Abstract
In the past few years, there has been an increasing neuroscientific interest in understanding the function of mammalian chromodomains helicase DNA-binding (CHD) proteins due to their association with severe developmental syndromes. Mammalian CHDs include nine members (CHD1 to CHD9), grouped into subfamilies according to the presence of specific functional domains, generally highly conserved in evolutionary terms. Mutations affecting these domains hold great potential to disrupt protein function, leading to meaningful pathogenic scenarios, such as embryonic defects incompatible with life. Here, we analysed the evolution of CHD proteins by performing a comparative study of the functional domains of CHD proteins between orthologous and paralogous protein sequences. Our findings show that the highest degree of inter-species conservation was observed at Group II (CHD3, CHD4, and CHD5) and that most of the pathological variations documented in humans involve amino acid residues that are conserved not only between species but also between paralogs. The parallel analysis of both orthologous and paralogous proteins, in cases where gene duplications have occurred, provided extra information showing patterns of flexibility as well as interchangeability between amino acid positions. This added complexity needs to be considered when the impact of novel mutations is assessed in terms of evolutionary conservation.
Keywords: chromodomains helicase DNA-binding protein, neurodevelopment, chromatin remodelling, transcription regulation, evolutionary conservation
1. Introduction
Mammalian chromodomains helicase DNA-binding (CHDs) proteins represent a family of nine proteins involved in chromatin remodelling and transcription regulation during mammalian development [1,2,3,4,5,6,7,8,9]. All these proteins present two chromodomains and two helicase domains and yet other specific functional domains permit to distinguish three groups of CHDs: I (CHD1 and CHD2), II (CHD3, CHD4, and CHD5), and III (CHD6, CHD7, CHD8, and CHD9) [1]. Group II proteins possess two additional plant homeodomains (PHD), responsible for the physical binding of CHDs to the Nucleosome Remodelling Deacetylase (NuRD) chromatin-remodelling complex, which plays a critical role in embryonic development [10,11,12]. Group III proteins present a Brahma and Kismet (BRK) domain with unclear functions [1,8,13]. Additionally, both group I and III proteins have smaller domains such as the SWI3, ADA2, N-COR, and TFIIB (SANT), whereas group I and II CHDs retain domains of unknown significance (DUF) [1,8,13].
Regardless of the mammalian model used, the central roles of CHDs in embryonic viability and in utero development have been firmly demonstrated either by in vitro or in vivo studies. As such, CHD1 affects cell proliferation and growth during mouse embryonic development [14]; CHD2 is associated with murine neurogenesis and memory formation [3,15]; CHD3 is essential for brain development [16]; CHD4 orchestrates neuronal connectivity and embryonic vascular development [17,18]; CHD5 regulates murine spermiogenesis [19]; CHD6 is associated with mouse coordination and ataxia [20]; CHD7 is involved in mice cell proliferation and neuronal differentiation [21]; CHD8 is connected to murine neurodevelopment and autistic-like features [22]; and CHD9 is involved in mouse oocyte chromatin remodelling, possibly being associated with epigenetic reprogramming [23]. In humans, defective CHDs have been linked to severe phenotypes such as Pilarowski-Bjornsson Syndrome [24], Epileptic Encephalopathy, childhood-onset [25,26], Snijders Blok-Campeau Syndrome [27], Sifrim-Hitz-Weiss Syndrome [28,29], CHARGE Syndrome [30,31,32,33,34] and autism spectrum disorder [35,36,37,38,39]. Noteworthy, a significant proportion of mutations associated with to these pathologies were mapped into conserved functional domains [24,27,28,31,34,35,40,41,42].
Given the impact of CHDs in human disease, in vivo studies using model organisms are crucial to obtain insights into the cascade of pathophysiological events that leads to the disorder. Lin and colleagues [43] reported several differences between mice and human transcriptional landscapes, partially explained by the metabolic and physiological differences among the species. If murine CHDs have been the focus of most studies performed until now [2,3,16,17,19,20,21,44,45,46,47], the near prospects are to extend the strategy to other non-human mammals because they are assumed to be the most suitable models for brain research. Evolutionary constraints are crucial for understanding the history of these proteins and to predict the impact that new mutations will eventually have on the protein. However, the current knowledge on the degree of similarity in structure and sequence between protein members of the CHDs family in those model organisms is still very limited.
As a better understanding of the genetic diversity associated with the functional domains of CHDs is essential for future research on neurodevelopmental and neurological disorders, we performed a comparative analysis of the functional domains of CHD proteins between Homo sapiens and five non-human mammals by compilingCHD pathogenic mutations to investigate their degree of conservation in orthologous as well as in paralogous sequences.
2. Materials and Methods
2.1. Sequence Retrieval and Alignment of CHD Proteins
Human canonical transcripts for each nine CHD protein were retrieved from the UniProt database [48]. Six mammalian species were selected—human (Homo sapiens), chimpanzee (Pan troglodytes), Rhesus macaque (Macaca mulatta), domestic cat (Felis catus), rat (Rattus norvegicus), and mouse (Mus musculus). Orthologous sequences were searched in the HomoloGene database from National Centre for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/homologene. Accessed on 2 November 2020) [49]. In cases where more than one transcript existed for the target protein, Position-Specific Iterated BLAST (PSI-BLAST) algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed on 2 November 2020) [50] was used to determine the highest homology transcript to each canonical human transcript retrieved from UniProt [48]. The reference for each sequence used in this work is displayed in Table S1. Additionally, we retrieved three protein sequences of the invertebrate Caenorhabditis elegans (UniProt: O61845, Q22516, and O17909) referred in the work of Flanagan and colleagues [51]. A global alignment including all nine sets of CHD proteins and their orthologues (54 sequences) was performed using the Clustal Omega tool (Max HMM Iterations = 5) (https://www.ebi.ac.uk/Tools/msa/clustalo/. Accessed on 2 November 2020) [52]. The resulting alignment file was refined using the default options of BMGE software [53] available at (https://ngphylogeny.fr/tools/. Accessed on 2 November 2020) [54,55] (BLOSUM 62 Matrix, Maximum Entropy Threshold = 0.5, Gap Rate Cut-off = 0.5 and Minimum Block Size = 5).
2.2. Phylogenetic Analysis of CHD Proteins
The refined sequence alignment file from BMGE was used to calculate a phylogenetic tree using the built it SMS [56] option in PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/. Accessed on 9 July 2021) [57] resulting in JTT+G+I. Posterior probabilities were determined using aBayes [58]. The tree analysis was performed until the average standard deviation of split frequencies approximated to 0. The final phylogenetic tree was then midpoint rooted and edited with the online software iTOL (https://itol.embl.de/shared_projects.cgi. Accessed on 9 July 2021) [59].
2.3. Alignment of CHD Functional Domains
We identified the amino acid sequences for each functional domain of human CHD proteins from the UniProt database (Chromo 1, Chromo 2, Helicase ATP-binding, Helicase C-terminal, PHD I, and PHD II) [48]. The amino acid sequences of SANT, DUF, and BRK domains were retrieved from the Conserved Domain Database (CDD) from NCBI (https://www.ncbi.nlm.nih.gov/cdd/. Accessed on 16 March 2021) [60] and Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de. Accessed on 16 March 2021) [61]. Sizes and coordinates of each domain were retrieved from the aforementioned databases. We used Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo. Accessed on 16 March 2021) [52] to compare the functional domains between: (a) H. sapiens and the five orthologue species P. troglodytes, M. mulatta, F. catus, R. norvegicus, and M. musculus (interspecific analysis); and (b) Paralogues of the same group (I, II, III) in H. sapiens (intraspecific analysis). MView (https://www.ebi.ac.uk/Tools/msa/mview. Accessed on 2 November 2020) [62] was used to analyze the results.
2.4. Retrieval of Pathogenic Missense Variants
On 2 March 2021, we retrieved the pathogenic missense variants affecting CHDs domains from the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar. Accessed on 2 March 2021) [63]. The filters used for our query were (1) Clinical significance—pathogenic and (2) Molecular consequence—missense. We only considered pathogenic variants previously reported in the literature. The pathogenic missense variants were located in the alignments of the functional domains.
3. Results
3.1. Phylogenetic Analysis of CHD Proteins
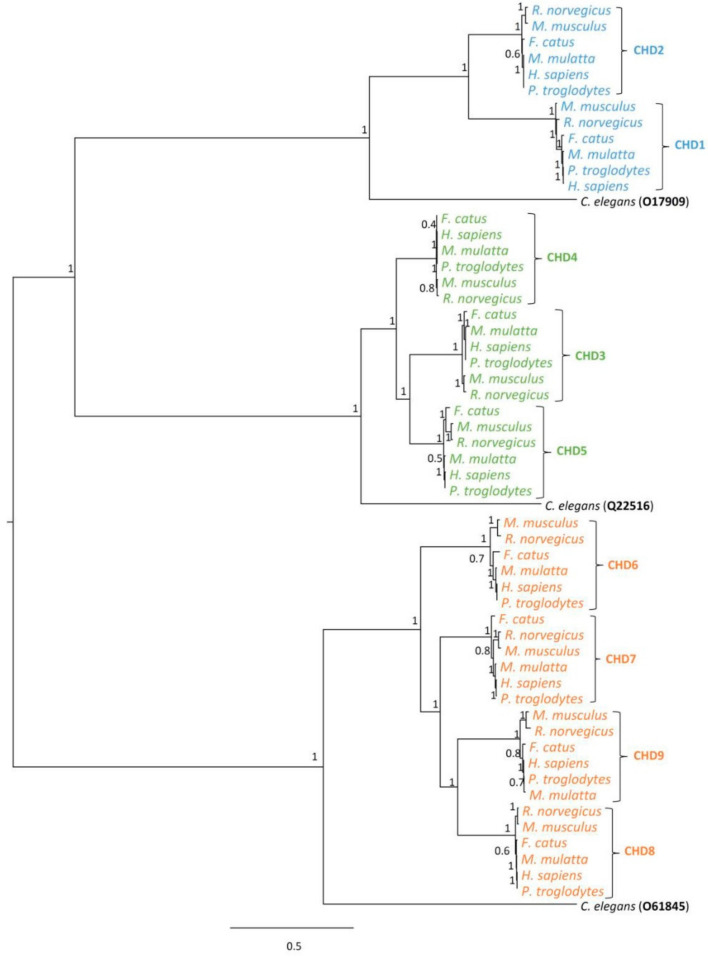
The phylogenetic analysis of CHD proteins considering the six mammalian species under investigation (Figure 1) showed that CHDs cluster in three clades that match the three groups of proteins from the CHD family based on the types of functional domains: I (CHD1 and CHD2), II (CHD3, CHD4, and CHD5), and III (CHD6, CHD7, CHD8, and CHD9). According to Flanagan and colleagues [51], each group of CHDs derived from one of three different ancestral proteins. In line with this and taking into account the position of the sequences from C. elegans, our analysis suggests that at least three ancestral CHDs existed before the divergence of vertebrates [6,51], an indication that the expansion of the CHD gene family by gene duplication events occurred after the divergence between invertebrates and vertebrates. Flanagan and colleagues [51] further showed that invertebrates, vertebrates and plants have three CHD classes whereas some unicellular organisms have one CHD group. As such, the question can be raised on whether the proliferation of different paralogues for CHD proteins might have occurred in the transition from unicellular to multicellular organisms due to the demanding needs of specialized functions that emerged with multicellularity [64,65,66]. The phylogenetic topology for group III CHDs shows that CHD6 places basally to CHD7, CHD8, and CHD9, indicating that the CHD6 sequence is more divergent than CHD7, CHD8, or CHD9. This result is consistent with those provided by the biochemical studies of Manning and Yusufzai [7] which suggest that CHD6 lacks nucleosome sliding activity and might have a different mechanism to disrupt chromatin, in contrast to other group III CHDs.
Figure 1.
Phylogenetic tree for the CHD family of proteins. Evolutionary model was determined automatically using the built it SMS [56] option in PhyML 3.0 [57] resulting in JTT+G+I. Branch support values shown at tree nodes correspond to posterior probabilities determined using aBayes [58]. The highlighting represents each protein group (blue: group I; green: group II and orange: group III).
3.2. Analysis of Domains between Human CHDs
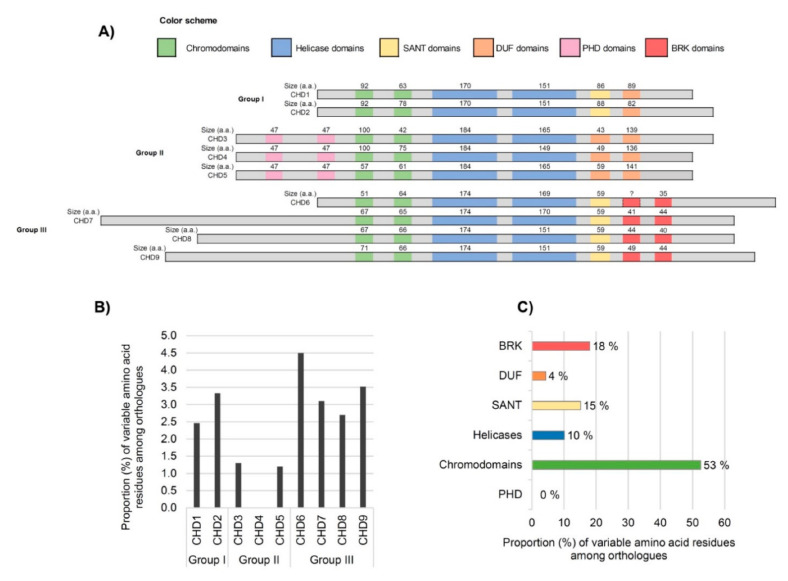
Next, to dissect the domain architecture of human CHDs, we performed alignments of the amino acid sequences (Figure 2A). The number of amino acids of the shared domains was similar among members of the CHD family group, with only a few exceptions, such as the Chromo 1 domain between Group II proteins (Figure 2A). The largest Chromo 1 domain is present in Group II (CHD3 and CHD4) and the smallest in Group III (CHD6). This domain shows high intra- and inter-group heterogeneity in terms of segment size. The helicase domain of CHDs, consisting of two modules: one Helicase ATP-binding and one Helicase C-terminal, is of extreme importance because it provides energy for nucleosome remodeling through its ATPase activity [13,27]. The Helicase ATP-binding has remained invariable in terms of amino acid content between all CHD proteins belonging to the same group, whereas the other helicase domain (C-terminal domain) reveals heterogeneity not only between groups but also considerably among members of groups II and III. In what concerns the group-specific domains DUF and BRK, our comparative analyses revealed some differences among members of the same group.
Figure 2.
Schematic representation of the architecture of CHD proteins. (A) Amino acid (a.a.) size of the conserved domains for each group of CHD paralogues in Homo sapiens. (B) Percentage of variable amino acid residues within the functional domains of CHD proteins among orthologues distributed by groups I, II, and III (n = 139). (C) Proportion of variable amino acid residues within the functional domains of CHD proteins among orthologues distributed by categories—Chromodomains, Helicases, SWI3, ADA2, N-COR, and TFIIB (SANT), Domains of Unknown Significance (DUF), Brahma and Kismet (BRK), and Plant Homeodomains (PHD) (n = 139).
Regarding the BRK domain(s) of the CHD6 protein, there is still contradictory information on whether it contains one BRK domain or two tandem BRK domains, as is reflected in the published schematic representations of the human CHD family [1,7,8,13]. For this reason, the first CHD6′s BRK domain is signalized with a question mark in Figure 2A (for sequence details see Figure S1) showing the conserved residues. For the second BRK domain, the SMART analysis retrieved a BRK domain of 35 a.a. in CHD6, as is represented in Figure 2A. The SANT domain of group III CHDs is shorter than group I. However, the size is the same in all four members (59 a.a.) and members of group I only differ by 2 amino acids; reinforcing the importance of this domain. The PHD domains, specific to group II CHDs, are well conserved in terms of size.
3.3. Inter-Species Comparison of CHD Domains
The comparative analysis of the functional domains’ sequences between humans and non-human primates, revealed 100% identity among H. sapiens, P. troglodytes, and M. mulatta (Figure S2), indicating that CHD domains are extremely conserved in primates. Concerning the comparison between humans and the other non-primate mammalian species (F. catus, R. norvegicus, and M. musculus), a total of 139 variable amino acid positions were detected within the Chromodomains, Helicases, SANT, DUF, BRK, and PHD of all nine CHDs (Figure 2B,C). The highest percentage of variable residues (4.5%) is observed on CHD6, whereas CHD4 displays no variability within functional domains between the species analyzed. The lowest levels of inter-species heterogeneity were observed at Group II (CHD3, CHD4, and CHD5). Since all members of this group are important components of the NuRD complex [10], the observed high level of conservation of this group might be an indication that the maintenance of the domain structure of the group is important to preserve the binding affinities to the NuRD complex, contributing to its functionality and allowing it to orchestrate the biological processes in which is involved. Besides, there are lines of evidence that CHD4 also plays fundamental NuRD-independent functions, including the proliferation of neural precursors and regulation of neuronal connectivity, whereas CHD3 and CHD5 have been associated with neuron differentiation and migration [17,18,67]. The degree of conservation of Group II CHDs domain among mammalian species, clearly indicates the relevance of the biological pathways, many still unknown, in which these CHDs participate.
The highest average proportion of inter-species heterogeneity was found among the members of group III. One of these proteins is CHD7 which was associated with CHARGE syndrome, the most well-established syndrome connected with the CHD family of proteins. Different mouse models were already reported for this syndrome exhibiting phenotypes that mimic human symptoms [68]. According to our data, 3.3% of the CHD7 domain sequence is variable among the orthologues from F. catus, R. norvegicus, and M. musculus. CHD9 also shows a high number of variable residues among orthologues (Figure 2B). Although CHD9 continues to be the less well-characterized member of the subfamily III [47], its role in the regulation of expression of osteogenic cells [69,70] has been well documented. In contrast, a recent study revealed that mice with depleted CHD9 survived and were fertile, indicating that CHD9 is dispensable for murine embryonic development [47]. The same study demonstrated that acute depletion of CHD9 in human cancer cells elicited more robust gene expression changes, suggesting that CHD9 is a highly context-dependent chromatin regulator. Thus, further studies are needed to elucidate if the amino acid differences between human and murine CHD9 influence the functions of this protein and its associated pathological phenotypes. Additionally worth mentioning, the proportion of amino acid differences among orthologues peaked in CHD6 (4.5%), a finding that suggests that the selective constraints related to this protein are more relaxed. Fittingly, mouse lines with deletion of part of the ATPase domain of CHD6 were reported to be viable [20].
The distribution of amino acid differences by domain category (Figure 2C) revealed that most of the variations fall within the chromodomains category (53%) whereas the PHD domains do not accommodate any variation in the six species of mammals herein considered. In eukaryotes, there are various protein families containing chromodomains, which are modules 40–100 amino acids long that despite being evolutionarily conserved, are diversified enough to allow their classification into different groups [51,71]. While the most well-known function of chromodomains is to recognize and bind to lysine-methylated histone tails, facilitating recruitment to chromatin [72,73], other important functions have been proposed, such as direct nucleic acid recognition and binding [72,73].
The analysis of the structure of chromodomains revealed that the most conserved region is limited to 21 residues that form a conserved core in each chromodomain, referred to as a chromobox, which has motifs differing between the double chromodomains as well as between modules of the nine CHD proteins [51]. Chromoboxes harbour critical aromatic residues for methyllysine recognition, namely at position 5 where there is always an aromatic residue, although other aromatic residues at positions 8 and/or 12 are required for methyllysine binding capability of chromodomains [74]. The observation that the chromodomains accumulate the highest proportion of amino acid differences among orthologues is rather puzzling, but meaningfully most of the differences detected were outside the chromoboxes (Figure S2), and among those inside a chromobox, none involved positions 5, 8 or 12.
CHD7 presented the highest proportion of variable residues at chromoboxes, with four positions varying among orthologues located there: p.Ile843Val (M. mulatta, F. catus, R. norvegicus and M. musculus), p.Arg920Leu (all non-primates), p.Arg921Lys (M. musculus), p.Ile924Leu (R. norvegicus). Comparatively to the chromodomains, the helicase domains present a much lower proportion of variable amino acid residues among orthologues (10%) despite having on average twice or more the size of the chromodomains. Many pathogenic mutations implicated in neurodevelopmental disorders were reported to cluster on these domains whose disruption activates pathogenic mechanisms associated with ATP hydrolysis [13,27,35].
The extreme conservation of the PHDs is quite interesting. These domains are uniquely present in the group II CHDs, where they are featured in two tandem modules that share very high sequence homology in the three human proteins. PHDs are central “readers” of histone post-translational modifications that control gene expression cascades by recruiting multiprotein complexes consisting of chromatin regulators and transcription factors. The majority of the PHDs characterized to date bind to unmodified or methylated states of histone H3 lysine 4 [75,76]. Different subclasses of PHD fingers are present in distinct PHD-containing proteins (over 100 human proteins containing this module), in which mutations disrupting PHD fingers have been associated with a wide range of human diseases, including immunological disorders, neurological syndromes (e.g., Sotos syndrome, Rubenstein-Taybi Syndrome, etc.), and cancer [75,77].
3.4. Analyses of the Pathological Diversity Associated with CHDs
Overall, although our previous data revealed that the six different domains harboured by CHDs are in general highly conserved among humans and the mammalian species here addressed, some changes seem to be tolerated, particularly concerning the chromodomains. This opened the challenge of further dissecting how our findings could provide insights into the genetic basis of CHD’s associated neurodevelopmental disorders. So, we extended the analysis towards mapping the described CHDs pathogenic variants in the context of the level of conservation of the different domains either among paralogues or orthologues. Viewing that, we used the NIH open-access database ClinVar to obtain the data set of CHDs pathogenic variants implying one of the six domains, being of note that all retrieved variants were classified as de novo mutations in the platform. This is not surprising since it is well documented that de novo mutations, including germinal and postzygotic mutations, are important players in the genetic architecture of neurodevelopmental disorders [78], and specifically involving CHDs de novo mutations in CHD8 have been recurrently found in autism [36,38].
The filtered CHDs variants are present in Table 1 whereas Figure 3 displays their location in the specific human sequences that are aligned with the paralogue regions. The overwhelming majority of these pathogenic variants are located at one of the two tandem helicase domains.
Table 1.
Collection of the described human deleterious mutations (de novo) affecting conserved domains of CHD proteins. Data obtained from the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar. Accessed on 2 November 2020) [63] and also described in the literature.
| Protein | Mutation | Domain | Phenotype | Inheritance | Orthologue Variability | Paralogue Variability |
References |
|---|---|---|---|---|---|---|---|
| CHD1 | p.Arg618Gln | Helicase ATP-binding (493-663 a.a.) |
Pilarowski-Bjornsson Syndrome (α) | Autosomal Dominant | No | No | Pilarowski et al. (2018) [24] |
| CHD3 | p.Leu915Phe p.His886Asp |
Helicase ATP-binding (748–932 a.a.) |
Snijders Blok-Campeau Syndrome, Intellectual disability (β) | Autosomal Dominant | No | No | Snijders Blok et al. (2018) [27] |
| p.Arg1121Pro, p.Trp1158Arg p.Asn1159Lys p.Arg1169Trp p.Arg1172Gln |
Helicase C-terminal (1064–1229 a.a.) |
Snijders Blok-Campeau Syndrome, Intellectual disability (β) | Autosomal Dominant | No | Yes (p.Arg1121Pro) |
Snijders Blok et al. (2018) [27] | |
| CHD4 | p.Ser851Tyr | Helicase ATP-binding (738–922 a.a.) |
Sifrim-Hitz-Weiss syndrome (γ) | Autosomal Dominant | No | Yes | Sifrim et al. (2016) [29] |
| p.Arg1068His p.Glu1094Lys p.Arg1127Gln p.Trp1148Leu p.Arg1173Leu |
Helicase C-terminal (1054–1203 a.a.) |
Sifrim-Hitz-Weiss syndrome (γ) | Autosomal Dominant | No | Yes (p.Arg1068His and p.Glu1094Lys) |
Sifrim et al. (2016) [29] Weiss et al. (2016) [28] Weiss et al. (2020) [80] Richards et al. (2015) [81] |
|
| CHD7 | p.Ser834Phe | Chromo 1 (800–867 a.a.) |
CHARGE association, Idiopathic hypogonadotropic hypogonadism (δ) | Autosomal Dominant | No | No | Delahaye et al. (2007) [33] Kim et al. (2008) [41] |
| p.Ile1028Val p.Cys1101Arg | Helicase ATP-binding (980-1154 a.a.) | CHARGE association (δ) | Autosomal Dominant | No | Yes | Vissers et al. (2004) [32] Bergman et al. (2011) [82] |
|
| p.Leu1294Pro p.Leu1302Pro |
Helicase C-terminal (1294–1464 a.a.) |
CHARGE association (δ) | Autosomal Dominant | No | Yes (p.Leu1302Pro) |
Hale et al. (2016) [83] Lalani et al. (2006) [84] Legendre et al. (2017) [42] |
Figure 3.
Scheme of CHD paralogues with pathogenic variants obtained from the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar. Accessed on 2 November 2020) [63] also described in the literature (Table 1) for domains Chromo 1, Helicase ATP-binding, and Helicase C-terminal. Letters α, β, γ, and δ represent the clinical phenotypes associated with each pathogenic variant (see Table 1). Chromobox and motifs I, Ia, Ib, Ic, II, III, IV, IVa, V, Va, and VI are displayed. The colour scheme represents the variable amino acid positions after using the multiple alignment viewer MView (COV—% coverage; ID—% identity). The mapping of the chromobox and the motifs was performed according to references [27,28,51,85,86].
The helicase domains of the superfamily 2 of helicases, to which CHDs belong, have 11 signature motifs (I, Ia, Ib, Ic, II, III, IV, IVa, V, Va, and VI, marked in boxes in Figure 3) crucial for protein activity [79]. Almost all mutations we are now addressing, cluster within or near one of these highly conserved motifs, especially motifs V and VI of the Helicase C-terminal, and motifs Ia, II and III of Helicase ATP-binding. The two mutations affecting the VI motif are located in the highly conserved arginine fingers.
The mutations residing in the Helicase C-terminal involve highly conserved residues among mammalian orthologues that, in general, are also invariable paralogue sites (seven in a total of ten mutations). One exception was p.Arg1121Pro in CHD3, which according to Snijders Blok and colleagues [27] changes a residue in a helix region that is predicted to affect the structure of the CHD3 protein and as demonstrated through functional studies to lower enzymatic activity. In human CHD1 and CHD2, the corresponding position is occupied by histidine (Figure S1), which is, like arginine, a positively charged amino acid, demonstrating the residue flexibility among paralogues. Contrarily, the pathological effects of the substitution of an arginine by a proline are quite predictable, given the very distinct properties of the two amino acids.
As a counterexample, we can take p.Trp1158Arg, again in CHD3, a variant that was previously shown to affect the chromatin remodelling ability of the enzyme [27]. In this case, the alteration involved a position where tryptophan was found to be highly conserved among paralogues, which suggests that the substitution of the residue in any of the paralogue proteins can be critical. Fittingly, Weiss and colleagues [80] performed a functional study on the variant p.Trp1148Leu in CHD4 that revealed its role in decreasing chromatin remodelling activity, a result rather similar to that reported for p.Trp1158Arg in CHD3. The obvious inference is that mutations affecting highly conserved positions among paralogue polypeptide segments will likely disturb the protein function of any of the paralogues. Snijders Blok and colleagues [27] also provided evidence that the arginines located at positions 1169 and 1172 of CHD3 are crucial for correct enzymatic function, further establishing that p.Arg1172Gln caused reduced ATPase activity in functional studies. We cannot omit to highlight that these two positions are highly conserved among orthologues and paralogues (Table 1 and Figure 3), reinforcing their importance for the function of all CHDs.
The pathogenic mutations in the Helicase ATP-binding domain also locate predominately in the highly conserved motifs of the core region (Figure 3). Whilst half of the mutations involved invariable residues in all paralogues, the remaining half are conserved only among paralogues belonging to the same group of CHDs. Among the latter is, for instance, the p.Cys1101Arg in CHD7, found in CHARGE patients [82]. Cysteine is present in all the members of group II and III CHDs (CHD3 to CHD9), but instead, CHD1 and CHD2 have phenylalanine in the homologous position. At this same residue of CHD7 Bergman and colleagues [82] identified in a patient also with CHARGE syndrome a tyrosine (p.Cys1101Tyr), an amino acid with similar properties to phenylalanine. Thus, cysteine at this position in CHD7 seems to be highly intolerant to change, leading to infer a crucial importance for the protein function and probably also for all members from group II and III (CHD3 to CHD9).
Out of the examined CHDs pathogenic mutations, only one changed the sequence of a chromodomain, which was p.Ser834Phe in CHD7, specifically located within the Chromo 1 domain. As mentioned, in each chromodomains of all CHDs there is a chromobox that represents the highly conserved core of the module [51]. It consists of 21-residues among which Ser834 that was here observed to be invariant in the comparison between orthologues or paralogues (Figure 3), further supporting the importance of this particular amino acid residue for the correct function of the chromodomains. In CHD7 this residue lies just next to one of the two aromatic amino acids that form an aromatic cage in chromodomains [73]. Since the aromatic cages of the chromodomains are thought to play a crucial role in the coordination of the recognition of histone methylated-lysine [73], it seems likely that the substitution p.Ser834Phe, given that it introduces an additional aromatic residue, might interfere with the aromatic cage itself, disrupting its function.
4. Discussion
The present work aimed to enrich the knowledge on CHD proteins, a family of enzymes associated with severe developmental disorders. The investigation of sequence variability residing in the evolutionarily conserved domains of the CHD family of proteins allowed us to consolidate and extend the current knowledge on these crucial proteins, highlighting some features that previously had been scarcely documented or addressed namely in what concerns the patterns of conservation of amino acid residues between orthologous sequences.
Firstly, the family of CHD proteins is of very ancient origin and the diversification of this protein family appears to have accompanied the increased requirements of multicellularity, bearing out the essentiality of a set of proteins known to be regulators of transcription and critical players during developmental processes.
Secondly, the mapping of deleterious variants in distinct CHDs, revealed they predominantly cluster in non-variable positions either among the nine human paralogues or among paralogues belonging to the same group of CHDs. Furthermore, most of the mutated positions laid in highly conserved motifs within specific catalytic domains, meaning that the disruption of those sequences results in severe disease phenotypes. In this analysis, we have only focused on CHD mutations causing Mendelian or quasi-mendelian neurodevelopmental disorders. Due to that, the search in the ClinVar only yielded results for missense mutations in CHD1, CHD2, CHD3, CHD4, and CHD7. However, a plethora of pathogenic mutations in these and other members of CHD proteins has been identified in more complex neurodevelopmental and psychiatric diseases and even in cancer. For instance, mutations in CHD8 have often been found in ASD patients [22,35,36,39], and very recently data provided by An and colleagues [35] indicated that mutations affecting the helicase domains of that protein were associated with more severe ASD phenotypes. CHD8 has also recently been implicated in Zahir Friedman syndrome (ZFS) [87]. The gene encoding CHD5 was also found to be a shared risk locus in ASD and two psychiatric disorders [88], fitting the observation that the effects of CHD5 deletion in mouse were consistent with the presentation of ASDs [89].
CHDs remodelers are also potent tumour suppressors and their disruption contribute to the development of a variety of cancers [90,91]. Among the members implicated in cancer, CHD6 is an illustrative example, as it was recently demonstrated to be a key regulator of the oxidative DNA damage response in a manner importantly dependent on the roles exerted by the double chromodomains and SANT domain of CHD6 [90]. A mutation at the CHD6 second SANT domain was identified in a patient clinically presenting Hallermann-Streiff syndrome, a rare premature aging disorder [92]. In a mouse model it was shown that depletion of part of the ATPase domain of CHD6 interferes with motor coordination, indicating that CHD6 could be responsible for some ataxia phenotypes [20].
The case of CHD6 illustrates not only how deficiencies in individual CHD proteins can lead to dissimilar diseases but also how disparate phenotypes can result from mutations in distinct domains or even in the same domain. The complexity of CHDs functions in is still far from being fully understood. Still, as Manning and Yusufzai [7] suggested, the huge diversity of dys/malfunctions associated with CHD paralogues may rely on their non-redundant remodelling activities, on the expression control of specific paralogs in a tissue/temporal dependent fashion, and on the interactions between the paralogs with different sets of sequence-specific transcription factors.
Taken together, all these observations indicate that the variability present among the catalytic domains of paralogues is important to the specialization of functions distributed by the different proteins. At the same time, it may also be noted that the conservation observed between paralogues in key amino acid residues implies that protein function is the result of a network of fine intra-molecular interactions [93,94,95] which can be dramatically impaired by a point alteration at any of these critical sites resulting in pathological conditions. Overall we show that the comparative analysis of both orthologous and paralogous proteins reveals patterns of tolerability and flexibility that represent valuable insights for an accurate prediction of the impact of mutations.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12111827/s1, Figure S1: Multiple sequence alignment scheme of the first Brahma and Kismet (BRK) domain among group III chromodomains helicase-DNA binding (CHD) protein orthologues (CHD6, CHD7, CHD8 and CHD9), Figure S2: Multiple sequence alignment of chromodomains helicase-DNA binding (CHD) protein orthologues (Hsap: Homo sapiens; Ptro: Pan troglodytes; Mmul: Macaca mulatta; Fcat: Felis catus; Rnor: Rattus norvegicus; Mmus: Mus musculus), Table S1: Accession numbers of CHD protein sequences used in the phylogenetic tree.
Author Contributions
Conceptualization, A.R.C., M.J.P. and L.A.; methodology, A.R.C., M.L.-M., M.O., M.J.P. and L.A.; formal analysis, A.R.C., M.J.P. and L.A.; investigation, A.R.C., M.L.-M., M.O., A.A., M.J.P. and L.A.; data curation, A.R.C.; writing—original draft preparation, A.R.C.; writing—review and editing, A.R.C., M.L.-M., M.O., A.A., M.J.P. and L.A.; supervision, M.J.P. and L.A.; funding acquisition, M.J.P. and L.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FEDER—Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020—Operacional Programme for Competitiveness and Internationalization (POCI), Portugal 2020, and by Portuguese funding through FCT—Fundação para a Ciência e a Tecnologia, within the framework of the Project POCI-01–0145-FEDER-007274 to i3S and by FCT research project POCI-01–0145-FEDER-29723. ARC holds a FCT PhD Fellowship (SFRH/BD/141702/2018).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this paper and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li W., Mills A.A. Architects of the genome: Chd dysfunction in cancer, developmental disorders and neurological syndromes. Epigenomics. 2014;6:381–395. doi: 10.2217/epi.14.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E., et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y.J., Khoshkhoo S., Frankowski J.C., Zhu B., Abbasi S., Lee S., Wu Y.E., Hunt R.F. Chd2 is necessary for neural circuit development and long-term memory. Neuron. 2018;100:1180–1193.e1186. doi: 10.1016/j.neuron.2018.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin D.M. Chromatin remodeling in development and disease: Focus on chd7. PLoS Genet. 2010;6:e1001010. doi: 10.1371/journal.pgen.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostapcuk V., Mohn F., Carl S.H., Basters A., Hess D., Iesmantavicius V., Lampersberger L., Flemr M., Pandey A., Thomä N.H., et al. Activity-dependent neuroprotective protein recruits hp1 and chd4 to control lineage-specifying genes. Nature. 2018;557:739–743. doi: 10.1038/s41586-018-0153-8. [DOI] [PubMed] [Google Scholar]
- 6.Thompson P.M., Gotoh T., Kok M., White P.S., Brodeur G.M. Chd5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 7.Manning B.J., Yusufzai T. The ATP-dependent chromatin remodeling enzymes chd6, chd7, and chd8 exhibit distinct nucleosome binding and remodeling activities. J. Biol. Chem. 2017;292:11927–11936. doi: 10.1074/jbc.M117.779470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Layman W.S., Hurd E.A., Martin D.M. Chromodomain proteins in development: Lessons from charge syndrome. Clin. Genet. 2010;78:11–20. doi: 10.1111/j.1399-0004.2010.01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann A., Spengler D. Chromatin remodeling complex nurd in neurodevelopment and neurodevelopmental disorders. Front. Genet. 2019;10:682. doi: 10.3389/fgene.2019.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basta J., Rauchman M. The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res. J. Lab. Clin. Med. 2015;165:36–47. doi: 10.1016/j.trsl.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung H., Kohnken R., Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (nurd) complex is required for peripheral nerve myelination. J. Neurosci. Off. J. Soc. Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marfella C.G., Imbalzano A.N. The chd family of chromatin remodelers. Mutat. Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman-Ayala M., Sachs M., Koh F.M., Onodera C., Bulut-Karslioglu A., Lin C.J., Wong P., Nitta R., Song J.S., Ramalho-Santos M. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015;142:118–127. doi: 10.1242/dev.114843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen T., Ji F., Yuan Z., Jiao J. Chd2 is required for embryonic neurogenesis in the developing cerebral cortex. Stem Cells. 2015;33:1794–1806. doi: 10.1002/stem.2001. [DOI] [PubMed] [Google Scholar]
- 16.Xie J., Gao S., Schafer C., Colijn S., Muthukumar V., Griffin C.T. The chromatin-remodeling enzyme CHD3 plays a role in embryonic viability but is dispensable for early vascular development. PLoS ONE. 2020;15:e0235799. doi: 10.1371/journal.pone.0235799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman J.V., Yamada T., Yang Y., Kong L., Wu D.Y., Zhao G., Gabel H.W., Bonni A. The chromatin remodeling enzyme Chd4 regulates genome architecture in the mouse brain. Nat. Commun. 2020;11:3419. doi: 10.1038/s41467-020-17065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram K.G., Curtis C.D., Silasi-Mansat R., Lupu F., Griffin C.T. The nurd chromatin-remodeling enzyme chd4 promotes embryonic vascular integrity by transcriptionally regulating extracellular matrix proteolysis. PLoS Genet. 2013;9:e1004031. doi: 10.1371/journal.pgen.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang T., Hess R.A., Kolla V., Higashi M., Raabe T.D., Brodeur G.M. CHD5 is required for spermiogenesis and chromatin condensation. Mech. Dev. 2014;131:35–46. doi: 10.1016/j.mod.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathrop M.J., Chakrabarti L., Eng J., Rhodes C.H., Lutz T., Nieto A., Liggitt H.D., Warner S., Fields J., Stöger R., et al. Deletion of the Chd6 exon 12 affects motor coordination. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2010;21:130–142. doi: 10.1007/s00335-010-9248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micucci J.A., Layman W.S., Hurd E.A., Sperry E.D., Frank S.F., Durham M.A., Swiderski D.L., Skidmore J.M., Scacheri P.C., Raphael Y., et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of charge syndrome. Hum. Mol. Genet. 2014;23:434–448. doi: 10.1093/hmg/ddt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama Y., Nishiyama M., Shoji H., Ohkawa Y., Kawamura A., Sato T., Suyama M., Takumi T., Miyakawa T., Nakayama K.I. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- 23.Ooga M., Funaya S., Hashioka Y., Fujii W., Naito K., Suzuki M.G., Aoki F. Chd9 mediates highly loosened chromatin structure in growing mouse oocytes. Biochem. Biophys. Res. Commun. 2018;500:583–588. doi: 10.1016/j.bbrc.2018.04.105. [DOI] [PubMed] [Google Scholar]
- 24.Pilarowski G.O., Vernon H.J., Applegate C.D., Boukas L., Cho M.T., Gurnett C.A., Benke P.J., Beaver E., Heeley J.M., Medne L., et al. Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J. Med. Genet. 2018;55:561–566. doi: 10.1136/jmedgenet-2017-104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvill G.L., Heavin S.B., Yendle S.C., McMahon J.M., O’Roak B.J., Cook J., Khan A., Dorschner M.O., Weaver M., Calvert S., et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suls A., Jaehn J.A., Kecskés A., Weber Y., Weckhuysen S., Craiu D.C., Siekierska A., Djémié T., Afrikanova T., Gormley P., et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijders Blok L., Rousseau J., Twist J., Ehresmann S., Takaku M., Venselaar H., Rodan L.H., Nowak C.B., Douglas J., Swoboda K.J., et al. CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat. Commun. 2018;9:4619. doi: 10.1038/s41467-018-06014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss K., Terhal P.A., Cohen L., Bruccoleri M., Irving M., Martinez A.F., Rosenfeld J.A., Machol K., Yang Y., Liu P., et al. De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am. J. Hum. Genet. 2016;99:934–941. doi: 10.1016/j.ajhg.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sifrim A., Hitz M.P., Wilsdon A., Breckpot J., Turki S.H., Thienpont B., McRae J., Fitzgerald T.W., Singh T., Swaminathan G.J., et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hittner H.M., Hirsch N.J., Kreh G.M., Rudolph A.J. Colobomatous microphthalmia, heart disease, hearing loss, and mental retardation–A syndrome. J. Pediatric Ophthalmol. Strabismus. 1979;16:122–128. doi: 10.3928/0191-3913-19790301-10. [DOI] [PubMed] [Google Scholar]
- 31.Jongmans M.C., Admiraal R.J., van der Donk K.P., Vissers L.E., Baas A.F., Kapusta L., van Hagen J.M., Donnai D., de Ravel T.J., Veltman J.A., et al. Charge syndrome: The phenotypic spectrum of mutations in the CHD7 gene. J. Med. Genet. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 33.Delahaye A., Sznajer Y., Lyonnet S., Elmaleh-Bergès M., Delpierre I., Audollent S., Wiener-Vacher S., Mansbach A.L., Amiel J., Baumann C., et al. Familial CHARGE syndrome because of CHD7 mutation: Clinical intra- and interfamilial variability. Clin. Genet. 2007;72:112–121. doi: 10.1111/j.1399-0004.2007.00821.x. [DOI] [PubMed] [Google Scholar]
- 34.Vuorela P., Ala-Mello S., Saloranta C., Penttinen M., Pöyhönen M., Huoponen K., Borozdin W., Bausch B., Botzenhart E.M., Wilhelm C., et al. Molecular analysis of the CHD7 gene in CHARGE syndrome: Identification of 22 novel mutations and evidence for a low contribution of large CHD7 deletions. Genet. Med. Off. J. Am. Coll. Med. Genet. 2007;9:690–694. doi: 10.1097/GIM.0b013e318156e68e. [DOI] [PubMed] [Google Scholar]
- 35.An Y., Zhang L., Liu W., Jiang Y., Chen X., Lan X., Li G., Hang Q., Wang J., Gusella J.F., et al. De novo variants in the Helicase-C domain of CHD8 are associated with severe phenotypes including autism, language disability and overgrowth. Hum. Genet. 2020;139:499–512. doi: 10.1007/s00439-020-02115-9. [DOI] [PubMed] [Google Scholar]
- 36.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T., et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L., et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D., et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson B., Grepo N., Thompson B.L., Kim J., Wang K., Evgrafov O.V., Lu W., Knowles J.A., Campbell D.B. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding rnas and autism-related genes. Transl. Psychiatry. 2015;5:e568. doi: 10.1038/tp.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eising E., Carrion-Castillo A., Vino A., Strand E.A., Jakielski K.J., Scerri T.S., Hildebrand M.S., Webster R., Ma A., Mazoyer B., et al. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol. Psychiatry. 2019;24:1065–1078. doi: 10.1038/s41380-018-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H.G., Kurth I., Lan F., Meliciani I., Wenzel W., Eom S.H., Kang G.B., Rosenberger G., Tekin M., Ozata M., et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and kallmann syndrome. Am. J. Hum. Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legendre M., Abadie V., Attié-Bitach T., Philip N., Busa T., Bonneau D., Colin E., Dollfus H., Lacombe D., Toutain A., et al. Phenotype and genotype analysis of a french cohort of 119 patients with CHARGE syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 2017;175:417–430. doi: 10.1002/ajmg.c.31591. [DOI] [PubMed] [Google Scholar]
- 43.Lin S., Lin Y., Nery J.R., Urich M.A., Breschi A., Davis C.A., Dobin A., Zaleski C., Beer M.A., Chapman W.C., et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc. Natl. Acad. Sci. USA. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni S., Nagarajan P., Wall J., Donovan D.J., Donell R.L., Ligon A.H., Venkatachalam S., Quade B.J. Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am. J. Med. Genet. Part A. 2008;146a:1117–1127. doi: 10.1002/ajmg.a.32178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whittaker D.E., Riegman K.L., Kasah S., Mohan C., Yu T., Pijuan-Sala B., Hebaishi H., Caruso A., Marques A.C., Michetti C., et al. The chromatin remodeling factor CHD7 controls cerebellar development by regulating reelin expression. J. Clin. Investig. 2017;127:874–887. doi: 10.1172/JCI83408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sood S., Weber C.M., Hodges H.C., Krokhotin A., Shalizi A., Crabtree G.R. CHD8 dosage regulates transcription in pluripotency and early murine neural differentiation. Proc. Natl. Acad. Sci. USA. 2020;117:22331–22340. doi: 10.1073/pnas.1921963117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alendar A., Lambooij J.P., Bhaskaran R., Lancini C., Song J.Y., van Vugt H., Snoek M., Berns A. Gene expression regulation by the Chromodomain helicase DNA-binding protein 9 (CHD9) chromatin remodeler is dispensable for murine development. PLoS ONE. 2020;15:e0233394. doi: 10.1371/journal.pone.0233394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The UniProt Consortium Uniprot: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flanagan J.F., Blus B.J., Kim D., Clines K.L., Rastinejad F., Khorasanizadeh S. Molecular implications of evolutionary differences in CHD double chromodomains. J. Mol. Biol. 2007;369:334–342. doi: 10.1016/j.jmb.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Criscuolo A., Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemoine F., Correia D., Lefort V., Doppelt-Azeroual O., Mareuil F., Cohen-Boulakia S., Gascuel O. Ngphylogeny.Fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47:W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., et al. Phylogeny.Fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefort V., Longueville J.-E., Gascuel O. SMS: Smart model selection in phyml. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of phyml 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Anisimova M., Gil M., Dufayard J.-F., Dessimoz C., Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I., Bork P. Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 60.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown N.P., Leroy C., Sander C. MView: A web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- 63.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crow K.D., Wagner G.P. What is the role of genome duplication in the evolution of complexity and diversity? Mol. Biol. Evol. 2005;23:887–892. doi: 10.1093/molbev/msj083. [DOI] [PubMed] [Google Scholar]
- 65.Niklas K.J., Newman S.A. The origins of multicellular organisms. Evol. Dev. 2013;15:41–52. doi: 10.1111/ede.12013. [DOI] [PubMed] [Google Scholar]
- 66.Trachana K., Jensen L.J., Bork P. Evolution and regulation of cellular periodic processes: A role for paralogues. EMBO Rep. 2010;11:233–238. doi: 10.1038/embor.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodman J.V., Bonni A. Regulation of neuronal connectivity in the mammalian brain by chromatin remodeling. Curr. Opin. Neurobiol. 2019;59:59–68. doi: 10.1016/j.conb.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bosman E.A., Penn A.C., Ambrose J.C., Kettleborough R., Stemple D.L., Steel K.P. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 69.Shur I., Socher R., Benayahu D. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J. Cell. Physiol. 2006;207:374–378. doi: 10.1002/jcp.20586. [DOI] [PubMed] [Google Scholar]
- 70.Newton A.H., Pask A.J. CHD9 upregulates RUNX2 and has a potential role in skeletal evolution. BMC Mol. Cell Biol. 2020;21:27. doi: 10.1186/s12860-020-00270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eissenberg J.C. Structural biology of the chromodomain: Form and function. Gene. 2012;496:69–78. doi: 10.1016/j.gene.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 72.Flanagan J.F., Mi L.Z., Chruszcz M., Cymborowski M., Clines K.L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 73.Yap K.L., Zhou M.M. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry. 2011;50:1966–1980. doi: 10.1021/bi101885m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blus B.J., Wiggins K., Khorasanizadeh S. Epigenetic virtues of chromodomains. Crit. Rev. Biochem. Mol. Biol. 2011;46:507–526. doi: 10.3109/10409238.2011.619164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jain K., Fraser C.S., Marunde M.R., Parker M.M., Sagum C., Burg J.M., Hall N., Popova I.K., Rodriguez K.L., Vaidya A., et al. Characterization of the plant homeodomain (PHD) reader family for their histone tail interactions. Epigenetics Chromatin. 2020;13:3. doi: 10.1186/s13072-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paul S., Kuo A., Schalch T., Vogel H., Joshua-Tor L., McCombie W.R., Gozani O., Hammell M., Mills A.A. Chd5 requires PHD-mediated histone 3 binding for tumor suppression. Cell Rep. 2013;3:92–102. doi: 10.1016/j.celrep.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baker L.A., Allis C.D., Wang G.G. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat. Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alonso-Gonzalez A., Calaza M., Amigo J., González-Peñas J., Martínez-Regueiro R., Fernández-Prieto M., Parellada M., Arango C., Rodriguez-Fontenla C., Carracedo A. Exploring the biological role of postzygotic and germinal de novo mutations in ASD. Sci. Rep. 2021;11:319. doi: 10.1038/s41598-020-79412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding H., Guo M., Vidhyasagar V., Talwar T., Wu Y. The Q motif is involved in DNA binding but not atp binding in ChlR1 helicase. PLoS ONE. 2015;10:e0140755. doi: 10.1371/journal.pone.0140755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss K., Lazar H.P., Kurolap A., Martinez A.F., Paperna T., Cohen L., Smeland M.F., Whalen S., Heide S., Keren B., et al. The CHD4-related syndrome: A comprehensive investigation of theclinical spectrum, genotype–phenotype correlations, and molecular basis. Genet. Med. 2020;22:389–397. doi: 10.1038/s41436-019-0612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergman J.E., Bocca G., Hoefsloot L.H., Meiners L.C., van Ravenswaaij-Arts C.M. Anosmia predicts hypogonadotropic hypogonadism in CHARGE syndrome. J. Pediatrics. 2011;158:474–479. doi: 10.1016/j.jpeds.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 83.Hale C.L., Niederriter A.N., Green G.E., Martin D.M. Atypical phenotypes associated with pathogenic CHD7 variants and a proposal for broadening CHARGE syndrome clinical diagnostic criteria. Am. J. Med. Genet. Part A. 2016;170a:344–354. doi: 10.1002/ajmg.a.37435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lalani S.R., Safiullah A.M., Fernbach S.D., Harutyunyan K.G., Thaller C., Peterson L.E., McPherson J.D., Gibbs R.A., White L.D., Hefner M., et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am. J. Hum. Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farnung L., Vos S.M., Wigge C., Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–542. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yasin H., Gibson W.T., Langlois S., Stowe R.M., Tsang E.S., Lee L., Poon J., Tran G., Tyson C., Wong C.K., et al. A distinct neurodevelopmental syndrome with intellectual disability, autism spectrum disorder, characteristic facies, and macrocephaly is caused by defects in CHD8. J. Hum. Genet. 2019;64:271–280. doi: 10.1038/s10038-019-0561-0. [DOI] [PubMed] [Google Scholar]
- 88.Rylaarsdam L., Guemez-Gamboa A. Genetic causes and modifiers of autism spectrum disorder. Front. Cell. Neurosci. 2019;13:385. doi: 10.3389/fncel.2019.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pisansky M.T., Young A.E., O’Connor M.B., Gottesman I.I., Bagchi A., Gewirtz J.C. Mice lacking the chromodomain helicase DNA-binding 5 chromatin remodeler display autism-like characteristics. Transl. Psychiatry. 2017;7:e1152. doi: 10.1038/tp.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moore S., Berger N.D., Luijsterburg M.S., Piett C.G., Stanley F.K.T., Schräder C.U., Fang S., Chan J.A., Schriemer D.C., Nagel Z.D., et al. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat. Commun. 2019;10:241. doi: 10.1038/s41467-018-08111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mills A.A. The Chromodomain Helicase DNA-binding chromatin remodelers: Family traits that protect from and promote cancer. Cold Spring Harb. Perspect. Med. 2017;7:a026450. doi: 10.1101/cshperspect.a026450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kargapolova Y., Rehimi R., Kayserili H., Brühl J., Sofiadis K., Zirkel A., Palikyras S., Mizi A., Li Y., Yigit G., et al. Overarching control of autophagy and DNA damage response by CHD6 revealed by modeling a rare human pathology. Nat. Commun. 2021;12:3014. doi: 10.1038/s41467-021-23327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suriano G., Azevedo L., Novais M., Boscolo B., Seruca R., Amorim A., Ghibaudi E.M. In vitro demonstration of intra-locus compensation using the ornithine transcarbamylase protein as model. Hum. Mol. Genet. 2007;16:2209–2214. doi: 10.1093/hmg/ddm172. [DOI] [PubMed] [Google Scholar]
- 94.Bridgham J.T., Ortlund E.A., Thornton J.W. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Serrano C., Teixeira C.S.S., Cooper D.N., Carneiro J., Lopes-Marques M., Stenson P.D., Amorim A., Prata M.J., Sousa S.F., Azevedo L. Compensatory epistasis explored by molecular dynamics simulations. Hum. Genet. 2021;140:1329–1342. doi: 10.1007/s00439-021-02307-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in this paper and supplementary material.