Abstract
The discrimination between Staphylococcus epidermidis strains that contaminate and infect blood cultures is a daily challenge for clinical laboratories. The results of PCR detection of putative virulence genes were compared for contaminating strains, sepsis-related strains, catheter strains, and saprophytic strains. Multiplex PCR was used to explore the atlE gene, which is involved in initial adherence, the intercellular adhesion gene cluster (ica), which mediates the formation of the biofilm, and the agrA, sarA, and mecA genes, which might contribute to the pathogenicity of S. epidermidis. Whereas the atlE, agrA, and sarA genes were almost ubiquitously amplified, the ica and mecA genes were detected significantly more in infecting strains than in contaminating strains (P ≤ 0.02) and thus appeared to be related to the potential virulence of S. epidermidis.
During the last decade, Staphylococcus epidermidis and other coagulase-negative staphylococci have emerged as a major cause of nosocomial infections. These organisms, which constitute a major component of the normal skin and mucosal microflora, are particularly responsible for catheter- and medical device-related sepsis (13). They also frequently contaminate blood cultures, making their interpretation a major concern for clinicians and for analytical laboratories. Although the decision for therapy relies mostly on the observation of sepsis symptoms, other criteria are often considered, such as the number of positive blood cultures and the similarity of their antibiotic resistance profiles (15). However, these criteria may be controversial in many cases: antibiotic resistance profiles may differ for isogenic strains, whereas infections may involve strains that are isolated only once. Moreover, numeric criteria may not be available for anemic or pediatric patients, who cannot undergo multiple venous punctures. The purpose of this study was to assess whether the person making the clinical decision may benefit from genetic data, such as the detection of genes encoding putative virulence factors.
The pathogenesis of S. epidermidis catheter-related infections mostly relies on adherence to polymer surfaces (4). The bacterial biofilm is produced in a two-step manner: the initial bacterial attachment to the surface is followed by biofilm formation, consisting of bacterial proliferation, intercellular adhesion, and extracellular slime substance production (9). A recent study demonstrated that the primary attachment of S. epidermidis to a polystyrene surface is related to a cell surface protein exhibiting vitronectin-binding activity. This protein is encoded by the chromosomal atlE gene and exhibits a high similarity to the major autolysin of Staphylococcus aureus (11). In other respects, investigation of the second stage of biofilm formation demonstrated that cell aggregation and biofilm accumulation were mediated by the products of the chromosomal ica gene locus, which comprises three intercellular adhesion genes (icaA, icaB, and icaC) organized in an operon structure and which leads to the biosynthesis of polysaccharide intercellular adhesin (10). Besides virulence factors that are involved in adherence and biofilm formation, homologs of the sar and agr loci of S. aureus have recently been characterized for S. epidermidis (7, 17, 20; W. J. B. Van Wamel, J. Verhoef, and A. C. Fluid, Abstr. 96th Gen. Meet. Am. Soc. Microbiol., abstr. B-338, p. 213, 1996). Whereas the products of these genes mediate the production of major virulence factors in S. aureus (12, 18), the functions of their homologs in S. epidermidis are still unknown. Indeed, the agr and sar loci might also be implicated in the regulation of the expression of the chromosomal mecA gene, which is responsible for methicillin resistance (16). Based on the lack of mecA transcription in phase variants (16) and on the absence of mutation in the mecI gene and mecA promoter or operator region in methicillin-resistant S. epidermidis (14), one can speculate on the role of the agr and sar loci in the possible coregulation of resistance and of virulence.
The pathogenicity of S. epidermidis may rely on the presence or absence of candidate genes that are involved in the virulence process. This was recently shown for the ica gene locus, which proved to be almost exclusively present in sepsis-causing strains and not detectable in saprophytic isolates (21). The purpose of the present study was to investigate whether the presence of ica and also that of the atlE, agrA, sarA, and mecA genes might discriminate between virulent S. epidermidis strains that cause real sepsis and nonvirulent S. epidermidis strains that contaminate blood cultures. To address this question, multiplex PCR amplification was used to compare the genetic backgrounds of strains collected from presumed sepsis and from catheter-related infections with those of blood culture-contaminating strains and of healthy carriage strains. Between September 1998 and March 1999, 138 S. epidermidis isolates were collected from 122 patients hospitalized in the University Hospital of Rouen, France, and from 16 healthy volunteers. The strains were intentionally selected for inclusion into four clinical groups. Group S included 39 strains that were potentially involved in a sepsis because they infected at least three distinct blood cultures or at least two blood cultures and one concomitant entry site and because the different isolates of each patient shared the same antibiotic resistance profile. Group C included 39 strains that were considered contaminating strains because they were isolated from only one blood culture and because they were not associated with any other S. epidermidis-positive cultures from potential entry sites of patients. Group K comprised 44 strains that significantly colonized intravascular devices. In our hospital, quantitative cultures of catheters are performed by rinsing the distal 6-cm segment of the catheter with 1 ml of broth and inoculating 100 μl of the broth on blood agar, and the cultures are considered significant when the bacterial count is ≥103 CFU/ml (3). Finally, 40 healthy volunteers, who did not attend the hospital, were asked to place their fingers on blood agar in order to collect saprophytic strains. However, S. epidermidis strains were isolated from only 16 of these individuals; the remaining 24 were colonized by other species of coagulase-negative staphylococci. Therefore, 16 saprophytic strains were included in group H as control strains for subsequent molecular analysis. All the strains of the study were identified by colony morphology, Gram stain characteristics, and results of the Pastorex Staph Plus test (Sanofi Diagnostics Pasteur, Marnes la Coquette, France) and the APISTAPH system (bioMérieux, La Balme les Grottes, France), performed according to the manufacturers' recommendations. In addition to the 138 strains of the study, 8 clinical isolates belonging to other coagulase-negative staphylococci species were analyzed to assess the specificities of the PCR primers.
Four pairs of primers were designed for amplification of fragments of the atlE, icaA, icaB, sarA, and agrA genes of S. epidermidis, with the help of previously published sequences (7, 10, 12, 17). The ica primers were designed to amplify the icaA and icaB genes of the ica locus, while Ziebuhr et al. used a Southern blot probe specific to the icaAB portion of the locus in a previous study (21). For amplification of the mecA gene, primers previously designed by Geha et al. (8) were used. For amplification of an internal control, we used universal primers targeting 16S rRNA genes (19). The nucleotide sequences of the primers are presented in Table 1. For DNA extraction, 10 μl of a 2× McFarland standard suspension of staphylococcal cells was placed in the amplification tube and submitted to a cell lysis program of a GeneAmp PCR system 2400 (Perkin-Elmer Cetus, Norwalk, Conn.). Subsequently, 40 μl of the PCR reagent mixture was added to the PCR tube to initiate amplification. The PCR reagent mixture consisted of 200 μM (each) dATP, dTTP, dCTP, and dGTP; 10 mM Tris (pH 8.3); 50 mM KCl; 1.5 mM MgCl2; 1.25 U of Taq polymerase (Perkin-Elmer Cetus) and 0.5 to 1 μM each PCR primer. Multiplex PCR was performed for combined amplification of the (i) atlE, icaA, icaB, and 16S rRNA genes; (ii) agrA, sarA, and 16S rRNA genes; and (iii) mecA and 16S rRNA genes. Therefore, each PCR included amplification of the 16S rRNA gene as an internal control. Each PCR was performed twice for confirmation of the results, and each experiment included a PCR-positive control strain and a negative control, consisting of the PCR mixture without bacterial DNA. The optimal primer concentration for multiplex amplification was 0.5 μM, with the exception of that for the agrA and 16S rRNA primers, which required a concentration of 1 μM. The optimal annealing temperature for all multiplex amplifications appeared to be 55°C. Finally, DNA amplifications were carried out with the following thermal cycling profile: an initial denaturation at 94°C for 2 min, followed by 30 cycles of amplification (denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min), and ending with a final extension at 72°C for 5 min. Amplification products were analyzed by agarose gel electrophoresis. Examples of the different multiplex amplifications are shown in Fig. 1. All the strains were amplifiable. With the exception of the mecA and universal 16S rRNA primers, all the PCR primers used in this study appeared to be specific to the S. epidermidis species. Under the high-stringency conditions, the atlE, icaAB, agrA, and sarA primers did not generate any PCR product for the following species: Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus warneri, Staphylococcus capitis, Staphylococcus lugdunensis, Staphylococcus saprophyticus, Staphylococcus xylosus, and Staphylococcus simulans. Statistical analysis of the PCR results was performed by using the chi-square test and the Fisher exact test.
TABLE 1.
DNA sequences of amplification primers
| Target gene(s) | Primera | DNA sequence (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| atlE | atlE-F | CAA CTG CTC AAC CGA GAA CA | 682 | This work |
| atlE-R | TTT GTA GAT GTT GTG CCC CA | |||
| icaAB | icaAB-F | TTA TCA ATG CCG CAG TTG TC | 546 | This work |
| icaAB-R | GTT TAA CGC GAG TGC GCT AT | |||
| sarA | sarA-F | TGG TCA CTT ATG CTG ACA GAT T | 313 | This work |
| sarA-R | TTT GCT TCT GTG ATA CGG TTG | |||
| agrA | agrA-F | CAA CAA CGA AAC ATG GTG CT | 923 | This work |
| agrA-R | TGT CAT CGA AAA TGG TTA CTT TG | |||
| mecA | mecA-F | GTA GAA ATG ACT GAA CGT CCG ATA A | 310 | 8 |
| mecA-R | CCA ATT CCA CAT TGT TTC GGT CTA A | |||
| 16S rRNA | 91E-F | GGA ATT CAA A(T/G)G AAT TGA CGG GGG C | 478 | 19 |
| 13B-R | CGG GAT CCC AGG CCC GGG AAC GTA TTC AC |
F, forward primer; R, reward primer.
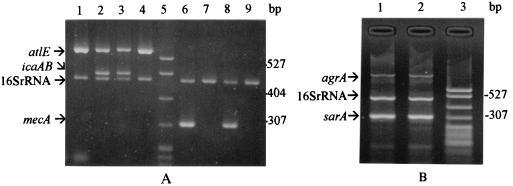
FIG. 1.
Multiplex amplifications of representative S. epidermidis strains. (A) Multiplex PCR of altE, icaAB, and 16S rRNA gene fragments (lanes 1 to 4) and of mecA and 16S rRNA gene fragments (lanes 6 to 9). Lanes 1, 4, 7, and 8, blood culture-contaminating isolates; lanes 2, 3, and 6, sepsis-related isolates; lane 5, molecular weight marker (pBR322 DNA-MspI digest); lane 9, healthy volunteer isolate. (B) Multiplex PCR of agrA, 16S rRNA, and sarA gene fragments. Lane 1, sepsis-related isolate; lane 2, blood culture-contaminating isolate; lane 3, molecular weight marker (pBR322 DNA-MspI digest).
The atlE gene, which encodes a vitronectin-binding cell surface protein involved in primary attachment (11), was ubiquitously amplified in S. epidermidis strains. The agrA and sarA genes may also be involved in the virulence of S. epidermidis, as was suggested by previous findings that insertional mutations in agr and mutations in both agr and sar respectively attenuate and nearly abolish the virulence of S. aureus in experimental endophthalmitis (2). Indeed, the agrA and sarA genes were amplified in almost all the infecting and contaminating strains, with the exception of only one catheter isolate lacking agrA, one sepsis isolate lacking sarA, and one contaminating isolate lacking both amplicons. As the great majority of strains harbor these genes, negative amplifications in a few isolates may be related to mutations of the annealing sequences and should be confirmed by Southern blot analysis. Although atlE, agrA, and sarA were quite ubiquitously found in the different groups of strains, the potential functions of their products in virulence cannot be excluded and neither can the possibility of point mutations or abnormal gene transcription in noninvasive strains.
In contrast, the amplification of the icaA, icaB, and mecA genes revealed striking differences between the different groups of strains. First, the ica locus was detected significantly more in infecting strains than in contaminating strains (P ≤ 0.003) (Table 2). These results are in agreement with those of a recent study in which S. epidermidis strains from clinical material were shown to differ from saprophytic strains by the presence of the icaA and icaB genes, their capacity for phase variation, their abilities to adhere to polymer and autoaggregate, and in their colony morphology on Congo red agar (21). Although phenotypic markers, such as culture on Congo red agar, also reflect the potential virulence of the strains and although phenotypic testing may be easier to perform than molecular analysis, the determination of phenotypes is hampered by the capacity of phase variants to change specific phenotypic features rapidly (5). Moreover, test tube adherence and in vitro slime production were shown to be of minor usefulness in guiding clinical decisions (15). Therefore, based on these previous findings, the detection of the ica gene locus is the most reliable means to address the discrimination of virulent and nonvirulent strains. In their study, Ziebuhr et al. demonstrated, by Southern hybridization with an icaAB probe, that the ica gene cluster was present in 44 of 52 (85%) blood culture isolates versus in only 2 of 36 (6%) saprophytic strains collected from healthy volunteers. This higher sensitivity for the detection of ica-positive infecting strains (85% versus 68.2 and 76.9% in our work) is relevant to the better sensitivity of Southern blot analysis, while PCR detection might be adversely affected by minor mutations. Incidentally, we found that the rate of strains carrying the ica locus among healthy volunteers (37.5%) was higher than the rate reported by Ziebuhr et al. (6%). Although the reason for this difference remains unclear, our result suggests that virulence factors can somehow be present in community strains and are not specific to nosocomial isolates. The innovative feature of the present study is the comparison of the ica PCR results between strains that potentially contaminate blood cultures and strains that potentially infect blood cultures or intravascular devices. The presence of the ica gene locus appeared to be statistically related to the potential virulence of the strains (Table 2). Although the detection of ica is neither sensitive nor specific enough to guide fully the clinical decision, it might be helpful when associated with other clinical and biological criteria of septicemia (15). In this aspect, further prospective investigations are needed and should include genetic, phenotypic, and clinical data. Moreover, as the PCR primers used in the present study appeared specific to S. epidermidis, further investigations are needed to establish the presence and determine the DNA sequences of ica homologs in various coagulase-negative species, such as S. haemolyticus, that are increasingly involved in sepsis (13).
TABLE 2.
Results of icaAB and mecA gene amplification grouped by the origins of the strains
| Origin of strainsa | No. of strains | % Positiveb for:
|
||
|---|---|---|---|---|
| icaAB | mecA | icaAB + mecA | ||
| S | 39 | 30 (76.9) A | 30 (76.9) C | 25 (64.1) E |
| K | 44 | 30 (68.2) B | 37 (84.0) D | 29 (65.9) F |
| C | 39 | 11 (28.2) A, B | 19 (48.7) C, D | 8 (20.5) E, F |
| H | 16 | 6 (37.5) | 4 (25.0) | 1 (6.2) |
Group S contains sepsis strains infecting at least three blood cultures or two blood cultures and one concomitant entry site and displaying the same antibiotic resistance profile for each patient. Group K contains strains significantly colonizing intravascular devices. Group C contains contaminating strains isolated from only one blood culture and not associated with positive culture of entry sites. Group H contains strains sampled from the hands of healthy volunteers.
Values with the same letter are statistically significantly different from each other as follows: A, P = 10−5; B, P = 0.003; C, P = 0.02; D, P = 10−3; E, P = 5 × 10−4; and F, P = 5 × 10−5.
Finally, the last candidate gene of the present work for discrimination between contaminating and invasive strains was the mecA gene, which controls the synthesis of the additional penicillin-binding protein PBP2′ in methicillin-resistant staphylococci. It is known that methicillin resistance is documented more often in disease-causing isolates than in colonizing isolates (1). Moreover, the lack of mecA transcription in slime-negative phase variants of methicillin-resistant S. epidermidis has suggested the possible implication of mecA gene regulation in pathogenicity (16). In the present study, mecA was found in almost half of the blood culture-contaminating strains and in more than 75% of the invasive strains. Despite the wide distribution of mecA among nosocomial staphylococci, the difference between the contaminating and invasive groups of strains was statistically significant for the presence of the mecA gene (P ≤ 0.02), in agreement with the results of a previous study reporting a higher rate of methicillin resistance in disease-causing strains than in colonizing isolates (1). However, although the presence of mecA was concordant with that of the ica locus for most of the invasive strains, the presence of one or both of these genes in 22 (56%) contaminating strains hampered the interest in a combined ica-mecA PCR. In any case, although the detection of mecA does not augment the interest in the detection of ica for the diagnosis of infection, it remains significant information for empiric antibiotic therapy. Unexpectedly, the mecA gene was amplified from four (25%) saprophytic strains sampled from the hands of healthy volunteers who did not attend the hospital. These data suggest the presence of methicillin-resistant S. epidermidis in the general population and might be somehow related to the increased incidence of methicillin-resistant S. aureus in the community (6). Whether the presence of the mecA gene in S. epidermidis strains of the healthy population reflects the dissemination of hospital strains or the role of antibiotics in food remains to be elucidated.
In conclusion, this study demonstrates the ability of the detection of the ica and mecA gene loci to discriminate between contaminating and infecting S. epidermidis strains. Although the ica and mecA PCRs lack sensitivity and specificity and cannot be considered biological tests, they may potentiate the clinical criteria used for the diagnosis of septicemia or catheter-related infections.
Acknowledgments
We gratefully thank Jean-François Menard for the performance of the statistical tests.
REFERENCES
- 1.Archer G L. Alteration of cutaneous staphylococcal flora as a consequence of antimicrobial prophylaxis. Rev Infect Dis. 1991;13(Suppl. 10):805–809. doi: 10.1093/clinids/13.supplement_10.s805. [DOI] [PubMed] [Google Scholar]
- 2.Booth M C, Cheung A L, Hatter K L, Jett B D, Callegan M C, Gilmore M S. Staphylococcal accessory gene regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65:1550–1556. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun-Buisson C, Abrouk F, Legrand P, Huet Y, Larabi S, Rapin M. Diagnosis of central venous catheter-related sepsis. Critical level of quantitative tip cultures. Arch Intern Med. 1987;147:873–877. [PubMed] [Google Scholar]
- 4.Christensen G D, Simpson W A, Bisno A L, Beachey E H. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen G D, Baddour L M, Madison B M, Parisi J T, Abraham S N, Hasty D L, Lowrance J H, Josephs J A, Simpson W A. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to β-lactam antibiotics, and virulence. J Infect Dis. 1990;161:1153–1159. doi: 10.1093/infdis/161.6.1153. [DOI] [PubMed] [Google Scholar]
- 6.Collignon P. Increased incidence of methicillin-resistant strains of Staphylococcus aureus in the community. J Infect Dis. 1999;179:1592. doi: 10.1086/314788. [DOI] [PubMed] [Google Scholar]
- 7.Fluckiger U, Wolz C, Cheung A L. Characterization of a sar homolog of Staphylococcus epidermidis. Infect Immun. 1998;66:2871–2878. doi: 10.1128/iai.66.6.2871-2878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geha D J, Uhl J R, Gustaferro C A, Persing D H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilmann C, Gerke C, Perdreau-Remington F, Götz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 11.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 12.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi N, Taniguchi K, Arasawa S. Analysis of diversity of mutations in the mecI gene and mecA promoter/operator region of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:717–720. doi: 10.1128/aac.42.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotilainen P. Association of coagulase-negative staphylococcal slime production and adherence with the development and outcome of adult septicemias. J Clin Microbiol. 1990;28:2779–2785. doi: 10.1128/jcm.28.12.2779-2785.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mempel M, Feucht H, Ziebuhr W, Endres M, Laufs R, Grüter L. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob Agents Chemother. 1994;38:1251–1255. doi: 10.1128/aac.38.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto M, Suusmuth R, Jung G, Götz F. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 1998;424:89–94. doi: 10.1016/s0014-5793(98)00145-8. [DOI] [PubMed] [Google Scholar]
- 18.Peng H L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1998;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;327:293–301. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 20.Vandenesch F, Projan S J, Kreiswirth B, Etienne J, Novick R P. Agr-related sequences in Staphylococcus lugdunensis. FEMS Microbiol Lett. 1993;111:115–122. doi: 10.1111/j.1574-6968.1993.tb06370.x. [DOI] [PubMed] [Google Scholar]
- 21.Ziebuhr W, Heilmann C, Götz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]