period. All cells were immunostained for Cx43 (red) with nuclear counterstaining using 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar = 50 µm. Images were taken with a 40× objective. A representative image is shown.
Unlike Cx32 and Cx26, Cx43 formed a delineated pattern in various liver cancer cell lines, which suggests predominant localization at the cell plasma membrane surface (Figure 6).
2.5. Gap Junctional Intercellular Communication in Liver Cancer Cell Lines
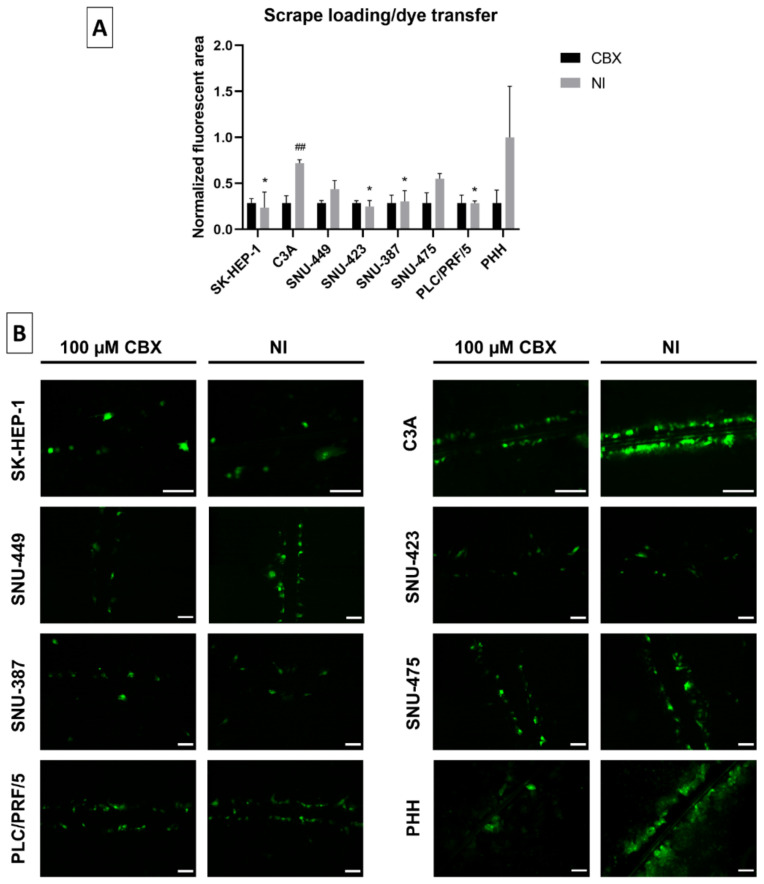
A scrape loading/dye transfer assay was used to assess GJIC. A scratch was hereby made with a needle into the confluent cell layer in the presence of the fluorescent dye Lucifer Yellow (LY). The latter was taken up by the damaged cells along the scrape. When functional, LY was transferred to undamaged neighboring cells through gap junctions. The fluorescent area was used as a measure to assess GJIC. Throughout these experiments, carbenoxolone disodium salt (CBX), a well-known inhibitor of gap junctions [50], was used as a control. C3A cells displayed functional GJIC, as shown by the significant difference between the CBX-inhibited and non-inhibited (NI) cells (Figure 7A,B).
Figure 7.
Gap junctional intercellular communication (GJIC) in liver cancer cell lines. All cell lines (n = 1–2, N = 1 (multiple images per well)) were grown to 100% confluence when a scrape was made across the well in presence of Lucifer Yellow (LY). Cells were washed and subjected to fluorescence microscopy analysis. Images were taken with a 10× or 20× objective (B). GJIC was either not inhibited (NI) or inhibited by 100 µM carbenoxolone disodium salt (CBX) for 10 min. The fluorescent area was measured with ImageJ. Data were normalized to the inhibited control and expressed as a ratio to the mean NI fluorescent area of primary human hepatocytes (PHH). Data are expressed as mean ± standard deviation (A). Significant differences compared to NI PHH are indicated with * p ≤ 0.05, while differences to the respective CBX control are indicated with ## p ≤ 0.01. Scale bar = 100 µm.
Although PHH displayed functional GJIC along certain scrapes, the higher variation in these cultures ensured that the overall GJIC was not significant compared to the inhibited control. In most cell lines, the NI fluorescent area was similar to the fluorescent area of the inhibited CBX control (Figure 7B). This indicated the absence of functional GJIC, which was also reflected by their significant lack of fluorescent dye transfer compared to NI PHH.
3. Discussion
Cell lines are popular experimental settings in scientific research because of a number of reasons, including ease of use and high reproducibility of testing results [5,51]. Many cell lines have a carcinogenic background and hence are frequently relied upon to test candidate anti-cancer drugs, as well as to elucidate mechanisms of carcinogenesis [51]. Among such mechanisms is cellular communication mediated by gap junctions [13,52]. Modifications in the expression of connexins in human and rodent liver cancer tissue have been reported on many occasions [21,23,24,38,53,54,55]. Several studies describe a reduction in Cx26 and Cx32 levels and a concomitant upregulation of Cx43 expression in various chronic liver diseases, most of which ultimately lead to HCC development [17]. The tumor-promoting role of Cx43 in HCC has been indicated on different occasions [56,57]. A knockdown of Cx43 production in human hepatoma cells triggers cell cycle arrest, boosts the differentiated status [56] and suppresses invasion, migration and metastasis [57], whereas inverse observations are seen in their Cx43-overexpressing counterparts [56].
To gain a broader insight into changes in connexin expression and gap junction functionality in HCC in vitro, this study subjected seven different human cell lines, commonly applied to liver cancer research, to an analysis of gene and protein expression, as well as of subcellular localization of Cx26, Cx32 and Cx43, along with testing of GJIC responses. Reminiscent of the in vivo situation [19,21,22], Cx43 de novo expression was found in various liver cancer cell lines when compared to PHH. The increase in protein abundance was hereby paralleled by almost identical changes at the mRNA level. This suggests regulation by the transcriptional machinery. In this respect, the proto-oncogenes c-fos and c-jun are known to be induced upon hepatocarcinogenesis [58]. These proteins form the transcription factor activator protein 1, which controls Cx43 expression [59]. In myometrial cells, activator protein 1 has been found to activate Cx43 gene transcription in stress conditions [60]. This mechanism could possibly underlie Cx43 production in liver cancer cells as well. Although most cell lines supported a tumor-promoting role for Cx43, this connexin species was not detected in SK-HEP-1 and C3A cells during immunoblot analysis. Indeed, contradicting studies about the role of Cx43 in HCC have been published. Various studies proposed a downregulation or even absence of Cx43 in human HCC samples [38,53,61], in vivo rat studies [62] and human [38] and rat liver cancer cell lines [63]. Another reason for the absence of Cx43 protein expression in C3A cells could be the fact that this cancer cell line has retained a more differentiated status compared to other liver cancer cell lines and therefore does not express the Cx43 protein [27,28]. The SK-HEP-1 cell line, on the other hand, is known to originate from endothelial cells and was more recently proposed to serve as a model for liver sinusoidal cells rather than HCC, which they have been widely used for in the past [64]. While liver endothelial cells normally express Cx43 [16], SK-HEP-1 does not, according to various studies [65,66,67], which is in line with the results of the present study.
Downregulation of Cx26 gene expression, as seen in the present study, has been equally observed in rat [68] and human HCC tissue [23] and complies with results from others using human liver cancer cell lines [38]. This has been associated with hypermethylation of the Cx26 gene promotor [68]. Cx26 and particularly Cx32 are known to act as liver tumor suppressors [17]. In this regard, Cx32-knockout animals display elevated susceptibility for the development of liver tumors [69,70]. At the same time, overexpression of Cx32 inhibits metastasis and proliferation of liver cancer cells [71]. Cx32 gene or protein expression has been repeatedly reported to be decreased in HCC in vivo [21,24,72,73], in vitro [38,49] and ex vivo [19,23,38,49,74]. This was also found by RT-qPCR analysis in the current study. However, with the exception of Cx32 expression in SNU-423 and PLC/PRF/5 cells, these findings were not reflected and actually contradictory at the protein level. Upregulated [53] or even unchanged [19,20,22] Cx32 protein expression has also been observed in human HCC samples. Such discrepancy between Cx32 mRNA and protein expression has been equally seen in non-alcoholic steatohepatitis, which often leads to HCC [75] and may be associated with shortening of the poly(A) tail in Cx32 mRNA [76,77].
GJIC deterioration is commonly observed in (chronic) liver diseases [78]. In HCC, GJIC has been inversely correlated with Cx43 expression and cell line malignancy levels [39,56]. Reduction of GJIC in HCC was supported by the results of the present study. This could represent an escape from homeostasis, this being a hallmark of cancer [13]. The lack of GJIC in SK-HEP-1 cells has been previously reported [79]. Nevertheless, gap junction activity could be detected in some liver cancer cell lines, in particular C3A cells and to a lesser extent in SNU-449 and SNU-475 cells.
In summary, the results of this study provide for the first time a full characterization of in vitro modifications in connexin expression as well as gap junction activity in liver cancer cell lines. PLC/PRF/5 is the cell line that stands out the most for displaying a decrease in Cx26 and Cx32 on both the transcriptional and translational levels, together with an increase in Cx43 on both levels and a reduction of GJIC. Although the presented data are based on single extractions and should be confirmed in follow-up studies, these findings may be of great relevance for the selection of liver cancer cell lines for future mechanistic investigation and testing of anti-cancer drugs that target connexins and their channels.
4. Materials and Methods
4.1. Reagents
Dimethyl sulfoxide (DMSO), CBX and LY were supplied by Sigma-Aldrich (St. Louis, MO, USA). All other reagents were obtained from various suppliers at the highest analytical grade possible.
4.2. Liver Cancer Cell Lines and Primary Human Hepatocytes
PHH were purchased from KaLy-cell (Plobsheim, France) and cultured in a sandwich culture for scrape loading/dye transfer analysis and immunocytochemistry analysis. PHH were thawed and transferred to a tube with prewarmed cryopreserved hepatocyte recovery medium (CHRM) (70001, APSciences, Columbia, MD, USA). After gentle mixing, PHH were pelleted by centrifugation at 100× g for 10 min at room temperature. Cell culture medium was discarded, and the PHH pellet was resuspended in 2–5 mL cold cryopreserved hepatocyte plating medium (CHPM) (70002, APSciences, Columbia, MD, USA) depending on the pellet size. A total of 0.36 × 106 cells were seeded per well in a 24-well plate coated with rigid rat tail collagen I (Corning, NY, USA). An amount of 0.1 mg/mL collagen was dissolved in 0.02 N acetic acid (Sigma-Aldrich, St. Louis, MO, USA) for coating of the plates before cell seeding. Plates were incubated at 37 °C with 5% CO2 and 95% humidity. At least 4 h after seeding, the cell culture medium was replaced with warm cryopreserved hepatocyte plating medium. Twenty-four hours after seeding, the monolayer was washed with Dulbecco’s phosphate-buffered saline (PBS) (Gibco, Waltham, MA, USA), and a Matrigel® (Corning, NY, USA) overlay was added. Matrigel® was dissolved in maintenance medium [Williams E medium (A12176-01, Gibco, Waltham, MA, USA), 1% L-glutamine–penicillin–streptomycin (Sigma-Aldrich, St. Louis, MO, USA), 1% insulin–transferrin–selenium (Gibco, Waltham, MA, USA) and 0.01% dexamethasone (Sigma-Aldrich, St. Louis, MO, USA) dissolved at 1 mM in DMSO] at a concentration of 0.25 mg/mL. Maintenance medium was replaced 2 h later, after gelatinization of the Matrigel® layer at 37 °C, and again 2 days after addition of the Matrigel® layer. PHH sandwich cultures were maintained for 4 days after seeding.
The Liver Cancer Panel (ATCC® TCP-1011™) was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and contained 7 human liver cancer cell lines, namely SK-HEP-1, C3A, SNU-449, SNU-423, SNU-387, SNU-475 and PLC/PRF/5 (Table 1). All cell lines were cultured at 37 °C with 5% CO2 and 95% humidity. Cells were grown according to the supplier’s recommendations and were used at 100% confluence unless specified otherwise. Cells were grown in Roswell Park Memorial Institute 1640 Medium (ATCC 30-2001™, Manassas, VA, USA or A1049101, Gibco, Waltham, MA, USA) or Eagle’s Minimum Essential Medium (EMEM)(ATCC 30-2003™, Manassas, VA, USA or Minimum Essential Medium (MEM) (11095080, Gibco, Waltham, MA, USA) with addition of 1% MEM Non-Essential Amino Acids Solution (11140035, Gibco, Waltham, MA, USA) and 1% Sodium Pyruvate (11360039, Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (ATCC, USA) (heat-inactivated for SNU-475, SNU-449 and SNU-423) and 100 units/mL penicillin and 100 µg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA).
4.3. Phase-Contrast Microscopy
Microscopy pictures were taken during the exponential growth phase of the cell lines and at 100% confluence with a Nikon Eclipse Ti-S microscope (Nikon, Tokyo, Japan) using a 10× objective. PHH sandwich cultures were imaged daily on the same microscope with a 20× objective. Scale bars were added with ImageJ software (version 1.52p) (Bethesda, MD, USA).
4.4. Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction Analysis
mRNA expression analysis was carried out as previously described with minor modifications [44]. In essence, total RNA was extracted from cultured cell lines and freshly thawed PHH using a GenEluteTM Mammalian Total RNA Miniprep Kit (Sigma-Aldrich, St. Louis, MO, USA) combined with an On-Column DNase I Digestion Set (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. RNA yield and purity were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Next, cDNA was generated with an iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) on a MiniAmp Plus Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) followed by cDNA purification using a GenElute™ PCR Clean-Up Kit (Sigma-Aldrich, St. Louis, MO, USA). RT-qPCR analysis was performed on an Applied Biosystems QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Samples were tested in triplicate and amplified with TaqMan® Gene Expression Assays (Applied Biosystems, Waltham, MA, USA) targeted towards 5 housekeeping genes, namely actin beta (ACTB), beta-2-microglobulin (B2M), GAPDH, hydroxymethylbilane synthase (HMBS) and ubiquitin C (UBC), as well as towards Cx26, Cx32 and Cx43 (Table 2).
Table 2.
Primers and probes for real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis. Assay identification (ID), accession number, assay location, amplicon size and exon boundaries are listed (GJA1, Cx43; GJB1, Cx32; GJB2, Cx26; ACTB, actin beta; B2M, beta-2-microglobulin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HMBS, hydroxymethylbilane synthase; UBC, ubiquitin C).
| Gene Symbol | Assay ID | Accession Number | Assay Location | Amplicon Size (Base Pairs) | Exon Boundary |
|---|---|---|---|---|---|
| GJA1 ( Cx43 ) | Hs00748445-s1 | NM_000165.4 | 1031 | 142 | 2 |
| GJB1 ( Cx32 ) | Hs00939759-s1 |
NM_000166.5 NM_001097642.2 |
1547 1496 |
63 | 2 |
| GJB2 ( Cx26 ) | Hs00269615-s1 | NM_004004.5 | 715 | 123 | 2 |
| ACTB | Hs01060665-g1 | NM_001101.3 | 208 | 63 | 2–3 |
| B2M | Hs00187842-m1 | NM_004048.2 | 134 | 64 | 1–2 |
| GAPDH | Hs02786624-g1 |
NM_001256799.2 NM_001289745.1 NM_001289746.1 NM_002046.5 |
870 928 822 836 |
157 | 7 8 7 8 |
| HMBS | Hs00609296-g1 |
NM_000190.3 NM_001024382.1 NM_001258208.1 NM_001258209.1 |
1070 972 950 1041 |
69 | 13–14 13–14 12–13 12–13 |
| UBC | Hs01871556-s1 | M26880.1 | 2173 | 135 | – |
The reaction mix contained 2 µL cDNA, 1 µL primer, 7 µL nuclease-free water and 10 µL of TaqMan® Fast Advanced Master Mix (Applied Biosystems, Waltham, MA, USA). Efficiency was estimated based on a serial dilution of pooled cDNA mix from all samples and 3 no-template controls. Relative alterations compared to PHH were calculated according to the Pfaffl method [80] in qbase+ (Biogazelle, Gent, Belgium).
4.5. Immunoblot Analysis
Immunoblot analysis was performed as described previously with small modifications [81]. Cell lines were washed twice with ice-cold PBS and harvested by scraping in the presence of radio-immunoprecipitation assay buffer (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 1% ethylenediaminetetraacetic acid (Thermo Fisher Scientific, Waltham, MA, USA) and 1% protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA). PHH were not cultured for protein extraction but were freshly thawed and washed twice with ice-cold PBS. After centrifugation for 5 min at 4 °C at 2060× g, the resulting pellet was resuspended in the same protein lysis buffer as the cell lines. Next, lysates were transferred to a pre-cooled tube and put on ice for 30 min, during which the lysate was vortexed every 5 min. Supernatants were collected by centrifugation for 20 min at 4 °C and 14,000× g and stored at −80 °C. Protein concentrations were determined with the PierceTM BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). For immunoblotting, proteins were separated on 10% (Cx43) or 12% (Cx26 and Cx32) Mini-PROTEAN TGX Stain-Free™ precast gels (Bio-Rad, Hercules, CA, USA). Samples for detection of Cx26 were heated for 5 min at 95 °C. Gels were blotted onto a nitrocellulose membrane (Cx43 and Cx32) (Bio-Rad, Hercules, CA, USA) or a polyvinylidene difluoride membrane (Cx26) (Bio-Rad, Hercules, CA, USA). Blocking of the membranes was performed in 5% non-fatty milk in Tris-buffered saline solution (20 mM Tris and 135 mM sodium chloride) with 0.1% Tween-20 for 1 h at room temperature on an orbital shaker. Membranes were incubated overnight at 4 °C with primary antibodies raised against Cx26, Cx32 or Cx43 (Table 3) diluted in blocking buffer.
Table 3.
Overview of primary antibodies. Specifications regarding the primary antibodies used in this study, including target, product code, dilution used in immunoblot analysis and immunostaining analysis and the supplier.
| Target | Product Code | Dilution Primary Antibody | Supplier | |
|---|---|---|---|---|
| Immunoblot | Immunocytochemistry | |||
| Cx26 | 51–2800 | 1:250 | 1:250 | Invitrogen, Waltham, MA, USA |
| Cx32 | C3470 | 1:600 | 1:500 | Sigma-Aldrich, St. Louis, MO, USA |
| Cx43 | C6219 | 1:4000 | 1:1000 | Sigma-Aldrich, St. Louis, MO, USA |
Membranes were washed 3 times for 10 min and incubated with a secondary antibody (Cx26 and Cx32: 1:500; Cx43: 1:2000, P0448, Dako, Næstved, Denmark) diluted in blocking buffer for 1 h at room temperature. Membranes were washed and visualized by using the Pierce™ ECL Western Blotting Substrate kit (Thermo Fisher Scientific, Waltham, MA, USA) on a ChemiDocTM MP imaging system (Bio-Rad, Hercules, CA, USA). Densiometric analysis was performed with Image Lab 6.0.1 software (Bio-Rad, Hercules, CA, USA). Signals were normalized against total protein loading and by sum of all data points in a replicate [42]. Data are expressed as fold change relative to the corresponding signals in PHH, unless mentioned otherwise.
4.6. Immunocytochemistry Analysis
Cell lines and PHH cultures were washed 3 times with ice-cold PBS for 5 min and fixed at −20 °C for 10 min with an acetone–ethanol (1:1) mixture that permeabilized simultaneously. Cells were then washed 3 times for 5 min with ice-cold PBS. Cells were blocked at room temperature with 1% bovine serum albumin (BSA)/5% donkey serum for 45 min and incubated overnight at 4 °C with primary antibodies. Primary antibodies targeted against Cx26, Cx32 and Cx43 (Table 3) were diluted in 1% BSA dissolved in PBS. Excess of antibody was washed away 3 times with PBS for 5 min. This was followed by incubation with a secondary antibody (1:500, Alexa Fluor 594-AffiniPure Donkey Anti-Rabbit IgG, 711-585-152, Jackson ImmunoResearch, West Grove, PA, USA) diluted in 1% BSA dissolved in PBS for 1 h at room temperature. Cells were again washed 3 times for 5 min with PBS and 1 time with double-distilled water. Nuclei were stained by adding VECTASHIELD Antifade Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA). Detection was performed on a Nikon Eclipse Ti microscope (Nikon, Tokyo, Japan) with a 40× objective. Image editing was done with ImageJ software (version 1.52p) (Bethesda, MD, USA).
4.7. Scrape Loading/Dye Transfer Assay
Cell lines and PHH were washed 3 times with scrape loading/dye transfer (SL/DT) buffer (137 mM NaCl, 5.36 mM KCl, 0.8 mM MgCl2, 5.55 mM glucose and 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES); pH 7.4) or CBX buffer (100 µM CBX in SL/DT buffer). Next, a scratch was made with a fine needle in the presence of LY (1 mg/mL) dissolved in SL/DT or CBX buffer. This was incubated for 10 min at room temperature. Then, cells were washed 4 times with Hank’s Balanced Salt Solution (HBSS)/ HEPES buffer (0.95 mM CaCl2.2H20, 0.81 mM MgSO4.7H2O, 137 mM NaCl, 5.36 mM KCl, 5.55 mM glucose and 25 mM HEPES; pH 7.4) and fixed for 10 min in 4% paraformaldehyde solution. Cells were washed again 4 times with HBSS/HEPES buffer and visualized on a Nikon Eclipse Ti-S microscope (Nikon, Tokyo, Japan) with a 10× or 20× objective. The fluorescent area of the dye spread was measured via ImageJ software (version 1.52p) (Bethesda, MD, USA). Data were normalized to the area of the inhibited cells and expressed as a ratio to the non-inhibited signals in PHH.
4.8. Statistical Analysis
One batch was used for PHH and all cell lines (n = 1). The number of technical replicates (N) is specified in the figure legends. Statistical analysis was performed in GraphPad Prism v9.0 software (GraphPad Software, San Diego, CA, USA), and data are presented as mean ± standard deviation. Normality was assessed by means of a Shapiro–Wilk normality test. Normally distributed RT-qPCR and immunoblot data were analyzed via an ordinary 1-way analysis of variance, followed by a Dunnett’s multiple comparisons test. Scrape loading/dye transfer data were analyzed in the same way when comparing the dye transfer of the cell lines to the dye transfer of the PHH. For comparing the inhibited to the non-inhibited fluorescent area within each cell line, multiple unpaired t-tests were performed with a Holm–Šidák correction for multiple comparisons. Significance levels are indicated by to the following symbols: * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 and **** p ≤ 0.0001.
Acknowledgments
The authors are grateful to Dinja De Win and Yasmin Dahdouh-Guebas for their excellent technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222212187/s1.
Author Contributions
Conceptualization, B.C. and M.V.; methodology, K.L. and A.P.; data analysis, K.L.; writing—original draft preparation, K.L. and M.V.; writing—review and editing, K.L., C.J.S.C., A.P., B.d.S.R., A.C., R.V.C., A.T., B.C. and M.V.; supervision, B.C. and M.V.; project administration, B.C. and M.V.; funding acquisition, B.C. and M.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Fund for Scientific Research-Flanders (FWO Vlaanderen), the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP-FWO Grant 18/10953-9), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grant 1270 310557/2019-4), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)—Brasil Finance code 001, the Alternatives Research and Development Foundation (ARDF), the Center for Alternatives to Animal Testing at Johns Hopkins University-USA (CAAT), the Methusalem program of the Flemish government and the University Hospital of the Vrije Universiteit Brussel-Belgium (Willy Gepts Fonds UZ-Brussel).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Koumbi L. Dietary factors can protect against liver cancer development. World J. Hepatol. 2017;9:119–125. doi: 10.4254/wjh.v9.i3.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sia D., Villanueva A., Friedman S.L., Llovet J.M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 4.Alqahtani A., Khan Z., Alloghbi A., Said Ahmed T.S., Ashraf M., Hammouda D.M. Hepatocellular carcinoma: Molecular mechanisms and targeted therapies. Medicina. 2019;55:526. doi: 10.3390/medicina55090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins S.D., Yuen G., Tu T., Budzinska M.A., Spring K., Bryant K., Shackel N.A. In vitro models of the liver: Disease modeling, drug discovery and clinical applications. In: Tirnitz-Parker J.E.E., editor. Hepatocellular Carcinoma. Codon Publications; Brisbane, Australia: 2019. [PubMed] [Google Scholar]
- 6.Hirschfield H., Bian C.B., Higashi T., Nakagawa S., Zeleke T.Z., Nair V.D., Fuchs B.C., Hoshida Y. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp. Mol. Med. 2018;50:e419. doi: 10.1038/emm.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlessi R., Kohn-Gaone J., Olynyk J.K., Tirnitz-Parker J.E.E. Mouse models of hepatocellular carcinoma. In: Tirnitz-Parker J.E.E., editor. Hepatocellular Carcinoma. Codon Publications; Brisbane, Australia: 2019. [PubMed] [Google Scholar]
- 8.Bagi C.M., Andresen C.J. Models of hepatocellular carcinoma and biomarker strategy. Cancers. 2010;2:1441–1452. doi: 10.3390/cancers2031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Febbraio M.A., Reibe S., Shalapour S., Ooi G.J., Watt M.J., Karin M. Preclinical models for studying NASH-driven HCC: How useful are they? Cell Metab. 2019;29:18–26. doi: 10.1016/j.cmet.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schachtschneider K.M., Schwind R.M., Darfour-Oduro K.A., De A.K., Rund L.A., Singh K., Principe D.R., Guzman G., Ray C.E., Jr., Ozer H., et al. A validated, transitional and translational porcine model of hepatocellular carcinoma. Oncotarget. 2017;8:63620–63634. doi: 10.18632/oncotarget.18872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q., Li J., Middleton A., Bhattacharya S., Conolly R.B. Bridging the data gap from in vitro toxicity testing to chemical safety assessment through computational modeling. Front. Public Health. 2018;6:261. doi: 10.3389/fpubh.2018.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willebrords J., Crespo Yanguas S., Maes M., Decrock E., Wang N., Leybaert L., da Silva T.C., Veloso Alves Pereira I., Jaeschke H., Cogliati B., et al. Structure, regulation and function of gap junctions in liver. Cell Commun. Adhes. 2015;22:29–37. doi: 10.3109/15419061.2016.1151875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zefferino R., Piccoli C., Gioia S.D., Capitanio N., Conese M. Gap junction intercellular communication in the carcinogenesis hallmarks: Is this a phenomenon or epiphenomenon? Cells. 2019;8:896. doi: 10.3390/cells8080896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neveu M.J., Hully J.R., Babcock K.L., Vaughan J., Hertzberg E.L., Nicholson B.J., Paul D.L., Pitot H.C. Proliferation-associated differences in the spatial and temporal expression of gap junction genes in rat liver. Hepatology. 1995;22:202–212. [PubMed] [Google Scholar]
- 15.Eugenin E.A., Gonzalez H.E., Sanchez H.A., Branes M.C., Saez J.C. Inflammatory conditions induce gap junctional communication between rat Kupffer cells both in vivo and in vitro. Cell Immunol. 2007;247:103–110. doi: 10.1016/j.cellimm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer R., Reinehr R., Lu T.P., Schonicke A., Warskulat U., Dienes H.P., Haussinger D. Intercellular communication via gap junctions in activated rat hepatic stellate cells. Gastroenterology. 2005;128:433–448. doi: 10.1053/j.gastro.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez-Guerra M., Hadjihambi A., Jalan R. Gap junctions in liver disease: Implications for pathogenesis and therapy. J. Hepatol. 2019;70:759–772. doi: 10.1016/j.jhep.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Maes M., Crespo Yanguas S., Willebrords J., Cogliati B., Vinken M. Connexin and pannexin signaling in gastrointestinal and liver disease. Transl. Res. 2015;166:332–343. doi: 10.1016/j.trsl.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krutovskikh V., Mazzoleni G., Mironov N., Omori Y., Aguelon A.M., Mesnil M., Berger F., Partensky C., Yamasaki H. Altered homologous and heterologous gap-junctional intercellular communication in primary human liver tumors associated with aberrant protein localization but not gene mutation of connexin 32. Int. J. Cancer. 1994;56:87–94. doi: 10.1002/ijc.2910560116. [DOI] [PubMed] [Google Scholar]
- 20.Oyamada M., Krutovskikh V.A., Mesnil M., Partensky C., Berger F., Yamasaki H. Aberrant expression of gap junction gene in primary human hepatocellular carcinomas: Increased expression of cardiac-type gap junction gene connexin 43. Mol. Carcinog. 1990;3:273–278. doi: 10.1002/mc.2940030507. [DOI] [PubMed] [Google Scholar]
- 21.Neveu M.J., Hully J.R., Babcock K.L., Hertzberg E.L., Nicholson B.J., Paul D.L., Pitot H.C. Multiple mechanisms are responsible for altered expression of gap junction genes during oncogenesis in rat liver. J. Cell Sci. 1994;107:83–95. doi: 10.1242/jcs.107.1.83. [DOI] [PubMed] [Google Scholar]
- 22.Wilgenbus K.K., Kirkpatrick C.J., Knuechel R., Willecke K., Traub O. Expression of Cx26, Cx32 and Cx43 gap junction proteins in normal and neoplastic human tissues. Int. J. Cancer. 1992;51:522–529. doi: 10.1002/ijc.2910510404. [DOI] [PubMed] [Google Scholar]
- 23.Sheen I.S., Jeng K.S., Wang P.C., Shih S.C., Chang W.H., Wang H.Y., Chen C.C., Shyung L.R. Are gap junction gene connexins 26, 32 and 43 of prognostic values in hepatocellular carcinoma? A prospective study. World J. Gastroenterol. 2004;10:2785–2790. doi: 10.3748/wjg.v10.i19.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krutovskikh V.A., Oyamada M., Yamasaki H. Sequential changes of gap-junctional intercellular communications during multistage rat liver carcinogenesis: Direct measurement of communication in vivo. Carcinogenesis. 1991;12:1701–1706. doi: 10.1093/carcin/12.9.1701. [DOI] [PubMed] [Google Scholar]
- 25.Fukuyama K., Asagiri M., Sugimoto M., Tsushima H., Seo S., Taura K., Uemoto S., Iwaisako K. Gene expression profiles of liver cancer cell lines reveal two hepatocyte-like and fibroblast-like clusters. PLoS ONE. 2021;16:e0245939. doi: 10.1371/journal.pone.0245939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eun J.R., Jung Y.J., Zhang Y., Zhang Y., Tschudy-Seney B., Ramsamooj R., Wan Y.J., Theise N.D., Zern M.A., Duan Y. Hepatoma SK Hep-1 cells exhibit characteristics of oncogenic mesenchymal stem cells with highly metastatic capacity. PLoS ONE. 2014;9:e110744. doi: 10.1371/journal.pone.0110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson L.J., Morgan K., Treskes P., Samuel K., Henderson C.J., LeBled C., Homer N., Grant M.H., Hayes P.C., Plevris J.N. Human hepatic HepaRG cells maintain an organotypic phenotype with high intrinsic CYP450 activity/metabolism and significantly outperform standard HepG2/C3A cells for pharmaceutical and therapeutic applications. Basic Clin. Pharmacol. 2017;120:30–37. doi: 10.1111/bcpt.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruso S., Calatayud A.L., Pilet J., La Bella T., Rekik S., Imbeaud S., Letouze E., Meunier L., Bayard Q., Rohr-Udilova N., et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology. 2019;157:760–776. doi: 10.1053/j.gastro.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Bartlett D.C., Hodson J., Bhogal R.H., Youster J., Newsome P.N. Combined use of N-acetylcysteine and Liberase improves the viability and metabolic function of human hepatocytes isolated from human liver. Cytotherapy. 2014;16:800–809. doi: 10.1016/j.jcyt.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montalbano M., Rastellini C., Wang X., Corsello T., Eltorky M.A., Vento R., Cicalese L. Transformation of primary human hepatocytes in hepatocellular carcinoma. Int. J. Oncol. 2016;48:1205–1217. doi: 10.3892/ijo.2015.3312. [DOI] [PubMed] [Google Scholar]
- 31.Maes M., Decrock E., Cogliati B., Oliveira A.G., Marques P.E., Dagli M.L., Menezes G.B., Mennecier G., Leybaert L., Vanhaecke T., et al. Connexin and pannexin (hemi)channels in the liver. Front. Physiol. 2014;4:405. doi: 10.3389/fphys.2013.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooreman A., Van Campenhout R., Ballet S., Annaert P., Van Den Bossche B., Colle I., Cogliati B., Vinken M. Connexin and pannexin (hemi)channels: Emerging targets in the treatment of liver disease. Hepatology. 2019;69:1317–1323. doi: 10.1002/hep.30306. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg E., Spray D.C., Reid L.M. Transcriptional and posttranscriptional control of connexin mRNAs in periportal and pericentral rat hepatocytes. Eur. J. Cell Biol. 1992;59:21–26. [PubMed] [Google Scholar]
- 34.Berthoud V.M., Iwanij V., Garcia A.M., Saez J.C. Connexins and glucagon receptors during development of rat hepatic acinus. Am. J. Physiol. 1992;263:650–658. doi: 10.1152/ajpgi.1992.263.5.G650. [DOI] [PubMed] [Google Scholar]
- 35.Greenwel P., Rubin J., Schwartz M., Hertzberg E.L., Rojkind M. Liver fat-storing cell clones obtained from a Cci4-cirrhotic rat are heterogeneous with regard to proliferation, expression of extracellular-matrix components, interleukin-6, and connexin-43. Lab. Investig. 1993;69:210–216. [PubMed] [Google Scholar]
- 36.Saez J.C. Intercellular gap junctional communication is required for an optimal metabolic response of the functional units of liver. Hepatology. 1997;25:775–776. doi: 10.1002/hep.510250348. [DOI] [PubMed] [Google Scholar]
- 37.Gingalewski C., Wang K., Clemens M.G., De Maio A. Posttranscriptional regulation of connexin 32 expression in liver during acute inflammation. J. Cell Physiol. 1996;166:461–467. doi: 10.1002/(SICI)1097-4652(199602)166:2<461::AID-JCP25>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y., Zhu J., Zhang N., Zhao Y., Li W.Y., Zhao F.Y., Ou Y.R., Qin S.K., Wu Q. Impaired gap junctions in human hepatocellular carcinoma limit intrinsic oxaliplatin chemosensitivity: A key role of connexin 26. Int. J. Oncol. 2016;48:703–713. doi: 10.3892/ijo.2015.3266. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki Y., Kubomoto A., Yamasaki H. Control of intracellular localization and function of Cx43 by SEMA3F. J. Membr. Biol. 2007;217:53–61. doi: 10.1007/s00232-007-9051-y. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y., Qin S.K., Wu Q., Wang Z.S., Zheng R.S., Tong X.H., Liu H., Tao L., He X.D. Connexin-dependent gap junction enhancement is involved in the synergistic effect of sorafenib and all-trans retinoic acid on HCC growth inhibition. Oncol. Rep. 2014;31:540–550. doi: 10.3892/or.2013.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ambrosi C., Boassa D., Pranskevich J., Smock A., Oshima A., Xu J., Nicholson B.J., Sosinsky G.E. Analysis of four connexin26 mutant gap junctions and hemichannels reveals variations in hexamer stability. Biophys. J. 2010;98:1809–1819. doi: 10.1016/j.bpj.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degasperi A., Birtwistle M.R., Volinsky N., Rauch J., Kolch W., Kholodenko B.N. Evaluating strategies to normalise biological replicates of Western blot data. PLoS ONE. 2014;9:e87293. doi: 10.1371/journal.pone.0087293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spray D.C., Bai S., Burk R.D., Saez J.C. Regulation and function of liver gap junctions and their genes. Prog. Liver Dis. 1994;12:1–18. [PubMed] [Google Scholar]
- 44.Maes M., McGill M.R., da Silva T.C., Abels C., Lebofsky M., Maria Monteiro de Araujo C., Tiburcio T., Veloso Alves Pereira I., Willebrords J., Crespo Yanguas S., et al. Involvement of connexin43 in acetaminophen-induced liver injury. Biochim. Biophys. Acta. 2016;1862:1111–1121. doi: 10.1016/j.bbadis.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traub O., Look J., Paul D., Willecke K. Cyclic adenosine monophosphate stimulates biosynthesis and phosphorylation of the 26 kDa gap junction protein in cultured mouse hepatocytes. Eur. J. Cell Biol. 1987;43:48–54. [PubMed] [Google Scholar]
- 46.Chu F.F., Doyle D. Turnover of plasma membrane proteins in rat hepatoma cells and primary cultures of rat hepatocytes. J. Biol. Chem. 1985;260:3097–3107. doi: 10.1016/S0021-9258(18)89478-9. [DOI] [PubMed] [Google Scholar]
- 47.Traub O., Druge P.M., Willecke K. Degradation and resynthesis of gap junction protein in plasma membranes of regenerating liver after partial hepatectomy or cholestasis. Proc. Natl. Acad. Sci. USA. 1983;80:755–759. doi: 10.1073/pnas.80.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yano T., Hernandez-Blazquez F.J., Omori Y., Yamasaki H. Reduction of malignant phenotype of HEPG2 cell is associated with the expression of connexin 26 but not connexin 32. Carcinogenesis. 2001;22:1593–1600. doi: 10.1093/carcin/22.10.1593. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y., Yao J.H., Du Q.Y., Zhou Y.C., Yao T.J., Wu Q., Liu J., Ou Y.R. Connexin 32 downregulation is critical for chemoresistance in oxaliplatin-resistant HCC cells associated with EMT. Cancer Manag. Res. 2019;11:5133–5146. doi: 10.2147/CMAR.S203656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Z.C., Oberheim N., Kettenmann H., Ransom B.R. Pharmacological “cross-inhibition” of connexin hemichannels and swelling activated anion channels. Glia. 2009;57:258–269. doi: 10.1002/glia.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raju K.L., Augustine D., Rao R.S., SV S., Haragannavar V.C., Nambiar S., Prasad K., Awan K.H., Patil S. Biomarkers in tumorigenesis using cancer cell lines: A systematic review. Asian Pac. J. Cancer Prev. 2017;18:2329–2337. doi: 10.22034/APJCP.2017.18.9.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schalper K.A., Carvajal-Hausdorf D., Oyarzo M.P. Possible role of hemichannels in cancer. Front. Physiol. 2014;5:237. doi: 10.3389/fphys.2014.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang Y., Wang Q., Guo Y., Ge H., Fu Y., Wang X., Tao L. Cx32 exerts anti-apoptotic and pro-tumor effects via the epidermal growth factor receptor pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:145. doi: 10.1186/s13046-019-1142-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neveu M.J., Hully J.R., Paul D.L., Pitot H.C. Reversible alteration in the expression of the gap junctional protein connexin 32 during tumor promotion in rat liver and its role during cell proliferation. Cancer Commun. 1990;2:21–31. doi: 10.3727/095535490820874731. [DOI] [PubMed] [Google Scholar]
- 55.Krutovskikh V.A., Mesnil M., Mazzoleni G., Yamasaki H. Inhibition of rat liver gap junction intercellular communication by tumor-promoting agents in vivo. Association with aberrant localization of connexin proteins. Lab. Invest. 1995;72:571–577. [PubMed] [Google Scholar]
- 56.Zhang D., Kaneda M., Nakahama K., Arii S., Morita I. Connexin 43 expression promotes malignancy of HuH7 hepatocellular carcinoma cells via the inhibition of cell-cell communication. Cancer Lett. 2007;252:208–215. doi: 10.1016/j.canlet.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 57.Ogawa K., Pitchakarn P., Suzuki S., Chewonarin T., Tang M., Takahashi S., Naiki-Ito A., Sato S., Takahashi S., Asamoto M., et al. Silencing of connexin 43 suppresses invasion, migration and lung metastasis of rat hepatocellular carcinoma cells. Cancer Sci. 2012;103:860–867. doi: 10.1111/j.1349-7006.2012.02228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elaut G., Henkens T., Papeleu P., Snykers S., Vinken M., Vanhaecke T., Rogiers V. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr. Drug Metab. 2006;7:629–660. doi: 10.2174/138920006778017759. [DOI] [PubMed] [Google Scholar]
- 59.Oyamada M., Oyamada Y., Takamatsu T. Regulation of connexin expression. Biochim. Biophys. Acta. 2005;1719:6–23. doi: 10.1016/j.bbamem.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell J.A., Lye S.J. Differential activation of the connexin 43 promoter by dimers of activator protein-1 transcription factors in myometrial cells. Endocrinology. 2005;146:2048–2054. doi: 10.1210/en.2004-1066. [DOI] [PubMed] [Google Scholar]
- 61.Shen Y.M., Li Y.X., Ma X.F., Wan Q.H., Jiang Z.M., Liu Y.X., Zhang D.Y., Liu X.Z., Wu W.H. Connexin 43 SUMOylation improves gap junction functions between liver cancer stem cells and enhances their sensitivity to HSVtk/GCV. Int. J. Oncol. 2018;52:872–880. doi: 10.3892/ijo.2018.4263. [DOI] [PubMed] [Google Scholar]
- 62.Tork O.M., Khaleel E.F., Abdelmaqsoud O.M. Altered cell to cell communication, autophagy and mitochondrial dysfunction in a model of hepatocellular carcinoma: Potential protective effects of curcumin and stem cell therapy. Asian Pac. J. Cancer Prev. 2015;16:8271–8279. doi: 10.7314/APJCP.2015.16.18.8271. [DOI] [PubMed] [Google Scholar]
- 63.Piechocki M.P., Burk R.D., Ruch R.J. Regulation of connexin32 and connexin43 gene expression by DNA methylation in rat liver cells. Carcinogenesis. 1999;20:401–406. doi: 10.1093/carcin/20.3.401. [DOI] [PubMed] [Google Scholar]
- 64.Tai Y., Gao J.H., Zhao C., Tong H., Zheng S.P., Huang Z.Y., Liu R., Tang C.W., Li J. SK-Hep1: Not hepatocellular carcinoma cells but a cell model for liver sinusoidal endothelial cells. Int. J. Clin. Exp. Pathol. 2018;11:2931–2938. [PMC free article] [PubMed] [Google Scholar]
- 65.Hermans M.M., Kortekaas P., Jongsma H.J., Rook M.B. pH sensitivity of the cardiac gap junction proteins, Connexin 45 and 43. Pflugers Arch. 1995;431:138–140. doi: 10.1007/BF00374389. [DOI] [PubMed] [Google Scholar]
- 66.Moreno A.P., Saez J.C., Fishman G.I., Spray D.C. Human Connexin43 gap junction channels—Regulation of unitary conductances by phosphorylation. Circ. Res. 1994;74:1050–1057. doi: 10.1161/01.RES.74.6.1050. [DOI] [PubMed] [Google Scholar]
- 67.Moreno A.P., Laing J.G., Beyer E.C., Spray D.C. Properties of gap junction channels formed of connexin-45 endogenously expressed in human hepatoma (Skhep1) cells. Am. J. Physiol. Cell Physiol. 1995;268:C356–C365. doi: 10.1152/ajpcell.1995.268.2.C356. [DOI] [PubMed] [Google Scholar]
- 68.Tsujiuchi T., Shimizu K., Itsuzaki Y., Onishi M., Sugata E., Fujii H., Honoki K. CpG site hypermethylation of E-cadherin and Connexin26 genes in hepatocellular carcinomas induced by a choline-deficient L-Amino Acid-defined diet in rats. Mol. Carcinog. 2007;46:269–274. doi: 10.1002/mc.20268. [DOI] [PubMed] [Google Scholar]
- 69.Hokaiwado N., Asamoto M., Futakuchi M., Ogawa K., Takahashi S., Shirai T. Both early and late stages of hepatocarcinogenesis are enhanced in cx32 dominant negative mutant transgenic rats with disrupted gap junctional intercellular communication. J. Membr. Biol. 2007;218:101–106. doi: 10.1007/s00232-007-9053-9. [DOI] [PubMed] [Google Scholar]
- 70.Kato H., Naiki-Ito A., Naiki T., Suzuki S., Yamashita Y., Sato S., Sagawa H., Kato A., Kuno T., Takahashi S. Connexin 32 dysfunction promotes ethanol-related hepatocarcinogenesis via activation of Dusp1-Erk axis. Oncotarget. 2016;7:2009–2021. doi: 10.18632/oncotarget.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao B., Zhao W., Wang Y., Xu Y., Xu J., Tang K., Zhang S., Yin Z., Wu Q., Wang X. Connexin32 regulates hepatoma cell metastasis and proliferation via the p53 and Akt pathways. Oncotarget. 2015;6:10116–10133. doi: 10.18632/oncotarget.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shoda T., Mitsumori K., Onodera H., Toyoda E., Uneyama C., Takada K., Hirose H. Liver tumor-promoting effect of beta-naphthoflavone, a strong CYP 1A1/2 inducer, and the relationship between CYP 1A1/2 induction and Cx32 decrease in its hepatocarcinogenesis in the rat. Toxicol. Pathol. 2000;28:540–547. doi: 10.1177/019262330002800406. [DOI] [PubMed] [Google Scholar]
- 73.Shoda T., Mitsumori K., Onodera H., Toyoda K., Uneyama C., Imazawa T., Hirose M. The relationship between decrease in Cx32 and induction of P450 isozymes in the early phase of clofibrate hepatocarcinogenesis in the rat. Arch. Toxicol. 1999;73:373–380. doi: 10.1007/s002040050676. [DOI] [PubMed] [Google Scholar]
- 74.Yu M., Zou Q., Wu X., Han G., Tong X. Connexin 32 affects doxorubicin resistance in hepatocellular carcinoma cells mediated by Src/FAK signaling pathway. Biomed. Pharmacother. 2017;95:1844–1852. doi: 10.1016/j.biopha.2017.09.065. [DOI] [PubMed] [Google Scholar]
- 75.Willebrords J., Cogliati B., Pereira I.V.A., da Silva T.C., Crespo Yanguas S., Maes M., Govoni V.M., Lima A., Felisbino D.A., Decrock E., et al. Inhibition of connexin hemichannels alleviates non-alcoholic steatohepatitis in mice. Sci. Rep. 2017;7:8268. doi: 10.1038/s41598-017-08583-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Maio A., Gingalewski C., Theodorakis N.G., Clemens M.G. Interruption of hepatic gap junctional communication in the rat during inflammation induced by bacterial lipopolysaccharide. Shock. 2000;14:53–59. doi: 10.1097/00024382-200014010-00010. [DOI] [PubMed] [Google Scholar]
- 77.Theodorakis N.G., De Maio A. Cx32 mRNA in rat liver: Effects of inflammation on poly(A) tail distribution and mRNA degradation. Am. J. Physiol. 1999;276:1249–1257. doi: 10.1152/ajpregu.1999.276.5.R1249. [DOI] [PubMed] [Google Scholar]
- 78.Crespo Yanguas S., Willebrords J., Maes M., da Silva T.C., Veloso Alves Pereira I., Cogliati B., Zaidan Dagli M.L., Vinken M. Connexins and pannexins in liver damage. EXCLI J. 2016;15:177–186. doi: 10.17179/excli2016-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Rijen H.V., van Veen T.A., Hermans M.M., Jongsma H.J. Human connexin40 gap junction channels are modulated by cAMP. Cardiovasc. Res. 2000;45:941–951. doi: 10.1016/S0008-6363(99)00373-9. [DOI] [PubMed] [Google Scholar]
- 80.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cooreman A., Van Campenhout R., Crespo Yanguas S., Gijbels E., Leroy K., Pieters A., Tabernilla A., Van Brantegem P., Annaert P., Cogliati B., et al. Cholestasis differentially affects liver connexins. Int. J. Mol. Sci. 2020;21:6534. doi: 10.3390/ijms21186534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request.