Abstract
Hat1p and Hat2p are the two subunits of a type B histone acetyltransferase from Saccharomyces cerevisiae that acetylates free histone H4 on lysine 12 in vitro. However, the role for these gene products in chromatin function has been unclear, as deletions of the HAT1 and/or HAT2 gene displayed no obvious phenotype. We have now identified a role for Hat1p and Hat2p in telomeric silencing. Telomeric silencing is the transcriptional repression of telomere-proximal genes and is mediated by a special chromatin structure. While there was no change in the level of silencing on a telomeric gene when the HAT1 or HAT2 gene was deleted, a significant silencing defect was observed when hat1Δ or hat2Δ was combined with mutations of the histone H3 NH2-terminal tail. Specifically, when at least two lysine residues were changed to arginine in the histone H3 tail, a hat1Δ-dependent telomeric silencing defect was observed. The most dramatic effects were seen when one of the two changes was in lysine 14. In further analysis, we found that a single lysine out of the five in the histone H3 tail was sufficient to mediate silencing. However, K14 was the best at preserving silencing, followed by K23 and then K27; K9 and K18 alone were insufficient. Mutational analysis of the histone H4 tail indicated that the role of Hat1p in telomeric silencing was mediated solely through lysine 12. Thus, in contrast to other histone acetyltransferases, Hat1p activity was required for transcriptional repression rather than gene activation.
The DNA of eukaryotic cells is packaged in a nucleoprotein complex known as chromatin. The fundamental unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around a protein core of histones H2A, H2B, H3, and H4. The core histones are important not only for the structural packaging of DNA in the nucleus but also for regulating many cellular processes that use DNA as a substrate. Much of the regulatory potential of the histones lies in their NH2 termini. The NH2-terminal tails, the first ∼30 amino acids of each histone, are largely unstructured and contain high concentrations of lysine and arginine residues (37). The physical characteristics of the NH2-terminal tails are regulated by extensive posttranslational modifications, which include phosphorylation, methylation, ubiquitination, ADP-ribosylation, and acetylation (69).
Acetylation of core histone NH2-terminal tails was discovered more than 30 years ago and has been the most extensively studied histone modification (3). Histone acetylation occurs on lysine residues, neutralizing their positive charge and changing their structure. As such, this modification is likely to affect the interaction of histones with both DNA and other proteins. The acetylation of the core histones is a dynamic process, with the acetylation state of a given histone determined by the actions of enzymes that add acetyl groups (histone acetyltransferases) and enzymes that remove them (histone deacetylases) (20, 27).
Histone acetyltransferases have traditionally been divided into two types, A and B. Type A histone acetyltransferases are nuclear enzymes that acetylate histones in a chromatin context. It was originally proposed that these enzymes were responsible for the increased levels of histone acetylation that are correlated with transcriptional activation. There is ample experimental evidence to support this idea, beginning with the discovery that the GCN5 gene product of Saccharomyces cerevisiae, a transcriptional coactivator of several genes, is a histone acetyltransferase (12). A number of other proteins known to be involved in transcriptional regulation have also been identified as type A histone acetyltransferases, including P/CAF, CBP, P300, TAFII250, ACTR, SRC-1, Tip60, Esa1p, and Elp3 (14, 16, 41, 44, 58, 61, 72–74). Thus, the acetylation of histones plays an important role in the activation of transcription.
Type B histone acetyltransferases are cytoplasmic enzymes that acetylate histones not associated with DNA. These enzymes are thought to be involved in the acetylation of newly synthesized histones (primarily H3 and H4) (11). When histone H4 is synthesized, it is rapidly acetylated in a specific, evolutionarily conserved pattern (52). There are four lysine residues in the histone H4 NH2 terminus, located at positions 5, 8, 12, and 16. In all eukaryotes that have been examined, newly synthesized histone H4 is acetylated at positions 5 and 12, with little or no acetylation seen at positions 8 and 16 (15, 60). It should be noted that the acetylation state of newly synthesized histone H4 in yeast has not been determined. In many organisms, newly synthesized histone H3 is also acetylated, but not in a strictly conserved pattern (33, 60).
It seems likely that acetylation of histones H3 and H4 plays an early role in chromatin assembly. Pulse-chase experiments indicate that this acetylation occurs rapidly after the synthesis of the histones (33, 52, 60). In addition, diacetylated histone H4 is complexed with histone H3 in the cytoplasm (13). Two protein complexes, chromatin assembly factor 1 (CAF-1) and replication-coupling assembly factor (RCAF), mediate replication-dependent chromatin assembly in vitro. Both of these activities preferentially interact with H3-H4 tetramers that contain species of H4 that are acetylated in patterns similar to those of the newly synthesized protein (30, 59, 68, 71). The population of histone H4 that associates with CAF-1 is acetylated at lysines 5, 8, and 12, while that associated with RCAF is acetylated at only lysines 5 and 12 (30, 68, 71). CAF-1 and RCAF have homologs in S. cerevisiae (CAC1, CAC2, and MSI1 for CAF-1 and ASF1 for RCAF), and genetic analyses suggest that these complexes are both involved in some aspect of chromatin assembly in vivo (18, 31, 34, 42, 56, 68).
The Hat1p-Hat2p complex isolated from S. cerevisiae is the quintessential type B histone acetyltransferase (26, 32, 46, 70). Hat1p, the catalytic subunit of the enzyme, when expressed in bacteria, acetylates histone H4 at the same residues that are modified on newly synthesized histone H4, lysines 5 and 12 (32, 46). However, its activity in vivo may be restricted, as the native enzyme acetylates H4 only at lysine 12 (46). Hat2p is a regulatory subunit of the enzyme; it is not required for catalytic activity but increases specific activity 10-fold. Hat2p appears to function by mediating the interaction between Hat1p and histone H4 (46, 70). Hat2p is an ortholog of two nearly identical human proteins, Rbap48 and Rbap46 (47, 48). Proteins in the Hat2p/Rbap48 family are subunits of protein complexes that modulate chromatin structure, including CAF-1, the nucleosome remodeling factor, and several histone deacetylase and transcriptional corepressor complexes (24, 40, 64, 71, 75). Thus, these proteins seem to play a central role in the communication between histones and chromatin-modifying activities.
Simple genetic analysis of HAT1 did not uncover an obvious role for type B histone acetyltransferases. Deletion of the gene does not affect cell growth or result in any other observable phenotype (32, 46). The lack of a hat1Δ phenotype suggested that type B histone acetyltransferases are not essential, but conservation of the acetylation pattern of newly synthesized histone H4 and of the Hat1p-Hat2p enzyme over a wide range of eukaryotes argues that this histone modification is important (15, 26, 60, 70). The lack of a phenotype may be explained by an acetyltransferase activity that can modify histone H4 just as Hat1p does or by structural redundancy in the histones that obviate the acetylation of histone H4 by Hat1p (35, 38).
In an effort to identify an in vivo role for the Hat1p-Hat2p histone acetyltransferase, we sought a sensitive genetic assay that monitored subtle changes in chromatin structure. Specifically, we explored whether Hat1p-Hat2p might play a role in telomeric silencing. Genes located near telomeres are transcriptionally repressed, or silenced, due to a special chromatin structure that is assembled over telomere-proximal DNA (19). Critical components of telomeric silent chromatin include the SIR proteins (Sir2p, Sir3p, and Sir4p), Rap1p (a telomere DNA-binding protein), and the histone H3 and H4 tails. It appears that the Sir proteins are recruited to telomeres through their interactions with Rap1p and one another and then “polymerize” along telomere-adjacent DNA by binding the NH2-terminal tails of histone H3 and H4 of the associated nucleosomes (4, 22, 51, 55). Telomeric silent chromatin shares many of the hallmarks of heterochromatin in other eukaryotes, including a distinct acetylation pattern on the histone H4 NH2-terminal tail. Histone H4 found in yeast silent chromatin is almost entirely unacetylated at positions 5, 8, and 16 but highly acetylated at position 12, just as it is in heterochromatin (9, 10). While it is not known whether this acetylation pattern plays a role in the formation of silent chromatin, the fact that Hat1p can specifically acetylate histone H4 at position 12 suggests that Hat1p may be necessary for silencing.
MATERIALS AND METHODS
Plasmid construction.
A plasmid containing wild-type HHT2 and HHF2 was constructed from pRM200 (39). First, pRM200 was digested with EcoRI and XbaI. The 4.6-kbp fragment containing HHT2 and HHF2 was isolated and blunt-end ligated into the HincII site of pUC9 to generate pMP1. Then 1.4 kbp of HHT2 downstream sequence was removed from pMP1 by digestion with HincII and SalI, followed by blunt-end religation (pMP2). The 2.7-kbp PstI fragment of pMP2 was ligated into the PstI site of pRS314 to generate pMP3, which was used as the wild-type HHT2-HHF2 plasmid in these studies.
A second plasmid containing the same fragment of HHT2 and HHF2 but with HHT2 lysines 9, 14, 18, and 23 changed to arginines (K9,14,18,23R) was constructed by ligating the 2.7-kbp PstI fragment of pRM253 into the PstI site of pRS314 (pMP6) (39). The H3 K9,14,18,23,27R allele was constructed from pMP6 by PCR amplification of a 1.4-kbp fragment extending from the NcoI site downstream of HHF2 to the PinAI site in the HHT2 open reading frame using the following primers: HHF2 NcoI, GGATTCCATGGGTTTCTGCG, and H3 K27R, CACCACCGGTAGATGGGGCGGATCTTCTGGCAGCTCTGGAGGC. The H3 K27R primers incorporate a mutation that changes HHT2 lysine 27 to arginine. This PCR fragment was digested with NcoI and PinAI and ligated into pMP6 that had been digested with the same enzymes to generate pMP8. The HHT2 K27R allele was confirmed by DNA sequencing.
All other HHT2 and HHF2 alleles were generated incrementally from pMP3, pMP6, and pMP8 by site-directed mutagenesis (Quik-Change site-directed mutagenesis kit; Stratagene) using appropriate combinations of the following PCR primers: H3 R9K A, CTAAACAAACAGCTAGAAAATCCACTGGTGG; H3 R9K B, CCACCAGTGGATTTTCTAGCTGTTTGTTTAG; H3 R14K A, CCACTGGTGGTAAAGCCCCAAGAA; H3 R14K B, TTCTTGGGGCTTTACCACCAGTGG; H3 R18K A, GCCCCAAGAAAACAATTAGCC; H3 R18K B, GGCTAATTGTTTTCTTGGGGC; H3 R23K A, CAATTAGCCTCCAAAGCTGCCAGAA; H3 R23K B, TTCTGGCAGCTTTGGAGGCTAATT; H4 K5R A, GCCTGGTAGAGGTAGAGGTGGTAAAGGTCTAGG; H4 K5R B, CCTAGACCTTTACCACCTCTACCTCTACCTGGC; H4 K8R A, GAGGTAAAGGTGGTAGAGGTCTAGGAAAAGG; H4 K8R B, CCTTTTCTCAGACCTCTACCACCTTGACCTC; H4 K12R A, GGTCTAGGAAGAGGTGGTGCC; H4 K12R B, GGCACCACCTCTTCCTAGACC; H4 K16R A, GGAAAAGGTGGTGCCAGACGTCACAGAAAGATTC; and H4 K16R B, GAATCTTTCTGTGACGTCTGGCACCACCTTTTCC.
Following site-directed mutagenesis, the coding sequences were checked by DNA sequencing to confirm that no PCR artifacts were incorporated into the plasmids.
The 2.7-kbp PstI fragment from pMP2 was ligated into the PstI site of pRS317 to generate pMP9. The same PstI fragment was also inserted into the PstI site of pRS412 to generate pRS412/HHT2-HHF2.
pHAT1::LYS2 was made by first deleting the BamHI-BglII fragment from the HAT1 open reading frame in pT7Sc-HAT1 (46). A SalI-SmaI fragment from YDpK containing the LYS2 coding sequence was then blunt-end ligated into the BamHI and BglII sites in pT7Sc-HAT1 (6).
Yeast strain construction.
Standard yeast culture and genetic methods were used (1, 23). Gene deletions were confirmed by Southern blotting or colony PCR.
Strain UCC1111 [MATα ade2::his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2 pRS412 (ADE2 CEN ARS) - HHF2-HHT2] was constructed as follows. URA3 was placed at the left arm of chromosome VII in strain BY4705 as described previously to generate UCC1091 (8, 19). The HHT2-HHF2 gene pair was replaced by MET15 using PCR-mediated gene disruption (UCC1095) (5). pMP9 was transformed into UCC1095, followed by replacement of the HHT1-HHF1 gene pair with HIS3 (UCC1098). The LEU2 gene was then inserted in place of the HIS3 gene by PCR-mediated gene disruption. pRS412-HHT2-HHF2 was then swapped with pMP9 to generate UCC1111.
The HAT1 and HAT2 genes were each disrupted in UCC1111 with HIS3 using PCR-mediated gene disruption to generate strains MPY1 and TKY101, respectively. HAT1 was disrupted with LYS2 in TKY101 by transformation with plasmid pHAT1::LYS2 that had been digested with EcoRI and HaeII to generate TKY104.
Plasmids containing wild-type or mutant HHT2-HHF2 alleles were transformed into these strains and selected on plates lacking tryptophan. Colonies that had lost the pRS412-HHT2-HHF2 plasmid, and which were thus ade2, were identified by their red color.
BY4705a (MATa ade2::hisΔ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0) was produced by changing the mating type of BY4705 by two-step replacement.
UCC6580 [MATα ade2::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 adh4::URA3-TEL (VII-L) sir3::HIS3] was generated by PCR-mediated disruption of the SIR3 gene in UCC1091.
Telomeric silencing assays.
Telomeric silencing was assayed essentially as described previously (19). Briefly, individual colonies of the indicated strains were resuspended in 200 μl of water. Tenfold serial dilutions of the cell suspensions were made, and 10 μl of each dilution was spotted onto synthetic complete plates (HC) and synthetic complete plates containing 0.1% 5-fluoroorotic acid (HC+5-FOA). The plates were then incubated for 3 days at 30°C unless otherwise indicated. In each case, at least three individual colonies of each strain were tested.
RT-PCR assays.
Total RNA was isolated as described (54). Reverse transcription (RT)-PCRs were performed using Ready-To-Go RT-PCR beads (Pharmacia) according to the manufacturer's instructions. The primers and cycling parameters for the amplification of MATa1 RNA were identical to those described (57). Reaction products were resolved on 2% agarose gels and visualized with ethidium bromide.
RESULTS
Hat1p and histone H3 are redundantly required for telomeric silencing.
Yeast telomeric chromatin is capable of repressing the transcription of genes placed within several kilobases of the end of a chromosome (50). The integrity of this silent chromatin structure can be sensitively assayed using the URA3 gene as a marker. Normal expression of the URA3 gene from an internal chromosomal locus causes yeast cells to be sensitive to 5-FOA due to conversion of the 5-FOA into a toxin by the URA3 gene product (7). When the URA3 gene is placed near a telomere, expression of the gene is repressed in a high percentage of cells in the population (30 to 50%), allowing them to grow in the presence of 5-FOA. When telomeric silencing is disrupted, the URA3 gene is expressed and the 5-FOA in the medium kills the cells (19).
The HAT1 gene was deleted in a strain containing a telomeric URA3 gene. As shown in Fig. 1, the absence of Hat1p had no effect on the level of telomeric silencing.
FIG. 1.
Effects of hat1Δ and histone H3 single-lysine-to-arginine mutations on telomeric silencing. The indicated histone H3 alleles were introduced into UCC1111 (HAT1) and MPY1 (hat1Δ). Telomeric silencing was measured by spotting 10-fold serial dilutions of cells on synthetic complete plates (HC) and synthetic complete plates containing 5-FOA (HC+5-FOA) (Materials and Methods). Plates were photographed after 3 days of growth at 30°C. The relevant genotypes of the strains are indicated on the left. WT, wild type. Assays were performed on at least three separate colonies from each strain.
We next asked whether Hat1p had a role in telomeric silencing that was masked by functional redundancies. One candidate for such an effect is the histone H3 tail, which is required for telomeric silencing. While the role of the lysines in the H3 tail has not been examined in depth, mutating several of these residues has been shown to cause modest decreases in telomeric silencing that are synergistic with a mutation that alters lysines 5, 8, and 12 of the histone H4 tail (65).
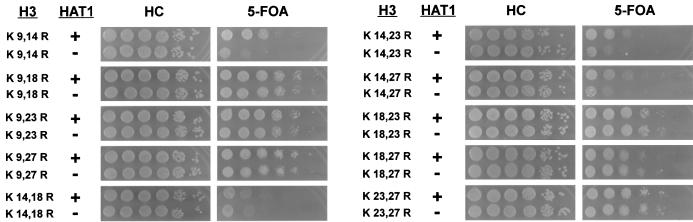
We began our analysis by changing each of the five acetylatable lysines in the histone H3 NH2-terminal tail to arginine. These changes preserve the basic charge of the residue but prevent it from being acetylated. Each allele was then introduced into wild-type and hat1Δ strains, and telomeric silencing was assayed as described above (Fig. 1). A number of observations can be made from this experiment. First, none of the lysine residues in the H3 tail was essential for telomeric silencing. Mutations at four of the histone H3 tail lysines (9, 18, 23, and 27) had little or no effect on telomeric silencing. Mutating the lysine at position 14 had the largest effect, causing a ≥10-fold reduction in silencing, as indicated by the decrease in the number of 5-FOA-resistant colonies. This is in contrast to the histone H4 tail, where mutation of lysine 16 completely abolishes telomeric silencing (4, 65). Also, while mutating individual lysine residues in the H3 tail did not uncover a significant hat1Δ phenotype, telomeric silencing was reduced further when hat1Δ mutations were combined with the H3 K14R allele (Fig. 1).
Next, we combined pairs of lysine-to-arginine mutations in the histone H3 tail with hat1Δ mutations. Five of the 10 histone H3 double-lysine-to-arginine mutations had little effect on telomeric silencing, even when they were combined with hat1Δ (Fig. 2). Two of the double histone H3 mutants showed decreased levels of silencing, independent of HAT1. However, three of the histone H3 double-lysine-to-arginine mutants had a much greater defect in telomeric silencing when the mutations were combined with a hat1Δ mutation (K9,14R, K14,23R, and K14,27R). In each case there was an approximately 100-fold effect on silencing. Interestingly, the hat1Δ phenotype required mutation of lysine 14 along with either lysine 9, 23, or 27. In each case when lysine 14 was unchanged, deletion of the HAT1 gene had no effect. Therefore, pairs of lysine residues in the histone H3 tail that include lysine 14 appear to be functionally redundant with Hat1p for the formation of telomeric silent chromatin.
FIG. 2.
Function of Hat1p in telomeric silencing is uncovered by particular histone H3 double-lysine-to-arginine mutations. The indicated histone H3 alleles were introduced into UCC1111 (HAT1) and MPY1 (hat1Δ). Telomeric silencing was assayed as described in the legend to Fig. 1. The genotypes are indicated on the left.
The hat1Δ-dependent decrease in growth on 5-FOA medium is specifically due to defects in telomeric silencing. hat1Δ/histone H3 mutants that lack a telomeric URA3 gene grow normally on 5-FOA, indicating that these mutants are not hypersensitive to this drug. In addition, the hat1Δ/histone H3 mutants increased the expression of the ADE2 gene when it was placed near the right arm of chromosome V, as determined by changes in colony color, demonstrating that this phenotype was neither telomere nor gene specific (data not shown) (19).
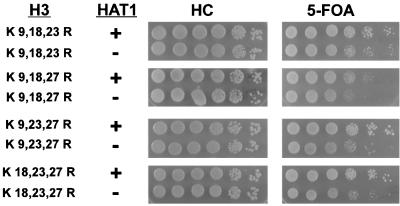
Each histone H3 triple-lysine-to-arginine mutant combination was also assayed for telomeric silencing. In each case when lysine 14 was changed to arginine, there was a hat1Δ effect on telomeric silencing similar to that shown in Fig. 2 (data not shown). The major difference between the double and triple mutants was a general reduction in the overall level of silencing. A different phenotype was observed when deletions of HAT1 were combined with H3 triple-lysine-to-arginine mutants that retain a lysine residue at position 14 (Fig. 3). While the number of 5-FOA-resistant colonies was unchanged, the size of the colonies was reduced dramatically in the hat1Δ background. This small-colony phenotype on HC+5-FOA plates was also observed for the H3 K14R mutant, independent of hat1Δ. The small-colony phenotype is not due to a general growth defect, as these strains grow normally in the absence of 5-FOA, but may be indicative of defects in the maintenance of the silent chromatin state (17, 42). Thus, a HAT1-dependent effect on telomeric silencing is observed when any three lysines are mutated to arginine in the histone H3 tail. However, the effect is modest if lysine 14 is not mutated.
FIG. 3.
hat1Δ causes a decrease in the size of 5-FOA-resistant colonies when combined with histone H3 triple-lysine-to-arginine mutations that retain lysine 14. The indicated histone H3 alleles were introduced into UCC1111 (HAT1) and MPY1 (hat1Δ). Telomeric silencing was assayed as in Fig. 1. The genotypes of the strains are indicated.
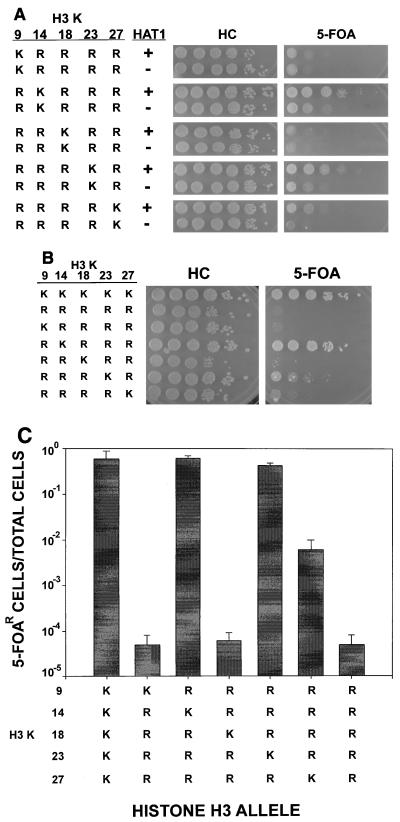
Next we examined the effect on telomeric silencing when four of five lysines in the histone H3 tail were mutated to arginine. In two cases (K14,18,23,27R and K9,14,23,27R) telomeric silencing was virtually abolished (Fig. 4A). This precluded an evaluation in these strains of Hat1p in telomeric silencing. In the other three mutants, silencing was still evident and was reduced when HAT1 was mutated. When lysine 14 remained (H3 K9,18,23,27R), mutating HAT1 did not decrease the number of 5-FOA-resistant colonies but significantly reduced their size (the small hat1Δ colonies were more readily visible after 7 days of growth). When lysine 14 was changed to arginine (H3 K9,14,18,27R and H3 K9,14,18,23R), hat1Δ decreased both the number and size of the 5-FOA-resistant colonies.
FIG. 4.
Single histone H3 lysine residues can be sufficient to support telomeric silencing. (A) The indicated histone H3 alleles were introduced into UCC1111 (HAT1) and MPY1 (hat1Δ). Telomeric silencing was assayed as in Fig. 1. (B) Analysis of the level of telomeric silencing of histone H3 alleles containing four or five lysine-to-arginine mutations. The indicated histone H3 alleles were introduced into UCC1111. Telomeric silencing was assayed as in Fig. 1. (C) Telomeric silencing was quantitated from three to five repetitions of the assays shown in panels A and B by counting the number of colonies that grew on HC and HC+5-FOA plates. The mean has been plotted, with the error bars representing the standard deviation. Plates were incubated for 7 days to aid in the counting of small colonies.
An unexpected aspect of the results presented in Fig. 4A is that histone H3 alleles that contain only a single lysine residue at position 14, 23, or 27 retained a high level of telomeric silencing. We directly compared the level of telomeric silencing observed with wild-type histone H3 to the level of silencing seen with alleles that change either four or five lysines to arginine (Fig. 4B and C). Mutating all five lysines in the H3 tail reduced telomeric silencing to the detection limits of our assay. This level of silencing was comparable to that found with an allele that changes histone H4 lysine 16 to arginine, which has been well documented to cause a complete loss of telomeric silencing (4, 65). This demonstrated that, collectively, the acetylatable lysines in the histone H3 tail were essential for telomeric silencing. In addition, lysine 14 alone was sufficient to support a nearly wild-type level of telomeric silencing. When lysine 23 was the sole unchanged residue, the number of 5-FOA-resistant colonies was similar to that of wild-type histone H3, but the size of these colonies was greatly reduced. Lysine 27 was capable of supporting a moderate level of telomeric silencing, while lysines 9 and 18 were incapable of supporting telomeric silencing. Therefore, there is a hierarchy to the requirement for the acetylatable lysines in the histone H3 tail, where lysine 14 is the most effective, lysine 23 is a little less effective, lysine 27 is weakly effective, and lysines 9 and 18 are completely ineffective.
Hat1p does not influence HMR silencing.
Telomeric heterochromatin involves many but not all of the factors involved in the transcriptional repression of the silent mating loci (4). The silent mating loci, HML and HMR, contain copies of yeast mating type-specific genes (a and α) that are expressed only when present at the MAT locus. The mechanism of transcriptional silencing at HML and HMR appears to be similar to the silencing of telomere-proximal genes (4, 51). However, HML and HMR silencing is stronger than telomeric silencing due to the presence of multiple cis-acting silencing elements and the participation of Sir1p in silencing at mating loci but not at telomeres (36).
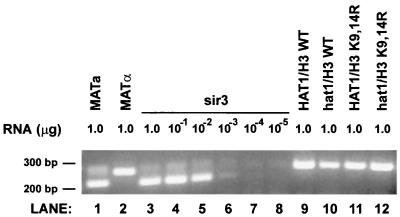
We tested whether histone H3 and hat1Δ mutations derepress the MATa1 gene at the HMRa locus. RT-PCR, using primers that flank a small intron, can be used to monitor expression of MATa1. The RT-PCR product produced from mature MATa1 mRNA is approximately 50 bp smaller than the product derived from genomic DNA (57). MATa1 is normally expressed at the MAT locus in MATa cells and repressed in MATα cells. This is visualized in the RT-PCR reactions shown in Fig. 5 (lanes 1 and 2), where the faster-migrating, mRNA-derived band is only generated from MATa RNA. As expected, MATa1 expression is completely derepressed in a MATα sir3Δ cell (Fig. 5, lane 3). This RT-PCR has a 1,000-fold range of sensitivity, as determined by assaying 10-fold serial dilutions of the MATα sir3Δ RNA (Fig. 5, lanes 3 to 8). In a MATα strain, hat1Δ and H3 K9,14R mutations do not cause any apparent derepression of the MATa1 gene when it is silenced at HMRa. Thus, as seen for the chromatin assembly factor Cac1p, the effects of Hat1p are more pronounced on telomeric silencing than on silent mating locus repression (17, 29, 42).
FIG. 5.
HMR silencing is unaffected by hat1Δ/histone H3 mutations. RT-PCRs were performed on total RNA (without removal of genomic DNA) isolated from the MATa strain BY4705a (lane 1) and MATα strains BY4705 (lane 2), UCC6580 (lanes 3 to 8), UCC1111 (lanes 9 and 11), and MPY1 (lanes 10 and 12). UCC1111 and MPY1 also contained the histone H3 alleles indicated. The amount of total RNA in each reaction is indicated. The migration of DNA molecular size standards is given on the left.
Hat1p affects telomeric silencing through histone H4 lysine 12.
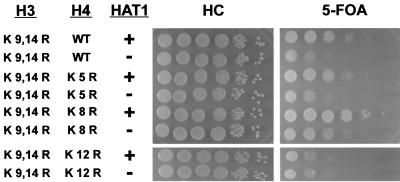
If the role of Hat1p in telomeric silencing is due to the acetylation of the histone H4 tail, then mutating lysine residues in the histone H4 tail might mimic the hat1Δ phenotypes. Like the hat1Δ mutation, individually mutating histone H4 lysines 5, 8, and 12 has no effect on telomeric silencing (data not shown). However, when the histone H4 mutations were combined with the histone H3 K9,14R allele (this allele produced the most pronounced hat1Δ phenotype with the fewest mutations in histone H3), only the K12R mutant of histone H4 mimicked the hat1Δ effect on telomeric silencing (Fig. 6). Importantly, combining hat1Δ with the H3 K9,14R and H4 K12R alleles did not result in further decreases in telomeric silencing, suggesting that Hat1p is functioning through histone H4 lysine 12. In contrast, mutating histone H4 lysine 5 or 8 to arginine did not result in a decrease in telomeric silencing in the H3 K9,14R background. In fact, changing lysine 8 to arginine increased the level of telomeric silencing. This is consistent with the recent finding that deletion of GCN5, whose gene product acetylates histone H4 lysine 8, results in an increase in telomeric silencing (63). In addition, altering histone H4 lysines 5 and 8 did not affect the hat1Δ phenotype normally seen in combination with the H3 K9,14R allele. Taken together, these results suggest that Hat1p functions through histone H4 lysine 12.
FIG. 6.
Role of Hat1p in telomeric silencing is mediated through histone H4 lysine 12. The indicated histone H3-H4 alleles were introduced into UCC1111 (HAT1) and MPY1 (hat1Δ). Telomeric silencing was assayed as in Fig. 1.
Hat2p is involved in telomeric silencing.
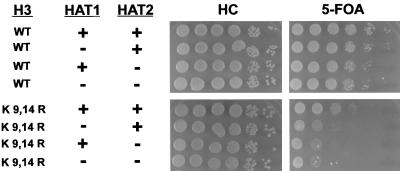
Hat2p was originally characterized biochemically as a positive regulatory subunit of the yeast histone H4-specific type B histone acetyltransferase complex (46). We took advantage of the hat1Δ telomeric silencing phenotype to test whether the regulation of Hat1p activity by Hat2p was also physiologically relevant. If Hat2p is required for the in vivo activity of Hat1p, deletion of the HAT2 gene should cause a telomeric silencing defect similar to that of hat1Δ when combined with histone H3 mutations. In the presence of wild-type histone H3, hat2Δ caused little if any decrease in telomeric silencing (Fig. 7). However, combining the histone H3 K9,14R allele with hat2Δ resulted in a decrease in telomeric silencing similar to that seen with hat1Δ. Consistent with the idea that Hat1p and Hat2p function together, further decreases in the level of telomeric silencing were not observed in the hat1Δ hat2Δ double mutant. These results indicate that the Hat1p-Hat2p interactions identified in vitro are functionally relevant in vivo.
FIG. 7.
Hat2p is involved in telomeric silencing. The indicated histone H3 alleles were introduced into UCC1111 (HAT1), MPY1 (hat1Δ), TKY101 (hat2Δ), and TKY104 (hat1Δhat2Δ). The strains were then assayed for telomeric silencing as in Fig. 1.
DISCUSSION
We have identified a role for the Hat1p-Hat2p type B histone acetyltransferase complex in telomeric silencing. The function of this enzyme is redundant with the histone H3 NH2-terminal tail, as the telomeric silencing defect of hat1Δ and hat2Δ strains is only apparent when particular lysine residues in histone H3 are also mutated.
In vivo characterization of Hat1p.
Up to this point, our understanding of Hat1p has been based solely on in vitro biochemical analyses, due to a lack of obvious phenotypes in hat1Δ cells. A role for Hat1p in telomeric silencing now allows in vivo characterization of this enzyme. The fundamental question about the function of Hat1p, whether histone H4 is an in vivo substrate of this enzyme, can now be answered affirmatively. When the histone H3 gene is appropriately mutated, changing lysine 12 on histone H4 mimics the hat1Δ phenotype. This is completely consistent with earlier work in which Hat1p isolated from yeast cytoplasmic extracts specifically acetylates histone H4 only on lysine 12 (32, 46). Recombinant Hat1p isolated from Escherichia coli acetylates lysine 5 and lysine 12 of histone H4 as well as histone H2A. This relaxation in specificity may be a fortuitous trait of recombinant Hat1p or may reflect the fact that Hat1p specificity is regulated in yeast. Our present study on telomeric silencing is consistent with Hat1p acting only on lysine 12 of histone H4.
Furthermore, the current study indicates that Hat2p is critical to Hat1p activity in vivo; hat1Δ, hat2Δ, and combined hat1Δ hat2Δ mutations caused the same loss of telomeric silencing. These results are best explained by Hat2p's ability to stabilize the interaction of histone H4 with Hat1p, thus increasing the specific activity of Hat1p-mediated acetylation (46, 70).
Histone acetylation and heterochromatin.
Histone H4 located in regions of silent chromatin in yeast or heterochromatin in Drosophila cells is acetylated in a distinct pattern. Lysines 5, 8, and 16 are hypoacetylated, and lysine 12 is acetylated at normal levels (10, 22, 67). It is unclear whether this acetylation pattern plays an active role in heterochromatin function, or whether this pattern is merely a consequence of the histones in heterochromatin being isolated from the histone acetyltransferases and deacetylases that normally act on euchromatin (22). Mutational analysis of the lysine residues of the H4 tail has produced contradictory results in terms of addressing the importance of this acetylation state for heterochromatic function. Mutating lysines 5, 8, and 12 to arginine, which is thought to mimic the constitutively unacetylated state, has little or no effect on transcriptional repression at HML and HMR, as measured by mating efficiency, or at telomeres (28, 45, 65). This suggests that these residues do not play a role in transcriptional silencing.
However, Braunstein et al. found that converting lysine 5 or 12 to glutamine, which has been suggested to mimic acetylated lysine, suppresses the mating defect seen when the other three histone H4 tail lysines are mutated to arginine (10). Enomoto and Berman found that converting lysines 5, 8, and 12 to arginine results in the formation of shmoo clusters when MATa cells are exposed to α-factor, indicative of subtle defects in HMLα silencing (17). In addition, Hecht et al., using an in vitro assay to detect interactions between histone H4 tail peptides and Sir3p, found that mutating lysine 16 to glutamine, which eliminates silencing in vivo, has no effect on the histone H4-Sir3p interaction. It is only when lysine 12 (or 5 and 12) is also changed to glutamine that the histone H4-Sir3p interaction is disrupted, suggesting that lysine 5 or 12 provides part of the interaction of histone H4 with Sir3p (25). Our findings help to clarify these results and strongly support the idea that acetylation of lysine 12 plays an important role in the formation or maintenance of silent chromatin (heterochromatin). Until now, the role of this modification has been obscured by redundancy with the histone H3 tail. This “redundancy” may be explained by binding of silencing proteins, such as Sir3p, to both histone H3 and H4 tails.
The fact that Hat1p promoted a repressive chromatin structure runs counter to the action of other known histone acetyltransferases, which are uniformly involved in the activation of transcription. This may reflect a fundamental in vivo difference between type B and type A histone acetyltransferases, which were originally distinguished based solely on in vitro biochemical properties. One explanation for these results is that lysine 12 acetylation, which is carried out by Hat1p, is important for the interaction between histone H4 and a structural component of silent chromatin, such as Sir3p. The fact that the acetylation of a histone, which is generally thought to loosen the interaction between histones and DNA, can promote a repressive chromatin structure underscores the idea that the acetylation of histones can act as a signal that modulates protein-protein interactions (62).
In regions of silent chromatin, histone H3 is hypoacetylated (10). However, it is not known whether the hypoacetylation of the histone H3 lysine residues is uniform or whether there is a specific pattern of acetylation. Our demonstration that lysine-to-arginine mutations at specific sites can have dramatic effects on telomeric silencing suggests that the acetylation of histone H3 might play an important role in silent chromatin function. Alternatively, lysine per se at specific sites in histone H3 may be important for silencing. Whichever mechanism is at work, lysine 14 is a key player in silencing, and lysines 23 and 27 contribute to a lesser extent.
Definitive evidence for the involvement of histone H3 acetylation in silent chromatin will require an analysis such as we have carried out here for histone H4 and Hat1p-Hat2p. That is, it will require identification of a histone H3-specific acetyltransferase that is required for silent chromatin function. An obvious candidate is Gcn5p, which can acetylate lysine 14 of histone H3 in vitro (21, 33, 66). However, it was recently shown that gcn5Δ cells have increased levels of telomeric silencing (63). Other candidate acetyltransferases include Sas2p and Sas3p, which have been shown to be important for full telomeric silencing (49). Both proteins show limited homology to the GNAT superfamily of acetyltransferases (43).
Chromatin assembly and heterochromatin.
As suggested above, Hat1p activity may be important in telomeric silencing because it acetylates lysine 12 of histone H4 and because acetylated lysine 12 is required for binding by silencing proteins (e.g., Sir3p). An alternative and mutually inclusive hypothesis is that acetylation of lysine 12 is required for interaction with a chromatin assembly factor, which ultimately leads to deposition of histone H4 for silent chromatin. One such potential chromatin assembly factor is CAF-1, which binds cytosolic histone H3-H4 tetramers and deposits them onto newly replicated DNA in vitro (30, 59). The histone H4 that is preferentially complexed with CAF-1 is acetylated on lysines 5, 8, and 12 (71). The fact that type B histone acetyltransferases, such as Hat1p, can acetylate free histone H4 on lysines 5 and 12 has led to the suggestion that Hat1p and CAF-1 operate in a common pathway, with Hat1p being at least partly responsible for acetylating the histone H4 that interacts with CAF-1 (2). Consistent with this model, mutants lacking the yeast CAF-1 subunits (CAC1, CAC2, and/or MSI1) display defects in telomeric silencing similar to those of the hat1Δ/histone H3 double mutants.
The pattern of acetylation of the histone H4 tail found in silent chromatin partially overlaps the acetylation pattern of newly synthesized histone H4. This has led to the idea that the modification present on histone H4 in silent chromatin is a result of the assembly of newly synthesized histones. However, if this is the case, why is heterochromatic histone H4 not acetylated at lysines 5 and 8? There are a number of possible explanations. CAF-1 may be capable of selectively interacting with and targeting populations of histones that have distinct acetylation states. In this way, CAF-1 may selectively assemble histone H4 acetylated at lysine 12 in regions of silent chromatin. Alternatively, histone deacetylases specific for lysines 5 and 8 may act in regions of silent chromatin to produce histones with the proper acetylation pattern. A third possibility is that the complete deacetylation of histone H4 following chromatin assembly that is seen in euchromatin also occurs in regions of silent chromatin. Hat1p may then reestablish the characteristic silent chromatin histone H4 acetylation pattern. This possibility is supported by the fact that Hat1p appears to be located in the nucleus as well as the cytoplasm (26, 46, 53, 70).
Thus, while we have demonstrated that the Hat1p-Hat2p histone acetyltransferase is important for telomeric silencing, the next challenge will be to determine when it plays its role in silent chromatin formation.
ACKNOWLEDGMENTS
We thank R. Mann and M. Grunstein for plasmids pRM200 and pRM253 and J. Boeke for strain BY4705. We also thank J. Basa for assistance in constructing some histone H3 alleles and P. Wade, R. Kamakaka, and A. Sklenar for critical reading of the manuscript.
This work was supported by an Ohio State University seed grant to M.R.P. and grant GM43893 from the National Institutes of Health to D.E.G.
REFERENCES
- 1.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 2.Adams C R, Kamakaka R T. Chromatin assembly: biochemical identities and genetic redundancy. Curr Opin Genet Dev. 1999;9:185–190. doi: 10.1016/S0959-437X(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 3.Allfrey V G, Faulkner R M, Mirsky A E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berben G, Dumont J, Gilliquet V, Bolle P A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 7.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 10.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 12.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 13.Chang L, Loranger S S, Mizzen C, Ernst S G, Allis C D, Annunziato A T. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 15.Chicoine L G, Schulman I G, Richman R, Cook R G, Allis C D. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena: evidence for functionally distinct H4 acetylation sites. J Biol Chem. 1986;261:1071–1076. [PubMed] [Google Scholar]
- 16.Clarke A S, Lowell J E, Jacobson S J, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 19.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 20.Grant P A, Berger S L. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- 21.Grant P A, Eberharter A, John S, Cook R G, Turner B M, Workman J L. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie C, Fink G R. Guide to yeast genetics and mlecular biology. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 24.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 25.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 26.Imhof A, Wolffe A P. Purification and properties of the xenopus hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry. 1999;38:13085–13093. doi: 10.1021/bi9912490. [DOI] [PubMed] [Google Scholar]
- 27.Johnson C A, Turner B M. Histone deacetylases: complex transducers of nuclear signals. Semin Cell Dev Biol. 1999;10:179–188. doi: 10.1006/scdb.1999.0299. [DOI] [PubMed] [Google Scholar]
- 28.Johnson L M, Kayne P S, Kahn E S, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman P D, Cohen J L, Osley M A. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 32.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 33.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 34.Le S, Davis C, Konopka J B, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Ling X, Harkness T A, Schultz M C, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 36.Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 38.Ma X J, Wu J, Altheim B A, Schultz M C, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mann R K, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Balbas M A, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 42.Monson E K, de Bruin D, Zakian V A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 44.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 45.Park E C, Szostak J W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 47.Qian Y W, Lee E Y. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 48.Qian Y W, Wang Y C, Hollingsworth R E, Jr, Jones D, Ling N, Lee E Y. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 49.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 50.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 51.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz-Carillo A, Wangh L J, Allfry V. Processing of newly synthesized histone molecules. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 53.Ruiz-Garcia A B, Sendra R, Galiana M, Pamblanco M, Perez-Ortin J E, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273:12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 56.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- 58.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith S, Stillman B. Purification and characterization of CAF-1, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 60.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 62.Sun Z W, Hampsey M. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999;152:921–932. doi: 10.1093/genetics/152.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 64.Thompson J S, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 65.Tse C, Georgieva E I, Ruiz-Garcia A B, Sendra R, Hansen J C. Gcn5p, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J Biol Chem. 1998;273:32388–32392. doi: 10.1074/jbc.273.49.32388. [DOI] [PubMed] [Google Scholar]
- 66.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 67.Tyler J K, Adams C R, Chen S R, Kobayashi R, Kamakaka R T, Kadonaga J T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 68.van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1989. [Google Scholar]
- 69.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 70.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 71.Wittschieben B O, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 73.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]