Abstract
In recent years, vaccine development using ribonucleic acid (RNA) has become the most promising and studied approach to produce safe and effective new vaccines, not only for prophylaxis but also as a treatment. The use of messenger RNA (mRNA) as an immunogenic has several advantages to vaccine development compared to other platforms, such as lower coast, the absence of cell cultures, and the possibility to combine different targets. During the COVID-19 pandemic, the use of mRNA as a vaccine became more relevant; two out of the four most widely applied vaccines against COVID-19 in the world are based on this platform. However, even though it presents advantages for vaccine application, mRNA technology faces several pivotal challenges to improve mRNA stability, delivery, and the potential to generate the related protein needed to induce a humoral- and T-cell-mediated immune response. The application of mRNA to vaccine development emerged as a powerful tool to fight against cancer and non-infectious and infectious diseases, for example, and represents a relevant research field for future decades. Based on these advantages, this review emphasizes mRNA and self-amplifying RNA (saRNA) for vaccine development, mainly to fight against COVID-19, together with the challenges related to this approach.
Keywords: RNA vaccines, mRNA vaccines, saRNA vaccines, COVID-19, SARS-CoV-2
1. Introduction
Vaccines are considered one of the most effective strategies to prevent, control, and eliminate infectious diseases [1,2]. The emergence of the new coronavirus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease of 2019 (COVID-19) [3], has highlighted the importance of vaccination for global health [4]. The virus was first reported in December 2019 in the city of Wuhan in China and, by the end of October 2021, its rapid spread caused the infection of [5] nearly 245 million people worldwide, leading to major economic and social impacts [6,7]. The high transmissibility of SARS-CoV-2 can be attributed to its unique characteristics, mainly human-to-human transmission that occurs by various mechanisms, even from asymptomatic carriers [8,9,10]. Although most COVID-19 patients present mild symptoms such as dry cough, fever, and fatigue [11], the hospitalization and mortality rates of the disease are higher when compared to those of diseases caused by other respiratory viruses, such as influenza [12,13]. The sheer number of patients and the rapid progression to life-threatening conditions has resulted in devastating overloads of health care systems in numerous parts of the world [14].
SARS-CoV-2 is a single-chain positive-sense RNA virus of the Betacoronavirus genera. The SARS-CoV-2 viral genome has 29.8 kilobases, with a G+C content of less than 40%, and is composed of six large open reading frames (ORFs) common to coronaviruses and two untranslated regions (UTRs) at the 5′ and 3′ ends [15]. Four structural proteins—membrane (M), envelope (E), spike (S), and nucleocapsid (N)—and sixteen non-structural proteins (nsp1-16) form the RNA genome of SARS-CoV-2 [16]. Among them, the S glycoprotein is an important target of therapies since it is responsible for entry into host cells via its interaction with the angiotensin-converting enzyme 2 (ACE2) cell receptor [17,18].
Early sequencing of the SARS-CoV-2 genome allowed for the prompt determination of its sequence identity/similarity with the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and SARS-CoV (both previously responsible for concerning outbreaks), and routine sequencing has facilitated the identification of new mutated SARS-CoV-2 variants-of-concern [19]. Numerous SARS-CoV-2 variants-of-concern have been identified, most notably, the B.1.1.7 (known as 501Y.V1), B.1.351 (known as 501Y.V2), and P.1 (known as 501Y.V3) variants that were first detected and identified in the United Kingdom, South Africa, and Brazil, respectively [20,21]. On May 31, 2021, the WHO (World Health Organization) decided to simplify the names of these variants-of-concern with Greek letters. Therefore, these four variants-of-concern are now called Alpha, Beta, Gamma, and Delta, respectively [22]. Variants-of-interest, with the potential to rise in status to variants-of-concern, continue to emerge. Sequencing of the SARS-CoV-2 genome patients has allowed rapid advances in basic research as well as product development, most notably with innovation in vaccine development [23,24,25,26].
International efforts to end the current pandemic have been unprecedented in terms of resource allocation, scientific focus, and the pace of innovation [27]. Given the potential to provide the population with the necessary immunity against the virus, the widespread use of a safe and effective vaccine has become the primary goal for controlling the SARS-CoV-2 pandemic [28]. Since the beginning of the pandemic, more than 100 clinical trials of COVID-19 vaccine candidates have been conducted, involving over 150 research groups [29]. The development of vaccines for COVID-19 has been supported by significant financial investment; for example, the U.S. government has provided more than USD 10.5 billion to vaccine companies to accelerate the delivery of their products [30]. Companies have developed vaccine candidates across a variety of technological platforms, including virus-like particle, recombinant protein, inactivated virus, live attenuated virus, viral vector (replicating and non-replicating), and nucleic acid (DNA and RNA) approaches [31,32].
RNA-based vaccines were among the first to emerge and have become prominent in national immunization programs. RNA vaccine technology builds on the central dogma of molecular biology, in which messenger RNA (mRNA) is the intermediate step between the translation of the encoding DNA and the production of its respective protein. It is a technology that enables the carriage of genetic information directly into the cell, allowing endogenous protein expression instead of administering protein (antigen) as an exogenous entity such as killed or defined subunit platforms [33]. Moreover, due to its capacity to activate various pattern-recognition receptors, RNA can be very immunogenic [34]. Another logistical advantage of RNA-based vaccines is that the RNA can be produced in a cell-free environment by in vitro transcription (IVT), removing the need for cultured cells in the manufacturing process and removing the associated quality and safety issues associated with them [35]. In this way, it is possible to perform simple downstream purification to provide rapid and cost-effective manufacturing relative to other vaccine platforms [36]. Several other advantages, such as scalability, flexibility in manipulating antigens of interest, and the induction of both cellular and humoral immunity [31,37], are inherent to RNA vaccines. These characteristics previously enabled RNA-based vaccines to be evaluated against non-infectious diseases such as cancer and allowed their manufacturers to rapidly respond to emerging infectious agents such as SARS-CoV-2 [38]. For this reason, RNA-based vaccines have become attractive in the pandemic situation. Therefore, considering the potential presented by RNA-based vaccines, in this review, we provide a brief history and evaluate RNA-based vaccines within the context of the COVID-19 pandemic, describing the prospects and challenges related to their use for immunization of large populations against SARS-CoV-2.
2. A Brief History of RNA-Based Vaccines
As with other platforms, the development of RNA-based vaccines involves antigen discovery and analysis of the nucleotide sequence that will be translated into the protein of interest. For RNA-based vaccines, screening of modified nucleotides can optimize expression, as can the selection of an appropriate method [39]. Various methods have been used, such as modified nucleosides and the development of nanoparticles capable of stabilizing the RNA and/or improving its cellular uptake and, consequently, improving the overall bioavailability of the RNA cargo [40].
Studies that served as a basis for the development of synthetic RNA vaccines were reported from the end of the twentieth century and were motivated by the 1961 discovery of mRNA [41]. An important milestone for the use of RNA in the pharmaceutical industry occurred in 1989, when researchers from Vical Incorporated and the Salk Institute demonstrated that mRNA introduced by a liposomal nanoparticle could transfect different types of eukaryotic cells [42]. In 1990, Wolff et al. [43] reported that the injection of mRNA without a protective complex could induce protein expression within a few days. The first studies of mRNA and self-amplifying (saRNA) as vaccines demonstrated cell-mediated and humoral-adaptive immune responses, respectively, against the influenza A virus [44,45]. This was followed by studies of mRNA as immunotherapies focused on oncology with in vitro and in vivo assays, using both protected or unprotected (“naked”) mRNA [46,47,48]. It is important to note that the basic difference between these two types of RNA is associated with the number of replications and, consequently, the expression of the antigen. By presenting additional sequences in the coding region, saRNA has a self-amplification mechanism, which can result in an increase in the transcription process when compared to mRNA [49]. In addition, mRNA or saRNA are produced by IVT and, besides the advantages mentioned above, the RNA sequence can be easily modified to improve the protein synthesis, such as the addition of a 30 poly(A) tail, a capping approach, and methylated nucleosides or pseudouridine [50].
The amount of protein (antigen) expressed is directly related to the content of mRNA that can enter the intracellular environment. The presence of well-defined mRNA in the cytosol allows the presentation of exogenous and endogenous antigens to occur and can provide activation of the immune system via different pathways [51,52]. However, it has been shown that naked mRNA is more susceptible to enzymatic hydrolysis (especially by omnipresent ribonucleases), which can directly compromise its potency; since any degradation in its structure can result in the incomplete expression of the antigen [53,54]. After this discovery, different studies were conducted to develop delivery vehicles for the mRNA that would protect the molecule from degradation and improve the induction of the immune system [55,56,57]. Thus, several in vivo studies using different types of delivery vehicles have been performed in an attempt to develop a safe and effective RNA vaccine [58,59]. However, some strategies have demonstrated a lack of immunogenicity in primates and humans in contrast to small animal models (also called a “primate barrier”), making the choice of the delivery vehicle an important bottleneck in the clinical use of RNA vaccines [60]. Recently, the use of lipid nanoparticles (LNPs) as a delivery system has been the main focus for RNA vaccine development [61]. In 2007, de Jong et al. [62] demonstrated that LPN-encapsulated antigen can induce a stronger immune response and enhance immune efficacy. Since then, the importance of this delivery system in the development of safe and effective vaccines in different routes of administration has been reported, directly contributing to the advancement in the use of mRNA vaccines.
These studies were of great importance in establishing benchmarks for further evaluation and, consequently, assisting in the process of optimizing these vaccines, providing essential safety and immunogenicity, and providing a basis for the production of RNA vaccines in accordance with good manufacturing practices [63,64]. In this context, the most important innovations in RNA-based vaccine technology in recent years have been directed toward the use of optimized RNA sequencing, the application of methods that allow large-scale cGMP production, and the development of efficient and safe materials for RNA delivery [38,65].
2.1. RNA Vaccines against Cancer
RNA-based cancer vaccines have been designed to express tumor-associated antigens, resulting in the stimulation of T-cell-mediated immune responses [66]. Acute myeloid leukemia, brain cancer, colorectal cancer, liver metastases, esophagus cancer, glioblastoma, prostate cancer, and melanoma are all examples of clinical conditions targeted by RNA-based cancer vaccine candidates [67], with most applied in a therapeutic rather than a prophylactic manner [68,69]. To date, most RNA-based cancer vaccines have used non-replicating mRNA technology [70,71]. One of the first phase I/II studies involved the direct injection of mRNA into melanoma patients: while a 200-microgram injection of naked mRNA was safe, clinical efficacy was not demonstrated [72]. Soon after, the same group published a phase I/II trial of a protamine-protected mRNA vaccine in metastatic melanoma patients (NCT00204607), demonstrating that protamine-protected mRNA was not only safe but generated more promising clinical efficacy [73]. These trials thereby addressed the reported weakness of RNA based-vaccines being easily degraded by omnipresent ribonucleases and demonstrated the importance of the delivery format in related outcomes, prompting research regarding novel encapsulation/delivery strategies, such as the use of biopolymers, liposomes, or dendritic cells (DCs) [65,74].
Before studies in humans, in vitro and in vivo studies using DCs electroporated with mRNA assessed their ability to trigger potent immune responses against tumor antigens [47]. Boczkowski et al. showed the capacity of RNA-pulsed DC-based vaccines to reduce lung metastases in rats. Early human trials to evaluate mRNA delivery used monocyte-derived DCs transfected ex vivo with antigen-encoding mRNA by electroporation to provide a cell-based vaccine approach, with the cells being re-infused into patients [75]. Since then, many phase I/II clinical trials using this delivery strategy have been published, with prostate cancer, melanoma [76], B-cell lymphoma [77], adenocarcinoma [78], and pancreatic cancer [79] being among the examples of the therapeutic targets.
The RNActive® vaccine platform (WO2002098443, WO2012019780), designed by CureVac, uses an mRNA complex with protamine and naked mRNA, where protamine assists in the process of inducing cellular immunity [80]. Furthermore, it is important to note that the ability to express the antigen within this complex is directly associated with the relationship between the mRNA and the protamine [81]. The platform has proven to be highly versatile, allowing the translation of different antigens of interest. Two cancer vaccines based on this technology have undergone phase 1/2 clinical trials: CV-9104, tested in patients with castration-resistant prostate cancer (NCT01817738) [82] and CV-9201, tested in patients with non-small-cell lung carcinoma (NCT00923312) [83]. Table 1 presents examples of clinical trials involving RNA-based vaccines against different cancers.
Table 1.
Clinical trials with RNA-based vaccines against cancer. Data collected from the studies registered at ClinicalTrials.gov as of 8 August 2021.
| Sponsor | NCT-Number | Country | Vaccine Type | Targets | Trial Phase | Status | Reference |
|---|---|---|---|---|---|---|---|
| Argos Therapeutics | NCT00087984 | USA and Canada | DCs electroporated with autologous tumor mRNA | Metastatic renal cell carcinoma | 1/2 | Completed | NA |
| NCT00664482 | USA | Pancreatic cancer | Not Applicable | ||||
| University Hospital Tuebingen | NCT00204516 | Germany | Naked mRNA | Melanoma | 1/2 | Completed | NA |
| Radboud University | NCT00228189 | Netherlands | DCs electroporated with tumor-associated antigen mRNA | Colorectal cancer | 1/2 | Completed | [84] |
| Medigene AG | NCT02405338 | Norway | Autologous DCs with tumor-associated antigen mRNA | Acute myeloid leukemia | 1/2 | Completed | [85] |
| BioNTech RNA Pharmaceuticals | NCT02035956 | Austria and Germany | Naked RNA | Melanoma | 1 | Completed | [86] |
| Universitair Ziekenhuis Brussel | NCT01302496 | Belgium | DCs electroporated with tumor-associated antigen and TriMix (CD70, CD40L, TLR4) mRNA | Melanoma | 2 | Completed | [87] |
| Herlev Hospital | NCT00978913 | Denmark | DCs loaded with tumor-associated antigen mRNA | Breast cancer, melanoma | 1 | Completed | [88] |
| NCT01446731 | Prostate cancer | 2 | [89] | ||||
| Duke Cancer Institute | NCT01890213 | USA | Alphavirus replicon encoding the tumor-associated antigen | Colon cancer | 1 | Completed | [90] |
| NCT00003433 | Antigen-RNA-pulsed DCs | IV colon cancer, liver metastases |
1/2 | NA | |||
| NCT00003432 | Breast cancer | Terminated (low accrual) | |||||
| University Hospital Erlangen | NCT01983748 | Germany | DCs loaded with autologous tumor mRNA | Melanoma | 3 | Recruiting | NA |
| University Hospital, Antwerp | NCT02649582 | Belgium | DCs electroporated with tumor-associated antigen mRNA | Glioblastoma | 1/2 | Recruiting | NA |
| Changhai Hospital | NCT03468244 | China | Naked mRNA | Advanced esophageal squamous carcinoma, gastric, colorectal, and pancreatic adenocarcinomas |
Not Applicable | Recruiting | NA |
| University of Campinas | NCT03083054 | Brazil | DCs electroporated with tumor-associated antigen mRNA | Acute myeloid leukemia | 1/2 | Active, not recruiting | NA |
| Guangdong 999 Brain Hospital | NCT02808416 | China | DCs pulsed with tumor-associated antigen mRNA | Brain metastases | 2 | Active, not recruiting | NA |
| NCT02709616 | Glioblastoma | 1 | |||||
| Life Research Technologies | NCT01456065 | Hungary and Austria | DCs loaded with tumor-associated antigen mRNA | Ovarian cancer | 1 | Unknown | NA |
DCs, dendritic cells; NA, not available; USA, United States.
2.2. RNA Vaccines against Non-Infectious Diseases
In addition to the applications in cancer and non-infection diseases, it is important to mention that RNA-based vaccines also have a great potential to be applied to the treatment and prophylaxis of non-infectious diseases, such as autoimmune and allergic diseases [91,92]. These diseases, while apparently diverse, share a common characteristic of an undesired and inappropriate immune response. Therefore, in a different way than the approach used for non-infectious disease and cancer, which is based on the principle that RNA, together with its formulation, provides immune stimulations of T-cell and antibody responses [65], in the case of non-infectious diseases, the goal is to suppress an immune response.
Companies specializing in therapeutics with mRNA have sought solutions to a wide range of health conditions. When it comes to autoimmune disorders, previous studies have shown that mRNA therapy has a high potential application in the treatment of these diseases [93,94,95]. It is important to note that there are over 100 distinct autoimmune disorders, and they are highly complex, where for each one, there may be a different treatment approach [96]. In general, for autoimmune disease applications, an mRNA-based vaccine acts by suppressing antigen-specific immune responses [91]. A recent study performed by BioNTech RNA Pharmaceuticals researchers [91] described the disease-suppressing effects of a non-inflammatory mRNA vaccine in mice models of multiple sclerosis. It was demonstrated that the delivery of the autoimmune target antigen by the mRNA vaccine candidate into antigen-presenting cells in the lymph nodes resulted in the prevention of disease symptoms, a reduction in the disease progression, and the restoration of motor functions [91]. This study highlights that mRNA therapy has the potential to treat autoimmune diseases by increasing immune cell tolerance and consequently reducing damage without jeopardizing the immune system functions. Another important approach for mRNA-based vaccines in the autoimmune disease context relies on the application of mRNA to encode the immunomodulation of signal molecules such as cytokines. Veiga et al. demonstrated the application of mRNA encoding IL-10 as an alternative to traditional recombinant protein therapies in inflammatory bowel diseases. The expression of IL-10 in target cells resulted in a significant decrease in pathological symptoms and in the severity of intestinal inflammation. It is important to mention Moderna’s potential mRNA medicine, mRNA-6231, which was designed to trigger peripheral tolerance pathways to restore immune homeostasis and reduce autoimmune pathology by buffering autoimmune activation. mRNA-6231 encodes for IL-2, which mutein designs to activate and expand regulatory T cells, buffering the immune response [97]. The clinical trial to evaluate the safety and tolerability of mRNA-6231 is underway (NCT04916431).
When it comes to RNA-based vaccine applications for allergic diseases therapy, in contrast to cancer or autoimmune diseases applications, vaccination against allergy does not involve the administration of self-antigens [96]. It is generally accepted that allergic reactions are triggered by recurring exposure to allergens that will lead to the production of allergen-specific IgE antibodies and the subsequent activation of inflammatory cell responses by allergen–IgE immune complexes [98]. Therefore, allergic diseases are treated with allergen-specific immunotherapy since this is the treatment that can alter the immunological basis of allergic diseases with long-term effects, and no preventive vaccination against type I allergy is available [99,100,101]. mRNA-based vaccines are studied as an alternative for allergen-specific immunotherapy since, through this technology, it is possible to deliver the allergen in a pure form and in lower doses, therefore decreasing the risk of anaphylactic side effects caused by pre-existing IgE and the occurrence of the production of any novel allergen-specific to IgE. Roesler et al. demonstrated that mRNA vaccine expression of important 29 allergens could protect the induction of IgE in animal models. In this study, the benefits of mRNA vaccination were also seen in the downregulation of inflammatory lung parameters [102]. Moreover, Hattinger et al. [103] demonstrated that vaccination with mRNA vaccines was responsible for preventing an allergen-specific response by immunomodulating the TH2-type response by suppressing TH2 cytokines, eosinophils, and IgE expression, and increasing TH1-type parameters.
Although mRNA vaccines were in the spotlight during the COVID-19 pandemic, it is important to note that much work has already been completed by using this technology platform in the treatment and prophylaxis of non-infectious diseases. Such studies demonstrate the high potential of RNA vaccine application.
2.3. RNA Vaccines against Infectious Diseases
In addition to the push for RNA-based vaccines to fight cancer, in recent years, the use of these platforms has gained prominence against infectious agents, especially against emerging infectious diseases such as those caused by the Zika virus, Zaire Ebolavirus, and coronavirus [49]. In general, RNA-based vaccines against pathogens are developed through four main steps: (1) construction of an optimized sequence (capable of enhancing immunogenicity) of antigen-encoding mRNA based on the selected antigen(s) of the target pathogen; (2) determination of the delivery material, in either the presence or absence of adjuvant molecules, and influenced by the route of administration; (3) demonstration of the in vivo expression of the encoded antigen); and (4) evaluation of immune induction [67,104]. Unlike the predominance of the conventional mRNA approach against cancer vaccines, saRNA technology has been more widely evaluated against infectious diseases [105].
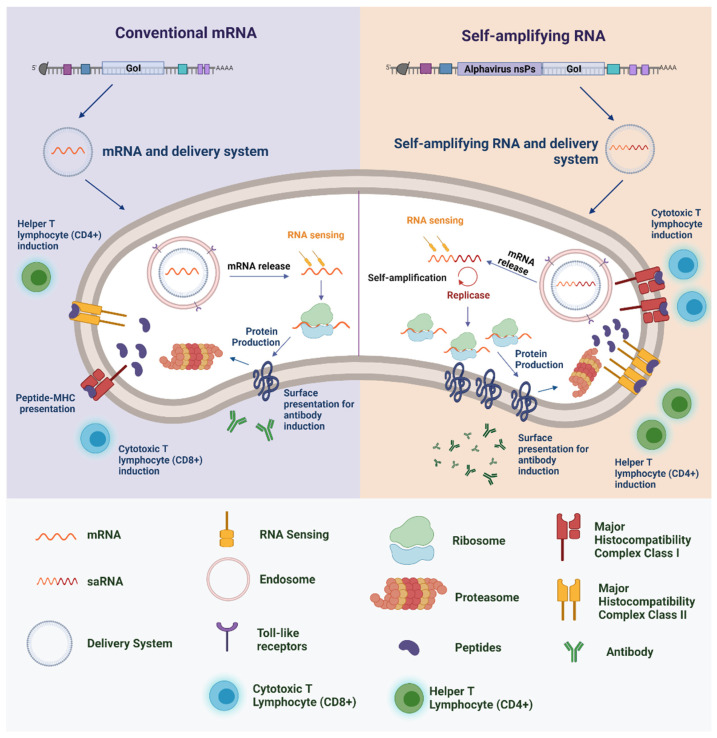
saRNA replicons are created by replacing the structural genes of, typically, an alphavirus (such as Semliki Forest virus (SFV), Sindbis virus (SINV), or Venezuelan equine encephalitis virus (VEEV) with the gene for the antigen of interest. It is important to note that the absence of endogenous viral structural genes in the replicons means that the production of infectious virions or virus-like vesicles in individuals after vaccination is negated, increasing the safety profile when compared to vaccine technologies such as attenuated virus [104]. When delivered into the cytoplasm of target cells, saRNA becomes capable of amplifying the mRNA to express the target antigen at very high levels [104,106,107]. Through this self-amplification system, it has already been estimated that 200,000 copies of RNA can be made from a single saRNA molecule, resulting in higher levels of protein expression relative to those achieved by conventional mRNA (Figure 1) [108]. Thus, vaccines based on saRNA technology can induce high levels of immunity even when administered in low amounts [109].
Figure 1.
Overview of mRNA and saRNA-based vaccine mechanisms for protein production. Adapted from Maruggi et al. [104]. GoI, gene of interest; MHC, major histocompatibility complex; nsPs, nonstructural proteins. Created with BioRender.com (accessed on 30 September 2021).
RNA-based vaccines have been used to deliver bacterial and parasite genes but, except for vaccine candidates for Chlamydia trachomatis [110], most of these remain in pre-clinical or early clinical stages of development (e.g., those against the protozoan Toxoplasma gondii [111], Plasmodium [112], and Leishmania donovani [113]). Recently, Raj et al. [114] developed an RNA vaccine for the expression of the glutamic-acid-rich protein (PfGARP) of Plasmodium falciparum and showed that it induced antibody formation in in vitro assays and in a non-human primate challenge model. Maruggi et al. [115] reported that saRNA vaccines based on bacterial antigens from Group A (GAS) and Group B (GBS) Streptococci induced protective efficacy in mice through the induction of functional antibodies.
RNA-based vaccines have advanced further in the context of viral infections, most notably for SARS-CoV-2 but also including other respiratory viruses (SARS-CoV-1) [116], insect-transmitted viruses (Zika, dengue, and Chikungunya) [117,118,119], animal-transmitted viruses (rabies) [120], as well as viruses transmitted by direct contact with humans or their fluids (e.g., Ebola [121], herpes (herpes simplex virus and human cytomegalovirus) [122,123], and HIV [124]) (Table 2). A pioneering study involving RNA vaccines for infectious agents was conducted by Fleeton et al. [125] against three viruses: influenza A virus, a tick-borne flavivirus (louping ill virus), and respiratory syncytial virus (RSV). In vivo experiments with a mouse model showed that the RNA vaccine encoding the envelope proteins of the three viruses afforded protection against each virus evaluated [125]. Just over a decade later, Petsch et al. [126] published important preclinical data on RNActive® platform-based influenza A virus infection vaccines, demonstrating that the vaccines promoted balanced, long-lived, and protective immunity against infection with influenza A virus in both small animals (mice and ferrets) and large animals (pigs) [126]. RNA vaccines can induce protective immunity against several influenza viruses and prolong the immune responses [50,127], and RNA-based vaccines are now being considered as alternatives that could overcome the bottlenecks typically faced by the conventional influenza vaccine.
Table 2.
Clinical trials with RNA-based vaccines against infectious diseases. Data collected from the studies registered at ClinicalTrials.gov as of 8 August 2021.
| Sponsor | NCT-Number | Country | Vaccine Type | Targets | Trial Phase | Status | Reference |
|---|---|---|---|---|---|---|---|
| Argos Therapeutics | NCT00672191 | United States and Canada | DCs loaded with autologous viral antigen and CD40L mRNAs | HIV | 2 | Completed | [128] |
| Massachusetts General Hospital | NCT00833781 | United States | Autologous DCs loaded with viral antigen mRNA | HIV | 2 | Completed | [129] |
| CureVac AG | NCT02241135 | Germany | RNActive viral Ag mRNA | Rabies | 1 | Completed | [130] |
| Moderna Therapeutics | NCT03392389 | United States | Lipid-nanoparticle-encapsulated, chemically modified viral antigen mRNA | Human Metapneumovirus and Human Parainfluenza | 1 | Completed | [131] |
| NCT03014089 | Zika | NA | |||||
| NCT03325075 | Chikungunya | [132] | |||||
| NCT03382405 | Cytomegalovirus | NA | |||||
| NCT03076385 | Germany | Influenza | [133] | ||||
| Fundacion Clinic per a la Recerca Biomédica | NCT02413645 | Spain | Naked Viral antigen and TriMix (CD40, CD70 and IL2) mRNA | HIV | 1 | Completed | [134] |
| AlphaVax | NCT00440362 | United States | Alphavirus replicon vaccine expressing a viral hemagglutinin protein | Influenza | 1/2 | Completed | NA |
| NCT00439803 | Alphavirus replicon vaccine expressing viral genes | Cytomegalovirus | 1 | [135] | |||
| NCT00097838 | United States, Botswana, and South Africa | Alphavirus replicon vaccine expressing viral protein | HIV | [136] |
DCs, dendritic cells; HIV, human immunodeficiency virus; NA, not available.
3. RNA Vaccines in the Context of COVID-19
The response to the COVID-19 pandemic has delivered many important milestones in vaccine development, especially for RNA-based platforms. Numerous vaccine candidates were proposed in record time, with the rapid-development process guided by the knowledge acquired from coronavirus targets that had already been used in successful vaccines for humans [137]. Almost seven months after initiating a clinical development program, the BNT162b2 vaccine produced by Pfizer in partnership with BioNTech became the first vaccine approved by United States FDA for emergency use approval (EUA) against COVID-19 [138,139]. Shortly after, Moderna’s mRNA-1273 vaccine, developed in collaboration with the National Institute Allergy and Infectious Diseases (NIAID)/National Institutes of Health (NIH), became the second COVID-19 vaccine to receive EUA in the United States [140] and represented the first approval of RNA-based vaccines.
3.1. The Main RNA-Based Vaccines for COVID-19
RNA-based vaccines were among the first candidates to enter preclinical and clinical development against COVID-19. According to the WHO, as of October 1, there were at least 194 preclinical studies of COVID-19 vaccine candidates, of which 24 included RNA-based vaccines [141]. These studies highlight the versatility of the RNA platform and have been characterized by the use of mRNA or saRNA, as well as the use of naked or nanostructured lipid-carrier delivery strategies (Table 3). Another important point is that the translated protein structure can also be modified through mRNA or saRNA ribonucleotides’ alteration, aiming to improve protein stability, immunogenicity, and the generation of neutralizing antibodies. Pallesen et al. [142] showed the structure-based design of S protein from MERS-CoV to generate a more efficient immunogenic, which was the Spike protein stabilized in the prefused protein. Their study demonstrated that genetic vaccination can be easily modified and adapted to a specific prophylactic use.
Table 3.
Overview of preclinical studies of RNA-based COVID-19 vaccine candidates.
| Developer | Vaccine Name | Active Substance (Antigen Type) and Formulation Details | Animal Model | Main Findings and/or Conclusions Considering | Reference |
|---|---|---|---|---|---|
| CureVac AG | CVnCoV | Lipid-nanoparticle-encapsulated mRNA that encodes full-length, prefusion-stabilized SARS-CoV-2 S protein | Mice (BALB/c) | Mice: innate immune response (systemic IL-6 and IFNα and robust cellular and humoral immune responses Hamsters: protected against challenge with wild-type SARS-CoV-2 NHP: those vaccinated with 8 µg were protected from challenge with wild-type SARS-CoV-2 (Victoria/1/2020) |
[146] |
| Syrian Hamsters (Mesocricetus auratus) | |||||
| Rhesus macaques (Macaca mulatta) | [148] | ||||
| Arcturus Therapeutics | ARCT-021 (LUNAR-COV19) |
Lipid-nanoparticle-encapsulated saRNA that encodes an alphavirus-based replicon and the SARS-CoV-2 full-length S glycoprotein (saRNA) | Mice (C57BL/6 (human ACE2 transgenic mouse model) and BALB/c) | Mice: single vaccination led to robust antibody responses Human ACE2 transgenic mice: protected from mortality and measurable infection following wild-type SARS-CoV-2 challenge |
[149] |
| Imperial College London | COVAC1 (LNP-nCoVsaRNA) | saRNA encoding SARS-CoV-2 S protein encapsulated within a cationic liposom | Mice (BALB/c) | Mice: high cellular responses (IFN-γ production), specific IgG | [143] |
| Pfizer and BioNTech | BNT162b1 | Nucleoside-modified mRNA that encodes immunogens derived from the S protein of SARS-CoV-2 stabilized in the prefusion conformation, formulated in lipid nanoparticle | Mice (BALB/c) | Mice: one intramuscular dose of either elicited dose-dependent antibody responses, strong CD4+, and CD8+ T-cell response NHP: protected macaques against challenge with wild-type SARS-CoV-2 |
[144] |
| BNT162b2 (known commercially as Comirnaty) |
Rhesus macaques (Macaca mulatta) | ||||
| HDT Bio | HDT-301 (LION/repRNA-CoV2S) |
saRNA that encodes SARS-CoV-2 full S protein formulated with alipid inorganic nanoparticle (LION) emulsion | Mice (BALB/c and C57BL/6) | Mice: single immunization triggered robust IgG and Th1 cells Aged mice: induced S-specific IgG and Th1 responses NHP: antibody responses persisted for at least 70 days, neutralized SARS-CoV-2 at titers comparable to those of convalescent plasma |
[145] |
| Pigtail macaque (Macaca nemestrina) | |||||
| Moderna | mRNA-1273 (known commercially as Spikevax) | Nucleoside-modified mRNA that encodes prefusion-stabilized SARS-CoV-2 encapsulated in a lipid nanoparticle | Rhesus macaques (Macaca mulatta) | NHP: induced robust neutralizing activity against SARS-CoV-2, rapid protection in the upper and lower airways without pathological changes in the lung after challenge | [150] |
| PLA Academy of Military Sciences, Abogen and Walvax | ARCoV | Lipid-nanoparticle-encapsulated mRNA encoding the receptor-binding domain of SARS-CoV-2 S protein | Mice (BALB/c) | Mice: neutralizing antibodies against SARS-CoV-2, cellular responses (Th1 cells) Two doses conferred complete protection against the challenge (wild-type SARS-CoV-2) NHP: Th1 responses, neutralizing antibodies against SARS-CoV-2 Note: manufactured as a liquid formulation that can be stored at room temperature for at least 1 week |
[147] |
| Cynomolgus monkeys (Macaca fascicularis) |
LUNAR: Lipid-Enabled and Unlocked Nucleomonomer Agent-Modified RNA; PLA: People’s Liberation Army; Names in italics represent scientific names.
In general, preclinical studies involving RNA-based vaccines have demonstrated that this technology can induce both humoral and cellular immune responses, with sufficient data to support advancement to clinical development phases [143,144,145,146,147]. To evaluate the immunogenicity of COVID-19 vaccine candidates, the induction of antigen-specific immunity (both antigen-binding IgG and virus-neutralizing antibodies) has been determined in animal models such as mice and non-human primates (Table 3). Accompanying the induction of circulating antibodies, the cellular responses involving CD4+ and CD8+ T cells have also typically been evaluated.
Moderna’s mRNA-1273 was the first COVID-19 vaccine candidate to enter the clinical phase in the Western hemisphere. Its rapid development was made possible by the technological advances achieved through the earlier use of the company’s platform for vaccines against other viruses (Table 2), as well as the important partnership with NAID, which previously resulted in different patent applications for vaccines for other infectious diseases, including Chikungunya (WO2017070624), Zika (WO2018151816), and Ebola (WO2017015457) [151]. Table 4 provides an overview of the clinical development of RNA-based COVID-19 vaccine candidates.
Table 4.
Overview of clinical trials involving the most advanced COVID-19 RNA vaccines.
| Vaccine | Trial Phase | Location (NCT Number) |
Doses | Dose Level | Main Immunogenicity and/or Efficacy Findings | Main Safety Findings |
Reference |
|---|---|---|---|---|---|---|---|
| mRNA-1273 (Spikevax) |
1 | USA | 2 (28 days apart) |
25 μg, 100 μg, or 250 μg | Dose-dependent responses observed. Antibody GMT of 40,227, 109,209, and 213,526 in 25 μg, 100 μg, and 250 μg recipients, respectively. After second vaccination, titers increased to 299,751, 782,719, and 1,192,154 for 25 μg, 100 μg, or 250 μg, respectively. | AEs were commonly reported at the highest doses and after the second immunization. Systemic and local AEs occurred after both injections (fatigue, chills, headache, myalgia, and injection site pain). No AEs were noted; no prespecified halting rules were met. | [163] |
| 2 | USA | 50 μg or 100 μg | The 100 μg dose induced a > binding antibody concentrations than 50 μg. | The most common solicited AEs were injection site pain, headache, and fatigue. | [164] | ||
| 3 | USA | 100 μg | There was 94.1% efficacy (95% CI, 89.3 to 96.8%; p < 0.001) for the prevention of symptomatic SARS-CoV-2 infection within two weeks of second dose. | AEs were rare and occurred with the same incidence as the placebo. The most common treatment-related AEs were fatigue, headache, and injection-site pain. | [158] | ||
| BNT162b2 (Comirnaty) |
1/2 | Germany (NCT04380701) USA (NCT04380701) |
2 (21 days apart) |
10 µg, 20 µg, 30 µg | Dose-dependent nAbs > convalescent serum. (1.7 to 4.6-fold higher in 18–55-year-olds, 1.1–2.2-fold higher in 65-to-85-year-olds. SARS-CoV-2 antigen-specific CD4+ and CD8+ T cells. | Lower incidence and severity of systemic reactions, especially in older adults (65 to 85 years). Transient local reactions and systemic events were dose-dependent, greater after the second dose. | [165] |
| 3 | USA, Argentina, Brazil, Germany, South Africa, Turkey (NCT04368728) |
30 μg | There was 95% efficacy (95% CI 90.3–97.6) evaluated 7 days after second dose. Efficacy in participants reported comorbidity of 94.7%. | 16–55-year-olds experienced more systemic effects than >55-year-olds. Headache and fatigue were the most common systemic effects of local, injection-site pain. | [166] | ||
| CVnCoV | 1 | Germany (NCT04449276) |
2 (28 days apart) |
2 µg, 4 µg, 6 µg, 8 µg, or 12 µg | nAbs and IgG S protein or RBD-binding IgG two weeks after the second 12 μg dose comparable to convalescent plasma. | Dose-dependent increase in the frequency and severity of systemic AEs and local reactions; most were mild or moderate and transient. No vaccine-related SAEs were reported. | [167] |
| 2/3 | Argentina, Belgium, Colombia, Dominican Republic, Germany, Mexico, Netherlands, Panama, Peru, and Spain (NCT04652102) |
12 µg | Reported 47% efficacy versus disease. | Data not yet released. | [168] | ||
| ARCT-021 (LUNAR-COV19) |
1 | Singapore (NCT04480957) |
1 | 1 μg, 5 μg, 7.5 μg, and 10 μg | nAbs increased dose, similar following one 5 μg or 7.5 μg dose. S-binding IgG titers overlapped with levels in convalescent plasma (except1 μg dose). | The 10 μg dose was associated with more local and systemic solicited AEs; 7.5 μg was well tolerated. Fatigue, headache, myalgia, chills, and fever were the most common AEs. | [169] |
| 2 | 2 (28 days apart) |
3 μg and 5 μg | nAbs increased with dose. No increase in CD8 IFN-γ T cells was reported after the second dose. | Fatigue and arthralgia occurred in a single participant following the second 5 μg dose (56–80-year-old cohort). |
AEs: adverse events; GMT, geometric mean titer; nAb, SARS-CoV-2 neutralizing antibody RBD, receptor-binding domain.
mRNA-1273 contains a nucleoside-modified mRNA that encodes the stabilized prefusion SARS-CoV-2 spike protein (S-2P) based on the originally identified Wuhan lineage virus (GenBank accession number MN908947.3) that is encapsulated in lipid nanoparticles. mRNA-1273 is administered intramuscularly in two doses scheduled 28 days apart [152]. The phase 1 clinical trial of mRNA-1273 was an open-label, dose-ranging trial that evaluated the safety, reactogenicity, and immunogenicity of the vaccine in 45 healthy adults (18 to 55 years of age) (NCT04283461). This trial started on 16 March in the United States, only 5 days after the World Health Organization (WHO) classified COVID-19 as a pandemic [153] and only 66 days after the release of the genetic sequence of SARS-CoV-2 [154]. Subsequently, a further phase 1 study included 40 healthy adults aged >56 years, stratified according to age (56 to 70 years or ≥71 years) (NCT04283461), with immunological and safety profiles similar to those reported in the phase 1 study with younger adults [155]. The phase 2 (NCT04405076) and 3 (NCT04470427) trials started in sequence on May 29 and on July 20 in adults aged 18 years and older with 600 and 30,420 participants, respectively.
Given the rapid progression, the long-term safety and durability of the humoral immune response elicited by mRNA-1273 are only now becoming clear. Available data demonstrate that circulating neutralizing and binding antibody titers at day 119 after the first vaccination (90 days after the second vaccination) are still at concentrations capable of providing protection in participants of different ages [156]. In the first phase 3 trial of mRNA-1273 (NCT04470427), the inclusion of different races or ethnic groups was reported, although each of it is 99 research centers were located in the United States. Clinical trials in various regions of the world have become a common practice in recent years, allowing the analysis of safety and efficacy data within groups from different locations [157]. Moderna then progressed to two phase 2/3 studies in participants under 18 years of age, assessing the safety, reactogenicity, and effectiveness of mRNA-1273 in adolescents 12 to <18 years old (NCT04649151) and in healthy children between 6 months and <12 years of age (NCT04796896), respectively. Again, these trials are exclusive to the United States.
Although the phase 3 trial of mRNA-1273 included patients from several risk groups for developing severe COVID-19, such as diabetics, hypertensive individuals, individuals with chronic lung disease, liver disease, significant heart disease, people with HIV, and people with severe obesity, other conditions, such as patients with chronic kidney disease, were not evaluated [158]. Thus, Broseta et al. [159] analyzed the evolution of IgG antibodies after the first dose of mRNA-1273 in 78 dialysis-dependent chronic kidney disease patients, showing that there was an increase in patients with a detectable immune response as the weeks progressed (8 weeks of evaluation), indicating that mRNA-1273 is also likely effective in this group. It was noteworthy, however, that relative to the general population, patients who received a kidney transplant had a lower response even after administration of the two doses of mRNA-1273, with this likely being associated with the use of high doses of immunosuppressants [160,161]. A single dose of mRNA-1273 was found to be more likely to induce the antibody response than BNT162b2 in solid-organ transplant recipients [162].
With an immediate USD 400 million awarded in grants [170], Pfizer and BioNTech developed the BNT162 series of COVID-19 RNA-based vaccine candidates encapsulated in a lipid nanoparticle [171]. The BNT162 series is a group of four candidate RNA vaccines (BNT162b1, BNT162b2, BNT162a1, and BNT162c2) developed with different RNA formats and target antigens: nucleoside-modified RNA (BNT162b1 and BNT162b2), non-modified uridine RNA (BNT162a1), and saRNA (BNT162c2), all with capabilities to synthesize protein S in its prefusion conformation [172]. Based on the Wuhan lineage virus encapsulated in a lipid nanoparticle, the BNT162b1 and BNT162a1 vaccines encode a trimerized receptor-binding domain (RBD) of the S protein, while the BNT162b2 and BNT162c2 vaccines encode a full-length spike protein [171]. Two phase 1/2 umbrella trials in Germany (NCT04380701 and EudraCT 2020-001038-36) and in the United States (NCT04368728) evaluated multiple vaccines under a single trial protocol [154]. Results have been published from phase 1 trials of BNT162 vaccines in Germany [173], the United States [165,174], and China (NCT04523571) [175]. Initially, the phase 1 study conducted in Germany evaluated the four different vaccines (BNT162a1, BNT162b1, BNT162b2, and BNT162c2), with candidates BNT162b1 and BNT162b2 prioritized for further development.
Data from Sahin et al. [173] suggest that two doses of 1–50 μg of BNT162b1 could protect against COVID-19 through multiple mechanisms, including humoral and cellular responses. The trial conducted in the United States demonstrated that the cellular and humoral response induced by BNT162b1 was dose-dependent, as was the reactogenicity: the second dose of the highest concentration evaluated (100 μg) was not administered due to increased reactogenicity [174]. Phase 1 safety and immunogenicity results of BNT162b1 in younger and older Chinese adults indicated a safe profile, with AEs that were transient and resolved spontaneously or could be managed with a simple standard of care [175]. BNT162b1, however, generated a higher incidence and severity of systemic reactions than BNT162b2, particularly in older adults (65 to 85 years old), while eliciting SARS-CoV-2-neutralizing antibodies in a similar dose-dependent manner [165]. BNT162b2 was selected for expanded clinical evaluation to support an application for marketing authorization [176]. For phase 2/3 (NCT04368728), 43,548 people underwent randomization at >160 sites worldwide (including Brazil, Argentina, Turkey, and South Africa) (Table 4), with about 20% of participants reporting some comorbidity (most commonly diabetes mellitus, hypertension, or chronic lung disease). The safety and efficacy of BNT162b2 in pregnant women, an important risk group for COVID-19 [177], was not released alongside the interim analysis, but Goldshtein et al. [178] demonstrated that pregnant women (gestational age up to 5 weeks) who received a single dose of BNT162b2 had a lower risk of SARS-CoV-2 infection 28 days after immunization than unvaccinated pregnant women, and Bookstein et al. [179] demonstrated that two doses of BNT162b2 generated a humoral immune response in pregnant women at 2–40 weeks of gestation, although anti-Spike RBD IgG levels were lower than those observed in vaccinated, non-pregnant women.
The high efficacy demonstrated in the phase 3 trial of BNT162b2 supported its eagerly awaited application for emergency use authorization (EUA). On 2 December 2020, UK regulators granted EUA to BNT162b2, making it the world’s first approved COVID-19 vaccine [180]. Nine days later, the US Food and Drug Administration (FDA) also granted EUA, facilitating its approval by other regulatory agencies around the world [181]. The high efficacy observed in the phase 3 trial, however, raised questions as to whether the conditions evaluated would represent “real life” outside of clinical trials. In phase 3 studies, conditions such as the monitoring and maintaining of the cold chain required to preserve vaccine integrity and the administration of the vaccine doses within the expected interval were closely followed, preventing the occurrence of problems in maintaining the quality and stability of the vaccine [182]. However, these variables can be difficult to control as mass vaccination is implemented, which causes the results related to the level of protection given the investigational product to be overestimated in the “real-world” [183]. Normally, these conditions are only evaluated in phase 4 studies, where the wide deployment of the vaccine is considered [184]. When Chodick et al. [185] analyzed the effectiveness of BNT162b2 in adults in Israel, they found that the first dose was associated with a ~51% reduction in the risk of SARS-CoV-2 infections on days 13 to 24 after immunization and 54% efficacy against symptomatic COVID-19, a similar level to that reported after provision of the first dose in the phase 3 study [166,185]. In a prospective cohort study conducted in Mexico to evaluate the effectiveness of BNT162b2 among the high-risk group of healthcare professionals, efficacy against severe cases of COVID-19 among individuals with only a single dose was 100% [186]. Thus, the efficacy rates reported from trials have generally held up under real-world conditions (Table 5).
Table 5.
Effectiveness of approved RNA-based vaccines against COVID-19 from other studies (real-world conditions).
| Evaluated Vaccine | Doses | Country | Participants | COVID-19 Outcomes | Effectiveness (%) | Reference |
|---|---|---|---|---|---|---|
| BNT162b2 | 1 | England | 156,930 | Symptomatic infection | 61 | [187] |
| Hospitalization | 80 | |||||
| Deaths | 85 | |||||
| BNT162b2 | 1 | Scotland | 1,331,993 | Hospitalization | 91 | [188] |
| BNT162b2 and mRNA-1273 (Spikevax) | 2 | USA | 1212 | Hospitalization | 87.1 | [189] |
| BNT162b2 | 2 | Israel | 6286 | Symptomatic infection | 61 (first dose) to 89 (second dose) | [190] |
| BNT162b2 | 2 | Israel | 596,618 | Documented infection | 46 (first dose) and 92 (second dose) | [191] |
| Symptomatic infection | 57 (first dose) and 94 (second dose) | |||||
| Hospitalization | 74 (first dose) and 87 (second dose) | |||||
| Severe disease | 62 (first dose) and 92 (second dose) |
In addition to being the first vaccine approved within the context of the pandemic of COVID-19, BNT162b2 was the first vaccine to be licensed for use in adolescents between the ages of 12 and 15 in countries such as the United States [192], Canada [193], and Brazil [194], as well as the European Union [195]. The authorization for use in adolescents came after the release of the phase 3 study initiated in the United States that evaluated BNT162b2 in children aged 12 to 15 years (NCT04368728). The vaccine demonstrated 100% efficacy against confirmed COVID-19 and induced strong immunogenicity one month after the second dose [196]. In parallel, another phase 1/2/3 study is being conducted at over 101 sites to evaluate the safety, tolerability, and immunogenicity of BNT162b2 administered at three different dosages (10 μg, 20 μg, and 30 μg) in healthy children between 6 months and 12 years of age (NCT04816643). It is expected that, as with adults and the elderly, the worldwide population under the age of 12 will soon have access to vaccines against SARS-CoV-2.
CVnCoV, the COVID-19 vaccine candidate developed by the German company CureVac, is based on mRNA technology capable of encoding the prefusion-stabilized form of the full-length S protein of the Wuhan lineage virus (GenBank accession number YP_009724390.1) and formulated on the LNP-formulated RNActive® [197]. It is important to note that there has been an alteration in the vaccine development strategy by CureVac, as a lipid carrier has replaced the protamine-based carrier that had been commonly used by the company, as presented in Table 2. This change may have been motivated by the advantages presented by the lipid vehicles, such as the ability to induce neutralizing antibodies after administration of the vaccine via standard intramuscular injection, whereas this property has only been demonstrated for protamine-based formulations since their administration without needles (a less common route), as well as by the better storage conditions of the LNPs [198,199]. The development of CVnCoV was carried out with the expectation that, because it only required refrigerated storage, it could facilitate access to low-income countries [168]. The phase 1 clinical trial of CVnCoV (NCT04449276) started in July 2020 in Germany and Belgium, following preclinical studies indicating that low doses of CVnCoV induced high titers of neutralizing antibodies and a robust cellular immune response against SARS-CoV-2 [146,148]. The antibody responses were elicited in a dose-dependent manner, and a 12 μg dose was selected for phase 2/3 clinical investigation (NCT04652102, NCT04515147, and NCT04674189) in Argentina, Peru, Panama, Belgium, Mexico, the Netherlands, as well as other European and Latin American countries [167]. Preliminary phase 3 data showed that the vaccine was only 47% effective at preventing COVID-19 [200], almost two times less effective than vaccines BNT162b2 and mRNA-1273 and below the 50% target initially set by the WHO.
The lower performance of CVnCoV relative to mRNA-1273 and BNT162b2, which use the same technological development platform, was speculatively attributed to variants-of-concern found in the countries where the study was conducted (such as the Lambda variant, circulating in different Latin American and European countries) [201,202]. However, other RNA vaccines are effective in controlling new variants-of-concern of SARS-CoV-2 [203,204,205], and the main reason for the relatively poor performance of CVnCoV may relate to either the low doses employed or the mRNA sequence used (CVnCoV uses normal uridine and is thus “unmodified” mRNA, whereas mRNA-1273 and BNT162b2 use a modified mRNA that replaces uridine with N1-methylpseudouridine). mRNA modified with N1-methylpseudouridine can result in more durable protein expression and thus longer antigen availability and may result in more robust immune system responses [206,207]. CureVac recently announced a new COVID-19 vaccine candidate called CV2CoV, a second-generation mRNA vaccine developed through a partnership with GlaxoSmithKline (GSK). This candidate is based on the analysis of potential SARS-CoV-2 variants-of-concern in multivalent vaccine formats. Recent preclinical data for CV2CoV show that it induced complete protection against variant B.1.351 in a transgenic mouse model [208].
The first RNA vaccine approved for clinical trials in China was ARCoV (CN111333704), a vaccine developed by the People’s Liberation Army (PLA) Academy of Military Sciences in collaboration with Suzhou Abogen Biosciences and Walvax Biotechnology Co. ARCoV is a lipid-nanoparticle-encapsulated mRNA vaccine encoding the RBD domain of SARS-CoV-2 (Wuhan) S protein [209]. Phase 1 and phase 2 trials evaluating the safety and immunogenicity of different doses of ARCoV in Chinese adults aged 18–59 years began in June 2020 (ChiCTR2000034112) and January 2021 (ChiCTR2100041855), respectively, while Phase 3 evaluation is being conducted in China and Mexico (NCT04847102) [210]. A major advantage of ARCoV over other RNA vaccines lies in its thermal stability as a liquid formulation that can be stored at room temperature for at least 1 week, thus facilitating distribution [147].
Vaccines based on saRNA technology also rapidly emerged. Among them, ARCT-021 (LUNAR-COV19), developed by Arcturus Therapeutics (San Diego, California, US) and Duke–NUS Medical School in Singapore, uses the STARR™ (Self-Transcribing and Replicating RNA) system in combination with the LUNAR® (Lipid-Enabled and Unlocked Nucleomonomer-Agent-Modified RNA) delivery system. ARCT-021 carries a sequence that encodes alphavirus replicase and the full-length, unmodified S protein (Wuhan sequence) [211]. This system is designed to increase and extend the expression of the antigen of interest, allowing vaccination at lower doses than conventional mRNA vaccines [212]. A differentiating feature of ARCT-021 is that, unlike the other vaccines mentioned above that are distributed as frozen liquids, it is presented in a lyophilized form that negates the need for an ultra-cold chain, which needs consistent storage at extremely cold temperatures (storage at about <−70 °C) [211]. Phase 1 (NCT04480957) and phase 2 (NCT04480957) trials are summarized in Table 4. Data from 42 phase 1 and 64 phase 2 patients indicated that ARCT-021 was well tolerated in a single 7.5 μg dose regimen and in a two-dose 5 μg regimen at the concentrations shown in Table 4. Neutralizing antibody titers increased with increasing dose, in addition to triggering the T-cell response against the S protein of SARS-CoV-2. ARCT-021 at a dose of 7.5 μg also showed satisfactory tolerability, with no vaccine-related serious AEs reported [169]. These results supported the further clinical development of ARCT-021, with a phase 2 clinical trial being conducted in the United States and Singapore with 600 participants (NCT04480957) to provide guidance for a subsequent phase 3 clinical trial.
With funding support from the British Government, Imperial College London also used saRNA technology to encode the prefusion-stabilized SARS-CoV-2 S-glycoprotein (Wuhan sequence) along with RNA replicase from an alphavirus, encapsulated in a lipid nanoparticle (COVAC1; LNP-nCoVsaRNA). After preclinical results demonstrated that two doses of COVAC1 induced cellular and humoral responses [143], a phase 1 trial was initiated in the United Kingdom in June 2020 to evaluate the safety and immunogenicity of COVAC1 in 320 18–75-year-olds (ISRCTN17072692). The trial evaluated COVAC1 at doses of 0.1 µg, 0.3 µg, and 1.0 µg, administered with a 4-week interval between the first and second injection. [213]. Although efficacy data have not yet been reported, together with Morningside Venture, Imperial College London has backed the creation of VacEquity Global Health (VGH), which aims to develop and distribute the COVAC1 in the United Kingdom as well as low- and middle-income countries [214].
HDT Bio’s COVID vaccine program, HDT-301, pairs a saRNA with a lipid inorganic nanoparticle (LION) formulation. Preclinical studies have demonstrated that HDT-301 induces neutralizing antibodies as well as a Th1-mediated immune response in both mice (including aged animals) and non-human primates. The generated neutralizing antibodies persisted for at least 70 days post-vaccination at titers compared to convalescent human serum [145]. Following technology transfer to the Indian biopharmaceutical Gennova Biopharmaceuticals (Pune, India), GMP manufacture and clinical evaluation of the HGCO19 vaccine candidate (incorporating the D614G variant sequence) was initiated [215] in a phase 1/2 trial (CTRI/2021/04/032688) to assess the safety and immunogenicity of a two-dose schedule of immunization with the experimental vaccine at either 5, 10, or 25 µg in healthy seronegative Indian adults. In late August 2021, the Drugs Controller General of India approved the advancement of HGC019 to phase II/III trials after it was found to be safe, tolerable, and immunogenic during its phase 1/2 evaluation. Phase 1 trials of HDT’s saRNA alternative SARS-CoV-2 Spike sequences have been approved by the FDA in the United States and Anvisa in Brazil (NCT04844268), respectively, and are scheduled to begin soon.
3.2. Efficacy of RNA-Based Vaccines against SARS-CoV-2 Variants
Several analyses of both clinical trial data and samples have assessed the ability of various COVID-19 vaccines to protect or generate neutralizing antibodies against different variants-of-concern of SARS-CoV-2 [216,217,218]. It is important to mention that the spike is not the only protein to become mutated. Mutations in the ORF1ab, ORF8, and nucleocapsid protein also occur [219]. Cai et al. [220] reported a comprehensive analysis demonstrating that, in terms of efficacy, RNA-based vaccines are the most effective and are capable of reaching greater than 94% efficacy. The authors attribute this achievement to the strong immunogenicity and effective presentation of SARS-CoV-2 antigens to the immune system of these vaccines. However, with the world population still to be immunized against SARS-CoV-2, ensuring the effectiveness of vaccines against the new variants-of-concern become a major concern for health authorities and scientists [221]. Currently, most of the data relating to the efficacy of RNA vaccines against SARS-CoV-2 variants-of-concern came from laboratory studies [222,223] that, in general, reveal that the vaccines elicit lower levels of neutralizing antibodies against SARS-CoV-2 variants-of-concern than against older isolates [222,223].
The B.1.1.7 variant was demonstrated to be more infectious than the Wuhan strain, around 60%, and also more resistant to monoclonal antibodies targeting the N-terminal domain [224]. However, both Comirnaty and Spikevax vaccines remain robust against this variant, generating antibodies two-fold less efficient in neutralizing the B.1.1.7 strain [225,226]. The second relevant variant found first in South Africa, known as B.1.351, shares the B.1.1.7 mutations D614G and N501Y in the S protein, which confers to this variant’s high transmissibility. The B.1.351 variant is more resistant to neutralization by convalescent serum and by serum from Comirnaty- and Spikevax-vaccinated people by six-fold [227,228]. The P.1 mutant was first detected in Brazil and shares D614G and N501Y S mutations with the other variants. Moreover, K417N/T and E484K mutations in the S protein are also found in RBD from the B.1.351 variant [229]. The Spikevax vaccine also presented reduced effectivity in generating neutralizing antibodies against P.1 compared to the original strain [223].
It is important to highlight that researchers believe the antibodies produced still might be sufficient to protect against SARS-CoV-2 infection, or at least against severe cases of COVID-19, since they can still neutralize the virus overall [230]. Since RNA technology can be more easily adapted to new variants-of-concern, and due to the important levels of efficacy observed against previously circulating SARS-CoV-2, RNA vaccines have enormous potential against SARS-CoV-2 variants-of-concern [230]. Since RNA technology can be more easily adapted to new variants-of-concern, and due to the high levels of efficacy observed against previously circulating SARS-CoV-2, RNA vaccines have great potential against SARS-CoV-2 variants-of-concern.
3.3. Composition of Main RNA-Based Vaccines for COVID-19
Within a brief period between the disclosure of the genetic code of SARS-CoV-2 and the authorization for use by different regulatory agencies, a large amount of information on RNA-based vaccines has been made available, which allows for a better investigation of the characteristics and composition of each developed vaccine, as well as enabling one to assess how these factors may influence immunogenicity and, consequently, the resulting protection. Table 6 shows a comparison of the components found in the main RNA vaccines against COVID-19, considering the mRNA or saRNA aspect, type of antigen used, and the composition of their delivery platform.
Table 6.
Components of the major RNA-based vaccine against COVID-19: characteristics of the mRNA or saRNA and antigen used and its delivery system composition.
| Vaccine Name | mRNA or saRNA Construct |
Antigen | Delivery Platform Composition | Diluent |
|---|---|---|---|---|
| mRNA-1273 (Spikevax) | N1-methylpseudouridine Modified 5′ CAP1 structure (m7GpppNmN) Codon optimization (GC-enriched sequence) |
Full-length S protein with two proline substitutions (K986P and V987P) Wuhan-Hu-1 (GenBank: MN908947.3) |
SM-102 Cholesterol DSPC PEG2000-DMG |
Sodium chloride |
| BNT162b2 | N1-methylpseudouridine Modified 5′ CAP1 structure Codon optimization |
Full-length S protein with two proline substitutions (K986P and V987P) Wuhan-Hu-1 (GenBank: MN908947) |
ALC-0315 (proprietary to Acuitas) ALC-0159 DSPC Cholesterol |
Sodium chloride |
| CVnCoV | Modified 5′ CAP1 structure (m7GpppNmN) Codon optimization (GC-enriched sequence |
Full-length S protein with two proline substitutions (K986P and V987P) Wuhan-Hu-1 (GenBank: YP_009724390.1) |
Cholesterol DSPC PEG-ylated lipid Ionizable lipid (undisclosed/proprietary to Acuitas) |
Sodium chloride |
| ARCT-021 (LUNAR-COV19) |
N1-methylpseudouridine Modified 5′ CAP1 structure (m7GpppNmN) Codon optimization Self-replicating replicon of VEEV |
Full-length, unmodified S protein Wuhan-Hu-1 (GenBank: YP_009724390.1) |
Ionizable lipid (undisclosed) DSPC Cholesterol PEG2000-DMG |
Sodium chloride |
| COVAC1 (LNP-nCoVsaRNA) |
Modified 5′ CAP1 structure (m7GpppNmN) Self-replicating replicon of VEEV F318V amino acid substitution |
Full-length S protein with two proline substitutions (K986P and V987P) and GGGGSGGGGS linker Wuhan-Hu-1 (GenBank: QHD43416.1) |
Ionizable cationic lipid (undisclosed) Phosphatidylcholine (undisclosed) Cholesterol PEG-lipid (undisclosed) |
Phosphate-buffered saline |
| ARCoV | Modified 5′ CAP1 structure (m7GpppNmN) Codon optimization |
RBD region of S protein Wuhan-Hu-1 (GenBank: MN908947.3) |
Ionizable lipid (undisclosed/proprietary to Acuitas) DSPC Cholesterol PEG-lipid (undisclosed) |
Not reported |
| HDT-301 (LION/repRNA-CoV2S) |
Modified 5′ CAP1 structure Self-replicating replicon of VEEV |
Full-length, unmodified S protein Wuhan-Hu-1 (GenBank: MN908947.3) |
DOTAP Squalene Span 60 Polysorbate 80 oleic-acid-coated iron oxide nanoparticles |
Sodium citrate buffer |
ALC-0159, 4-Hydroxybutyl)azanediyl)bis(hexane-6,1-diyl) bis(2-hexyldecanoate); ALC-0315, 4-Hydroxybutyl)azanediyl)bis(hexane-6,1-diyl) bis(2-hexyldecanoate); DOTAP, lipid 1,2-dioleoyl-3-trimethylam-monium-propane; DSPC, 1,2-distearoyl-snglycero-3 phosphocholine; PEG2000-DMG, 1 monomethoxypolyethyleneglycol-2,3- dimyristylglycerol with polyethylene glycol of average molecular weight 2000; Span 60, sorbitan monostearate; VEEV, venezuelan equine encephalitis virus.
Naked RNA cannot pass through the cell membrane and efficiently leak into the cytoplasm due to its intrinsic properties such as size, charge, and degradability [231,232]. Therefore, RNA vaccines require effective and safe delivery methods. Most RNA vaccines evaluated have the same antigen delivery strategy: the use of LNPs. This prominence has been achieved due to critical advances that have been made in the LNPs’ formulation processes, primarily to achieve improvements in formulation stability, which increase the ability to induce cellular and humoral immune responses, improve endosomal escape, decrease reactogenicity, and increase the efficacy of RNA delivery into the cytosolic environment [107]. In general, LPNs provide a solid lipid structure for RNA protection and are basically composed of four components: (i) cationic or ionizable lipids that act in the RNA complexation to increase cellular uptake efficiency, as well as promote endosomal escape; (ii) cholesterol, which has the function of stabilizing the nanoparticle; (iii) auxiliary phospholipids that act by stabilizing the lipoplex and promoting membrane fusion; and (iv) PEGylated lipids to reduce non-specific interactions, preventing particle aggregation [233,234].
The development of an effective LNP platform for mRNA or saRNA delivery involves a crucial step, which is the choice of cationic or ionizable lipid. Considering the RNA vaccines developed against COVID-19, ionizable lipids were used by the vast majority of developers (Table 6). This choice may be associated with the advantages that ionizable lipids possess: even though they present the same capacity to complex RNA as cationic lipids, studies have shown that their use provides more safety to the formulations [235,236]. More specifically, ionizable lipids exhibit a positively charged pKa at a low (acidic) pH, which allows anionic RNA complexation. However, it is important to highlight that, in a neutral pH environment, the charge of ionizable lipids is neutral, which may reduce possible nonspecific interactions and hence their toxicity [198].
The vaccines mRNA-1273 (Spikevax) and BNT162b2 have the molecules SM-102 and ALC-0315 as ionizable lipids, respectively, in their formulations [237]. These lipidic molecules present a biodegradable character due to the presence of ester bonds in the lipid tails, resulting in a chemical similarity [238]. However, they exhibit structural differences that may have a bearing on the immunogenicity induced by these vaccines, in addition to the efficacy of mRNA delivery [206]. ALC-0315 is also estimated to be the ionizable lipid found in the carrier system of CVnCoV [238]. According to the developers of ARCT-021, the ionizable lipid molecule used on the LUNAR® platform is also biodegradable and has optimized efficiency for mRNA delivery [239] due to the ability of this lipid to be rapidly degraded under normal physiological conditions by ester bond breaking, resulting in rapid metabolization and an improved safety profile [240]. The saRNA from the COVAC1 vaccine (LNP-nCoVsaRNA) is delivered in an LNP system that is owned by Acuitas. One of the probable ionizable lipids used in this formulation is Lipid A9 [238], developed by Acuitas and described in patent document US10221127B2 (lipids and lipid nanoparticle formulations for delivery of nucleic acids), which has equivalent properties to ionizable lipids SM-102 and ALC-0315 [236]. In addition to ionizable lipids, it is important to note that the helper lipids used in the RNA vaccine formulations against COVID-19 are similar, with variations in their concentration [240]. The HDT-301 (LION/repRNA-CoV2S) COVID-19 vaccine candidate uses a delivery system based on lipid inorganic nanoparticles (LION), more specifically, nanoemulsions, where the molecule DOTAP acts as a cationic lipid capable of establishing an electrostatic association with RNA molecules [145]. Like LPNs, cationic nanoemulsions (CNE) are also considered as an important antigen delivery platform within the context of RNA vaccine development. In general, the emulsions used are typically a water-in-oil emulsion, composed of three more components in addition to the cationic lipid: (1) squalene, (2) sorbitan trioleate (or monostearate in the case of HDT-301), and (3) polysorbate 80 [61]. It is important to highlight that one of the differentials found in the LION formulation compared to conventional CNEs (cationic nanoemulsions) is the presence of superparamagnetic iron oxide (Fe3O4) nanoparticles (SPIO), which can act as the stability intensifier. Preclinical trials have shown good efficacy of CNEs in delivering antigens against HIV, RSV, and cytomegalovirus and in inducing immune responses [241]. A feature of this approach is the ability to store the carrier system (CNEs) and saRNA or mRNA in separate vials and only combine them at the time of vaccine administration [238].
In addition to the delivery system, the engineering involved in the mRNA construction process is one of the critical points for an RNA vaccine to achieve an acceptable safety and efficacy profile. Optimization of the mRNA sequence to improve the immunogenic and safety profile has been one of the main challenges in developing new vaccines. This optimization has focused on the following approaches: increasing mRNA half-life to increase the duration of protein expression and protein levels, the optimization of poly-(A)-tail length, the modification of the 5′ and 3′ untranslated regions (UTRs) to adjust immunogenicity, capping strategies to prevent rapid mRNA degradation, and the incorporation of modified nucleotides within the coding sequence [242,243,244]. Thus, even non-structural (or non-coding) elements have importance in achieving the key outcomes associated with producing a vaccine, since they can interfere with mRNA stability and translatability [245]. The incorporation of N1-methylpseudouridine (m1Ψ), RNA capping, and codon optimization were the main strategies adopted by the developers of the RNA-based COVID-19 vaccines (Table 6). The use of modified nucleotide m1Ψ has been one of the main discussions within the strategies for mRNA engineering for COVID-19 vaccines. The presence of modified uridine in the mRNA construct may promote increased immune evasion, as it can minimize the recognition of the mRNA by proteins involved in the innate immune response to exogenous mRNA, in addition to increased protein production due to its ability to facilitate the translation of the mRNA into proteins via the ribosome [246,247]. These features may increase the biological stability of the mRNA and may also reduce the reactogenicity of vaccines using this technology [248]. The low efficacy of CVnCoV compared to mRNA-1273 (Spikevax) and BNT162b2 has been mainly related to the absence of these modified uridines in the structure of the CVnCoV mRNA, since they may confer stability to the mRNA before administration [249].
The combination of the delivery system with improved properties and the optimized mRNA should promote the initiation of the translation process of the target antigen in vivo. During previous outbreaks caused by other coronaviruses, the S protein has been considered as the main antigen for the development of vaccine candidates and, similarly, all mRNA vaccines against SARS-CoV-2 were developed to induce immune responses against the S protein or the receptor-binding domain [172]. Preclinical studies with candidate vaccines against SARS-CoV-1 and MERS-CoV from different technology platforms have shown that the presence of a full-length S gene or the protein itself induces both neutralizing antibody responses and protective immunity [250,251,252], which may have been used as the basis for the widespread use of this antigen in SARS-CoV-2 vaccines, as shown in Table 6. An important optimization strategy widely used by the developers of the SARS-CoV-2 mRNA vaccines (such as Pfizer and BioNTech, CureVac, Moderna, and Imperial College London) was the use of a gene sequence with a minor change that allows for the translation of the full-length S protein with two proline (2P) substitutions (K986P and V987P mutations), leading to greater stability during pre-melting conformation of the glycoprotein. Consequently, it is estimated that this modification may lead to a greater induction of antibody formation [253]. This strategy was also based on previous lessons learned during the development of vaccines against other CoVs, such as SARS-CoV-1, MERS-CoV, and HKU1 [116,142]. Despite its importance, studies have shown that the full-length S protein may also be associated with the induction of harmful immune responses through antibody-dependent enhancement mediated by non-neutralizing antibodies [254,255,256]. This evidence may have motivated the developers of the ARCoV vaccine to use the RBD subunit of the SARS-CoV-2 S-protein as an antigen, which has been the region of the S-protein that has major neutralizing epitopes capable of inducing high titers of neutralizing antibodies and low levels of non-neutralizing antibodies when compared to the full-length S-protein [17,257,258].
Furthermore, it is important to note that the definition of the components of a vaccine has a direct bearing on the possible costs of manufacturing and making the vaccine available. One of the most important advantages of the RNA platform over conventional vaccine technologies is that its manufacture is simpler, since it does not contain steps such as cell culture, which require more time and investment [107]. Moreover, even mRNAs encoding different antigens have similar physical and chemical characteristics, which means that once the manufacturing process is established, no major adaptations in the facility are required [249]. However, it is important to note that some factors may influence the costs associated with the production of RNA vaccines for COVID-19, such as the materials required to produce the delivery system and mRNA, particularly if there is a need for the inclusion of modified nucleosides and capping in the mRNA sequence, as well as the costs with consumables, such as the use of single-use equipment. For example, the CleanCap reagent (TriLink Biotechnologies, Inc., San Diego, CA, USA) is a major cost component for the production of vaccines developed by Moderna, Pfizer and BioNTech, CureVac, and Imperial College London [259]. Within this expectation, saRNA-based vaccines are expected to have a lower production cost, since a lower concentration per dose of RNA is needed to induce the required immune response, which reduces the amount of materials needed for their manufacture.
4. Perspectives and Challenges Associated with COVID-19 Immunization Based on RNA Vaccines
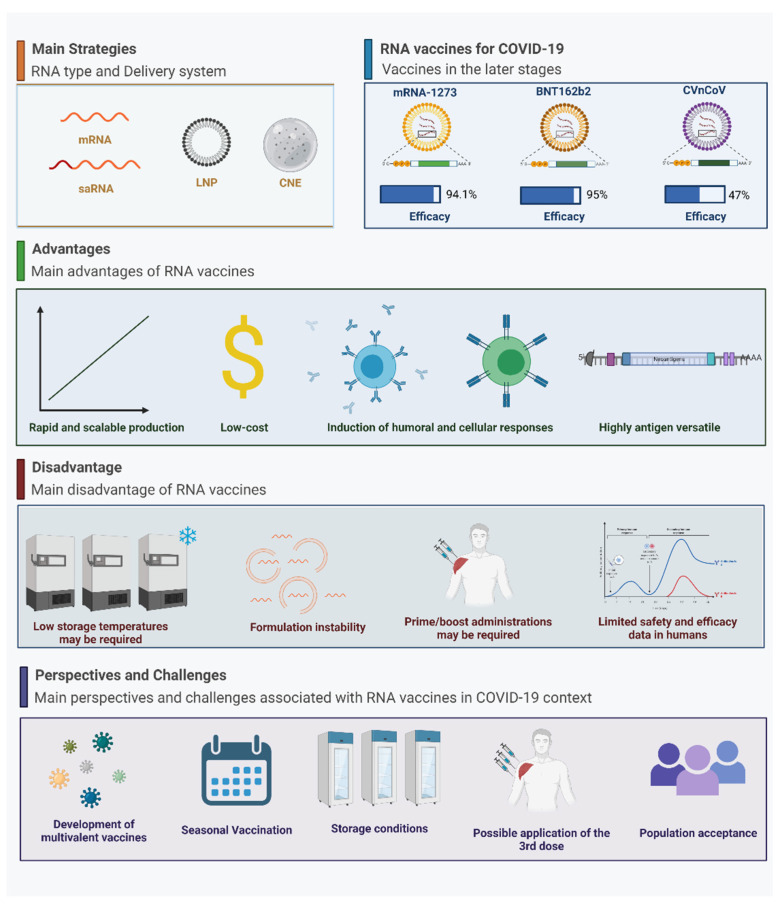
The COVID-19 pandemic has quickly transitioned RNA vaccines from a promising platform to becoming a major tool for COVID-19 prevention and a return to normal daily routines. Following their FDA emergency use authorization (EUA), by March 2021, more than 170 million doses of RNA-based vaccines had been made available worldwide, representing 43% of the total vaccines produced [260]. Within eight months of EUA, BNT162b2 (known as Comirnaty) has been approved in almost 90 countries, while mRNA-1273 (now named Spikevax) has been approved in more than 60 countries, making these two vaccines central within national immunization programs [261]. As observed in other COVID-19 vaccines based on different technology platforms, such as the adenoviral vectors vaccines that have been widely distributed, immunization with COVID-19 RNA vaccines still faces challenges associated with mass production, extended distribution chains, sustained efficacy in the face of emergent variants-of-concern, amongst others (Figure 2).
Figure 2.
Perspectives and challenges for RNA-based vaccines for COVID-19. Created with BioRender.com (accessed on 30 September 2021).
4.1. Perspectives and Challenges Associated with Widespread Production and Availability
One of the main challenges associated with RNA vaccines is related to the maintenance of the immune response over time against SARS-CoV-2 through neutralizing antibody titers. In a study conducted in Israel with 4868 participants, it was observed that in the third month after administration of the second dose of the Comirnaty vaccine, IgG antibody levels decreased at a consistent rate; in the same period, the level of neutralizing antibodies decreased rapidly, with an average reduction of more than 70% from initial values [262]. Another study analyzing the same vaccine found that 3 months after the second dose, there was a reduction of up to 44.7% in antibody titers [263]. The study by Khoury et al. [264] demonstrated that the concentration of neutralizing antibodies could predict immune protection from symptomatic SARS-CoV-2 infection, where antibody decay indicates a higher probability of infection. Importantly, the decline in antibody levels may be related to the reduced effectiveness of RNA vaccines in preventing SARS-CoV-2 infection. In a recent preprint, compared to the 96% observed efficacy two weeks after receipt of both doses, the efficacy of the BNT162b2 (Comirnaty) vaccine against symptomatic COVID-19 decreased to 84% 6 months after the second dose [265]. Studies by Benotmane et al. [266] and Hall et al. [267] indicated that the provision of a third mRNA-1273 (Spikevax) dose in transplant recipients increased the humoral immune response and antibody titers. A similar result was reported by Del Bello et al. [268], who analyzed the efficacy of BNT162b2 (Comirnaty) under the same conditions. To address this issue, booster immunizations are now being recommended for some at-risk individuals in the United States and some other countries [269]. This concern is accentuated by the possibility of the emergence of new variants-of-concern with a higher morbidity and mortality rate. Thus, the development of mRNA vaccines that protect for a longer period is a major challenge for developers.
However, because new variants-of-concern usually present with mutations in the S protein gene that can reduce the affinity of neutralizing antibodies, another strategy under consideration is the use of variant-specific and/or multivalent vaccines. In this context, as discussed earlier, the manufacturing flexibility of RNA-based vaccines affords an advantage over other vaccine platforms. Moderna has adjusted its mRNA vaccine to match the Spike gene sequence of variant 501Y.V2 [270], and clinical trials are already underway (NCT04785144). Thus, a major expectation is that, like other respiratory viruses such as influenza, SARS-CoV-2 will eventually become endemic and present seasonally, which will require developers to constantly update the antigen used [271].
The countries where COVID-19 vaccines have been incorporated into the routine of the health system have had to prepare strategic planning related to their storage, transport, manipulation, stock control, and distribution, mainly because approved mRNA vaccines currently require special storage conditions [272,273]. The stability of mRNA-1273 and BNT162b2 require extremely low temperatures (between −80 to −20 °C) for distribution within their approved specifications, restricting availability to countries and regions where ultra-cool equipment is not widely available [274]. This scenario is typical of many developing countries, where the purchase of refrigeration equipment costing upwards of USD 20000 per unit is prohibitive even before considering that many communities live without continuous electrical power capable of supporting such equipment [275]. It is important to note that outside of exceptional conditions, such as the pandemic caused by SARS-CoV-2, vaccines that require a storage temperature below −20 °C are not in compliance with the requirements placed on WHO prequalification [276]. These special storage conditions can result in not only logistical and economic difficulties but can also have a direct impact on environmental issues. The study by Santos et al. [277] demonstrated the possible impacts of this special storage condition on electricity consumption and pollutant emission, where it was reported that stock maintenance of Comirnaty and Spikevax vaccines is able to emit more CO2 and consume more electricity compared to vaccines developed from other technologies, such as CoronaVac and Sputnik V. Thus, the development of thermostable RNA-based vaccines that do not require expensive equipment and special storage conditions and distribution chain is of importance. The use of thermostable lipid nanoparticles in the RNA system and freeze-dried techniques are considered the technological advancements that could achieve this goal [211,278].
Another important challenge is associated with the production of RNA vaccines. Biopharmaceutical companies are estimated to be able to produce up to 12 billion doses by 2021; however, to have an equitable distribution of doses among countries, especially considering the need for booster doses, this amount is not believed to be sufficient [279]. However, a point of importance is that production sites for RNA vaccines are primarily concentrated in high-income countries, necessitating that middle- and low-income countries require importation and adaptations beyond the cold chain for internal distribution [259]. Given this need, countries such as South Africa and India have sought to establish partnerships to produce RNA vaccines in their territories [280,281]. This can be reflected in the low availability of RNA vaccines to the population, where, in India, more than 85% of its population has been immunized with a vaccine based on viral vector technology [261]. In addition, according to the study by Rosa et al. [50], the production of RNA vaccines in a sustainable and cost-effective way has important challenges and bottlenecks identified in the downstream and upstream production steps, such as the use of high-cost and limited availability reagents, as well as the use of methods with low stability. In this way, it is fundamental that new production methods are proposed so that the stage of availability, commercialization, and meeting the market demand can be supplied in a quick and equitable manner.
4.2. Perspectives and Challenges Associated with Acceptance of COVID-19 Vaccines
Although the approval of the first RNA vaccine during the COVID-19 pandemic was a milestone, its uniqueness and rapid development led to public concerns about the safety of RNA-based vaccines [282]. This skepticism has strengthened anti-vaccine movements, raising questions about the new technology and its “real” performance [283]. Solís Arce et al. [284] showed that concerns about possible side effects are the most common reasons for the reluctance of people to receive these vaccines, despite an ever-building safety profile and regulatory scrutiny that skepticism has brought. Public perceptions may be incorrect; through a real-world safety and reactogenicity study, Mathioudakis et al. [285] demonstrated that recipients of mRNA vaccines reported milder and less frequent systemic side effects than recipients of viral vector-based vaccines. Data such as these have been reflected in countries such as Brazil, where the level of acceptance for BNT162b2 (the only RNA vaccine currently approved in the country) is higher than those developed through other technological platforms, with lower reporting of post-vaccine AEs [286]. Health authorities are promoting critical awareness campaigns identifying the benefits of immunization with approved and safe vaccines. Large clinical (phase 3 trials) and real-life studies (after EUA) have shown the safe and effective profile of RNA-based vaccines in protecting against COVID-19, and the data-based incorporation of RNA-based vaccines in national immunization programs represents an important step to increase vaccine acceptance levels.
5. Conclusions
Despite being studied over the past few years and having initial studies conducted in the context of cancer vaccines [126,241,287], RNA-based vaccines first became available on a population level in response to the SARS-CoV-2 pandemic, highlighting the importance of having a highly safe, flexible, and scalable vaccine available for the prevention of not only for epidemic outbreaks, but also important diseases such as cancer, autoimmune diseases, and allergic disease. In addition, the many advantages of this technology when compared to conventional approaches, such as allowing the delivery of the genetic material directly to the cells, without the need to administer exogenous antigens, high immunogenicity, and production in a cell-free environment, allows RNA-based vaccines to have good potential in the future of biopharmaceuticals. Therefore, there is an increased interest from the industry in the development of this technological platform.
In the context of COVID-19, it is important to highlight not only the rapid development and availability of mRNA vaccines but also their high efficacy. Thus far, clinical trials and the subsequent expansive use of COVID-19 vaccines demonstrate not only the safety but also the efficacy of the RNA vaccine platforms. As demonstrated in clinical studies, mRNA vaccines, particularly Pfizer’s and Moderna’s mRNA vaccines, have been shown to be highly safe and effective. However, it is important to mention that, even with high efficacy, such vaccines need to be applied in two doses, and the application of a booster dose (third dose) is already being considered for risk groups (the elderly and people with immunodeficiencies). A promising alternative to resolve this issue is the application of saRNA vaccines, which, thanks to their self-replicative capacity, have the potential to confer an immune and long-lasting response with just a single dose. However, clinical immunogenicity data for these vaccines are not yet available.
It is important to highlight that many strategies need to be to evaluated when developing RNA-based vaccines. Among them are the delivery technologies applied to it since naked RNA cannot enter cells due to its intrinsic properties. Knowledge of the delivery platform composition is especially important since it impacts the formulation stability, the ability to induce an immune response, decreased reactogenicity, and the efficacy of RNA delivery. LNPs are the most used platform, and their composition provides a solid structure that protects RNA from degradation. It is also important to note that the components of a vaccine directly influence the planning and costs of manufacturing, key factors for making the immunizer available.
Despite the rapid response capability and flexibility of RNA vaccine technology demonstrated as these vaccines have become central to efforts to contain SARS-CoV-2, tied to their high safety and efficacy, RNA-based vaccines face significant challenges, mostly related to the scale of production, supply chains, and long-term efficacy, the latter of which is accentuated with the emergence of new variants. Moreover, RNA-based vaccines still face skepticism and strengthening anti-vaccine movements. However, thanks to its versatility, RNA technology is changing the vaccine industry and providing hope that it may be used to generate candidates with the potential to protect against a wide variety of cancers and infectious diseases [65].
Acknowledgments
The authors thank SENAI CIMATEC and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) (BAMS is a Technological fellow from CNPq 315351/2018-7).
Author Contributions
Conceptualization, B.A.S.M., L.A.B.M., L.P.C.d.S.A., M.B.P.S., S.G.R. and R.B.; methodology, B.A.S.M., K.V.S.H., L.M.d.S.F., L.A.B.M., L.P.C.d.S.A. and R.B.; validation, B.A.S.M., L.A.B.M., L.P.C.d.S.A., M.B.P.S., P.B., M.S.D., S.G.R. and R.B.; writing—original draft preparation, B.A.S.M., K.V.S.H., L.M.d.S.F., L.A.B.M., L.P.C.d.S.A., V.P.C.R., M.B.P.S., P.B., M.S.D., S.G.R. and R.B.; writing—review and editing, B.A.S.M., K.V.S.H., L.M.d.S.F., M.B.P.S., P.B., M.S.D. and R.B.; visualization, B.A.S.M., P.B. and M.S.D.; supervision, B.A.S.M., L.A.B.M., L.P.C.d.S.A. and R.B.; project administration, B.A.S.M. and L.A.B.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
P.B., M.S.D. and S.G.R. have an equity interest in HDT Bio. B.A.S.M., K.V.S.H.; L.M.d.S.F., L.A.B.M., L.P.C.d.S.A., V.P.C.R., M.B.P.S. and R.B. declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plotkin S.A. Vaccines: Past, present and future. Nat. Med. 2005;11:S5–S11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang N., Li C., Zhu Y., Li K., Liang J., Wang L., Du L., Jiang S., Yang F., He X., et al. Current development of COVID-19 diagnostics, vaccines and therapeutics. Microbes Infect. 2019;22:231–235. doi: 10.1016/j.micinf.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dodd R.H., Pickles K., Nickel B., Cvejic E., Ayre J., Batcup C., Bonner C., Copp T., Cornell S., Dakin T., et al. Concerns and motivations about COVID-19 vaccination. Lancet Infect. Dis. 2021;21:161–163. doi: 10.1016/S1473-3099(20)30926-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu H., Stratton C.W., Tang Y.-W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker P.G.T., Whittaker C., Watson O.J., Baguelin M., Winskill P., Hamlet A., Djafaara B.A., Cucunubá Z., Mesa D.O., Green W., et al. The impact of COVID-19 and strategies for mitigation and suppression in low- and middle-income countries. Science. 2020;369:413–422. doi: 10.1126/science.abc0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO WHO Coronavirus Disease (COVID-19) Dashboard. [(accessed on 28 October 2021)]. Available online: https://covid19.who.int/
- 8.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meselson M. Droplets and Aerosols in the Transmission of SARS-CoV-2. N. Engl. J. Med. 2020;382:2063. doi: 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arslan M., Xu B., El-Din M.G. Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020;743:140709. doi: 10.1016/j.scitotenv.2020.140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Chen X., Deng X., Chen Z., Gong H., Yan H., Wu Q., Shi H., Lai S., Ajelli M., et al. Disease burden and clinical severity of the first pandemic wave of COVID-19 in Wuhan, China. Nat. Commun. 2020;11:5411. doi: 10.1038/s41467-020-19238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piroth L., Cottenet J., Mariet A.-S., Bonniaud P., Blot M., Tubert-Bitter P., Quantin C. Comparison of the characteristics, morbidity, and mortality of COVID-19 and seasonal influenza: A nationwide, population-based retrospective cohort study. Lancet Respir. Med. 2021;9:251–259. doi: 10.1016/S2213-2600(20)30527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara P., Albano L. COVID-19 and healthcare systems: What should we do next? Public Health. 2020;185:1–2. doi: 10.1016/j.puhe.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malick M.S.S., Fernandes H. The Genomic Landscape of Severe Acute Respiratory Syndrome Coronavirus 2. Adv. Mol. Pathol. 2021;4:231–235. doi: 10.1016/j.yamp.2021.06.006. [DOI] [Google Scholar]
- 20.Wang G.-L., Wang Z.-Y., Duan L.-J., Meng Q.-C., Jiang M.-D., Cao J., Yao L., Zhu K.-L., Cao W.-C., Ma M.-J. Susceptibility of Circulating SARS-CoV-2 Variants to Neutralization. N. Engl. J. Med. 2021;384:2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernal J.L., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO Tracking SARS-CoV-2 Variants. [(accessed on 13 August 2021)]. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 23.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seib K., Dougan G., Rappuoli R. The Key Role of Genomics in Modern Vaccine and Drug Design for Emerging Infectious Diseases. PLoS Genet. 2009;5:e1000612. doi: 10.1371/journal.pgen.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty S., Mallajosyula V., Tato C.M., Tan G.S., Wang T.T. SARS-CoV-2 vaccines in advanced clinical trials: Where do we stand? Adv. Drug Deliv. Rev. 2021;172:314–338. doi: 10.1016/j.addr.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Peng Y., Xu H., Cui Z., Williams R. The COVID-19 Vaccine Race: Challenges and Opportunities in Vaccine Formulation. AAPS PharmSciTech. 2020;21:225. doi: 10.1208/s12249-020-01744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen A. For Billion-Dollar COVID Vaccines, Basic Government-Funded Science Laid the Groundwork. [(accessed on 26 January 2021)]. Available online: https://www.scientificamerican.com/article/for-billion-dollar-covid-vaccines-basic-government-funded-science-laid-the-groundwork/
- 31.Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 32.Conte C., Sogni F., Affanni P., Veronesi L., Argentiero A., Esposito S. Vaccines against Coronaviruses: The State of the Art. Vaccines. 2020;8:309. doi: 10.3390/vaccines8020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradley E.S., McNeel D.G. Cancer Therapeutic Targets. Volume 1–2. Springer; New York, NY, USA: 2017. DNA Vaccines; pp. 183–198. [Google Scholar]
- 34.Pardi N., Weissman D. Methods in Molecular Biology. Volume 1499. Humana Press Inc.; Totowa, NJ, USA: 2017. Measuring the Adjuvant Activity of RNA Vaccines; pp. 143–153. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Zhang Z., Luo J., Han X., Wei Y., Wei X. mRNA vaccine: A potential therapeutic strategy. Mol. Cancer. 2021;20:33. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iavarone C., O’Hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 37.Ho W., Gao M., Li F., Li Z., Zhang X., Xu X. Next-Generation Vaccines: Nanoparticle-Mediated DNA and mRNA Delivery. Adv. Healthc. Mater. 2021;10:e2001812. doi: 10.1002/adhm.202001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Jahanafrooz Z., Baradaran B., Mosafer J., Hashemzaei M., Rezaei T., Mokhtarzadeh A., Hamblin M.R. Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today. 2019;25:552–560. doi: 10.1016/j.drudis.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuller D.H., Berglund P. Amplifying RNA Vaccine Development. N. Engl. J. Med. 2020;382:2469–2471. doi: 10.1056/NEJMcibr2009737. [DOI] [PubMed] [Google Scholar]
- 41.Brenner S., Jacob F., Meselson M. An Unstable Intermediate Carrying Information from Genes to Ribosomes for Protein Synthesis. Nature. 1961;190:576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 42.Malone R.W., Felgner P.L., Verma I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 44.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.-G., Lévy J.-P., Meulien P. Induction of virus-specific cytotoxic T lymphocytesin vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 45.Zhou X., Berglund P., Rhodes G., Parker S., Jondal M., Liljeström P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine. 1994;12:1510–1514. doi: 10.1016/0264-410X(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 46.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- 47.Boczkowski D., Nair S.K., Snyder D., Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J. Exp. Med. 1996;184:465–472. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoerr I., Obst R., Rammensee H.G., Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur. J. Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Bloom K., Berg F.V.D., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosa S.S., Prazeres D.M., Azevedo A.M., Marques M.P. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39:2190–2200. doi: 10.1016/j.vaccine.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deering R.P., Kommareddy S., Ulmer J.B., Brito L.A., Geall A.J. Nucleic acid vaccines: Prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014;11:885–899. doi: 10.1517/17425247.2014.901308. [DOI] [PubMed] [Google Scholar]
- 52.Liu M.A. Immunologic Basis of Vaccine Vectors. Immunity. 2010;33:504–515. doi: 10.1016/j.immuni.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Beelman C.A., Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 54.Pogocki D., Schöneich C. Chemical stability of nucleic acid–derived drugs. J. Pharm. Sci. 2000;89:443–456. doi: 10.1002/(SICI)1520-6017(200004)89:4<443::AID-JPS2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 55.Mintzer M.A., Simanek E.E. Nonviral Vectors for Gene Delivery. Chem. Rev. 2008;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 56.Martin M.E., Rice K.G. Peptide-guided gene delivery. AAPS J. 2007;9:E18–E29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pack D.W., Hoffman A.S., Pun S., Stayton P. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 58.Lee H., Lytton-Jean A., Chen Y., Love K.T., Park A.I., Karagiannis E., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M., et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perche F., Benvegnu T., Berchel M., Lebegue L., Pichon C., Jaffrès P.-A., Midoux P. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomed. Nanotechnol. Biol. Med. 2011;7:445–453. doi: 10.1016/j.nano.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Thalhamer J., Weiss R., Scheiblhofer S., editors. Gene Vaccines. Springer; Vienna, Austria: 2012. [Google Scholar]
- 61.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Jong S., Chikh G., Sekirov L., Raney S., Semple S., Klimuk S., Yuan N., Hope M., Cullis P., Tam Y. Encapsulation in liposomal nanoparticles enhances the immunostimulatory, adjuvant and anti-tumor activity of subcutaneously administered CpG ODN. Cancer Immunol. Immunother. 2007;56:1251–1264. doi: 10.1007/s00262-006-0276-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pascolo S. The messenger’s great message for vaccination. Expert Rev. Vaccines. 2015;14:153–156. doi: 10.1586/14760584.2015.1000871. [DOI] [PubMed] [Google Scholar]
- 64.WHO Evaluation of the Quality, Safety and Efficacy of RNA-Based. [(accessed on 28 January 2021)]. Available online: https://www.who.int/docs/default-source/biologicals/ecbs/reg-considerations-on-rna-vaccines_1st-draft_pc_tz_22122020.pdf?sfvrsn=c13e1e20_3.
- 65.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coulie P.G., Van den Eynde B.J., Van Der Bruggen P., Boon-Falleur T. Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat. Rev. Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 67.Xu S., Yang K., Li R., Zhang L. mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection. Int. J. Mol. Sci. 2020;21:6582. doi: 10.3390/ijms21186582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang R., Billingsley M., Mitchell M.J. Biomaterials for vaccine-based cancer immunotherapy. J. Control. Release. 2018;292:256–276. doi: 10.1016/j.jconrel.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McNamara M.A., Nair S.K., Holl E.K. RNA-Based Vaccines in Cancer Immunotherapy. J. Immunol. Res. 2015;2015:1–9. doi: 10.1155/2015/794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 72.Weide B., Carralot J.-P., Reese A., Scheel B., Eigentler T.K., Hoerr I., Rammensee H.-G., Garbe C., Pascolo S. Results of the First Phase I/II Clinical Vaccination Trial With Direct Injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 73.Weide B., Pascolo S., Scheel B., Derhovanessian E., Pflugfelder A., Eigentler T., Pawelec G., Hoerr I., Rammensee H.-G., Garbe C. Direct Injection of Protamine-protected mRNA: Results of a Phase 1/2 Vaccination Trial in Metastatic Melanoma Patients. J. Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 74.Kowalski P., Rudra A., Miao L., Anderson D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilboa E., Vieweg J. Cancer immunotherapy with mRNA-transfected dendritic cells. Immunol. Rev. 2004;199:251–263. doi: 10.1111/j.0105-2896.2004.00139.x. [DOI] [PubMed] [Google Scholar]
- 76.Bol K.F., Figdor C., Aarntzen E.H., Welzen M.E., Van Rossum M.M., Blokx W., Van De Rakt M.W., Scharenborg N.M., De Boer A.J., Pots J.M., et al. Intranodal vaccination with mRNA-optimized dendritic cells in metastatic melanoma patients. OncoImmunology. 2015;4:e1019197. doi: 10.1080/2162402X.2015.1019197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Timmerman J.M., Czerwinski D.K., Davis T.A., Hsu F.J., Benike C., Hao Z.M., Taidi B., Rajapaksa R., Caspar C.B., Okada C.Y., et al. Idiotype-pulsed dendritic cell vaccination for B-cell lymphoma: Clinical and immune responses in 35 patients. Blood. 2002;99:1517–1526. doi: 10.1182/blood.V99.5.1517. [DOI] [PubMed] [Google Scholar]
- 78.Loveland B.E., Zhao A., White S., Gan H., Hamilton K., Xing P.-X., Pietersz G.A., Apostolopoulos V., Vaughan H., Karanikas V., et al. Mannan-MUC1–Pulsed Dendritic Cell Immunotherapy: A Phase I Trial in Patients with Adenocarcinoma. Clin. Cancer Res. 2006;12:869–877. doi: 10.1158/1078-0432.CCR-05-1574. [DOI] [PubMed] [Google Scholar]
- 79.Yanagisawa R., Koizumi T., Koya T., Sano K., Koido S., Nagai K., Kobayashi M., Okamoto M., Sugiyama H., Shimodaira S. WT1-pulsed Dendritic Cell Vaccine Combined with Chemotherapy for Resected Pancreatic Cancer in a Phase I Study. Anticancer Res. 2018;38:2217–2225. doi: 10.21873/anticanres.12464. [DOI] [PubMed] [Google Scholar]
- 80.Kallen K.-J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., Baumhof P., Scheel B., Koch S.D., Fotin-Mleczek M. A novel, disruptive vaccination technology. Hum. Vaccines Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rauch S., Lutz J., Kowalczyk A., Schlake T., Heidenreich R. In: RNA Vaccines. Kramps T., Elbers K., editors. Volume 1499. Springer; New York, NY, USA: 2017. Methods in Molecular Biology. [Google Scholar]
- 82.Kübler H., Scheel B., Gnad-Vogt U., Miller K., Schultze-Seemann W., Dorp F.V., Parmiani G., Hampel C., Wedel S., Trojan L., et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: A first-in-man phase I/IIa study. J. Immunother. Cancer. 2015;3:26. doi: 10.1186/s40425-015-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sebastian M., Schröder A., Scheel B., Hong H.S., Muth A., von Boehmer L., Zippelius A., Mayer F., Reck M., Atanackovic D., et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019;68:799–812. doi: 10.1007/s00262-019-02315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lesterhuis W.J., De Vries I.J.M., Schreibelt G., Schuurhuis D.H., Aarntzen E.H., De Boer A., Scharenborg N.M., Van De Rakt M., Hesselink E.J., Figdor C., et al. Immunogenicity of dendritic cells pulsed with CEA peptide or transfected with CEA mRNA for vaccination of colorectal cancer patients. Anticancer. Res. 2010;30:5091–5098. [PubMed] [Google Scholar]
- 85.Eckl J., Raffegerst S., Schnorfeil F., Prinz-Schulz P., Fingerhut C., Floisand Y., Josefsen D., Bigalke I., Pinkernell K., Kvalheim G., et al. DC Vaccination Induces Antigen Specific Immune Responses in AML Patients: A 1-Year Interim Assessment. Blood. 2019;134:3923. doi: 10.1182/blood-2019-129749. [DOI] [Google Scholar]
- 86.Sahin U., Derhovanessian E., Miller M., Kloke B.-P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 87.Wilgenhof S., Van Nuffel A., Benteyn D., Corthals J., Aerts C., Heirman C., Van Riet I., Bonehill A., Thielemans K., Neyns B. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann. Oncol. 2013;24:2686–2693. doi: 10.1093/annonc/mdt245. [DOI] [PubMed] [Google Scholar]
- 88.Borch T.H., Engell-Noerregaard L., Iversen T.Z., Ellebaek E., Met Ö., Hansen M., Andersen M.H., Straten P.T., Svane I.M. mRNA-transfected dendritic cell vaccine in combination with metronomic cyclophosphamide as treatment for patients with advanced malignant melanoma. OncoImmunology. 2016;5:e1207842. doi: 10.1080/2162402X.2016.1207842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kongsted P., Borch T.H., Ellebaek E., Iversen T.Z., Andersen R., Met Ö., Hansen M., Lindberg H., Sengeløv L., Svane I.M. Dendritic cell vaccination in combination with docetaxel for patients with metastatic castration-resistant prostate cancer: A randomized phase II study. Cytotherapy. 2017;19:500–513. doi: 10.1016/j.jcyt.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 90.Morse M.A., Hobeika A., Gwin W., Osada T., Gelles J., Rushing C., Niedzwiecki D., Lyerly H. Phase I study of alphaviral vector (AVX701) in colorectal cancer patients: Comparison of immune responses in stage III and stage IV patients. J. Immunother. Cancer. 2015;3:P444. doi: 10.1186/2051-1426-3-S2-P444. [DOI] [Google Scholar]
- 91.Krienke C., Kolb L., Diken E., Streuber M., Kirchhoff S., Bukur T., Akilli-Öztürk Ö., Kranz L.M., Berger H., Petschenka J., et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 92.Scheiblhofer S., Thalhamer J., Weiss R. DNA and mRNA vaccination against allergies. Pediatr. Allergy Immunol. 2018;29:679–688. doi: 10.1111/pai.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sahin U., Karikó K., Türeci Ö. MRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 94.Steinman L., Ho P.P., Robinson W.H., Utz P.J., Villoslada P. Antigen-specific tolerance to self-antigens in protein replacement therapy, gene therapy and autoimmunity. Curr. Opin. Immunol. 2019;61:46–53. doi: 10.1016/j.coi.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 95.Veiga N., Goldsmith M., Granot Y., Rosenblum D., Dammes N., Kedmi R., Ramishetti S., Peer D. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L., Wang F.-S., Gershwin M.E. Human autoimmune diseases: A comprehensive update. J. Intern. Med. 2015;278:369–395. doi: 10.1111/joim.12395. [DOI] [PubMed] [Google Scholar]
- 97.Steinle H., Weber J., Stoppelkamp S., Große-Berkenbusch K., Golombek S., Weber M., Canak-Ipek T., Trenz S.-M., Schlensak C., Avci-Adali M. Delivery of synthetic mRNAs for tissue regeneration. Adv. Drug Deliv. Rev. 2021;179:114007. doi: 10.1016/j.addr.2021.114007. [DOI] [PubMed] [Google Scholar]
- 98.Curin M., Khaitov M., Karaulov A., Namazova-Baranova L., Campana R., Garib V., Valenta R. Next-Generation of Allergen-Specific Immunotherapies: Molecular Approaches. Curr. Allergy Asthma Rep. 2018;18:39. doi: 10.1007/s11882-018-0790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barnes P.J. Therapeutic strategies for allergic diseases. Nature. 1999;402:31–38. doi: 10.1038/35037026. [DOI] [PubMed] [Google Scholar]
- 100.Linhart B., Valenta R. Vaccines for allergy. Curr. Opin. Immunol. 2012;24:354–360. doi: 10.1016/j.coi.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steveling-Klein E.H., Durham S.R. Encyclopedia of Respiratory Medicine. Elsevier; Amsterdam, The Netherlands: 2022. Immunotherapy for Allergy; pp. 491–502. [Google Scholar]
- 102.Roesler E., Weiss R., Weinberger E.E., Fruehwirth A., Stoecklinger A., Mostböck S., Ferreira F., Thalhamer J., Scheiblhofer S. Immunize and disappear—Safety-optimized mRNA vaccination with a panel of 29 allergens. J. Allergy Clin. Immunol. 2009;124:1070–1077.e11. doi: 10.1016/j.jaci.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 103.Hattinger E., Scheiblhofer S., Roesler E., Thalhamer T., Weiss R. Prophylactic mRNA Vaccination against Allergy Confers Long-Term Memory Responses and Persistent Protection in Mice. J. Immunol. Res. 2015;2015:1–12. doi: 10.1155/2015/797421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019;27:757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blakney A., Ip S., Geall A. An Update on Self-Amplifying mRNA Vaccine Development. Vaccines. 2021;9:97. doi: 10.3390/vaccines9020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ljungberg K., Liljestrom P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines. 2014;14:177–194. doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- 107.Ballesteros-Briones M.C., Silva-Pilipich N., Herrador-Cañete G., Vanrell L., Smerdou C. A new generation of vaccines based on alphavirus self-amplifying RNA. Curr. Opin. Virol. 2020;44:145–153. doi: 10.1016/j.coviro.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lundstrom K. Self-Amplifying RNA Viruses as RNA Vaccines. Int. J. Mol. Sci. 2020;21:5130. doi: 10.3390/ijms21145130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2017;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blakney A.K., McKay P.F., Christensen D., Yus B.I., Aldon Y., Follmann F., Shattock R.J. Effects of cationic adjuvant formulation particle type, fluidity and immunomodulators on delivery and immunogenicity of saRNA. J. Control. Release. 2019;304:65–74. doi: 10.1016/j.jconrel.2019.04.043. [DOI] [PubMed] [Google Scholar]
- 111.Luo F., Zheng L., Hu Y., Liu S., Wang Y., Xiong Z., Hu X., Tan F. Induction of Protective Immunity against Toxoplasma gondii in Mice by Nucleoside Triphosphate Hydrolase-II (NTPase-II) Self-amplifying RNA Vaccine Encapsulated in Lipid Nanoparticle (LNP) Front. Microbiol. 2017;8:605. doi: 10.3389/fmicb.2017.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garcia A.B., Siu E., Sun T., Exler V., Brito L., Hekele A., Otten G., Augustijn K., Janse C.J., Ulmer J.B., et al. Neutralization of the Plasmodium-encoded MIF ortholog confers protective immunity against malaria infection. Nat. Commun. 2018;9:2714. doi: 10.1038/s41467-018-05041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duthie M., Van Hoeven N., Macmillen Z., Picone A., Mohamath R., Erasmus J., Hsu F.-C., Stinchcomb D.T., Reed S.G. Heterologous Immunization With Defined RNA and Subunit Vaccines Enhances T Cell Responses That Protect Against Leishmania donovani. Front. Immunol. 2018;9:2420. doi: 10.3389/fimmu.2018.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raj D.K., Das Mohapatra A., Jnawali A., Zuromski J., Jha A., Cham-Kpu G., Sherman B., Rudlaff R.M., Nixon C.E., Hilton N., et al. Anti-PfGARP activates programmed cell death of parasites and reduces severe malaria. Nature. 2020;582:104–108. doi: 10.1038/s41586-020-2220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maruggi G., Chiarot E., Giovani C., Buccato S., Bonacci S., Frigimelica E., Margarit I., Geall A., Bensi G., Maione D. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. 2017;35:361–368. doi: 10.1016/j.vaccine.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 116.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhong Z., Catani J.P.P., Mc Cafferty S., Couck L., Broeck W.V.D., Gorlé N., Vandenbroucke R.E., Devriendt B., Ulbert S., Cnops L., et al. Immunogenicity and Protection Efficacy of a Naked Self-Replicating mRNA-Based Zika Virus Vaccine. Vaccines. 2019;7:96. doi: 10.3390/vaccines7030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Roth C., Cantaert T., Colas C., Prot M., Casadémont I., Levillayer L., Thalmensi J., Langlade-Demoyen P., Gerke C., Bahl K., et al. A Modified mRNA Vaccine Targeting Immunodominant NS Epitopes Protects Against Dengue Virus Infection in HLA Class I Transgenic Mice. Front. Immunol. 2019;10:1424. doi: 10.3389/fimmu.2019.01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Szurgot I., Ljungberg K., Kümmerer B.M., Liljeström P. Infectious RNA vaccine protects mice against chikungunya virus infection. Sci. Rep. 2020;10:21076. doi: 10.1038/s41598-020-78009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aldrich C., Leroux–Roels I., Huang K.B., Bica M.A., Loeliger E., Schoenborn-Kellenberger O., Walz L., Leroux-Roels G., von Sonnenburg F., Oostvogels L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: A phase 1 trial. Vaccine. 2021;39:1310–1318. doi: 10.1016/j.vaccine.2020.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-Based Vaccines Elicit Robust Immune Responses and Protect Guinea Pigs From Ebola Virus Disease. J. Infect. Dis. 2017;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Egan K.P., Hook L.M., Naughton A., Pardi N., Awasthi S., Cohen G.H., Weissman D., Friedman H.M. An HSV-2 nucleoside-modified mRNA genital herpes vaccine containing glycoproteins gC, gD, and gE protects mice against HSV-1 genital lesions and latent infection. PLoS Pathog. 2020;16:e1008795. doi: 10.1371/journal.ppat.1008795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nelson C.S., Jenks J.A., Pardi N., Goodwin M., Roark H., Edwards W., McLellan J.S., Pollara J., Weissman D., Permar S.R. Human Cytomegalovirus Glycoprotein B Nucleoside-Modified mRNA Vaccine Elicits Antibody Responses with Greater Durability and Breadth than MF59-Adjuvanted gB Protein Immunization. J. Virol. 2020;94:e00186-20. doi: 10.1128/JVI.00186-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.D’Haese S., Lacroix C., Garcia F., Plana M., Ruta S., Vanham G., Verrier B., Aerts J.L. Off the beaten path: Novel mRNA-nanoformulations for therapeutic vaccination against HIV. J. Control. Release. 2020;330:1016–1033. doi: 10.1016/j.jconrel.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 125.Fleeton M.N., Chen M.S., Berglund P., Rhodes G., Parker S.E., Murphy M., Atkins G.J., Liljestrom P. Self-Replicative RNA Vaccines Elicit Protection against Influenza A Virus, Respiratory Syncytial Virus, and a Tickborne Encephalitis Virus. J. Infect. Dis. 2001;183:1395–1398. doi: 10.1086/319857. [DOI] [PubMed] [Google Scholar]
- 126.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.-J., Stitz L., et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 127.Wei C.-J., Crank M.C., Shiver J., Graham B.S., Mascola J.R., Nabel G.J. Next-generation influenza vaccines: Opportunities and challenges. Nat. Rev. Drug Discov. 2020;19:239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jacobson J.M., Routy J.-P., Welles S., DeBenedette M., Tcherepanova I., Angel J.B., Asmuth D.M., Stein D.K., Baril J.-G., McKellar M., et al. Dendritic Cell Immunotherapy for HIV-1 Infection Using Autologous HIV-1 RNA. JAIDS J. Acquir. Immune Defic. Syndr. 2016;72:31–38. doi: 10.1097/QAI.0000000000000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gandhi R.T., Kwon D.S., Macklin E., Shopis J.R., McLean A.P., McBrine N., Flynn T., Peter L., Sbrolla A., Kaufmann D.E., et al. Immunization of HIV-1-Infected Persons With Autologous Dendritic Cells Transfected With mRNA Encoding HIV-1 Gag and Nef. JAIDS J. Acquir. Immune Defic. Syndr. 2016;71:246–253. doi: 10.1097/QAI.0000000000000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 131.Shaw C., Lee H., Knightly C., Kalidindi S., Zaks T., Smolenov I., Panther L. 2754. Phase 1 Trial of an mRNA-Based Combination Vaccine Against hMPV and PIV3. Open Forum Infect. Dis. 2019;6:S970. doi: 10.1093/ofid/ofz360.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shaw C., Panther L., August A., Zaks T., Smolenov I., Bart S., Watson M. Safety and immunogenicity of a mRNA-based chikungunya vaccine in a phase 1 dose-ranging trial. Int. J. Infect. Dis. 2019;79:17. doi: 10.1016/j.ijid.2018.11.058. [DOI] [Google Scholar]
- 133.Feldman R.A., Fuhr R., Smolenov I., Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ö., Pujar H.S., et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 134.Leal L., Guardo A.C., Morón-López S., Salgado M., Mothe B., Heirman C., Pannus P., Vanham G., Ham H.J.V.D., Gruters R., et al. Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS. 2018;32:2533–2545. doi: 10.1097/QAD.0000000000002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bernstein D.I., Reap E.A., Katen K., Watson A., Smith K., Norberg P., Olmsted R.A., Hoeper A., Morris J., Negri S., et al. Randomized, double-blind, Phase 1 trial of an alphavirus replicon vaccine for cytomegalovirus in CMV seronegative adult volunteers. Vaccine. 2009;28:484–493. doi: 10.1016/j.vaccine.2009.09.135. [DOI] [PubMed] [Google Scholar]
- 136.Wecker M., Gilbert P., Russell N., Hural J., Allen M., Pensiero M., Chulay J., Chiu Y.-L., Karim S.A., Burke D.S. Phase I Safety and Immunogenicity Evaluations of an Alphavirus Replicon HIV-1 Subtype CgagVaccine in Healthy HIV-1-Uninfected Adults. Clin. Vaccine Immunol. 2012;19:1651–1660. doi: 10.1128/CVI.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Padron-Regalado E. Vaccines for SARS-CoV-2: Lessons from Other Coronavirus Strains. Infect. Dis. Ther. 2020;9:255–274. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.The Lancet Respiratory Medicine Realising the potential of SARS-CoV-2 vaccines—A long shot? Lancet Respir. Med. 2021;9:117. doi: 10.1016/S2213-2600(21)00045-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Forni G., Mantovani A. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021;28:626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ledford H. Moderna COVID vaccine becomes second to get US authorization. Nature. 2020 doi: 10.1038/d41586-020-03593-7. [DOI] [PubMed] [Google Scholar]
- 141.WHO Draft Landscape and Tracker of COVID-19 Candidate Vaccines. [(accessed on 1 October 2021)]. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- 142.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3–9. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 145.Erasmus J.H., Khandhar A.P., O’Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., et al. An Alphavirus -derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rauch S., Roth N., Schwendt K., Fotin-Mleczek M., Mueller S.O., Petsch B. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. npj Vaccines. 2021;6:1–9. doi: 10.1038/s41541-021-00311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rauch S., Gooch K., Hall Y., Salguero F.J., Dennis M.J., Gleeson F.V., Harris D., Ho C., Humphries H.E., Longet S., et al. mRNA vaccine CVnCoV protects non-human primates from SARS-CoV-2 challenge infection. bioRxiv. 2020 doi: 10.1101/2020.12.23.424138. [DOI] [Google Scholar]
- 149.De Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., Yau C., Low J.G., Kalimuddin S., Matsuda D., et al. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021;29:1970–1983. doi: 10.1016/j.ymthe.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Neumann P.J., Cohen J.T., Kim D.D., Ollendorf D.A. Consideration Of Value-Based Pricing For Treatments And Vaccines Is Important, Even In The COVID-19 Pandemic. Health Aff. 2021;40:53–61. doi: 10.1377/hlthaff.2020.01548. [DOI] [PubMed] [Google Scholar]
- 152.Belete T.M. Review on Up-to-Date Status of Candidate Vaccines for COVID-19 Disease. Infect. Drug Resist. 2021;14:151–161. doi: 10.2147/IDR.S288877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mullard A. COVID-19 vaccine development pipeline gears up. Lancet. 2020;395:1751–1752. doi: 10.1016/S0140-6736(20)31252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shenoy P. Multi-regional clinical trials and global drug development. Perspect. Clin. Res. 2016;7:62–67. doi: 10.4103/2229-3485.179430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Broseta J.J., Rodríguez-Espinosa D., Soruco E., Maduell F. Weekly seroconversion rate of the mRNA-1273 SARS-CoV-2 vaccine in haemodialysis patients. Nephrol. Dial. Transplant. 2021;36:1754–1755. doi: 10.1093/ndt/gfab195. [DOI] [PubMed] [Google Scholar]
- 160.Ikizler T.A., Coates P.T., Rovin B.H., Ronco P. Immune response to SARS-CoV-2 infection and vaccination in patients receiving kidney replacement therapy. Kidney Int. 2021;99:1275–1279. doi: 10.1016/j.kint.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Caillard S., Chavarot N., Bertrand D., Kamar N., Thaunat O., Moal V., Masset C., Hazzan M., Gatault P., Sicard A., et al. Occurrence of severe COVID-19 in vaccinated transplant patients. Kidney Int. 2021;100:477–479. doi: 10.1016/j.kint.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boyarsky B.J., Werbel W.A., Avery R.K., Tobian A.A.R., Massie A.B., Segev D.L., Garonzik-Wang J.M. Immunogenicity of a Single Dose of SARS-CoV-2 Messenger RNA Vaccine in Solid Organ Transplant Recipients. JAMA. 2021;325:1784. doi: 10.1001/jama.2021.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chu L., McPhee R., Huang W., Bennett H., Pajon R., Nestorova B., Leav B. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine. 2021;39:2791–2799. doi: 10.1016/j.vaccine.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kremsner P.G., Mann P., Kroidl A., Leroux-Roels I., Schindler C., Gabor J.J., Schunk M., Leroux-Roels G., Bosch J.J., Fendel R., et al. Safety and immunogenicity of an mRNA-lipid nanoparticle vaccine candidate against SARS-CoV-2. Wien. Klin. Wochenschr. 2021;133:931–941. doi: 10.1007/s00508-021-01922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dolgin E. CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature. 2021;594:483. doi: 10.1038/d41586-021-01661-0. [DOI] [PubMed] [Google Scholar]
- 169.Low J.G., de Alwis R., Chen S., Kalimuddin S., Leong Y.S., Mah T.K.L., Yuen N., Tan H.C., Zhang S.L., Sim J.X.Y., et al. A phase 1/2 randomized, double-blinded, placebo controlled ascending dose trial to assess the safety, tolerability and immunogenicity of ARCT-021 in healthy adults. medRxiv. 2021 doi: 10.1101/2021.07.01.21259831. [DOI] [Google Scholar]
- 170.Liu K., Gu Z., Islam S., Scherngell T., Kong X., Zhao J., Chen X., Hu Y. Global landscape of patents related to human coronaviruses. Int. J. Biol. Sci. 2021;17:1588–1599. doi: 10.7150/ijbs.58807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Knezevic I., Liu M.A., Peden K., Zhou T., Kang H.-N. Development of mRNA Vaccines: Scientific and Regulatory Issues. Vaccines. 2021;9:81. doi: 10.3390/vaccines9020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li Y.-D., Chi W.-Y., Su J.-H., Ferrall L., Hung C.-F., Wu T.-C. Coronavirus vaccine development: From SARS and MERS to COVID-19. J. Biomed. Sci. 2020;27:104. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T-cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 174.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 175.Li J., Hui A., Zhang X., Yang Y., Tang R., Ye H., Ji R., Lin M., Zhu Z., Türeci Ö., et al. Safety and immunogenicity of the SARS-CoV-2 BNT162b1 mRNA vaccine in younger and older Chinese adults: A randomized, placebo-controlled, double-blind phase 1 study. Nat. Med. 2021;27:1062–1070. doi: 10.1038/s41591-021-01330-9. [DOI] [PubMed] [Google Scholar]
- 176.NIH A Trial Investigating the Safety and Effects of Four BNT162 Vaccines Against COVID-2019 in Healthy and Immunocompromised. Adults. [(accessed on 13 July 2021)]; Available online: https://clinicaltrials.gov/ct2/show/NCT04380701.
- 177.Subbaraman N. Pregnancy and COVID: What the data say. Nature. 2021;591:193–195. doi: 10.1038/d41586-021-00578-y. [DOI] [PubMed] [Google Scholar]
- 178.Goldshtein I., Nevo D., Steinberg D.M., Rotem R.S., Gorfine M., Chodick G., Segal Y. Association Between BNT162b2 Vaccination and Incidence of SARS-CoV-2 Infection in Pregnant Women. JAMA. 2021;326:728. doi: 10.1001/jama.2021.11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Peretz S.B., Regev N., Novick L., Nachshol M., Goffer E., Ben-David A., Asraf K., Doolman R., Levin E.G., Yochay G.R., et al. Short-term outcome of pregnant women vaccinated with BNT162b2 mRNA COVID-19 vaccine. Ultrasound Obstet. Gynecol. 2021;58:450–456. doi: 10.1002/uog.23729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ledford H., Cyranoski D., Van Noorden R. The UK has approved a COVID vaccine—Here’s what scientists now want to know. Nature. 2020;588:205–206. doi: 10.1038/d41586-020-03441-8. [DOI] [PubMed] [Google Scholar]
- 181.Ledford H. US authorization of first COVID vaccine marks new phase in safety monitoring. Nature. 2020;588:377–378. doi: 10.1038/d41586-020-03542-4. [DOI] [PubMed] [Google Scholar]
- 182.Kim J.H., Marks F., Clemens J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021;27:205–211. doi: 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 183.Clemens J., Brenner R., Rao M., Tafari N., Lowe C. Evaluating New Vaccines for Developing Countries. JAMA. 1996;275:390–397. doi: 10.1001/jama.1996.03530290060038. [DOI] [PubMed] [Google Scholar]
- 184.Victora C.G., Habicht J.-P., Bryce J. Evidence-Based Public Health: Moving Beyond Randomized Trials. Am. J. Public Health. 2004;94:400–405. doi: 10.2105/AJPH.94.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chodick G., Tene L., Patalon T., Gazit S., Ben Tov A., Cohen D., Muhsen K. Assessment of Effectiveness of 1 Dose of BNT162b2 Vaccine for SARS-CoV-2 Infection 13 to 24 Days After Immunization. JAMA Netw. Open. 2021;4:e2115985. doi: 10.1001/jamanetworkopen.2021.15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Murillo-Zamora E., Trujillo X., Huerta M., Riós-Silva M., Mendoza-Cano O. Effectiveness of BNT162b2 COVID-19 Vaccine in Preventing Severe Symptomatic Infection among Healthcare Workers. Medicina. 2021;57:746. doi: 10.3390/medicina57080746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bernal J.L., Andrews N., Gower C., Robertson C., Stowe J., Tessier E., Simmons R., Cottrell S., Roberts R., O’Doherty M., et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: Test negative case-control study. BMJ. 2021;373:n1088. doi: 10.1136/bmj.n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vasileiou E., Simpson C.R., Shi T., Kerr S., Agrawal U., Akbari A., Bedston S., Beggs J., Bradley D., Chuter A., et al. Interim findings from first-dose mass COVID-19 vaccination roll-out and COVID-19 hospital admissions in Scotland: A national prospective cohort study. Lancet. 2021;397:1646–1657. doi: 10.1016/S0140-6736(21)00677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tenforde M.W., Patel M.M., Ginde A.A., Douin D.J., Talbot H.K., Casey J.D., Mohr N.M., Zepeski A., Gaglani M., McNeal T., et al. Effectiveness of Severe Acute Respiratory Syndrome Coronavirus 2 Messenger RNA Vaccines for Preventing Coronavirus Disease 2019 Hospitalizations in the United States. Clin. Infect. Dis. 2021:ciab687. doi: 10.1093/cid/ciab687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zacay G., Shasha D., Bareket R., Kadim I., Sikron F.H., Tsamir J., Mossinson D., Heymann A.D. BNT162b2 Vaccine Effectiveness in Preventing Asymptomatic Infection With SARS-CoV-2 Virus: A Nationwide Historical Cohort Study. Open Forum Infect. Dis. 2021;8:ofab262. doi: 10.1093/ofid/ofab262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernán M.A., Lipsitch M., Reis B., Balicer R.D. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.FDA Coronavirus (COVID-19) Update: FDA Authorizes Pfizer-BioNTech COVID-19 Vaccine for Emergency Use in Adolescents in Another Important Action in Fight Against Pandemic. [(accessed on 9 August 2021)]; Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-pfizer-biontech-covid-19-vaccine-emergency-use.
- 193.Health Canada Health Canada Authorizes Use of the Pfizer-BioNTech COVID-19 Vaccine in Children 12 to 15 Years of Age. [(accessed on 9 August 2021)]. Available online: https://www.canada.ca/en/health-canada/news/2021/05/health-canada-authorizes-use-of-the-pfizer-biontech-covid-19-vaccine-in-children-12-to-15-years-of-age.html.
- 194.Agência Brasil Brazil Authorizes Pfizer Vaccine for Children Aged 12 and Older. [(accessed on 9 August 2021)]. Available online: https://agenciabrasil.ebc.com.br/en/saude/noticia/2021-06/brazil-authorizes-pfizer-vaccine-children-aged-12-and-older.
- 195.EMA First COVID-19 Vaccine Approved for Children Aged 12 to 15 in EU. [(accessed on 9 August 2021)]. Available online: https://www.ema.europa.eu/en/news/first-covid-19-vaccine-approved-children-aged-12-15-eu.
- 196.Frenck R.W., Klein N.P., Kitchin N., Gurtman A., Absalon J., Lockhart S., Perez J.L., Walter E.B., Senders S., Bailey R., et al. Safety, Immunogenicity, and Efficacy of the BNT162b2 Covid-19 Vaccine in Adolescents. N. Engl. J. Med. 2021;385:239–250. doi: 10.1056/NEJMoa2107456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Batty C.J., Heise M.T., Bachelder E.M., Ainslie K.M. Vaccine formulations in clinical development for the prevention of severe acute respiratory syndrome coronavirus 2 infection. Adv. Drug Deliv. Rev. 2020;169:168–189. doi: 10.1016/j.addr.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. npj Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Baker N., Dolgin E. Coronapod: CureVac disappoints in COVID vaccine trial. Nature. 2021 doi: 10.1038/d41586-021-01694-5. [DOI] [PubMed] [Google Scholar]
- 201.Slavov S.N., Patané J.S.L., Bezerra R.D.S., Giovanetti M., Fonseca V., Martins A.J., Viala V.L., Rodrigues E.S., Santos E.V., Barros C.R.S., et al. Genomic monitoring unveil the early detection of the SARS-CoV-2 B.1.351 (beta) variant (20H/501Y.V2) in Brazil. J. Med. Virol. 2021;93:6782–6787. doi: 10.1002/jmv.27190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Romero P.E., Dávila-Barclay A., Salvatierra G., González L., Cuicapuza D., Solis L., Marcos-Carbajal P., Huancachoque J., Maturrano L., Tsukayama P. The Emergence of SARS-CoV-2 Variant Lambda (C.37) in South America. medRxiv. 2021 doi: 10.1128/Spectrum.00789-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Woldemeskel B.A., Garliss C.C., Blankson J.N. SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J. Clin. Investig. 2021;131:e149335. doi: 10.1172/JCI149335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zani A., Caccuri F., Messali S., Bonfanti C., Caruso A. Serosurvey in BNT162b2 vaccine-elicited neutralizing antibodies against authentic B.1, B.1.1.7, B.1.351, B.1.525 and P.1 SARS-CoV-2 variants. Emerg. Microbes Infect. 2021;10:1241–1243. doi: 10.1080/22221751.2021.1940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jalkanen P., Kolehmainen P., Häkkinen H.K., Huttunen M., Tähtinen P.A., Lundberg R., Maljanen S., Reinholm A., Tauriainen S., Pakkanen S.H., et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat. Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: The COVID-19 case. J. Control. Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hoffmann D., Corleis B., Rauch S., Roth N., Mühe J., Halwe N.J., Ulrich L., Fricke C., Schön J., Kraft A., et al. CVnCoV and CV2CoV protect human ACE2 transgenic mice from ancestral B BavPat1 and emerging B.1.351 SARS-CoV-2. Nat. Commun. 2021;12:4048. doi: 10.1038/s41467-021-24339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li D.-D., Li Q.-H. SARS-CoV-2: Vaccines in the pandemic era. Mil. Med. Res. 2021;8:1–15. doi: 10.1186/s40779-020-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pinghui Z. Coronavirus: Late-Stage Trial of Chinese mRNA Vaccine Candidate to Begin in Mexico. [(accessed on 7 August 2021)]. Available online: https://www.scmp.com/news/china/science/article/3133200/coronavirus-late-stage-trial-chinese-mrna-vaccine-candidate.
- 211.Therapeutics A. Arcturus—A clinical-stage mRNA therapeutics and vaccines company. [(accessed on 30 September 2021)];Biopharma Deal. 2021 2682:B9. Available online: https://www.nature.com/articles/d43747-021-00073-3. [Google Scholar]
- 212.Zhao J., Zhao S., Ou J., Zhang J., Lan W., Guan W., Wu X., Yan Y., Zhao W., Wu J., et al. COVID-19: Coronavirus Vaccine Development Updates. Front. Immunol. 2020;11:3435. doi: 10.3389/fimmu.2020.602256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Scheuber A. COVID-19 Vaccine Secures New Government Investment. [(accessed on 10 August 2021)]. Available online: https://www.imperial.ac.uk/news/197573/covid-19-vaccine-secures-government-investment/
- 214.Scheuber A. Imperial Social Enterprise to Accelerate Low-Cost COVID-19 Vaccine. [(accessed on 10 August 2021)]. Available online: https://www.imperial.ac.uk/news/198053/imperial-social-enterprise-accelerate-low-cost-covid-19/
- 215.Cagigi A., Loré K. Immune Responses Induced by mRNA Vaccination in Mice, Monkeys and Humans. Vaccines. 2021;9:61. doi: 10.3390/vaccines9010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Abu-Raddad L.J., Chemaitelly H., Butt A.A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., Blane B., Bonsall D., Cicconi P., Charlton S., et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): An exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hitchings M.D., Ranzani O.T., Torres M.S.S., de Oliveira S.B., Almiron M., Said R., Borg R., Schulz W.L., de Oliveira R.D., da Silva P.V., et al. Effectiveness of CoronaVac among healthcare workers in the setting of high SARS-CoV-2 Gamma variant transmission in Manaus, Brazil: A test-negative case-control study. Lancet Reg. Health—Am. 2021;1:100025. doi: 10.1016/j.lana.2021.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G.-W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. Biol. 2021;4:228. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cai C., Peng Y., Shen E., Huang Q., Chen Y., Liu P., Guo C., Feng Z., Gao L., Zhang X., et al. A comprehensive analysis of the efficacy and safety of COVID-19 vaccines. Mol. Ther. 2021;29:2794–2805. doi: 10.1016/j.ymthe.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Blasi F., Gramegna A., Sotgiu G., Saderi L., Voza A., Aliberti S., Amati F. SARS-CoV-2 vaccines: A critical perspective through efficacy data and barriers to herd immunity. Respir. Med. 2021;180:106355. doi: 10.1016/j.rmed.2021.106355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 225.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M.E., Zhao Y., Duyvesteyn H.M., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29:529–539.e3. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 229.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Moore J.P., Offit P.A. SARS-CoV-2 Vaccines and the Growing Threat of Viral Variants. JAMA. 2021;325:821. doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 231.Guevara M.L., Persano F., Persano S. Advances in Lipid Nanoparticles for mRNA-Based Cancer Immunotherapy. Front. Chem. 2020;8:589959. doi: 10.3389/fchem.2020.589959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: Optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chen S., Tam Y.Y.C., Lin P.J., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 234.Park K.S., Sun X., Aikins M.E., Moon J.J. Non-viral COVID-19 vaccine delivery systems. Adv. Drug Deliv. Rev. 2020;169:137–151. doi: 10.1016/j.addr.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Habrant D., Peuziat P., Colombani T., Dallet L., Gehin J., Goudeau E., Evrard B., Lambert O., Haudebourg T., Pitard B. Design of Ionizable Lipids To Overcome the Limiting Step of Endosomal Escape: Application in the Intracellular Delivery of mRNA, DNA, and siRNA. J. Med. Chem. 2016;59:3046–3062. doi: 10.1021/acs.jmedchem.5b01679. [DOI] [PubMed] [Google Scholar]
- 236.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Messengers of hope. Nat. Biotechnol. 2020;39:1. doi: 10.1038/s41587-020-00807-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Buschmann M., Carrasco M., Alishetty S., Paige M., Alameh M., Weissman D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines. 2021;9:65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Arcturus Therapeutics Addressing Key Challenges of Lipid-Mediated Delivery Systems for mRNA through Innovation. [(accessed on 12 August 2021)]. Available online: https://endpts.com/sp/addressing-key-challenges-of-lipid-mediated-delivery-systems-for-mrna-through-innovation/
- 240.Thi T., Suys E., Lee J., Nguyen D., Park K., Truong N. Lipid-Based Nanoparticles in the Clinic and Clinical Trials: From Cancer Nanomedicine to COVID-19 Vaccines. Vaccines. 2021;9:359. doi: 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., et al. A Cationic Nanoemulsion for the Delivery of Next-generation RNA Vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Linares-Fernández S., Lacroix C., Exposito J.-Y., Verrier B. Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2019;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 244.Granados-Riveron J.T., Aquino-Jarquin G. Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed. Pharmacother. 2021;142:111953. doi: 10.1016/j.biopha.2021.111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Von Niessen A.G.O., Poleganov M.A., Rechner C., Plaschke A., Kranz L., Fesser S., Diken M., Löwer M., Vallazza B., Beissert T., et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2018;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA. 2019;116:24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2013;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Nance K.D., Meier J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Central Sci. 2021;7:748–756. doi: 10.1021/acscentsci.1c00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and Immunogenic Characterization of Recombinant Baculovirus-Expressed Severe Acute Respiratory Syndrome Coronavirus Spike Protein: Implication for Vaccine Design. J. Virol. 2006;80:5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Volz A., Kupke A., Song F., Jany S., Fux R., Shams-Eldin H., Schmidt J.C., Becker C., Eickmann M., Becker S., et al. Protective Efficacy of Recombinant Modified Vaccinia Virus Ankara Delivering Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein. J. Virol. 2015;89:8651–8656. doi: 10.1128/JVI.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2020;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wang S.-F., Tseng S.-P., Yen C.-H., Yang J.-Y., Tsao C.-H., Shen C.-W., Chen K.-H., Liu F.-T., Liu W.-T., Chen Y.-M.A., et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., et al. Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tseng C.-T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS Coronavirus Vaccines Leads to Pulmonary Immunopathology on Challenge with the SARS Virus. PLoS ONE. 2012;7:e35421. doi: 10.1371/annotation/2965cfae-b77d-4014-8b7b-236e01a35492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains Multiple Conformation-Dependent Epitopes that Induce Highly Potent Neutralizing Antibodies. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 259.Kis Z., Kontoravdi C., Shattock R., Shah N. Resources, Production Scales and Time Required for Producing RNA Vaccines for the Global Pandemic Demand. Vaccines. 2020;9:3. doi: 10.3390/vaccines9010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Irwin A. What it will take to vaccinate the world against COVID-19. Nature. 2021;592:176–178. doi: 10.1038/d41586-021-00727-3. [DOI] [PubMed] [Google Scholar]
- 261.WHO COVID19 Vaccine Tracker. [(accessed on 9 October 2021)]. Available online: https://covid19.trackvaccines.org/
- 262.Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Favresse J., Bayart J.-L., Mullier F., Elsen M., Eucher C., Van Eeckhoudt S., Roy T., Wieers G., Laurent C., Dogné J.-M., et al. Antibody titres decline 3-month post-vaccination with BNT162b2. Emerg. Microbes Infect. 2021;10:1495–1498. doi: 10.1080/22221751.2021.1953403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 265.Thomas S.J., Moreira E.D., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Polack F.P., Zerbini C., et al. Six Month Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. medRxiv. 2021 doi: 10.1101/2021.07.28.21261159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Benotmane I., Gautier G., Perrin P., Olagne J., Cognard N., Fafi-Kremer S., Caillard S. Antibody Response After a Third Dose of the mRNA-1273 SARS-CoV-2 Vaccine in Kidney Transplant Recipients With Minimal Serologic Response to 2 Doses. JAMA. 2021;326:1063. doi: 10.1001/jama.2021.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hall V.G., Ferreira V.H., Ku T., Ierullo M., Majchrzak-Kita B., Chaparro C., Selzner N., Schiff J., McDonald M., Tomlinson G., et al. Randomized Trial of a Third Dose of mRNA-1273 Vaccine in Transplant Recipients. N. Engl. J. Med. 2021;385:1244–1246. doi: 10.1056/NEJMc2111462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Del Bello A., Abravanel F., Marion O., Couat C., Esposito L., Lavayssière L., Izopet J., Kamar N. Efficiency of a boost with a third dose of anti-SARS-CoV-2 messenger RNA-based vaccines in solid organ transplant recipients. Am. J. Transplant. 2021 doi: 10.1111/ajt.16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Burki T. Booster shots for COVID-19—The debate continues. Lancet Infect. Dis. 2021;21:1359–1360. doi: 10.1016/S1473-3099(21)00574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Callaway E., Ledford H. How to redesign COVID vaccines so they protect against variants. Nature. 2021;590:15–16. doi: 10.1038/d41586-021-00241-6. [DOI] [PubMed] [Google Scholar]
- 271.Engelbrecht F.A., Scholes R.J. Test for Covid-19 seasonality and the risk of second waves. One Health. 2020;12:100202. doi: 10.1016/j.onehlt.2020.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Abdulla Z., Al-Bashir S., Al-Salih N., Aldamen A., Abdulazeez M. A Summary of the SARS-CoV-2 Vaccines and Technologies Available or under Development. Pathogens. 2021;10:788. doi: 10.3390/pathogens10070788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Sandbrink J.B., Shattock R.J. RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front. Immunol. 2020;11:3329. doi: 10.3389/fimmu.2020.608460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Crommelin D.J., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2020;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.The Lancet Infectious Diseases COVID-19 vaccine equity and booster doses. Lancet Infect. Dis. 2021;21:1193. doi: 10.1016/S1473-3099(21)00486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Choi E.M. COVID-19 vaccines for low- and middle-income countries. Trans. R. Soc. Trop. Med. Hyg. 2021;115:447–456. doi: 10.1093/trstmh/trab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Santos A., Gaspar P., de Souza H. Refrigeration of COVID-19 Vaccines: Ideal Storage Characteristics, Energy Efficiency and Environmental Impacts of Various Vaccine Options. Energies. 2021;14:1849. doi: 10.3390/en14071849. [DOI] [Google Scholar]
- 278.Ball R.L., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: Examining the effect of pH, temperature, and lyophilization. Int. J. Nanomed. 2016;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Maxmen A. The fight to manufacture COVID vaccines in lower-income countries. Nature. 2021;597:455–457. doi: 10.1038/d41586-021-02383-z. [DOI] [PubMed] [Google Scholar]
- 280.Hotez P.J., Narayan K.M.V. Restoring Vaccine Diplomacy. JAMA. 2021;325:2337. doi: 10.1001/jama.2021.7439. [DOI] [PubMed] [Google Scholar]
- 281.Kavanagh M.M., Gostin L.O., Sunder M. Sharing Technology and Vaccine Doses to Address Global Vaccine Inequity and End the COVID-19 Pandemic. JAMA. 2021;326:219. doi: 10.1001/jama.2021.10823. [DOI] [PubMed] [Google Scholar]
- 282.Machingaidze S., Wiysonge C.S. Understanding COVID-19 vaccine hesitancy. Nat. Med. 2021;27:1338–1339. doi: 10.1038/s41591-021-01459-7. [DOI] [PubMed] [Google Scholar]
- 283.Ball P. Anti-vaccine movement could undermine efforts to end coronavirus pandemic, researchers warn. Nature. 2020;581:251. doi: 10.1038/d41586-020-01423-4. [DOI] [PubMed] [Google Scholar]
- 284.Arce J.S.S., Warren S.S., Meriggi N.F., Scacco A., McMurry N., Voors M., Syunyaev G., Malik A.A., Aboutajdine S., Adeojo O., et al. COVID-19 vaccine acceptance and hesitancy in low- and middle-income countries. Nat. Med. 2021;27:1385–1394. doi: 10.1038/s41591-021-01454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Mathioudakis A.G., Ghrew M., Ustianowski A., Ahmad S., Borrow R., Papavasileiou L.P., Petrakis D., Bakerly N.D. Self-Reported Real-World Safety and Reactogenicity of COVID-19 Vaccines: A Vaccine Recipient Survey. Life. 2021;11:249. doi: 10.3390/life11030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hacknen D. Por que os Brasileiros Estão Preferindo Tomar a Vacina da Pfizer? [(accessed on 14 August 2021)]. Available online: https://jc.ne10.uol.com.br/brasil/2021/06/12133656-por-que-os-brasileiros-estao-preferindo-tomar-a-vacina-da-pfizer.html.
- 287.Pardi N., Hogan M., Pelc R., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.