Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that synthesizes and secretes a wide range of virulence factors. P. aeruginosa poses a potential threat to human health worldwide due to its omnipresent nature, robust host accumulation, high virulence, and significant resistance to multiple antibiotics. The pathogenicity of P. aeruginosa, which is associated with acute and chronic infections, is linked with multiple virulence factors and associated secretion systems, such as the ability to form and utilize a biofilm, pili, flagella, alginate, pyocyanin, proteases, and toxins. Two-component systems (TCSs) of P. aeruginosa perform an essential role in controlling virulence factors in response to internal and external stimuli. Therefore, understanding the mechanism of TCSs to perceive and respond to signals from the environment and control the production of virulence factors during infection is essential to understanding the diseases caused by P. aeruginosa infection and further develop new antibiotics to treat this pathogen. This review discusses the important virulence factors of P. aeruginosa and the understanding of their regulation through TCSs by focusing on biofilm, motility, pyocyanin, and cytotoxins.
Keywords: Pseudomonas aeruginosa, virulence, two-component system (tcs), biofilm, motility, pyocyanin, cytotoxins
1. Introduction
Antimicrobial resistance (AMR) has become a serious global health threat because the rapid emergence of AMR has led to considerable increase in morbidity and mortality across the world [1,2,3,4]. Consequently, it is critical to develop new antibiotics or alternative therapeutic strategies to address pathogen antimicrobial resistance [5,6,7]. The increase of multi-drug resistant (MDR), pan-drug resistant (PDR), and extensively drug-resistant (XDR) isolates of P. aeruginosa constitute a substantial therapeutic challenge [8]. P. aeruginosa has been included in the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp) pathogen list and identified as a critical priority pathogen [9,10]. According to the US Centers for Disease Control and Prevention, 32,600 cases and 2700 deaths caused by multi-drug resistant P. aeruginosa were recorded in 2019 [11].
P. aeruginosa is a Gram-negative, environmental pathogen that is present in diverse habitats [12]. P. aeruginosa is a clinically and epidemiologically important bacterium that causes both acute and chronic infections. Furthermore, this opportunistic pathogen is linked most often to infections in immunocompromised patients [13,14]. The pathogenesis of P. aeruginosa depends on the virulence factors, which have a crucial role in bacterial colonization, and on host tissue invasion, which can result in life-threatening infections. The important virulence factors of P. aeruginosa include biofilm formation, motility (pili, flagella), pigment (pyocyanin), cytotoxins, phospholipases, elastases, and proteases [15].
P. aeruginosa has both resistance and virulence traits, and the versatility is related to its large genome and core-essential genes [16,17]. Its genome is comprised of multiple two-component systems (TCSs) and several regulatory genes [18]. TCSs function via a signal-response coupling mechanism that allows bacteria to recognize and respond to the signals in a diverse environment. A previous report has suggested that various TCSs are involved in regulation of virulence factors of P. aeruginosa [19]. It is important to understand how P. aeruginosa senses and responds to environmental stimuli via TCSs throughout the infectious process to enhance our knowledge of its pathogenesis. Therefore, it is essential to understand the role of TCSs in virulence to develop novel therapeutics against AMR strains of P. aeruginosa. Moreover, these diverse TCS-based control mechanisms of virulence factors shape the adaptation and survival of P. aeruginosa in unfavorable environments.
Consequently, it is imperative to review recent advancements in the field of TCSs. This knowledge will help refine our understanding of the intricate regulatory network architecture that controls the virulence of P. aeruginosa. In this review, we explore the regulation of four important virulence factors: biofilm formation, motility, pyocyanin, and cytotoxins, by assessing various TCSs of P. aeruginosa, as depicted in Figure 1. However, in this review, input signals to TCSs have not been included nor mentioned in figures and text, since the focus has been placed mainly on cellular signaling through TCS and their effects on key virulence factors.
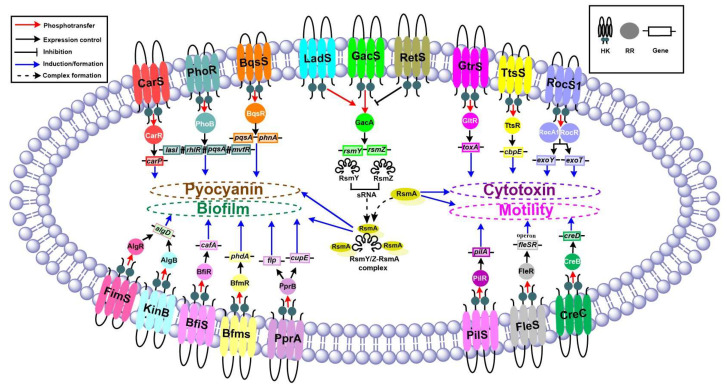
Figure 1.
Schematic representation of TCSs that regulate four important virulence factors of biofilm formation, motility, pyocyanin, and cytotoxins, in P. aeruginosa. The important TCSs and the genes responsible for producing the respective virulence factors are highlighted. Sensor HKs are shown in the cell membrane (different colors), while their respective round structures signify cognate RRs. The small rectangles represent the genes. The arrows and dotted lines represent various functions as indicated in the inset.
2. TCSs in P. aeruginosa
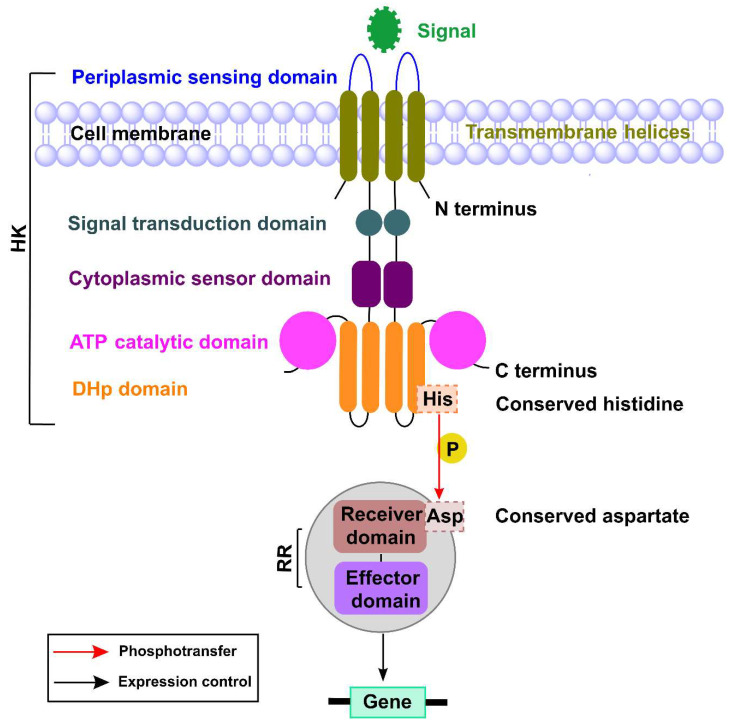
TCSs are key mediators of signal transduction in bacteria [20]. They also play an important role in sensing various external stimuli and responding to changes in environmental conditions. These TCSs are comprised primarily of a histidine kinase (HK) and its cognate response regulator (RR) that governs various signal transduction pathways [21]. The sensor histidine kinases are multidomain structures primarily composed of a periplasmic sensing domain that is responsible for identifying specific signals; the signal transduction domain; cytoplasmic sensor domain; adenosine triphosphate (ATP) catalytic domain; and dimerization histidine phosphotransfer domain (DHp) [21,22]. The sensor HK is responsible for autophosphorylation of the conserved histidine within the HK domain, and it transfers the phosphate from its conserved histidine residue to the aspartate residue of its cognate RR. The RR is responsible for intracellular responses, for which its effector domain undergoes a conformational change that allows it to bind to DNA, which triggers changes in gene expression [21] (Figure 2).
Figure 2.
The basic mechanisms of a two-component signaling system. The sensor HK senses the environmental signal using the sensing domain that is connected to the signal transduction domain, a cytoplasmic sensor domain, an ATP catalytic domain, and a dimerization histidine phosphotransfer domain (DHp). The conserved histidine residue of HK transfers the phosphate to the conserved aspartate residue of the receiver domain of its RR. The effector domain of the phosphorylated RR binds to its target and regulates gene expression.
In general, bacterial genomes have a diverse number of TCSs, and the overall number can differ from one bacterium to another [23]. Most bacterial species require an array of TCSs because of the varying input detection domains of HK, which allow the bacteria to receive multiple environmental stimuli. More than half the known TCSs are responsible for controlling virulence factors. The number of TCSs associated with virulence in P. aeruginosa is increasing rapidly due to the development of whole genome-based approaches [19]. When comparing the genome sizes of various bacteria, the P. aeruginosa PA14 strain has the largest sequenced genome of 6.54 million base pairs (Mbp) and 5973 genes, which is slightly larger than that of P. aeruginosa PA01, which has a genome size of 6.26 Mbp [24]. The genome of P. aeruginosa PA14 is larger than those of other known pathogenic bacteria such as Escherichia coli K12 (4.64 Mbp), Klebsiella pneumoniae HS11286 (5.33 Mbp) and Acinetobacter baumanni HX386 (4.09 Mbp) [25,26,27]. The number of TCS of P. aeruginosa is also higher than other Gram-negative bacteria; such as P. aeruginosa PA01 (63 HKs; 64 RRs), Escherichia coli K12 (28 HKs; 32 RRs), Klebsiella pneumoniae HS11286 (32 HKs; 32 RRs), and A. baumanni XH386 (14 HKs; 15 RRs) [18,23,28]. The detailed comparison is mentioned in Table 1.
Table 1.
Summary of genome size and TCSs of P. aeruginosa with other Gram-negative pathogens.
| Bacterial Strain | Genome Size (Mbp) | Number of Genes (NCBI Ref Seq No.) | Sensor Kinase | Response Regulator | Reference |
|---|---|---|---|---|---|
| Pseudomonas aeruginosa PA01 | 6.26 | 5697 (NC_002516.2) | 63 | 64 | [18,24] |
| Escherichia coli K12 | 4.64 | 4609 (NC_000913.3) | 28 | 32 | [25,28] |
| Klebsiella pneumoniae HS11286 | 5.33 | 5404 (NC_016845.1) | 32 | 32 | [23,26] |
| Acinetobacter baumanni XH386 | 4.09 | 4062 (CP010779.1) | 14 | 15 | [23,27] |
3. Virulence of P. aeruginosa
P. aeruginosa infection involves a series of stages starting from bacterial adherence and followed by colonization, invasion, dissemination, and finally severe systemic diseases. Its pathogenesis is determined by multiple virulence factors [29]. P. aeruginosa infection involves a series of steps, from bacterial adherence followed by colonization, invasion, and dissemination, which finally cause severe systemic infection [29]. In short, adhesion and colonization are the first two important steps of pathogenesis where the bacteria undergo initial attachment to the surface for colonization. In the third step of pathogenesis, the colonized bacteria start to invade the host tissues. P. aeruginosa then enters the fourth step for facilitating dissemination and causing systemic infection by damaging host tissues such as skin, blood, respiratory, and urinary tract. Each stage of infection is highly influenced and controlled by multiple virulence factors.
These virulence factors allow bacteria to escape from host defenses and trigger a variety of diseases such as respiratory, skin, blood, and urinary tract infections.
4. Key Virulence Factors in P. aeruginosa
The pathogenic profile of P. aeruginosa is connected to multiple virulence factors including, but not limited to, a protein secretion system, biofilm formation, cell surface components, quorum sensing system (QS), and exoenzymes, which play an important role in infection severity [30]. Various secreted products such as toxin, exopolysaccharide, and enzymes and cell surface components such as capsules, lipopolysaccharides, glycoproteins, and lipoproteins play a major role in pathogenesis [31]. Moreover, siderophores including pyochelin and pyoverdine are crucial virulence factors that allow bacteria to divide in the presence of ferrous ions and enhance bacterial metal resistance [32]. The Type III secretion systems (T3SS) of P. aeruginosa produce various toxins such as ExoU, ExoS, ExoT, and ExoY, which are responsible for ventilator-associated pneumonia [33]. ExoS and ExoT are hetero-bifunctional cytotoxins with amino-terminal Rho-GAP (GTP binding protein of rho - GTPase activating protein) and C-terminal ADP-ribosyltransferase activities, respectively. They play a role in interfering with the signal transduction in the host involved in phagocytic oxidation of NADPH (nicotinamide adenine dinucleotide phosphate) [34,35]. Moreover, ExoU is responsible for destruction of cellular physiology through its phospholipase activity [36]. ExoY is another important toxin that has a role in pathogenesis of P. aeruginosa based on adenylate cyclase activity [37]. These virulence factors are present in other pathogenic bacteria as well; however, several virulence factors such as pyocyanin, rhamnolipids, and cup fimbriae are specific to pseudomonas species [19,38,39]. Among various virulence factors in P. aeruginosa, biofilm formation, motility, pyocyanin, and secreted toxins are the four most important since they are responsible for acute and chronic infections. Although there are two types of regulation systems, TCS and a QS, which control the expression of these virulence factors, we focus on TCSs and their roles in controlling the four key virulence factors in P. aeruginosa (Table 2).
Table 2.
List of TCSs that contribute to virulence in P. aeruginosa.
| Virulence | Description | Gene | Virulence Factor | TCS | Reference |
|---|---|---|---|---|---|
| Biofilm | Reversible attachment/adherence | fleSR operon | Flagella | FleSR | [40] |
| pilA | Type IV pili | PilSR | [41]. | ||
| cupB, cupC | CupC fimbriae | RocS1-RocR-RocA1 | [42] | ||
| Irreversible attachment | algD | Alginate | FimS-AlgR, KinB-AlgB | [43,44,45,46] | |
| cafA | Rnase G | BfiSR | [47] | ||
| Microcolony | psl, pel | Exopolysaccharides (Pel, Psl) | GacSA, RetS | [23,48,49,50,51] | |
| cupD | CupD fimbriae | RcsCB, PvrSR | [52] | ||
| PA5330 | MifSR | [53] | |||
| Maturation | phdA | eDNA | BfmSR | [54] | |
| cupE | Cup E fimbriae | PprAB | [55] | ||
| Dispersion | rhlAB operon | Rhamnolipid | BqsSR | [56] | |
| Motility | Swimming/ Swarming |
fleSR operon | Flagella | FleSR | [40,57,58] |
| GacSA | [59] | ||||
| CreCB | [60] | ||||
| carP | CarSR | [19,61] | |||
| Twitching | pilA | Type IV pili | PilSR | [41,62,63] | |
| fimU, pilV, pilW, pilX, pilE, pilY1 | FimS-AlgR | [64,65,66] | |||
| PilT, pilU, fimX, pilB, pilZ | ChpA-PilG | [67,68] | |||
| Pigment | Pyocyanin | phz | Pyocyanin | GacSA-LadS-RetS | [59] |
| carP | CarSR | [61] | |||
| pqsA, phnA | BqsSR | [56] | |||
| PA2573-PA2572 | [69] | ||||
| lasI, rhlR, pqsA, mvfR | PhoRB | [70] | |||
| czcR | AlgZR | [71] | |||
| Toxin | Secreted by Type II, Type III secretion systems | cbpE | CbpE | TtsSR | [72] |
| toxA | ToxA | GtrS-GltR | [73] | ||
| Type III secretion | GacS-LadS-RetS, CsrA/RsmA | [74,75] | |||
| rhlR | Elastase | RsmA | [76] | ||
| exoU | ExoU | LadS | [77] | ||
| exoY, exoT, | ExoY, ExoT | RocS1-RocR-RocA1 | [78] | ||
| exoT, exoS | ExoT, ExoS | CbrAB | [79] | ||
| exoS | ExoS | PA2573-PA2572 | [69] | ||
| lasI, rhlR, pqsA, mvfR | Cytotoxicity | PhoRB | [70] | ||
| algD | FimS-AlgR | [19,46] |
4.1. Biofilm, a City of Microbes
P. aeruginosa cells are highly organized and are able to form a complex community called a “biofilm” or “a microbe city” [80]. The biofilm is enclosed inside the extracellular matrix and can adhere to both biotic and abiotic surfaces. The matrix is composed primarily of lipids, polysaccharides, and extracellular DNA (eDNA) [81]. Biofilm formation is an important trait attribute to chronic P. aeruginosa infections which allows the bacteria to evade the host immune response [82,83]. P. aeruginosa produces a robust biofilm that is one of the most critical virulence factors in its pathogenesis. P. aeruginosa is linked to device-associated infections characterized by formation of thick biofilms on the surface of implanted materials [84].
The development of a biofilm of P. aeruginosa occurs in five important phenotypic stages (Figure 3). The first step is initial adherence or reversible attachment, in which the free-living bacteria attach to appropriate surfaces, but can also be detached depending on the concentration of bacteria or environmental changes, such as in physical force. In the second step (irreversible attachment), bacteria can be attached irreversibly by lying flat along the surface to protect themselves from physical barriers [85]. After irreversible attachment, the bacteria form small colonies in the extra polymeric substance matrix. Next, microcolonies of the bacteria enlarge and converge with other microcolonies to produce an additional organized phenotype in a non-colonized space. A surface devoid of attached bacteria is filled by reproducing bacteria, which ultimately cover the entire surface to provide a mushroom-like appearance. Finally, in unfavorable conditions, the synthesis of matrix compounds declines, and the matrix is cleaved enzymatically, which can lead to biofilm dispersion [84].
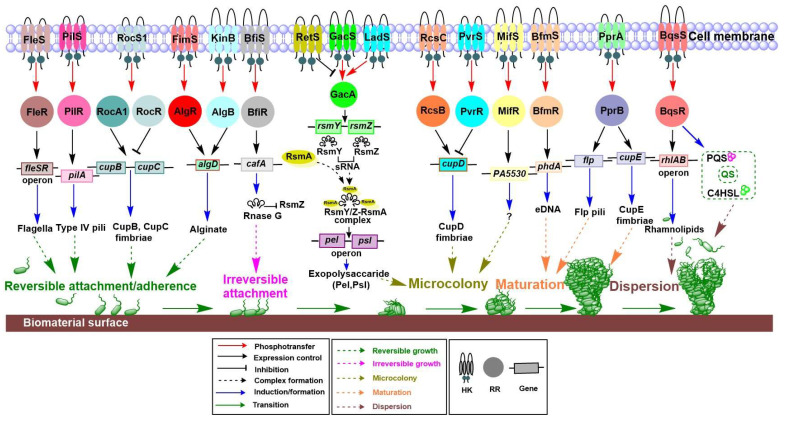
Figure 3.
The role of TCSs in biofilm formation. Important stages of P. aeruginosa biofilm formation are reversible attachment/adherence, irreversible attachment, microcolony formation, maturation, and dispersion. Many TCS are important for initial adherence. FleSR and PilSR are responsible for flagella and pili motility structures during initial attachment. RocS1-RocR and RocA1 play a role in production of the CupB and CupC fimbriae structure. FimS-AlgR and KinB-AlgB are responsible for alginate biosynthesis and help with adherence. During the initial stages, BfiSR has a role in irreversible growth. The GacSA acts in a parallel and antagonistic manner to LadS and RetS. The free RsmA regulatory proteins, together with sRNAs (RsmY and RsmZ) lead to the complex formation of RsmY/RsmZ-RsmA and play a role in the production of exopolysaccharides Pel and Psl. RcsCB and PvrSR are important for the CupD fimbriae structure. BfmSR and MifSR are essential for regulating microcolony formation and initial maturation during biofilm formation. PprAB is crucial for the CupE fimbriae structure and helps microcolonies to form and mature. BqsSR plays a role in the production of rhamnolipids and is responsible for dispersion of biofilm.
The extracellular components of P. aeruginosa play a significant role in the initial step of biofilm formation. Flagella and type IV pili are essential components of the matrix, have a role in adhesion to the surface, and are linked with initial attachment during biofilm production [86]. Previous studies confirmed that deletion of type IV pilus and flagellum genes resulted in deficient biofilm formation [86]. Therefore, both type IV pili and flagella-dependent motilities seem to have a role in the formation of P. aeruginosa biofilm [87,88]. A mutation study determined that the assembly of fimbriae subunits is regulated by a chaperone-usher pathway involved in biofilm formation [89].
Three types of polysaccharides (alginate, Pel, and Psl) are produced by P. aeruginosa and contribute to biofilm development by providing structural integrity [90]. The alginate is a linear, unbranched polymer made up of L-guluronic acid and D-mannuronic acid [91]. This provides structural support and protection and is responsible for nutrient and water retention in the biofilm [92]. The Pel polysaccharide is rich in glucose and is involved in pellicle formation [93], while Psl is made up of a pentasaccharide containing D-mannose, L-rhamnose, and D-glucose [94]. Both Pel and Psl provide a structured platform for biofilm production [92]. During the biofilm formation, a subpopulation of cells lyse and release eDNA, an important component of the P. aeruginosa biofilm matrix. The eDNA also contributes to cellular alignment, serves as a nutrient source and a cation chelator, and allows the biofilm environment to become acidic, which limits antimicrobial agent penetration [84].
4.1.1. Role of TCSs in Each Stage of Biofilm Formation
Biofilm formation in P. aeruginosa is controlled tightly by TCSs. TCSs control the production of key components of the biofilm in response to environmental stimuli and ultimately trigger the bacterium to change from planktonic to sessile life phases, and vice versa. Figure 3 illustrates the critical TCSs and genes responsible for each stage of biofilm formation.
Reversible or initial attachment is an important stage in biofilm formation. Many TCSs are responsible for surface attachment. Important surface components, including flagella, type IV pili, and fimbriae, are responsible for motility and initial attachment. The promoter fusions and microarray studies revealed that fleSR transcriptional activation is directly regulated by a master transcriptional regulator FleQ [40]. Furthermore, at least 26 genes in P. aeruginosa are regulated directly or indirectly by FleSR [40]. Among these genes, approximately 20 are responsible for flagellar formation, which eventually facilitates reversible attachment during biofilm formation. Consistently, P. aeruginosa carrying fleS and fleR mutations have shown a significant reduction in bacterial adherence to the substrate [95,96]. Interestingly, it has been noted that FleQ regulates the fleSR operon [40]. PilSR is another important TCS responsible for regulating the expression of the type IV pilus and facilitating initial attachment through twitching motility [41]. Earlier mutational studies have shown that pilR and fleR genes are necessary for twitching and swimming motility, respectively [97]. Roc is another TCS in P. aeruginosa and is comprised of RocS1 and RocA1, a sensor kinase and RR, respectively. Roc is another TCS in P. aeruginosa and comprised of RocS1, RocR and RocA1, which is a sensor HK, an antagonist of RocA1, and RR, respectively. The Roc stimulates cupB and cupC gene expression, which leads to CupB and CupC fimbriae production, responsible for adhesion. A two-hybrid assay revealed that RocS1 interacts with both RocA1 and RocR, which suggests that RocR modulates RocA1 activities by competing with RocA1 for the interaction with RocS1 [42]. These gene clusters in the PA14 strain are responsible for biofilm maturation [78]. Other TCSs, namely FimS-AlgR (AlgZ-AlgR) and KinB-AlgB, play a role in motility regulation and positively regulate alginate biosynthesis [43,44]. AlgB RR directly binds to the algD promoter, which is a key gene for alginate biosynthesis [45], which is crucial for P. aeruginosa biofilm development and has a role in adherence [98]. An earlier study showed that AlgR RR positively regulates the transcription of algD [99]. A previous study examined the role of BfiSR, another TCS, in biofilm formation and observed that it regulates the irreversible attachment of biofilm through transcriptional activation of cafA that encodes RNaseG [47]. Deactivation of the cafA gene leads to enhancement of the RsmZ level and inhibits biofilm formation. Rsm (repressor of secondary metabolism) system is a well-characterized small RNA (sRNA)-based regulatory system consisting of Rsm X, Y, and Z. Upregulation of cafA restored biofilm formation in the mutant bfiS background and decreased the level of rsmZ with respect to that of wild-type PA14 [47]. Among many TCSs in the P. aeruginosa genome, GacSA is responsible for switching from acute to chronic infection by controlling the expression of small RNAs (sRNAs), rsmY and rmsZ, through interaction with RetS and LadS [48,49]. It was suggested that LadS and RetS act agonistically and antagonistically, respectively, by forming a hetero complex with GacS. PA1611, a hybrid HK, also affects the biofilm formation by forming a heterocomplex with RetS [48]. A sensor HK GacS detects the unknown environmental signals which trigger the transfer of phosphate to RR GacA, which controls the production of sRNAs, such as RsmY and RsmZ. These RNAs regulate biofilm formation, motility, and T3SS via direct or indirect mechanisms [51]. RsmY and RsmZ bind to the RsmA regulatory protein and build an RsmY/Z-RsmA complex. This complex enhances the levels of pel and psl operons that are responsible for formation of microcolonies in the initial and later stages of biofilm formation in P. aeruginosa PA01 strain [23,49]. A previous report has suggested that PvrSR and RcsCB are similar to the Roc system and are responsible for cupD regulation [52]. Two additional TCS, PvrS and RcsC, are hybrid and unorthodox sensor histidine kinases, respectively. RcsB RR stimulates cupD expression, while PvrR RR has antagonistic activity to that of RcsB on cupD expression. PvrR is involved in the c-di-GMP degradation pathway through phosphodiesterase activity [52]. Pyruvate fermentation and a response regulator, MifR, support the formation of a microcolony [100]. Another study confirmed that MifSR senses the levels of α-ketoglutarate (α-KG) and regulates its transport and metabolism [53]. The activated MifR, along with sigma factor RpoN (σ54), initiates the transcription of the PA5530 gene, which is an α-KG-specific transporter gene. However, the exact mechanism of action to form the microcolonies is not known [53].
4.1.2. TCS Involvement in Controlling Biofilm Formation
Mutations in bfiS, bfmR, and mifR genes prevent biofilm formation, indicating involvement of these genes in various stages of biofilm formation. Interruptions in sequencing and assembly of these genes disturb the overall development and maintenance of the biofilm [101]. BfmR activates the phdA gene and has a role in the release of eDNA, which provides integrity in biofilm maturation [54]. PprAB is an another important TCS that plays a role in stimulating cupE gene cluster expression and producing CupE fimbriae that is responsible for cell-to-cell connections during microcolony formation, and thus is involved in colony formation in the early stages of biofilm formation and, also in 3D mushroom-like structure shaping during the biofilm maturation [55]. The production of Flp pilin, a major subunit of type IVb pili responsible for adhesion, can be observed in the late stationary phase. PprB is a RR that binds to three intergenic regions upstream of the flp–rcp, tadF–fppA, and pprB genes [102]. Post-maturation and biofilm dispersion are necessary stages for biofilm persistence. The detached cells migrate away, which accounts for reversible growth, and ultimately help bacteria to maintain the biofilm [84]. A mutation study on bqsS has demonstrated that BqsSR plays a critical role in biofilm degeneration by modulating the synthesis of rhamnolipids and signaling molecules such as 4-hydroxy-2-heptylquinoline (C4HSL) and pseudomonas quinolone signal (PQS) [56]. Production of rhamnolipids is controlled by the rhlAB operon.
4.2. Motility System
The motility system is an important virulence factor in many pathogenic bacteria because it is necessary for proliferation, colonization, and infection. Motility allows bacteria to adjust to diverse environmental conditions [103]. The three distinct types of motility in P. aeruginosa are swimming, swarming, and twitching [104].
4.2.1. Role of TCSs in Swimming and Swarming Motilities
P. aeruginosa possesses a single polar unsheathed flagellum that is crucial for both swimming and swarming motility [105,106]. Flagellar proteins are also responsible for adhesion, invasion, and biofilm formation [107]. Flagellin, a flagellar protein, facilitates the inflammatory response via the innate immune system and interacts specifically with a number of pattern recognition receptors (PRRs) of the host.
Several TCSs are engaged in the synthesis, assembly, and regulation of flagellar proteins, as shown in Figure 4. In P. aeruginosa, FleSR is responsible for the expression of many genes involved in flagellar biosynthesis [40]. It is well established that transcriptional and post-transcriptional events are controlled by the interlinked transcriptional regulatory circuits that consist of FleR RR, FleQ (TR), and sigma factor RpoN (σ54) [40]. Nonmotile mutants have been found in a large population of P. aeruginosa, isolated from CF patients [108]. Previous reports have suggested that restricted motility causes aggregated growth of bacteria, which strikingly increases the resistance of bacteria to macrophage ingestion. Furthermore, it was also found that the non-motile P. aeruginosa with aggregated growth has higher antibiotic tolerance as compared to the motile strains [108,109]. The σ factor AlgT has a role in P. aeruginosa motility, and its suppression decreases the flagellum expression by preventing the expression of flagellar regulator FleQ [110]. The flagella-impaired strains that have been isolated from CF patients are more pathogenic toward the host immune system than are other motile strains [110]. FleQ regulates the transcription of fleS-fleR, as well as a number of additional flagellar, adhesion, and biofilm-associated genes. In addition, several flagellar genes controlled by FleQ are antagonized by the cyclic diguanylate (c-di-GMP)-related pathway [57,58]. GacSA, which works in parallel to and antagonistic with LadS and RetS, also has an important role in motility. Swarming motility, a physiological phenomenon defined as multicellular, flagella-mediated migration of bacteria on a surface. Swarming is regulated by the GacSA signaling system. The sensor proteins and LadS activate the GacSA signaling system, while RetS represses its activation in response to external stimuli. Upon activation, GacSA stimulates the generation of two sRNAs, RsmY and RsmZ, which sequester the free RsmA by forming a RsmA-RsmY-RsmZ complex. Since RsmA has a role in repressing genes involved in chronic infection, such as pel and psl (biofilm formation), phz (pyocyanin), and genes in T6SS; as well as in activating genes associated with acute infection, such as gene sin swarming motility, lipAH (lipase), rhlAB (rhamnolipids), and genes in T3SS, the GacSA signal triggers the chronic infection via the production of RsmY and RsmZ [59]. Additionally, HptB, a histidine phosphotransfer protein, indirectly controls the expression of RsmA through a poorly-defined mechanism [111]. HptB also has a role in swarming since mutations in hptB genes lead to swarming deficiencies [112]. A prior study found that SuhB, a ribosome-associated protein, regulates multiple virulence factors that are responsible for swimming motility, biofilm production, type III secretion, and type VI secretion [113]. SuhB regulates the motile-sessile transition by inversely controlling the swimming motility and biofilm formation through the GacA-RsmY/Z-RsmA system [113]. The same study has revealed that the motility loss in suhB mutant strains can be recovered by mutations in gacA or rsmY/Z. Furthermore, yet another finding in this study was that the excessive production of RsmA protein in P. aeruginosa can also rescue the motility defect caused by a mutation in suhB. The mutational analysis has revealed that gacA or sRNAs rsmY/rsmZ or RsmA overproduction are rescued the motility defects in suhB mutant [113]. CreCB is an important conserved signaling system in many bacteria including P. aeruginosa and plays an important role in swarming and antibiotic resistance [60]. One study demonstrated that CarS HK is responsible for sensing external Ca2+ concentration and modulating Ca2+ homeostasis via CarR RR. CarSR also regulates swarming motility through a target gene carP [61]. PilSR, which plays a role in pilus-dependent twitching motility, is also associated with flagellum-dependent swimming motility [41].
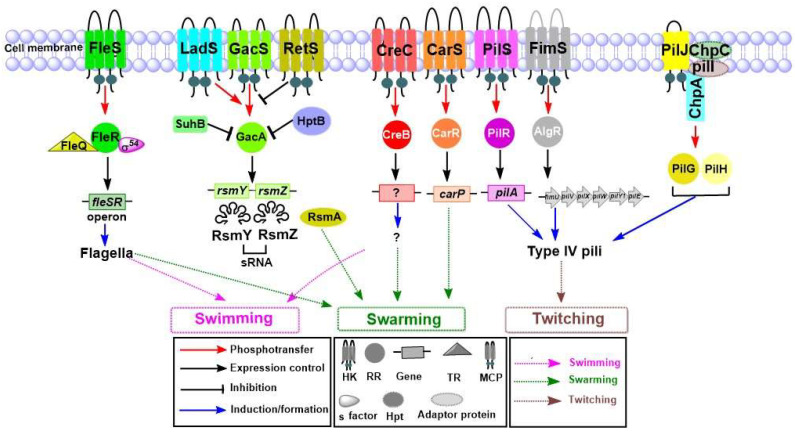
Figure 4.
Functional characterization of TCSs in P. aeruginosa and their involvement in motility-related virulence. The FleS sensor HK phosphorylates its cognate RR FleR and facilitates transcription of flagellar genes. The interconnected transcriptional regulatory circuits consisting of FleR, FleQ TR, and σ54 factors have a major role in flagellar biogenesis and are responsible for swimming and swarming motility. GacSA works along with hybrid sensor HKs LadS and RetS through a parallel and antagonistic mechanism. The free RsmA regulatory protein is involved in swarming motility. The SuhB regulator indirectly regulates motility through the GacSA, HptB also works along with GacSA and indirectly controls free RsmA. CreCB plays a major role in swarming motility by an unknown mechanism. CarSR is involved in swarming motility through its target carP. PilSR has a major role in controlling type IV pili and twitching motility. FimS-AlgR also is important for twitching motility. The Chp chemosensory system impacts type IV pili. The MCP receptor PilJ senses the environmental signal and phosphorylates ChpA using two adaptor proteins, ChpC and PilI. Phosphorylated PilG and PilH play a role in the extension and retraction of type IV pili.
4.2.2. Role of TCSs in Twitching Motility
P. aeruginosa uses hair-like appendages, known as type IV pili, which are an essential virulence factor [114]. Pili are surface organelles important for biofilm initiation, colonization, bacterial aggregation, twitching, and cellular invasion. Most type IV pili use cytoplasmic ATPase to elongate and retract, which are important features of motility [114]. Various TCSs are responsible for production, function, and control of type IV pili in P. aeruginosa. Previous research has shown the diversity of a type IV pilin allele based on P. aeruginosa strains collected from CF patients and the environment [115]. One report showed that more than 40 P. aeruginosa genes contribute to the function of type IV pili [66]. These genes encode major structural protein PilA and other minor proteins such as PilE, PilV, PilX, PilW, PilY1, PilY2, and FimT, which are responsible for formation of the tip and base of the pili [62]. Additionally, other proteins regulate the production of pili and are responsible for twitching motility in response to environmental signals, as shown in Figure 4. PilSR is the most important TCS responsible for expression of the gene pilA [63]. The role of FimS-AlgR in the regulation of twitching motility has been explained in earlier studies [65,66]. FimS-AlgR controls the twitching motility by positively regulating the expression of the genes involved in the assembly of minor pilins FimU-PilVWXE and the putative adhesin PilY1 prime pilin, which are known to mediate the twitching motility in P. aeruginosa [64]. The twitching motility of P. aeruginosa is controlled by a chemosensory system, Chp system [67]. The putative HK ChpA is coupled with a methyl-accepting chemotaxis protein (MCP) receptor, PilJ, which is controlled by two CheW-like adaptor proteins, ChpC and PilI. This complex senses currently unknown environmental signals, and facilitates the conformational change of PilJ, which causes ChpA autophosphorylation [67]. Moreover, it was proposed that PilJ also recognizes the major pilin subunit PilA as a sensor of mechanically induced conformational changes in the stretched type IV pili [116]. The phosphorylated RR PilG interacts with the motor complex of PilZ, ATPase PilB, and diguanylate cyclase FimX to facilitate pilus extension, while another RR PilH plays a role in retraction of type IV pili through interaction with ATPases PilT and PilU [68].
4.3. Pyocyanin
Pyocyanin is a blue-green pigment with a strong antibiotic effect against other bacterial species [117]. Several infections associated with pyocyanin cytotoxic effects have been reported, and they involve pro-inflammatory and free radical production resulting in cellular damage and necrosis [118,119,120]. Pyocyanin is produced in both the planktonic and biofilm states. However, since it plays a role in biofilm formation in P. aeruginosa [121], pyocyanin detection can be employed as a rapid approach for detecting P. aeruginosa infections in patients [122].
TCSs Responsible for Pyocyanin Production
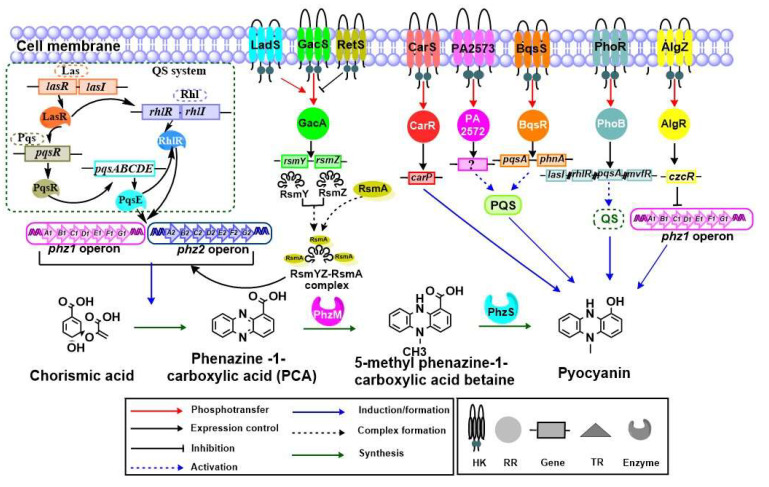
Like many other virulence factors, pyocyanin is controlled by a complex TCS network, as shown in Figure 5. Pyocyanin allows the bacterial population to coordinate a response to an environmental change [123]. Pyocyanin production is regulated by three interlinked QS systems las, rhl, and pqs [124,125,126]. Moreover, thioesterase (PqsE), 2-heptyl-4-hydroxyquinoline (HHQ), and 2-heptyl-3-hydroxy-4-quinolone (PQS) play essential roles in pyocyanin production [127,128]. In P. aeruginosa, the PQS QS system is regulated positively and negatively by Las and Rhl, respectively, and is responsible for pyocyanin synthesis [129,130]. A previous report has suggested that phz1 and phz2 operons play a central role in the biosynthesis of pyocyanin. Both phz1 and phz2 operons consist of functionally-associated genes, phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2, respectively, and each operon encodes a set of enzymes responsible for the synthesis of phenazine-1-carboxylic acid from its precursor chorismic acid. P. aeruginosa produces pyocyanin through multiple steps that begin with chorismic acid production via a complex phenazine biosynthetic route [131,132]. PCA is transferred to pyocyanin with the help of an adenosylmethionine-dependent methyltransferase (PhzM) and flavin-comprising hydroxylase (PhzS) [133,134].
Figure 5.
Biosynthesis and regulation of pyocyanin pigment by TCSs. The QS system (las, rhl, and pqs), the autoinducer synthase (PqsE), and RhlR TR are highlighted in a green dotted rectangle and play an important role in regulation of phz genes. Both phz1 and phz2 operons are responsible for biosynthesis of pyocyanin. They convert chorismic acid into phenazine-1-carboxylic acid (PCA), which is converted into 5-methyl phenazine-1-carboxylic acid betaine and finally pyocyanin pigment via phenazine enzymes (PhzM and PhzS). GacSA along with LadS and RetS hybrid HKs function in a parallel and antagonistic approach. The RsmY/Z-RsmA complex enhances the regulation of phz genes. CarSR is responsible for pyocyanin production by controlling the carP. PA2572-PA2573 and BqsSR also are responsible for pyocyanin production through PQS. PhoRB is another important TCS that is responsible for pyocyanin production through the QS network. AlgZR plays an antagonistic role in the production of pyocyanin.
LasR positively regulates the expression of rhlR and pqsR while RhlR inhibits the expression of pqsABCDE. PqsE activates the Rhl system by an unknown mechanism and this system directly controls the production of pyocyanin [135,136]. Pseudomonas quinolone system (PQS) system consists of five genes. pqsABCDE in the chromosome of P. aeruginosa, PqsA, an anthranilate-coenzyme A ligase, plays a role in the first step of PQS biosynthesis by triggering the synthesis of anthranilate-coenzyme A. PqsD controls the production of 2-aminobenzoylacetate (2-ABA) from anthraniloyl-coenzyme A and malonyl-coenzyme A. PqsB and PqsC form a heterodimer that catalyzes a reaction to produce 2-heptyl-4-quinolone (HHQ) through the condensation of octanoyl-coenzyme A and 2-ABA. Finally, PqsE thioesterase in alkylquinolone biosynthesis hydrolyzes the biosynthetic intermediate to generate 2-ABA [124].
GacSA in P. aeruginosa also works with RetS hybrid sensor HKs that are associated with the production of phenazine metabolites [137]. Previous reports suggested that CarSR is responsible for sensing the external Ca2+ concentration to modulate Ca2+ homeostasis [61]. Furthermore, the same studies also suggested that CarSR regulates the transcription of carP, which modulates the pyocyanin production and swarming motility in the high Ca2+ condition. Additionally, an orphan chemotaxis sensor, PA2573, regulates the production of pyocyanin in P. aeruginosa. Consistently, it was observed that the mutation in the PA2573 gene significantly reduces the production of pyocyanin [69]. Another study reported that pyocyanin production also is controlled by BqsSR that is positively regulated by the PQS system [56]. A gene expression study explained the role of BqsSR in modulating the transcriptional expression of pqsA and phnA, which are responsible for PQS biosynthesis [56]. Accordingly, it was reported that mutations in bqsS and bqsR significantly decrease pyocyanin production [56]. One study observed that the production of pyocyanin was affected by a RR and PhoB under phosphate-limited conditions [70]. PhoB is responsible for activation of genes involved in QS, such as lasI, rhlR, pqsA, and mvfR [70]. AlgZR is a key component that regulates pyocyanin production through CzcR, a repressor of pyocyanin. CzcR directly binds to the phz1 operon to repress the synthesis of pyocyanin [71].
4.4. Secretory System and Secreted Virulence Factors
The secretory system and secreted virulence factors of P. aeruginosa are considered to be essential. The different types of secretion systems (Types I, II, III, V, and VI) identified in P. aeruginosa participate in pathogenicity by secreting a variety of toxins and hydrolytic enzymes [138].
4.4.1. Secretory System
Of these secretion systems, type II and III (T2SS, and T3SS) are important. The T2SS of P. aeruginosa is responsible for secreting various secretory proteins such as lipase, phospholipase, exotoxin A, proteases, and alkaline phosphatase. It is well known that T2SS secretes proteins into the extracellular environment using a pilus-like structure [107]. Another type of T2SS, which is denoted as the third Xcp homolog (Txc), was discovered in P. aeruginosa strain PA7 [72]. A protein secreted through the Txc secretion system binds to chitin and thus it is known as chitin-binding protein E (CbpE). Apart from CbpE, other chitin-binding proteins such as CbpD (chitin-binding protein D) and elastase are also secreted through the T2SS system in P. aeruginosa [139]. Most of these secretory proteins promote the virulence of P. aeruginosa by damaging host cells and tissues. Among the proteins secreted by the T2SS, exotoxin A (also known as ToxA), is an important secretory protein. ToxA possesses ADP-ribosylation activity that helps bacteria to alter the protein synthesis of host cells [140]. Elastase is another virulence factor that belongs to the protease family. A previous review has extensively discussed the role of elastase in the modulation of initial defense mechanisms which eventually led to damaging host tissues [141]. Earlier research demonstrated that the LasA protease and LasB elastase play important a role in the pathogenesis of P. aeruginosa [142].
The T3SS is another important secretion system associated with higher mortality and cytotoxin delivery to the host cell [143]. Early research demonstrated that exoS, exoT, exoU, and exoY genes can be used as markers for chronic P. aeruginosa infection in hospitals [132]. T3SS plays a critical role in toxin secretion, host tissue destruction, and host immune response disruption. There are four major toxins or effector proteins controlled by the T3SS: ExoS, ExoT, ExoU, and ExoY [36]. ExoS exhibits GTPase-activating protein activity (GAP) and ADP ribosyl transferase activity (ADPRT), while ExoT exhibits N-terminal GAP activity and carboxy-terminal ADPRT activity [144]. ExoS and ExoT are considered bifunctional cytotoxins. ExoU has a role in acute infection and is associated with oxidative imbalance by acting as an important phospholipase [145]. ExoY is an adenylate cyclase that has a role in breaking down host microtubules and in disrupting the cell to cell junction in endothelial cells, which eventually leads to tissue edema [146].
4.4.2. TCSs That Influence the Secretion Systems and Their Substrates
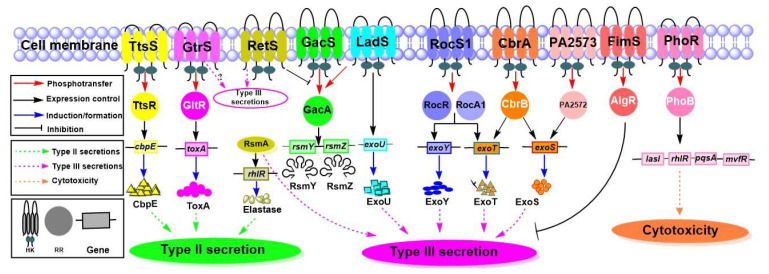
Many TCSs are involved in the regulation of the secreted proteins in T2SS and T3SS (Figure 6) [72]. An earlier study revealed that GltR regulates the expression of genes for glucose metabolism and transport [73], while sensor kinase GtrS facilitates bacterial host interaction and dissemination [147]. However, it was revealed that GtrS and GltR forms a TCS that is responsible for regulating toxA gene expression [73]. The role of GtrS HK in type III secretion has been elucidated in response to host cells. In the pneumonia infection model, GtrS was found critical for the host cell response during colonization and dissemination. It was also revealed that GtrS induces T3SS under microaerobic conditions [148].
Figure 6.
TCS-dependent regulation of secreted proteins. This figure summarizes important sensor HKs and their cognate RRs that are responsible for proteins secreted by type II, type III, and Txc secretion systems. The TtsSR is responsible for the Txc secretion and is involved in the production of CbpE. GtrS-GltR is responsible for the production of ToxA, which is an important toxin secreted by the type II secretion system. GacSA interacts with LadS and RetS in agonistic and antagonistic manners, respectively. GacSA controls both elastase and type III secretion systems by producing RsmY and RsmZ, which prevent the activities of RsmA by forming a RsmA-RsmY-RsmZ complex. LadS, specifically, is involved in regulation of ExoU. The RocS1-RocR-RocA1 is involved in the production of ExoY and ExoT toxins secreted by the type III secretion system. CbrAB has a role in the synthesis of ExoS and ExoT toxins. The PA2573-PA2572 is involved in the synthesis of the ExoS toxin, and the FimS-AlgR is responsible for suppressing type III secretion. The PhoRB is required for cytotoxicity under a phosphate-limited condition.
GacSA regulates type III secretion through RsmA activation via an unknown environmental stimulus [74]. The hybrid sensor kinase PA1611 interacts with RetS and facilitates type III secretion and biofilm formation [51,97]. A previous study showed that marine strain P. aeruginosa ID4365 modulates the production of elastase, rhamnolipids, and pyocyanin via RhlR and RsmA [76]. This report also suggested that inactivation of rsmA causes a surge in pyocyanin production but decreases elastase and rhamnolipid production via reduction of RhlR. Based on the mutational study of LadS, it was proposed that ExoU is regulated by LadS in P. aeruginosa at the transcriptional level, as well as the protein or phenotypic level via an unknown mechanism [77]. Another study explained that the SadARS signaling system, also called the RocS1−RocR−RocA1 system, contributes to production of secreted proteins through the type III secretion system [19,78]. A microarray study revealed that SadARS has a role in the synthesis of ExoY and ExoT [78]. CbrAB plays a role in the regulation of type III secretion and its effector exoenzymes ExoS and ExoT [79]. Another study explained the role of PA2573-PA2572 in the production of the ExoS toxin [69]. PhoRB is responsible for the expression of various virulence genes involved in cytotoxicity through QS system [70,148]. Among the key virulence factors regulated by QS, lasI, pqsA, mvfR, and rhlR are activated by PhoB in a phosphate-limiting condition [70]. A previous study indicated that FimS-AlgR plays an important role in the type III secretion since AlgR RR suppresses the type III secretion system in a mucoid background [149]. During the chronic P. aeruginosa CF infection, the strains switched into an alginate-overproducing mucoid phenotype, which protects them from host immune responses and oxidative burst [150,151]. TtsSR positively regulates the expression of txc gene clusters and the cbpE gene [72].
5. TCSs as Targets for Drug Development
Rapid emergence of antimicrobial resistance has become a major health threat worldwide. Therefore, developing new antibiotics and other treatment avenues are critical. However, there are limited numbers of targets for drug development. In this aspect, it is important to understand the structures and functions of TCSs to use them as new drug targets based on their role in production of virulence factors and antibiotic resistance [19]. Numerous studies have shown that TCSs are necessary for the coordinated expression of virulence determinants. They are important for the bacterial growth and survival. TCSs of P. aeruginosa have a complex signaling mechanism that is involved in host pathogenesis. The regulatory behavior of TCSs makes them excellent targets for antimicrobial therapy to overcome infection caused by drug-resistant bacteria [152,153]. Many studies have described that both antibiotics and anti-virulence drugs can be designed by targeting TCSs [154]. The wide dominance and functional variety of TCSs also favor the possibilities of screening novel small-molecule inhibitors against at least one of these TCSs [152].
TCSs play a central role not only in the coordination of numerous virulence factors, but also in bacterial growth and viability. Moreover, histidine phosphorylation acts as a key signaling mechanism in the bacterial TCSs while Ser/Thr/Tyr phosphorylation occurs during the cellular signal transduction in eukaryotic cells. Therefore, bacterial TCSs are considered to be promising targets for the development of novel antibiotics or antivirulence agents [152]. There have been reports on the development of antibiotics against P. aeruginosa, through the targeting of TCSs. For example, halogenated phenyl-thiazoles were developed to be used as small molecule inhibitors of AlgR2 TCS. However, these compounds also showed inhibitory activity on KinA, CheA, and NRII [155]. In another report, it was proposed that mucin glycans can be used for controlling the P. aeruginosa infection by inhibiting GacS-GacA via RetS-dependent signaling [156]. These promising outcomes signify the role of TCSs in developing anti-virulent or antimicrobial drugs. Despite such a significant role, there are several issues to be considered for the development of TCS-targeting inhibitors. For one, the development of a drug that is highly selective to TCSs may be difficult due to the high level of structural similarities among bacterial HKs and RRs [157,158,159]. Furthermore, the toxicity issue poses an additional barrier to the development of TCS-targeting drugs. Since the ATP-binding pocket in bacterial TCSs is sequentially and structurally similar to those in some human proteins, drugs targeting bacterial TCSs may show inhibitory activities on human proteins [160].
6. Conclusions
P. aeruginosa is a Gram-negative bacterium, and its virulence is associated with TCSs. Many TCSs directly regulate P. aeruginosa virulence, including biofilm formation, motility, toxin secretion, and pigment production. Control of important bacterial TCSs offers a new opportunity for treatment of bacterial infections. Many TCSs are responsible for regulation of biofilm formation in a stage-specific manner, leading to chronic infection. Moreover, a number of TCSs is involved in motility-related phenotypes, pyocyanin biosynthesis, and cytotoxin secretion. These complex TCSs allow bacteria to react suitably to diverse environmental stimuli. Additionally, TCSs undergo substantial selective pressure within hosts, mainly during acute and chronic infection. Therefore, it is essential to understand how TCSs detect environmental signals, transduce signals, and regulate gene expression during the infectious process to increase our knowledge on bacterial pathogenicity controlled by TCSs. Further studies on virulence factors and their cognate TCSs will contribute to developing innovative antibacterial and anti-virulence strategies to overcome AMR.
Author Contributions
The review was conceptualized by M.S., R.A., and K.K.K.; the editing was performed by R.A. and K.K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Research Foundation (NRF) of Korea to R.A. (2020R1I1A1A01070592) and K.K.K. (2017M3A9E4078553). The funders had no role in the study design, data collection, analysis, or the decision to submit the work for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Founou R.C., Founou L.L., Essack S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE. 2017;12:e0189621. doi: 10.1371/journal.pone.0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abadi A.T.B., Rizvanov A.A., Haertlé T., Blatt N.L. World Health Organization report: Current crisis of antibiotic resistance. BioNanoScience. 2019;9:778–788. doi: 10.1007/s12668-019-00658-4. [DOI] [Google Scholar]
- 3.De Oliveira D.M., Forde B.M., Kidd T.J., Harris P.N., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tillotson G.S., Zinner S.H. Burden of antimicrobial resistance in an era of decreasing susceptibility. Expert Rev. Anti-Infect. Ther. 2017;15:663–676. doi: 10.1080/14787210.2017.1337508. [DOI] [PubMed] [Google Scholar]
- 5.Mizar P., Arya R., Kim T., Cha S., Ryu K.-S., Yeo W.-S., Bae T., Kim D.W., Park K.H., Kim K.K., et al. Total Synthesis of Xanthoangelol B and Its Various Fragments: Toward Inhibition of Virulence Factor Production of Staphylococcus aureus. J. Med. Chem. 2018;61:10473–10487. doi: 10.1021/acs.jmedchem.8b01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeo W.-S., Arya R., Kim K.K., Jeong H., Cho K.H., Bae T. The FDA-approved anti-cancer drugs, streptozotocin and floxuridine, reduce the virulence of Staphylococcus aureus. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-20617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imdad S., Chaurasia A.K., Kim K.K. Identification and validation of an antivirulence agent targeting HlyU-regulated virulence in Vibrio vulnificus. Front. Cell. Infect. Microbiol. 2018;8:152. doi: 10.3389/fcimb.2018.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes M., Vira D., Medikonda R., Kumar N. Extensively and pan-drug resistant Pseudomonas aeruginosa keratitis: Clinical features, risk factors, and outcome. Graefe Arch. Clin. Exp. Ophthalmol. 2016;254:315–322. doi: 10.1007/s00417-015-3208-7. [DOI] [PubMed] [Google Scholar]
- 9.Rice L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. The University of Chicago Press; Chicago, IL, USA: 2008. [DOI] [PubMed] [Google Scholar]
- 10.Tacconelli E., Magrini N., Kahlmeter G., Singh N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Volume 27. World Health Organization; Geneva, Switzerland: 2017. pp. 318–327. [Google Scholar]
- 11.CDC . Antibiotic Resistance Threats in the United States, 2019. Department of Health and Human Services, CDC; Atlanta, GA, USA: 2019. pp. 1–150. [DOI] [Google Scholar]
- 12.Moradali M.F., Ghods S., Rehm B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migiyama Y., Yanagihara K., Kaku N., Harada Y., Yamada K., Nagaoka K., Morinaga Y., Akamatsu N., Matsuda J., Izumikawa K., et al. Pseudomonas aeruginosa bacteremia among immunocompetent and immunocompromised patients: Relation to initial antibiotic therapy and survival. Jpn. J. Infect. Dis. 2016;69:91–96. doi: 10.7883/yoken.JJID.2014.573. [DOI] [PubMed] [Google Scholar]
- 14.Turner K.H., Everett J., Trivedi U., Rumbaugh K.P., Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkowska K., Otlewska A., Guiamet P.S., Wrzosek H., Machnowski W. Pre-Columbian Archeological Textiles: A Source of Pseudomonas aeruginosa with Virulence Attributes. Appl. Sci. 2020;10:116. doi: 10.3390/app10010116. [DOI] [Google Scholar]
- 16.Stover C., Pham X., Erwin A., Mizoguchi S., Warrener P., Hickey M., Brinkman F., Hufnagle W., Kowalik D., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 17.Poulsen B.E., Yang R., Clatworthy A.E., White T., Osmulski S.J., Li L., Penaranda C., Lander E.S., Shoresh N., Hung D.T. Defining the core essential genome of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2019;116:10072–10080. doi: 10.1073/pnas.1900570116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigue A., Quentin Y., Lazdunski A., Méjean V., Foglino M. Two-component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/S0966-842X(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 19.Francis V.I., Stevenson E.C., Porter S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017;364:fnx104. doi: 10.1093/femsle/fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitrophanov A.Y., Groisman E.A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 22.Raghavan V., Groisman E.A. Orphan and hybrid two-component system proteins in health and disease. Curr. Opin. Microbiol. 2010;13:226–231. doi: 10.1016/j.mib.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhagirath A.Y., Li Y., Patidar R., Yerex K., Ma X., Kumar A., Duan K. Two component regulatory systems and antibiotic resistance in Gram-negative pathogens. Int. J. Mol. Sci. 2019;20:1781. doi: 10.3390/ijms20071781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D.G., Urbach J.M., Wu G., Liberati N.T., Feinbaum R.L., Miyata S., Diggins L.T., He J., Saucier M., Déziel E., et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blattner F.R., Plunkett G., 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 26.Liu P., Li P., Jiang X., Bi D., Xie Y., Tai C., Deng Z., Rajakumar K., Ou H.Y. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 2012;194:1841–1842. doi: 10.1128/JB.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Y., Quan J., Hua X., Feng Y., Li X., Wang J., Ruan Z., Shang S., Yu Y. Complete genome sequence of Acinetobacter baumannii XH386 (ST208), a multi-drug resistant bacteria isolated from pediatric hospital in China. Genom. Data. 2016;7:269–274. doi: 10.1016/j.gdata.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 29.Vasil M.L. Pseudomonas aeruginosa: Biology, mechanisms of virulence, epidemiology. J. Pediatrics. 1986;108:800–805. doi: 10.1016/S0022-3476(86)80748-X. [DOI] [PubMed] [Google Scholar]
- 30.Jurado-Martín I., Sainz-Mejías M., McClean S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021;22:3128. doi: 10.3390/ijms22063128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leitão J.H. Microbial Virulence Factors. Int. J. Mol. Sci. 2020;21:5320. doi: 10.3390/ijms21155320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braud A., Geoffroy V., Hoegy F., Mislin G.L., Schalk I.J. Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal tolerance. Environ. Microbiol. Rep. 2010;2:419–425. doi: 10.1111/j.1758-2229.2009.00126.x. [DOI] [PubMed] [Google Scholar]
- 33.Alonso B., Fernández-Barat L., Di Domenico E.G., Marín M., Cercenado E., Merino I., de Pablos M., Muñoz P., Guembe M. Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect. Dis. 2020;20:909. doi: 10.1186/s12879-020-05534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vareechon C., Zmina S.E., Karmakar M., Pearlman E., Rietsch A. Pseudomonas aeruginosa effector ExoS inhibits ROS production in human neutrophils. Cell Host Microbe. 2017;21:611–618. doi: 10.1016/j.chom.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbieri J., Sun J. Reviews of Physiology, Biochemistry and Pharmacology. Volume 152. Springer; Berlin/Heidelberg, Germany: 2004. Pseudomonas aeruginosa exos and exot; pp. 79–92. [DOI] [PubMed] [Google Scholar]
- 36.Foulkes D.M., McLean K., Haneef A.S., Fernig D.G., Winstanley C., Berry N., Kaye S.B. Pseudomonas aeruginosa toxin ExoU as a therapeutic target in the treatment of bacterial infections. Microorganisms. 2019;7:707. doi: 10.3390/microorganisms7120707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yahr T.L., Vallis A.J., Hancock M.K., Barbieri J.T., Frank D.W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eslami P., Hajfarajollah H., Bazsefidpar S. Recent advancements in the production of rhamnolipid biosurfactants by Pseudomonas aeruginosa. RSC Adv. 2020;10:34014–34032. doi: 10.1039/D0RA04953K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meirelles L.A., Newman D.K. Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol. Microbiol. 2018;110:995–1010. doi: 10.1111/mmi.14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dasgupta N., Wolfgang M.C., Goodman A.L., Arora S.K., Jyot J., Lory S., Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 2003;50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 41.Kilmury S.L., Burrows L.L. The Pseudomonas aeruginosa PilSR two-component system regulates both twitching and swimming motilities. Mbio. 2018;9:e01310-18. doi: 10.1128/mBio.01310-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulasekara H.D., Ventre I., Kulasekara B.R., Lazdunski A., Filloux A., Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 43.Mikkelsen H., Sivaneson M., Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 2011;13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- 44.Ma S., Wozniak D.J., Ohman D.E. Identification of the Histidine Protein Kinase KinB in Pseudomonas aeruginosa and Its Phosphorylation of the Alginate Regulator AlgB. J. Biol. Chem. 1997;272:17952–17960. doi: 10.1074/jbc.272.29.17952. [DOI] [PubMed] [Google Scholar]
- 45.Leech A.J., Sprinkle A., Wood L., Wozniak D.J., Ohman D.E. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J. Bacteriol. 2008;190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deretic V., Gill J.F., Chakrabarty A. Pseudomonas aeruginosa infection in cystic fibrosis: Nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987;15:4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrova O.E., Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development directly through CafA by its control over the small RNA rsmZ. J. Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambonnier G., Roux L., Redelberger D., Fadel F., Filloux A., Sivaneson M., de Bentzmann S., Bordi C. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet. 2016;12:e1006032. doi: 10.1371/journal.pgen.1006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L., Hu Y., Liu Y., Zhang J., Ulstrup J., Molin S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 2011;13:1705–1717. doi: 10.1111/j.1462-2920.2011.02503.x. [DOI] [PubMed] [Google Scholar]
- 50.Parkins M.D., Ceri H., Storey D.G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- 51.Kong W., Chen L., Zhao J., Shen T., Surette M.G., Shen L., Duan K. Hybrid sensor kinase PA 1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 2013;88:784–797. doi: 10.1111/mmi.12223. [DOI] [PubMed] [Google Scholar]
- 52.Mikkelsen H., Ball G., Giraud C., Filloux A. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS ONE. 2009;4:e6018. doi: 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatke G., Kumari H., Silva-Herzog E., Ramirez L., Mathee K. Pseudomonas aeruginosa MifS-MifR two-component system is specific for α-ketoglutarate utilization. PLoS ONE. 2015;10:e0129629. doi: 10.1371/journal.pone.0129629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrova O.E., Schurr J.R., Schurr M.J., Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol. Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giraud C., Bernard C.S., Calderon V., Yang L., Filloux A., Molin S., Fichant G., Bordi C., de Bentzmann S. The PprA–PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone–usher pathway system assembling fimbriae. Environ. Microbiol. 2011;13:666–683. doi: 10.1111/j.1462-2920.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- 56.Dong Y., Zhang X.-F., An S.-W., Xu J.-L., Zhang L.-H. A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol. 2008;1:88–96. doi: 10.4161/cib.1.1.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arora S.K., Ritchings B.W., Almira E.C., Lory S., Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiménez-Fernández A., López-Sánchez A., Jiménez-Díaz L., Navarrete B., Calero P., Platero A.I., Govantes F. Complex interplay between FleQ, cyclic diguanylate and multiple σ factors coordinately regulates flagellar motility and biofilm development in Pseudomonas putida. PLoS ONE. 2016;11:e0163142. doi: 10.1371/journal.pone.0163142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jimenez P.N., Koch G., Thompson J.A., Xavier K.B., Cool R.H., Quax W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moya B., Dötsch A., Juan C., Blázquez J., Zamorano L., Haussler S., Oliver A. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 2009;5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guragain M., King M.M., Williamson K.S., Pérez-Osorio A.C., Akiyama T., Khanam S., Patrauchan M.A., Franklin M.J. The Pseudomonas aeruginosa PAO1 two-component regulator CarSR regulates calcium homeostasis and calcium-induced virulence factor production through its regulatory targets CarO and CarP. J. Bacteriol. 2016;198:951–963. doi: 10.1128/JB.00963-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang B., Whitchurch C.B., Mattick J.S. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hobbs M., Collie E., Free P., Livingston S., Mattick J. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 1993;7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 64.Marko V.A., Kilmury S.L., MacNeil L.T., Burrows L.L. Pseudomonas aeruginosa type IV minor pilins and PilY1 regulate virulence by modulating FimS-AlgR activity. PLoS Pathog. 2018;14:e1007074. doi: 10.1371/journal.ppat.1007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitchurch C.B., Alm R.A., Mattick J.S. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattick J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 67.Nolan L.M., Cavaliere R., Turnbull L., Whitchurch C.B. Extracellular ATP inhibits twitching motility-mediated biofilm expansion by Pseudomonas aeruginosa. BMC Microbiol. 2015;15:55. doi: 10.1186/s12866-015-0392-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bertrand J.J., West J.T., Engel J.N. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J. Bacteriol. 2010;192:994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McLaughlin H.P., Caly D.L., McCarthy Y., Ryan R.P., Dow J.M. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS ONE. 2012;7:e42205. doi: 10.1371/journal.pone.0042205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng X., Ahator S.D., Zhang L.-H. Molecular mechanisms of phosphate stress activation of Pseudomonas aeruginosa quorum sensing systems. MSphere. 2020;5:e00119-20. doi: 10.1128/mSphere.00119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Little A.S., Okkotsu Y., Reinhart A.A., Damron F.H., Barbier M., Barrett B., Oglesby-Sherrouse A.G., Goldberg J.B., Cody W.L., Schurr M.J., et al. Pseudomonas aeruginosa AlgR phosphorylation status differentially regulates pyocyanin and pyoverdine production. MBio. 2018;9:e02318-17. doi: 10.1128/mBio.02318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cadoret F., Ball G., Douzi B., Voulhoux R. Txc, a new type II secretion system of Pseudomonas aeruginosa strain PA7, is regulated by the TtsS/TtsR two-component system and directs specific secretion of the CbpE chitin-binding protein. J. Bacteriol. 2014;196:2376–2386. doi: 10.1128/JB.01563-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daddaoua A., Molina-Santiago C., la Torre J.d., Krell T., Ramos J.-L. GtrS and GltR form a two-component system: The central role of 2-ketogluconate in the expression of exotoxin A and glucose catabolic enzymes in Pseudomonas aeruginosa. Nucleic Acids Res. 2014;42:7654–7665. doi: 10.1093/nar/gku496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vakulskas C.A., Potts A.H., Babitzke P., Ahmer B.M., Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. MMBR. 2015;79:193. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhagirath A.Y., Pydi S.P., Li Y., Lin C., Kong W., Chelikani P., Duan K. Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 2017;3:162–175. doi: 10.1021/acsinfecdis.6b00153. [DOI] [PubMed] [Google Scholar]
- 76.Cocotl-Yañez M., Soto-Aceves M.P., González-Valdez A., Servín-González L., Soberón-Chávez G. Virulence factors regulation by the quorum-sensing and Rsm systems in the marine strain Pseudomonas aeruginosa ID4365, a natural mutant in lasR. FEMS Microbiol. Lett. 2020;367:fnaa092. doi: 10.1093/femsle/fnaa092. [DOI] [PubMed] [Google Scholar]
- 77.Mikkelsen H., McMullan R., Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS ONE. 2011;6:e29113. doi: 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuchma S.L., Connolly J.P., O’toole G.A. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yeung A.T., Bains M., Hancock R.E. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J. Bacteriol. 2011;193:918–931. doi: 10.1128/JB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watnick P., Kolter R. Biofilm, city of microbes. J. Bacteriol. 2000;182:2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 82.Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents—how P. aeruginosa can escape antibiotics. Front. Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allesen-Holm M., Barken K.B., Yang L., Klausen M., Webb J.S., Kjelleberg S., Molin S., Givskov M., Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 84.Thi M.T.T., Wibowo D., Rehm B.H. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020;21:8671. doi: 10.3390/ijms21228671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hinsa S.M., Espinosa-Urgel M., Ramos J.L., O’Toole G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 86.Karygianni L., Ren Z., Koo H., Thurnheer T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 87.O’Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 88.Barken K.B., Pamp S.J., Yang L., Gjermansen M., Bertrand J.J., Klausen M., Givskov M., Whitchurch C.B., Engel J.N., Tolker-Nielsen T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 89.Vallet I., Olson J.W., Lory S., Lazdunski A., Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: Identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghafoor A., Hay I.D., Rehm B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chanasit W., Gonzaga Z.J.C., Rehm B.H. Analysis of the alginate O-acetylation machinery in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2020;104:2179–2191. doi: 10.1007/s00253-019-10310-6. [DOI] [PubMed] [Google Scholar]
- 92.Lee K., Yoon S.S. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 2017;27:1053–1064. doi: 10.4014/jmb.1611.11056. [DOI] [PubMed] [Google Scholar]
- 93.Maunders E., Welch M. Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol. Lett. 2017;364:fnx120. doi: 10.1093/femsle/fnx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Byrd M.S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A.B., Richardson S.H., Ma L., Ralston B., Parsek M.R., Anderson E.M., et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ritchings B.W., Almira E.C., Lory S., Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gellatly S.L., Bains M., Breidenstein E.B., Strehmel J., Reffuveille F., Taylor P.K., Yeung A.T., Overhage J., Hancock R.E. Novel roles for two-component regulatory systems in cytotoxicity and virulence-related properties in Pseudomonas aeruginosa. AIMS Microbiol. 2018;4:173. doi: 10.3934/microbiol.2018.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goodman A.L., Kulasekara B., Rietsch A., Boyd D., Smith R.S., Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 98.Moradali M.F., Rehm B.H. Extracellular Sugar-Based Biopolymers Matrices. Springer; Cham, Switzerland: 2019. The role of alginate in bacterial biofilm formation; pp. 517–537. [Google Scholar]
- 99.Mohr C., Leveau J., Krieg D., Hibler N., Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petrova O.E., Schurr J.R., Schurr M.J., Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 2012;86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petrova O.E., Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bernard C.S., Bordi C., Termine E., Filloux A., de Bentzmann S. Organization and PprB-dependent control of the Pseudomonas aeruginosa tad locus, involved in Flp pilus biology. J. Bacteriol. 2009;191:1961. doi: 10.1128/JB.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Behzadi P., Baráth Z., Gajdács M. It’s Not Easy Being Green: A Narrative Review on the Microbiology, Virulence and Therapeutic Prospects of Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics. 2021;10:42. doi: 10.3390/antibiotics10010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Khan F., Pham D.T.N., Oloketuyi S.F., Kim Y.-M. Regulation and controlling the motility properties of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2020;104:33–49. doi: 10.1007/s00253-019-10201-w. [DOI] [PubMed] [Google Scholar]
- 105.Jose R., Singh V. Swarming in Bacteria: A Tale of Plasticity in Motility Behavior. J. Indian Inst. Sci. 2020;100:515–524. doi: 10.1007/s41745-020-00177-2. [DOI] [Google Scholar]
- 106.Jyot J., Ramphal R. Pseudomonas: Model Organism, Pathogen, Cell Factory. Wiley-VCH; Weinheim, Germany: 2008. Flagella and pili of Pseudomonas aeruginosa; pp. 85–108. [Google Scholar]
- 107.Alhazmi A. Pseudomonas aeruginosa—Pathogenesis and pathogenic mechanisms. Int. J. Biol. 2015;7:44. doi: 10.5539/ijb.v7n2p44. [DOI] [Google Scholar]
- 108.Mahenthiralingam E., Campbell M.E., Speert D.P. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Staudinger B.J., Muller J.F., Halldórsson S., Boles B., Angermeyer A., Nguyen D., Rosen H., Baldursson Ó., Gottfreðsson M., Guðmundsson G.H. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 2014;189:812–824. doi: 10.1164/rccm.201312-2142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tart A.H., Wolfgang M.C., Wozniak D.J. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 2005;187:7955. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Robitaille S., de los Santos Y.L., Groleau M.-C., Jean-Pierre F., Doucet N., Perreault J., Déziel E. An experimentally evolved variant of RsmA confirms its central role in the control of Pseudomonas aeruginosa social motility. bioRxiv. 2020 doi: 10.1101/2020.07.15.203992. [DOI] [Google Scholar]
- 112.Hsu J.-L., Chen H.-C., Peng H.-L., Chang H.-Y. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li K., Yang G., Debru A.B., Li P., Zong L., Li P., Xu T., Wu W., Jin S., Bao Q. SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front. Microbiol. 2017;8:1045. doi: 10.3389/fmicb.2017.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hospenthal M.K., Costa T.R., Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat. Rev. Microbiol. 2017;15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 115.Kus J.V., Tullis E., Cvitkovitch D.G., Burrows L.L. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology. 2004;150:1315–1326. doi: 10.1099/mic.0.26822-0. [DOI] [PubMed] [Google Scholar]
- 116.Persat A., Inclan Y.F., Engel J.N., Stone H.A., Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2015;112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baron S.S., Rowe J.J. Antibiotic action of pyocyanin. Antimicrob. Agents Chemother. 1981;20:814–820. doi: 10.1128/AAC.20.6.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Britigan B.E., Roeder T.L., Rasmussen G.T., Shasby D.M., McCormick M.L., Cox C.D. Interaction of the Pseudomonas aeruginosa secretory products pyocyanin and pyochelin generates hydroxyl radical and causes synergistic damage to endothelial cells. Implications for Pseudomonas-associated tissue injury. J. Clin. Investig. 1992;90:2187–2196. doi: 10.1172/JCI116104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Denning G.M., Wollenweber L.A., Railsback M.A., Cox C.D., Stoll L.L., Britigan B.E. Pseudomonas pyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect. Immun. 1998;66:5777–5784. doi: 10.1128/IAI.66.12.5777-5784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lau G.W., Ran H., Kong F., Hassett D.J., Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 2004;72:4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alatraktchi F.A., Breum Andersen S., Krogh Johansen H., Molin S., Svendsen W.E. Fast selective detection of pyocyanin using cyclic voltammetry. Sensors. 2016;16:408. doi: 10.3390/s16030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu L., Cao X., Ma W., Chen L., Li S., Hu B., Xu Y. In-situ and continuous monitoring of pyocyanin in the formation process of Pseudomonas aeruginosa biofilms by an electrochemical biosensor chip. Sens. Actuators B Chem. 2021;327:128945. doi: 10.1016/j.snb.2020.128945. [DOI] [Google Scholar]
- 123.Jayaseelan S., Ramaswamy D., Dharmaraj S. Pyocyanin: Production, applications, challenges and new insights. World J. Microbiol. Biotechnol. 2014;30:1159–1168. doi: 10.1007/s11274-013-1552-5. [DOI] [PubMed] [Google Scholar]
- 124.Lin J., Cheng J., Wang Y., Shen X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 2018;8:230. doi: 10.3389/fcimb.2018.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Castañeda-Tamez P., Ramírez-Peris J., Pérez-Velázquez J., Kuttler C., Jalalimanesh A., Saucedo-Mora M.Á., Jiménez-Cortés J.G., Maeda T., González Y., Tomás M., et al. Pyocyanin restricts social cheating in Pseudomonas aeruginosa. Front. Microbiol. 2018;9:1348. doi: 10.3389/fmicb.2018.01348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.García-Reyes S., Soberón-Chávez G., Cocotl-Yanez M. The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein. J. Med. Microbiol. 2020;69:25–34. doi: 10.1099/jmm.0.001116. [DOI] [PubMed] [Google Scholar]
- 127.Gallagher L.A., McKnight S.L., Kuznetsova M.S., Pesci E.C., Manoil C., Couture-Tosi E., Delacroix H., Mignot T., Mesnage S., Chami M., et al. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Diggle S.P., Matthijs S., Wright V.J., Fletcher M.P., Chhabra S.R., Lamont I.L., Kong X., Hider R.C., Cornelis P., Cámara M., et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 129.Brint J.M., Ohman D.E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mukherjee S., Moustafa D., Smith C.D., Goldberg J.B., Bassler B.L. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017;13:e1006504. doi: 10.1371/journal.ppat.1006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mavrodi D.V., Bonsall R.F., Delaney S.M., Soule M.J., Phillips G., Thomashow L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Higgins S., Heeb S., Rampioni G., Fletcher M.P., Williams P., Cámara M. Differential regulation of the phenazine biosynthetic operons by quorum sensing in Pseudomonas aeruginosa PAO1-N. Front. Cell. Infect. Microbiol. 2018;8:252. doi: 10.3389/fcimb.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Parsons J.F., Greenhagen B.T., Shi K., Calabrese K., Robinson H., Ladner J.E. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Greenhagen B.T., Shi K., Robinson H., Gamage S., Bera A.K., Ladner J.E., Parsons J.F. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry. 2008;47:5281–5289. doi: 10.1021/bi702480t. [DOI] [PubMed] [Google Scholar]
- 135.Chincholkar S., Thomashow L. Microbial Phenazines: Biosynthesis, Agriculture and Health. Springer; Cham, Switzerland: 2013. [Google Scholar]
- 136.Bala A., Kumar L., Chhibber S., Harjai K. Augmentation of virulence related traits of pqs mutants by Pseudomonas quinolone signal through membrane vesicles. J. Basic Microbiol. 2015;55:566–578. doi: 10.1002/jobm.201400377. [DOI] [PubMed] [Google Scholar]
- 137.Goswami M., Espinasse A., Carlson E.E. Disarming the virulence arsenal of Pseudomonas aeruginosa by blocking two-component system signaling. Chem. Sci. 2018;9:7332–7337. doi: 10.1039/C8SC02496K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bleves S., Viarre V., Salacha R., Michel G.P., Filloux A., Voulhoux R. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int. J. Med Microbiol. 2010;300:534–543. doi: 10.1016/j.ijmm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 139.Galloway D. Pseudomonas aeruginosa elastase and elastolysis revisited: Recent developments. Mol. Microbiol. 1991;5:2315–2321. doi: 10.1111/j.1365-2958.1991.tb02076.x. [DOI] [PubMed] [Google Scholar]
- 140.Wolf P., Elsässer-Beile U. Pseudomonas exotoxin A: From virulence factor to anti-cancer agent. Int. J. Med Microbiol. 2009;299:161–176. doi: 10.1016/j.ijmm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 141.Pavlovskis O.R., Wretlind B. Assessment of protease (elastase) as a Pseudomonas aeruginosa virulence factor in experimental mouse burn infection. Infect. Immun. 1979;24:181–187. doi: 10.1128/iai.24.1.181-187.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ruffin M., Bilodeau C., Maillé E., LaFayette S.L., McKay G.A., Thu Ngan Trinh N., Beaudoin T., Desrosiers M.Y., Rousseau S., Nguyen D., et al. Quorum-sensing inhibition abrogates the deleterious impact of Pseudomonas aeruginosa on airway epithelial repair. FASEB J. 2016;30:3011–3025. doi: 10.1096/fj.201500166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarges E.d.S.N.F., Rodrigues Y.C., Furlaneto I.P., de Melo M.V.H., da Cunha Brabo G.L., Lopes K.C.M., Quaresma A.J.P.G., Lima L.N.G.C., Lima K.V.B. Pseudomonas aeruginosa type III secretion system virulotypes and their association with clinical features of cystic fibrosis patients. Infect. Drug Resist. 2020;13:3771. doi: 10.2147/IDR.S273759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hauser A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009;7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bagayoko S., Leon-Icaza S.-A., Pinilla M., Hessel A., Santoni K., Bordignon P.-J., Moreau F., Eren E., Boyance A., Naser E. Phospholipid peroxidation fuels ExoU phospholipase-dependent cell necrosis and supports Pseudomonas aeruginosa-driven pathology. bioRxiv. 2021 doi: 10.1101/2021.02.17.431580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morrow K.A., Frank D.W., Balczon R., Stevens T. Non-Canonical Cyclic Nucleotides. Springer; Cham, Switzerland: 2016. The Pseudomonas aeruginosa exoenzyme Y: A promiscuous nucleotidyl cyclase edema factor and virulence determinant; pp. 67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.O’Callaghan J., Reen F.J., Adams C., Casey P.G., Gahan C.G., O’Gara F. A novel host-responsive sensor mediates virulence and type III secretion during Pseudomonas aeruginosa–host cell interactions. Microbiology. 2012;158:1057–1070. doi: 10.1099/mic.0.056127-0. [DOI] [PubMed] [Google Scholar]
- 148.Bains M., Fernández L., Hancock R.E. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012;78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu W., Badrane H., Arora S., Baker H.V., Jin S. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2004;186:7575–7585. doi: 10.1128/JB.186.22.7575-7585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pedersen S.S., Møller H., Espersen F., Sørensen C.H., Jensen T., Høiby N. Mucosal immunity to Pseudomonas aemginosa alginate in cystic fibrosis. Apmis. 1992;100:326–334. doi: 10.1111/j.1699-0463.1992.tb00879.x. [DOI] [PubMed] [Google Scholar]
- 151.Lyczak J.B., Cannon C.L., Pier G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tiwari S., Jamal S.B., Hassan S.S., Carvalho P.V., Almeida S., Barh D., Ghosh P., Silva A., Castro T.L., Azevedo V. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: An overview. Front. Microbiol. 2017;8:1878. doi: 10.3389/fmicb.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yu W.L., Chen J.M., Sun Y., Geng X. Targeting two-component signal-transduction systems of bacteria for good potential drug design. J. Intensive Crit. Care. 2015;2 doi: 10.21767/2471-8505.100012. [DOI] [Google Scholar]
- 154.Fleitas Martínez O., Cardoso M.H., Ribeiro S.M., Franco O.L. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front. Cell. Infect. Microbiol. 2019;9:74. doi: 10.3389/fcimb.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Roychoudhury S., Zielinski N.A., Ninfa A.J., Allen N.E., Jungheim L.N., Nicas T.I., Chakrabarty A. Inhibitors of two-component signal transduction systems: Inhibition of alginate gene activation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 1993;90:965–969. doi: 10.1073/pnas.90.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang B.X., Wheeler K.M., Cady K.C., Lehoux S., Cummings R.D., Laub M.T., Ribbeck K. Mucin glycans signal through the sensor kinase RetS to inhibit virulence-associated traits in Pseudomonas aeruginosa. Curr. Biol. 2021;31:90–102. doi: 10.1016/j.cub.2020.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Barrett J.F., Hoch J.A. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob. Agents Chemother. 1998;42:1529–1536. doi: 10.1128/AAC.42.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Domagala J.M., Alessi D., Cummings M., Gracheck S., Huang L., Huband M., Johnson G., Olson E., Shapiro M., Singh R. Resolving the Antibiotic Paradox: Progress in Understanding Drug Resistance and Development of New Antibiotics. Volume 456. Springer; New York, NY, USA: 2012. Inhibition of NRII by the Diphenolic Methanes (Bisphenols) pp. 269–286. [Google Scholar]
- 159.Domagala J.M., Alessi D., Cummings M., Gracheck S., Huang L., Huband M., Johnson G., Olson E., Shapiro M., Singh R., et al. Resolving the Antibiotic Paradox. Springer; Boston, MA, USA: 1998. Bacterial Two-Component Signalling as a Therapeutic Target in Drug Design; pp. 269–286. [DOI] [PubMed] [Google Scholar]
- 160.Bem A.E., Velikova N., Pellicer M.T., Baarlen P.V., Marina A., Wells J.M. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem. Biol. 2015;10:213–224. doi: 10.1021/cb5007135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.