Abstract
A long-term assessment of quantitative hepatitis C virus (HCV) testing was performed at the University of Pittsburgh Medical Center. The Quantiplex HCV RNA 2.0 branched-chain DNA (bDNA) assay (Bayer Diagnostics) for hepatitis C viral load determination was used to test 3,471 specimens. bDNA-negative samples were also tested by an in-house qualitative reverse transcriptase (RT)-PCR assay with a measured sensitivity of fewer than 100 HCV genome equivalents per milliliter. Of 1,239 bDNA-negative specimens, 74.1% were negative and 25.9% were positive by RT-PCR, indicating the presence of viremia in a significant proportion of bDNA-negative samples. We discuss the medical and economic implications of these results and propose two alternatives for clinical laboratories to consider in approaching quantitative HCV testing. For laboratories able to perform a sensitive RT-PCR assay for ≤40% of the bDNA test cost, prescreening bDNA requests by RT-PCR may be the most cost-effective approach.
With an estimated 4 million Americans infected by hepatitis C virus (HCV), this agent has become an important cause of chronic liver disease (3, 30). Primary infection with HCV leads to persistent viremia in ∼85% of patients, with development of chronic liver disease in >60% of cases. Approximately 20% of individuals with chronic hepatitis C eventually develop medically significant sequelae, including cirrhosis, end stage liver disease, or hepatocellular carcinoma (3). Thus, therapeutic management of HCV infection has become an important medical problem. Recombinant interferon alone or in combination with ribavirin are current treatments used for chronic hepatitis C (5, 23).
Quantitation of HCV RNA in serum or plasma has become an important clinical assay for assessing and managing chronic HCV infection and is commonly used to assess prognosis and monitor the efficacy of antiviral therapy (6, 7, 10, 11, 13, 17, 21, 24, 33). Among several host-related and viral factors, greater treatment efficacy has been associated with lower hepatitis C viral loads before therapy or after commencing therapy, emphasizing the need for HCV quantitation (23, 31, 32). Several quantitative HCV assays are commercially available, employing primarily two technologies to assess HCV RNA levels: target amplification approaches, such as quantitative reverse transcriptase PCR (RT-PCR) assays, and signal amplification approaches, such as the branched-chain DNA assay. The performance characteristics of commercially available quantitative HCV assays have been extensively compared with each other and with RT-PCR, which is regarded as the most sensitive method, or “gold standard,” to detect low level viremia (1, 2, 4, 10–12, 14–16, 19, 20, 22, 25–28). The Quantiplex HCV RNA 2.0 assay (Bayer Diagnostics, Emeryville, Calif.), widely used throughout the United States and based on branched DNA (bDNA) technology, was found to be precise and reproducible in several independent studies, but considerably less sensitive than RT-PCR. Reported sensitivities for the bDNA assay, in comparison to those for various RT-PCR assays, ranged from 60 to 91% (4, 15, 16, 19, 29). Despite the availability of several commercial target amplification methods, many laboratories use the bDNA assay for HCV quantitation because of its precision and reproducibility, as well as its abundance of clinical experience. Because of the lower sensitivity of the bDNA assay, it has been recommended that routine RT-PCR testing of all bDNA-negative specimens be performed to rule out low-level viremia (10, 11). Our laboratory has tested HCV bDNA-negative specimens by a more sensitive in-house HCV RT-PCR assay for several years. The purpose of this article is (i) to communicate our experience with this approach and (ii) to give recommendations for the optimization of testing strategies for hepatitis C viral load testing based on our data.
The evaluation included all blood specimens referred to the Molecular Diagnostics Division at the University of Pittsburgh Medical Center for quantitative HCV testing between 18 March 1996 and 10 June 1999. The University of Pittsburgh is a major referral center for hepatic diseases, including HCV infection. During the study period, a total of 3,471 clinical specimens (serum or plasma) were referred for routine quantitative HCV testing to assess viral load. Serum or plasma specimens were stored at −80°C prior to analysis. The Quantiplex HCV RNA 2.0 assay (Bayer Diagnostics) was used to assess viral load according to the manufacturer's recommendations. The lower limit of detection for the bDNA assay is 200,000 RNA genome equivalents/ml (0.2 Meq/ml). All specimens with a result of <0.2 Meq/ml for the bDNA assay were analyzed by an in-house liquid hybridization-gel retardation RT-PCR method with primers targeting the conserved 5′ untranslated region (5′UTR) of HCV (4). The sensitivity of RT-PCR in comparison to that of the bDNA assay was determined by analyzing dilutions of RNA from samples with known bDNA values (data not shown). The limit of detection of the RT-PCR method was fewer than 100 HCV genome equivalents per milliliter, which is theoretically 3 orders of magnitude more sensitive than the bDNA assay. Although the RT-PCR assay was not designed to be quantitative, the relative intensity of the HCV-specific hybridizing signal was graded by utilizing a semiquantitative scale from 1+ (weakly positive) to 4+ (strongly positive) to obtain a relative estimate of the quantity of HCV RNA in each specimen. Testing serial dilutions of known positive samples was shown to result in a decrease in signal intensity (data not shown), thus validating the use of this semiquantitative scale. The results of the bDNA and RT-PCR assays were accessioned into a computer database program (RBase 6.0; Microrim Inc.) and later retrieved for data analysis.
Of the 3,471 specimens referred to the laboratory for quantitative HCV testing, 1,264 (36.4%) yielded a negative result (<0.2 Meq/ml) with the Quantiplex HCV RNA 2.0 assay. RT-PCR testing was performed for 1,239 of the 1,264 bDNA-negative specimens in order to detect viremia below the detection limit of the bDNA assay. The RT-PCR assay yielded a negative result for 918 of the bDNA-negative specimens. The predictive value of a negative bDNA result was therefore 74.1%. The more sensitive RT-PCR assay yielded positive results for 321 (25.9%) of the bDNA-negative specimens. In other words, 9.2% of all specimens referred to the laboratory for HCV quantitation were found to be negative by the bDNA assay, but tested positive with the RT-PCR assay. This is consistent with reported ranges of sensitivity for the bDNA assay, from 60 to 91% in comparison to RT-PCR assays (4, 15, 16, 19, 29). Approximately 80% of the specimens that tested negative by the bDNA assay and positive by RT-PCR had weak signal intensities (1+ or 2+), consistent with the hypothesis that these specimens contained relatively low levels of HCV. The other 20% of the bDNA-negative specimens exhibited stronger signal intensities (3+ and 4+), even though they were not detected by the bDNA assay.
Much of the data regarding bDNA assay characteristics has been derived from testing small numbers of patients or a selection of special patient populations (4, 11, 14–16, 19, 22, 25). Our study is the first long-term assessment of a quantitative HCV assay in a large and heterogenous population. These data indicate that ∼26% of all bDNA-negative patient specimens may show low-level viremia when tested with more-sensitive methods like RT-PCR. Not many laboratories offer automatic performance of RT-PCR for bDNA-negative cases, even though this approach has been previously recommended (10, 11) and appears desirable from our data. The clinical significance of bDNA-negative, RT-PCR-positive samples in our study is unknown. However, two recent large multicenter studies of chronic hepatitis C treatment with interferon alpha-2b alone or in combination with ribavirin were published. These studies showed a correlation between “sustained virologic response” (defined by a negative result for serum HCV by RT-PCR, with an assay sensitivity of 100 copies/ml) and clinical improvement as assessed by alanine aminotransferase (ALT) concentration in serum and hepatic histology (5, 23). In addition, HCV testing by RT-PCR has been recommended, in addition to serum ALT testing, as the basis on which to define end-of-treatment response and sustained response to treatment (8, 18). Thus, it is important to provide HCV RT-PCR status to clinicians monitoring treatment efficacy. However, clinicians and laboratories must recognize that the proportion of RT-PCR-positive cases will vary, depending on the sensitivity of the assay used. Sensitivities of “home-brew” RT-PCR assays may vary widely from laboratory to laboratory, and there is currently no defined quantitative HCV standard (34). Thus, assessments of virologic status must include information regarding the sensitivity and characteristics of the assay used. Ideally, any RT-PCR assay used to assess virologic status should have a calculated sensitivity of at least 100 copies/ml, since studies evaluating sustained virologic response have been defined by this number. Clinicians ordering testing for HCV quantitation may not always be familiar with the different test characteristics of bDNA and RT-PCR assays. Thus, the clinical laboratory has an important role in educating laboratory users as well as ensuring that more sensitive testing is performed on specimens below the detection limits of the bDNA assay.
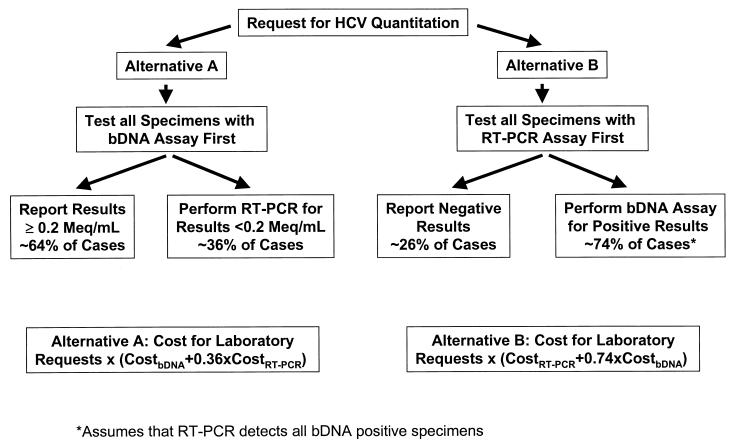
Cost constraints are increasingly a major factor in determining the extent of laboratory testing performed for many patients. The costs of performing the bDNA assay are considerable and are likely higher for most institutions than the cost for in-house RT-PCR assays. In our study, 26.4% of the specimens submitted for quantitative HCV testing were actually found to be negative by HCV RT-PCR. Thus, if we had prescreened requests for bDNA testing during the study period by first performing the RT-PCR assay, we could have eliminated 26% of the bDNA tests. The two major factors that are important in determining which test to perform first are (i) the frequency of negative versus positive test results produced by the two assays described (or similar assays) and (ii) the total cost of testing (direct and indirect costs) for each assay. Based on our experience and the assumption that the relative frequencies of bDNA negative and positive results in other institutions are comparable to ours, we suggest two alternative approaches to optimize the detection of low-level viremia in laboratories that use bDNA technology for HCV quantitation (Fig. 1). For laboratories able to perform a sensitive RT-PCR assay for ≤40% the bDNA test cost, prescreening bDNA requests by RT-PCR (alternative B) should be more cost-effective.
FIG. 1.
Strategies to optimize detection of low-level viremia for laboratories using the bDNA assay for HCV quantitation. Solving the equalized cost equations for alternatives A and B shows that the costs of both approaches would be equal if the cost of RT-PCR (CostRT-PCR) is approximately 0.4 times the cost of the bDNA assay (CostbDNA). Therefore, alternative B is more cost advantageous for a given laboratory whose CostRT-PCR is less than 0.4 times CostbDNA. Use of these algorithms assumes similar relative frequencies of positive versus negative test results to those obtained in our laboratory. The term Cost in this algorithm refers to the total costs per test (direct and indirect costs).
These algorithms do not take into account reimbursement practices for the various assays. The relative levels of reimbursement are higher for qualitative RT-PCR than for quantitative HCV testing from most third-party payers, even though the costs of testing may be less. An important additional consideration is that the Quantiplex HCV RNA 2.0 assay is a manufactured kit without Food and Drug Administration (FDA) approval for clinical diagnostic use. Medicare intermediaries and other third-party payers may well interpret such tests to be “investigational” (therefore, not “medically necessary”) and thus not reimbursable to clinical laboratories. On the other hand, the FDA has determined that FDA clearance or approval for home-brew tests is not necessary if performed in laboratories qualified to perform high complexity testing, as described in reference 9. Thus, clinical laboratories may realize greater reimbursement for in-house tests than for quantitative bDNA testing. Because of the cost savings and uncertainties in reimbursement for bDNA (or other quantitative HCV) testing, clinical laboratories performing a carefully validated in-house RT-PCR assay for molecular HCV testing may find it most prudent to prescreen samples for HCV positivity before performing quantitative testing.
Acknowledgments
We are thankful to technologists Ralph Anderson, Anke Bakker, Ronald Busch, Sandy Cicone, Yiping Chen, Bonnie Frye, James Klenner, Rick Nestler, and LeAnn Stringos for carrying out the HCV laboratory testing over this period.
REFERENCES
- 1.Albadalejo J, Alonso R, Antinozzi R, Bogard M, Bourgault A M, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J, Young C. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862–865. doi: 10.1128/jcm.36.4.862-865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter H J, Sanchez-Pescador R, Urdea M S, Wilber J C, Lagier R J, Di Bisceglie A M, Shih J W, Neuwald P D. Evaluation of branched DNA signal amplification for the detection of hepatitis C virus RNA. J Viral Hepat. 1995;2:121–132. doi: 10.1111/j.1365-2893.1995.tb00017.x. [DOI] [PubMed] [Google Scholar]
- 3.Alter M J. Epidemiology of hepatitis C. Hepatology. 1997;26:62S–65S. doi: 10.1002/hep.510260711. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Cooper D L, Ehrlich G D. Comparative analysis of three nucleic acid-based detection systems for hepatitis C virus RNA in plasma from liver transplant recipients. Mol Cell Probes. 1996;10:331–336. doi: 10.1006/mcpr.1996.0045. [DOI] [PubMed] [Google Scholar]
- 5.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 6.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diodati G, Bonetti P, Noventa F, Casarin C, Rugge M, Scaccabarozzi S, Tagger A, Pollice L, Tremolada F, Davite C, et al. Treatment of chronic hepatitis C with recombinant human interferon-alpha 2a: results of a randomized controlled clinical trial. Hepatology. 1994;19:1–5. [PubMed] [Google Scholar]
- 8.European Association for the Study of the Liver. EASL International Consensus Conference on Hepatitis C. Consensus Statement. J Hepatol. 1999;30:956–961. [PubMed] [Google Scholar]
- 9.Federal Register. Medical devices; classification/reclassification; restricted devices; analyte specific reagents—FDA. Final rule. Fed Regist. 1997;62:62243–62260. [PubMed] [Google Scholar]
- 10.Gretch D R. Diagnostic tests for hepatitis C. Hepatology. 1997;26:43S–47S. doi: 10.1002/hep.510260708. [DOI] [PubMed] [Google Scholar]
- 11.Gretch D R, dela Rosa C, Carithers R L, Jr, Willson R A, Williams B, Corey L. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann Intern Med. 1995;123:321–329. doi: 10.7326/0003-4819-123-5-199509010-00001. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi J, Ohmiya M, Kishihara Y, Tani Y, Kinukawa N, Ikematsu H, Kashiwagi S. A statistical analysis of predictive factors of response to human lymphoblastoid interferon in patients with chronic hepatitis C. Am J Gastroenterol. 1994;89:2151–2156. [PubMed] [Google Scholar]
- 14.Hayashi J, Yoshimura E, Kishihara Y, Yamaji K, Etoh Y, Ikematsu H, Kashiwagi S. Hepatitis C virus RNA levels determined by branched DNA probe assay correlated with levels assessed using competitive PCR. Am J Gastroenterol. 1996;91:314–318. [PubMed] [Google Scholar]
- 15.Ichijo T, Matsumoto A, Kobayashi M, Furihata K, Tanaka E. Quantitative measurement of HCV RNA in the serum: a comparison of three assays based on different principles. J Gastroenterol Hepatol. 1997;12:500–506. doi: 10.1111/j.1440-1746.1997.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 16.Jacob S, Baudy D, Jones E, Xu L, Mason A, Regenstein F, Perrillo R P. Comparison of quantitative HCV RNA assays in chronic hepatitis C. Am J Clin Pathol. 1997;107:362–367. doi: 10.1093/ajcp/107.3.362. [DOI] [PubMed] [Google Scholar]
- 17.Lau J Y, Davis G L, Kniffen J, Qian K P, Urdea M S, Chan C S, Mizokami M, Neuwald P D, Wilber J C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay K L. Therapy of hepatitis C: overview. Hepatology. 1997;26:71S–77S. doi: 10.1002/hep.510260713. [DOI] [PubMed] [Google Scholar]
- 19.Lu R H, Hwang S J, Chan C Y, Chang F Y, Lee S D. Quantitative measurement of serum HCV RNA in patients with chronic hepatitis C: comparison between Amplicor HCV monitor system and branched DNA signal amplification assay. J Clin Lab Anal. 1998;12:121–125. doi: 10.1002/(SICI)1098-2825(1998)12:2<121::AID-JCLA8>3.0.CO;2-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lunel F, Cresta P, Vitour D, Payan C, Dumont B, Frangeul L, Reboul D, Brault C, Piette J C, Huraux J M. Comparative evaluation of hepatitis C virus RNA quantitation by branched DNA, NASBA, and monitor assays. Hepatology. 1999;29:528–535. doi: 10.1002/hep.510290237. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto A, Tanaka E, Suzuki T, Ogata H, Kiyosawa K. Viral and host factors that contribute to efficacy of interferon-alpha 2a therapy in patients with chronic hepatitis C. Dig Dis Sci. 1994;39:1273–1280. doi: 10.1007/BF02093793. [DOI] [PubMed] [Google Scholar]
- 22.Mayerat C, Burgisser P, Lavanchy D, Mantegani A, Frei P C. Comparison of a competitive combined reverse transcription-PCR assay with a branched-DNA assay for hepatitis C virus RNA quantitation. J Clin Microbiol. 1996;34:2702–2706. doi: 10.1128/jcm.34.11.2702-2706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 24.Mita E, Hayashi N, Hagiwara H, Ueda K, Kanazawa Y, Kasahara A, Fusamoto H, Kamada T. Predicting interferon therapy efficacy from hepatitis C virus genotype and RNA titer. Dig Dis Sci. 1994;39:977–982. doi: 10.1007/BF02087547. [DOI] [PubMed] [Google Scholar]
- 25.Nolte F S, Thurmond C, Fried M W. Preclinical evaluation of AMPLICOR hepatitis C virus test for detection of hepatitis C virus RNA. J Clin Microbiol. 1995;33:1775–1778. doi: 10.1128/jcm.33.7.1775-1778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poljak M, Seme K, Koren S. Evaluation of the automated COBAS AMPLICOR hepatitis C virus PCR system. J Clin Microbiol. 1997;35:2983–2984. doi: 10.1128/jcm.35.11.2983-2984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichard O, Norkrans G, Fryden A, Braconier J H, Sonnerborg A, Weiland O. Comparison of 3 quantitative HCV RNA assays—accuracy of baseline viral load to predict treatment outcome in chronic hepatitis C. Scand J Infect Dis. 1998;30:441–446. doi: 10.1080/00365549850161395. [DOI] [PubMed] [Google Scholar]
- 28.Roth W K, Lee J H, Ruster B, Zeuzem S. Quantification of hepatitis C virus RNA by RT-PCR in comparison to the branched DNA method. Z Gastroenterol. 1998;36:5–11. [PubMed] [Google Scholar]
- 29.Shiratori Y, Kato N, Yokosuka O, Hashimoto E, Hayashi N, Nakamura A, Asada M, Kuroda H, Ohkubo H, Arakawa Y, Iwama A, Omata M. Quantitative assays for hepatitis C virus in serum as predictors of the long-term response to interferon. J Hepatol. 1997;27:437–444. doi: 10.1016/s0168-8278(97)80346-7. [DOI] [PubMed] [Google Scholar]
- 30.Tong M J, Blatt L M, McHutchison J G, Co R L, Conrad A. Prediction of response during interferon alfa 2b therapy in chronic hepatitis C patients using viral and biochemical characteristics: a comparison. Hepatology. 1997;26:1640–1645. doi: 10.1002/hep.510260637. [DOI] [PubMed] [Google Scholar]
- 31.Walsh K M, Good T, Cameron S, Thorburn D, McCruden E A, Mills P R, Morris A J. Viral kinetics can predict early response to alpha-interferon in chronic hepatitis C. Liver. 1998;18:191–195. doi: 10.1111/j.1600-0676.1998.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 32.Wiley T E, Briedi L, Lam N, Layden T J. Early HCV RNA values after interferon predict response. Dig Dis Sci. 1998;43:2169–2172. doi: 10.1023/a:1026613116942. [DOI] [PubMed] [Google Scholar]
- 33.Yamada G, Takatani M, Kishi F, Takahashi M, Doi T, Tsuji T, Shin S, Tanno M, Urdea M S, Kolberg J A. Efficacy of interferon alfa therapy in chronic hepatitis C patients depends primarily on hepatitis C virus RNA level. Hepatology. 1995;22:1351–1354. [PubMed] [Google Scholar]
- 34.Zaaijer H L, Cuypers H T, Reesink H W, Winkel I N, Gerken G, Lelie P N. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]