Abstract
Multidrug-resistant Salmonella enterica serotype Typhi isolates from four outbreaks of typhoid fever in southern Vietnam between 1993 and 1997 were compared. Pulsed-field gel electrophoresis, bacteriophage and plasmid typing, and antibiotic susceptibilities showed that independent outbreaks of multidrug-resistant typhoid fever in southern Vietnam are caused by single bacterial strains. However, different outbreaks do not derive from the clonal expansion of a single multidrug-resistant serotype Typhi strain.
Typhoid fever remains endemic in many developing countries. Multidrug resistance (MDR; resistance to chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole) and quinolone resistance in Salmonella enterica serotype Typhi are increasing (13). In Vietnam, since the initial outbreak of MDR serotype Typhi in Kien Giang in 1993 (12), MDR serotype Typhi has become the norm, with over 80% of isolates having an MDR phenotype by 1994 (21). MDR serotype Typhi could have become established in Vietnam in a number of ways. A single MDR bacterial clone could have spread from the original outbreak in 1993, or several different clones could have been introduced and spread. Alternatively, resistance may have spread by genetic exchange between serotype Typhi strains or between serotype Typhi and other members of the family Enterobacteriaceae in normal gut flora. A previous study in Vietnam, combining ribotyping, pulsed-field gel electrophoresis (PFGE), and bacteriophage typing, detected a variety of different strains and PFGE types in sporadic typhoid cases associated with relapse (23). In Asia in general, there have been reports describing many PFGE types in circulation (5, 9). In Malaysia, PFGE suggested that individual outbreaks were associated with closely related strains, whereas isolates of serotype Typhi from sporadic cases were very diverse (17). In Papua New Guinea, very few PFGE types are reported from sporadic cases, although this may be related to the relatively recent introduction of typhoid fever into this country (19). In Chile, serotype Typhi strains isolated between 1977 and 1986 were of multiple serotype Typhi ribotypes (2), suggesting multiple sources of infection. In this study, we have used PFGE and other typing methods to compare serotype Typhi clinical isolates from four outbreaks of MDR typhoid fever in southern Vietnam. It is important to understand both the causes of outbreaks and the mode of spread of MDR in individual areas of endemicity if rational strategies for prevention of MDR typhoid are to be implemented.
Seventy-five serotype Typhi isolates, from four MDR typhoid outbreaks, were studied (20, 21; C. Parry, J. Wain, N. T. Chinh, H. Vinh, and J. J. Farrar, Letter, Lancet 351:1289, 1998). Serotype Typhi clinical isolates were identified using Kligler iron agar slants, urea agar, Simmons citrate agar, SIM medium, and MR test medium (all from Oxoid, Basingstoke, United Kingdom) together with agglutination by antisera specific for O9 and Vi antigens (Murex, Dartford, United Kingdom). Antimicrobial sensitivity was determined on primary isolation by a modified Bauer-Kirby method. PFGE of restriction enzyme-cleaved serotype Typhi genomic DNA was performed using restriction enzymes XbaI and BlnI (Boehringer Mannheim, Lewes, United Kingdom) and the intron-encoded endonuclease I-CeuI (New England Biolabs, Hitchin, United Kingdom). These had previously been used to type serotype Typhi isolates from this region (23). DNA was prepared from isolates cultured from frozen beads onto nutrient agar (Oxoid) using the method of Liu and Sanderson (7, 8). PFGE of chromosomal DNA fragments was carried out in gels of 1% agarose (Boehringer Mannheim), at 6 V/cm, in 0.5× TBE buffer (0.045 M Tris-borate, 1 mM EDTA [pH 8.0]) at 4°C, using a Bio-Rad (Hemel Hempstead, United Kingdom) CHEF-DR II apparatus. The following conditions were used: pulse times ramped from 10 to 50 s over 12 h, then 20 to 25 s over 8 h, then 10 to 15 s over 8 h, and finally 2 to 10 s over 8 h (gel size, 13 by 20 cm). Similarity coefficients (F values) for PFGE patterns were calculated using the formula F = 2nxy/(nx + ny). nx is the total number of DNA fragments from isolate X, ny is the total number of DNA fragments from isolate Y, and nxy is the number of DNA fragments shared by the two isolates. We have used definitions from the work of Tenover et al. (16) to group PFGE patterns. The PFGE patterns were matched to a reference library of PFGE type patterns obtained (using the same gel conditions) from serotype Typhi strains isolated throughout Vietnam over the last 6 years.
All studies described in this work were approved by the Ethical and Scientific Committee of the Centre for Tropical Diseases, Ho Chi Minh City, Vietnam.
The Department of Enteric Pathogens, Central Public Health Laboratory, London, United Kingdom, performed phage typing. Plasmids were isolated using the method of Kado and Liu (6). Plasmid DNA was run on a 0.8% agarose gel (14), and the size was estimated by comparison with marker plasmids of known size.
Details of strains and results of antimicrobial susceptibility testing, plasmid analysis, phage typing, and PFGE patterns for the 75 serotype Typhi isolates representative of the four typhoid outbreaks described in this study are presented in Table 1.
TABLE 1.
Characteristics of the isolates used in this study
| Outbreak (yr) | No. of isolates with phenotype/total no.
|
Phage type (n) | PFGE type | |
|---|---|---|---|---|
| MDR | Nalr | |||
| KG (1993) | 23/25 | 0/25 | Untypeable (25) | VN1a |
| Type 52 (1) | ||||
| CB (1993–1994) | 9/9 | 6/9 | Untypeable (9) | VN2 |
| TT (1994) | 17/17 | 0/17 | Type E1 (17) | VN3 |
| D6 (1996–1997) | 23/24 | 19/24 | Untypeable (18) | VN3 |
| Type E2 (2) | ||||
| Type E3 (4) | ||||
One isolate was VN3.
The first outbreak was in Kien Giang province in the Mekong delta (KG) during April and May 1993; 3,049 people were infected (12, 20). Twenty-five serotype Typhi isolates were studied, of which 24 of 25 were MDR, all were nalidixic acid sensitive, and all were Vi phage untypeable, except one which was phage type 56. In the Cai Be outbreak (CB) in the Mekong delta between September 1993 and July 1994, some isolates were resistant to nalidixic acid (21). Nine serotype Typhi isolates were studied: all were MDR, six of nine were Nalr, and all were Vi phage untypeable. In the Thu Thiem outbreak (TT), in March and June 1994, 364 cases of typhoid fever were reported from this district on the outskirts of Ho Chi Minh City (HCMC) (21). Seventeen MDR but Nals isolates of serotype Typhi were studied, and all were Vi phage type E1. Finally, a prolonged outbreak of MDR and Nalr serotype Typhi occurred in districts 6 and 8 of HCMC (D6) between November 1996 and November 1997. Twenty-four MDR serotype Typhi isolates, 20 of which were Nalr, were studied (Parry et al., Letter, 1998). Eighteen isolates were Vi phage type untypeable, four were E3, and two were E2.
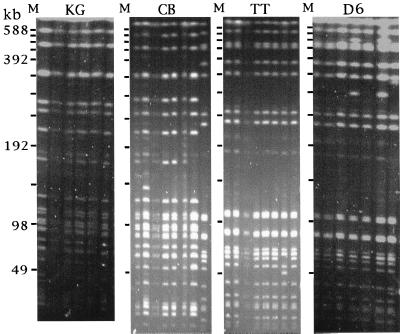
PFGE was carried out using BlnI and XbaI cleavage of chromosomal DNA from all of the 75 serotype Typhi strains. The BlnI restriction enzyme, which typically produced 17 to 25 resolvable DNA fragments, was found to be slightly more discriminating than XbaI, which generated 18 to 20 resolvable DNA fragments (results not shown). However, the data obtained using both enzymes allowed similar conclusions to be drawn, and consequently, only the BlnI data are presented here. PFGE patterns of representative isolates from each outbreak are shown in Fig. 1. This was prepared by combining data from several different PFGE gels to generate a consensus pattern.
FIG. 1.
PFGE (BlnI cleavage) patterns of representative isolates from each of the four typhoid outbreaks in southern Vietnam. Bacteriophage lambda concatemer molecular size markers (M; indicated by dashes with sizes of DNA fragments) are indicated to the left of each panel. KG isolates, from left to right, are KG 9, KG 24, KG 27, KG 57, KG 66, and KG 108. CB isolates, from left to right, are CT 1, CT 4, CT 5, CT 38, CT 47, CT 48, CT 53, and CT 88. TT isolates, from left to right, are CT 51, CT 64, CT 86, CT 90, CT 91, CT 92, CT 102, and CT 105. D6 isolates, from left to right, are 14076, 14113, 14222, 14231, 14132, 14182, and 14285.
For each outbreak, a particular PFGE pattern of serotype Typhi could be identified as predominant. Indeed, there was a remarkable homogeneity of PFGE patterns between the isolates from within each outbreak with, at most, two DNA fragment differences. The similarity coefficients (F values) from representative isolates were calculated. DNA fragments below 49 kb were not included in order to exclude plasmid-derived DNA fragments. For isolates within the same outbreak, the F values were between 0.97 and 1. Each outbreak was caused by a single clone (or very homogenous group of strains) of serotype Typhi, in terms of chromosomal DNA restriction pattern. F values from a representative isolate from each outbreak are presented in matrix form (Table 2). The predominant strains isolated in the two HCMC outbreaks (TT and D6) were indistinguishable by PFGE (F value of 1). This PFGE pattern was termed VN3. The predominant KG outbreak pattern, designated VN1, was closely related (three DNA fragments different) to the CB outbreak pattern, designated VN2, and could be derived from a common parent (F value of 0.91). One CB isolate had a VN3 PFGE pattern typical of the HCMC types. There was no close relationship between the rural (VN1 and VN2) and the urban (VN3) patterns (F values of 0.47 to 0.48). I-CeuI analysis (7) revealed that the VN1 and VN2 Mekong delta (KG and CB) patterns were Sanderson type 3 and that the VN3 HCMC (TT and D6) patterns were type 2. These are the most common types found worldwide (8). All but three of the MDR isolates harbored a large (140-MDa) plasmid, which transfers MDR to Escherichia coli (23). An 80- to 90-MDa plasmid was present in 23 of 25 KG isolates, 1 of 9 CB isolates, 0 of 17 TT isolates, and 22 of 24 D6 isolates.
TABLE 2.
Matrix showing F valuesa
| Outbreak |
F value for outbreak:
|
|||
|---|---|---|---|---|
| KG | CB | TT | D6 | |
| KG | 1.0 | |||
| CB | 0.91 | 1.0 | ||
| TT | 0.48 | 0.47 | 1.0 | |
| D6 | 0.48 | 0.47 | 1.0 | 1.0 |
An F value of 1 represents identity.
We have shown that different serotype Typhi isolates from within each of four outbreaks of MDR typhoid fever exhibited genetic homogeneity. These data suggest that each outbreak of MDR typhoid, in this region where typhoid is endemic, may have originated from a single source, allowing the spread of a single serotype Typhi strain. Furthermore, the four outbreaks were caused by at least three readily distinguishable PFGE types, although two of the patterns were highly related to each other. Studies have shown that the PFGE patterns associated with clearly defined typhoid outbreaks in regions where typhoid is not endemic are usually highly related and relatively stable (3, 17). As few as one or two DNA fragment differences on PFGE can occur, possibly because of the rearrangement of the serotype Typhi genome around the rrn operons (1, 11). In areas where typhoid fever is endemic, a local outbreak would be expected to occur against a diverse background of sporadic cases (2, 4, 9, 10, 17, 18), although if typhoid fever has been recently introduced into an area, the diversity may be more restricted (19). We have detected many different PFGE patterns for serotype Typhi strains isolated from southern Vietnam (23).
The data suggest that the KG and TT outbreaks were due to a common source. In KG, the strain may have originated either from a plasmid-carrying PFGE type imported with food or a case or carrier or from a local serotype Typhi strain acquiring the resistance via a plasmid from the normal gut flora. The uniform PFGE types of the D6 and CB isolates, however, could be distinguished by different phage types. Isolates in these outbreaks included a new Nalr phenotype, and selection for this phenotype may have occurred by the hospitalization of patients with a strain not responding to first-line therapy rather than by origin from a common source.
Is the HCMC PFGE type derived from the Mekong delta PFGE type? The differences between these strains give an F value of 0.47, which translates to eight to nine band differences on a gel. These differences may be due in part to a rearrangement of the genome as shown by having a different I-CeuI type, but this cannot explain the level of difference observed. It seems highly unlikely that the two outbreak strains are clonally related. The spread of MDR typhoid in Vietnam is therefore not due to a single strain. Why these two PFGE types are associated with outbreaks against a background of multiple types in sporadic cases is not clear. Higher transmission rates for MDR serotype Typhi than for sensitive strains have been demonstrated previously (22), and so it is possible that these two PFGE types have virulence-associated factors on the genome which enhance the transmission potential of these particular bacterial strains.
Acknowledgments
We thank the directors and staff of the Centre for Tropical Diseases, Dong Nai Paediatric Hospital, and Dong Thap Provincial Hospital for their support during this work.
This work was supported by The Wellcome Trust.
REFERENCES
- 1.Echeita M A, Usera M A. Chromosomal rearrangements in Salmonella enterica serotype Typhi affecting molecular typing in outbreak investigations. J Clin Microbiol. 1998;36:2123–2126. doi: 10.1128/jcm.36.7.2123-2126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fica A E, Prat Miranda S, Fernandez Ricci A, D'Ottone K, Cabello F C. Epidemic typhoid in Chile: analysis by molecular and conventional methods of Salmonella typhi strain diversity in epidemic (1977 and 1981) and nonepidemic (1990) years. J Clin Microbiol. 1996;34:1701–1707. doi: 10.1128/jcm.34.7.1701-1707.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruner E, Flepp M, Gabathuler U, Thong K L, Altwegg M. Outbreak of typhoid fever in a non-endemic area: comparison of three molecular typing methods. J Microbiol Methods. 1997;28:179–185. [Google Scholar]
- 4.Hampton M D, Ward L R, Rowe B, Threlfall E J. Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype Typhi. Emerg Infect Dis. 1998;4:317–319. doi: 10.3201/eid0402.980223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermans P W, Saha S K, van Leeuwen W J, Verbrugh H A, van Belkum A, Goessens W H. Molecular typing of Salmonella typhi strains from Dhaka (Bangladesh) and development of DNA probes identifying plasmid-encoded multidrug-resistant isolates. J Clin Microbiol. 1996;34:1373–1379. doi: 10.1128/jcm.34.6.1373-1379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kado C I, Liu S-L. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S-L, Hessel A, Sanderson K L. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S-L, Sanderson K L. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci USA. 1996;93:10303–10308. doi: 10.1073/pnas.93.19.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair S, Poh C L, Lim Y S, Tay L, Goh K T. Genome fingerprinting of Salmonella typhi by pulsed-field gel electrophoresis for subtyping common phage types. Epidemiol Infect. 1994;113:391–402. doi: 10.1017/s0950268800068400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nastasi A, Mammina C, Villafrate M R. rDNA fingerprinting as a tool in epidemiological analysis of Salmonella typhi infections. Epidemiol Infect. 1991;107:565–576. doi: 10.1017/s0950268800049268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navarro F, Llovet T, Echeita M A, Coll P, Aladuena A, Usera M A, Prats G. Molecular typing of Salmonella enterica serovar Typhi. J Clin Microbiol. 1996;34:2831–2834. doi: 10.1128/jcm.34.11.2831-2834.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen T A, Ha Ba K, Nguyen T D. La fievre typhoide au sud du Viet-Nam, 1990–1993. Bull Soc Pathol Exot. 1993;86:476–478. [PubMed] [Google Scholar]
- 13.Pang T, Levine M, Wain J, Finlay B. Typhoid fever—important issues still remain. Trends Microbiol. 1998;6:131–133. doi: 10.1016/s0966-842x(98)01236-0. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Shanahan P M A, Jesudason M V, Thomson C J, Amyes S G B. Molecular analysis of and identification of antibiotic resistance genes in clinical isolates of Salmonella typhi from India. J Clin Microbiol. 1998;36:1595–1600. doi: 10.1128/jcm.36.6.1595-1600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thong K L, Cheong Y M, Puthucheary S, Koh C L, Pang T. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1135–1141. doi: 10.1128/jcm.32.5.1135-1141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thong K L, Puthucheary S, Yassin R M, Sudarmono P, Padmidewi M, Soewandojo E, Handojo I, Sarasombath S, Pang T. Analysis of Salmonella enterica serovar Typhi isolates from Southeast Asia by pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1938–1941. doi: 10.1128/jcm.33.7.1938-1941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thong K L, Passey M, Clegg A, Combs B G, Yassin R M, Pang T. Molecular analysis of isolates of Salmonella typhi obtained from patients with fatal and nonfatal typhoid fever. J Clin Microbiol. 1996;34:1029–1033. doi: 10.1128/jcm.34.4.1029-1033.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran T H, Bethell D B, Nguyen T T, Wain J, To S D, Le T P, Bui M C, Nguyen M D, Pham T T, Walsh A L. Short course of ofloxacin for treatment of multidrug-resistant typhoid. Clin Infect Dis. 1995;20:917–923. [PubMed] [Google Scholar]
- 21.Vinh H, Wain J, Vo T N, Cao N N, Mai T C, Bethell D, Nguyen T T, Tu S D, Nguyen M D, White N J. Two or three days of ofloxacin treatment for uncomplicated multidrug-resistant typhoid fever in children. Antimicrob Agents Chemother. 1996;40:958–961. doi: 10.1128/aac.40.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wain J, Hoa N T, Chinh N T, Vinh H, Everett M J, Diep T S, Day N P, Solomon T, White N J, Piddock L J, Parry C M. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 23.Wain J, Hien T T, Connerton P, Ali T, Parry C, Chinh N T T, Vinh H, Phuong C X T, Diep T S, Farrar J J, White N J, Dougan G. Molecular typing of multiple-antibiotic-resistant Salmonella enterica serovar Typhi from Vietnam: application to acute and relapse cases of typhoid fever. J Clin Microbiol. 1999;37:2466–2472. doi: 10.1128/jcm.37.8.2466-2472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]