Abstract
A variety of Chlamydia species belonging to the Chlamydiaceae family have been reported in birds. Until recently, C. psittaci was considered to be the most common avian species, although found in both birds and mammals, while C. abortus has only been found in mammals. Recently, a new group of avian C. abortus strains with worldwide distribution in various wild bird families has been described. In this study, whole genome sequencing (WGS) of three of these strains (15-70d24, 15-49d3 and 15-58d44, representing genotypes G1, G2 and 1V, respectively) that were isolated from wild birds were analysed. Genome assemblies based on both short-read Illumina and long-read Nanopore data indicate that these avian C. abortus strains show features characteristic of both C. abortus and C. psittaci species, although phylogenetic analyses demonstrate a closer relationship with classical C. abortus strains. Currently, species classification established by the ICSP Subcommittee on the taxonomy of Chlamydiae, determines that these avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 should be classified as C. abortus. However, the authors of this study conclude that the current taxonomic definition of C. abortus is outdated and should be amended to include two subgroups, mammalian and avian, the latter of which would include all isolates so far referred to as atypical C. psittaci or C. psittaci/C. abortus intermediates.
Keywords: Chlamydia, avian C. abortus, WGS, Illumina, Nanopore
1. Introduction
Birds as well as other animals can be carriers of various pathogens, including the obligate intracellular bacteria of the family Chlamydiaceae. This family currently consists of the two genera: Chlamydia (C.) and recently described Chlamydiifrater [1]. Genus Chlamydia includes 14 recognised species: C. abortus, C. avium, C. buteonis, C. caviae, C. felis, C. gallinacea, C. muridarum, C. pecorum, C. pneumoniae, C. poikilothermis, C. psittaci, C. serpentis, C. suis, and C. trachomatis [2,3,4,5]. Moreover, there are four Candidatus species (Cand.): Cand. C. ibidis, Cand. C. sanzinia, Cand. C. corallus and Cand. C. testudinis [6,7,8,9]. Chlamydiae are widely distributed throughout the world causing a variety of diseases both in humans and animals, including zoonoses [10,11]. C. psittaci, which is encountered mainly in birds, has been considered for a long time as the sole species causing avian chlamydiosis [12,13,14]. However, it has been shown that other chlamydial species can also be harboured by birds. In addition to the classical C. abortus, C. pecorum, C. trachomatis, C. suis or C. muridarum species [15,16,17], new chlamydial species have been detected in birds within recent years: C. avium isolated from pigeons and psittacine birds, C. gallinacea carried by domestic poultry, C. buteonis in raptors and Cand. C. ibidis in the African sacred ibis [5,6,18]. Scientific reports show that the spectrum of chlamydial lineages occurring in birds is wide. Latterly, a new group of avian C. abortus strains with worldwide distribution in various wild bird families (Anatidae, Corvidae, Psittacidae, Rallidae) were described [19,20]. These non-classified chlamydial strains were previously referred as atypical C. psittaci or C. psittaci/C. abortus intermediates as they were demonstrating features of both C. psittaci and C. abortus. Most recently, the genome sequence of one of these strains, C. psittaci strain 84/2334 has been described and the strain reclassified as C. abortus species [21]. Availability of whole genome sequencing (WGS) provides a unique opportunity in understanding the genetic diversity and biology of the novel taxa in the genus Chlamydia, and hence to create a taxonomic definition. Recently, we have announced partial draft genome sequences of three avian Chlamydia abortus strains corresponding to genotypes G1, G2 and 1V (strains 15-70d24, 15-49d3 and 15-58d44, respectively), isolated from wild birds in Poland, based on Illumina sequencing [22,23]. As the genomes were incomplete, we decided to improve sequence data and perform confirmatory resequencing based on short-read Illumina and long-read Nanopore Technology to facilitate contig gap closure and conduct in-depth genome analysis. Here, we describe the results of this hybrid sequence analysis and phenotypic features of these three avian C. abortus strains.
2. Results
2.1. Genomes of Avian C. abortus (Strains 15-70d24, 15-49d3 and 15-58d44, Representing Genotypes G1, G2, 1V, Respectively) Based on Hybrid Sequencing
Sequencing resulted in single circular chromosomal contigs for each of the strains 15-70d24, 15-49d3, 15-58d44, with a length of 1,141,680, 1,132,330 and 1,151,406 bp, and average sequence coverage of 468×, 2437× and 7427×, respectively. G+C content for all three strains at 39.6–39.9% was more in keeping with that of classical C. abortus (39.9%; [21]) than C. psittaci (38.8–39.1%; [21]) strains. Single contig plasmid sequences for 15-70d24, 15-49d3 and 15-58d44 with lengths of 7553, 7556 and 7553 bp and sequence coverage of 1692×, 8017× and 9043×, respectively, were also extracted from the total whole genome raw sequence reads by comparison with other C. psittaci plasmid sequences. Detailed properties of the genomes in comparison to representatives of the family Chlamydiaceae are presented in Supplementary Table S1. The genome sequences of avian C. abortus (strains 15-70d24, 15-49d3 and 15-58d44) chromosomes and plasmids based on hybrid sequencing and Nanopore raw data obtained in this study have been deposited in European Nucleotide Archive (ENA) under the following accession numbers: LS450958.2, LS450956.2, OU508367.1 (chromosomes); LS450959.2, LS450957.2, OU508368.1 (plasmids); ERR6415086, ERR6415087, ERR6415088 (Nanopore), respectively. Accession numbers for Illumina raw data have been published previously [22,23]. All accession numbers related to WGS of avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 (G1, G2 and 1V, respectively) are included as a part of bioproject PRJEB26715.
2.2. Phenotypic Characterisation of Avian C. abortus Strains Corresponding to Genotypes G1, G2 and 1V (Strains 15-70d24, 15-49d3 and 15-58d44)
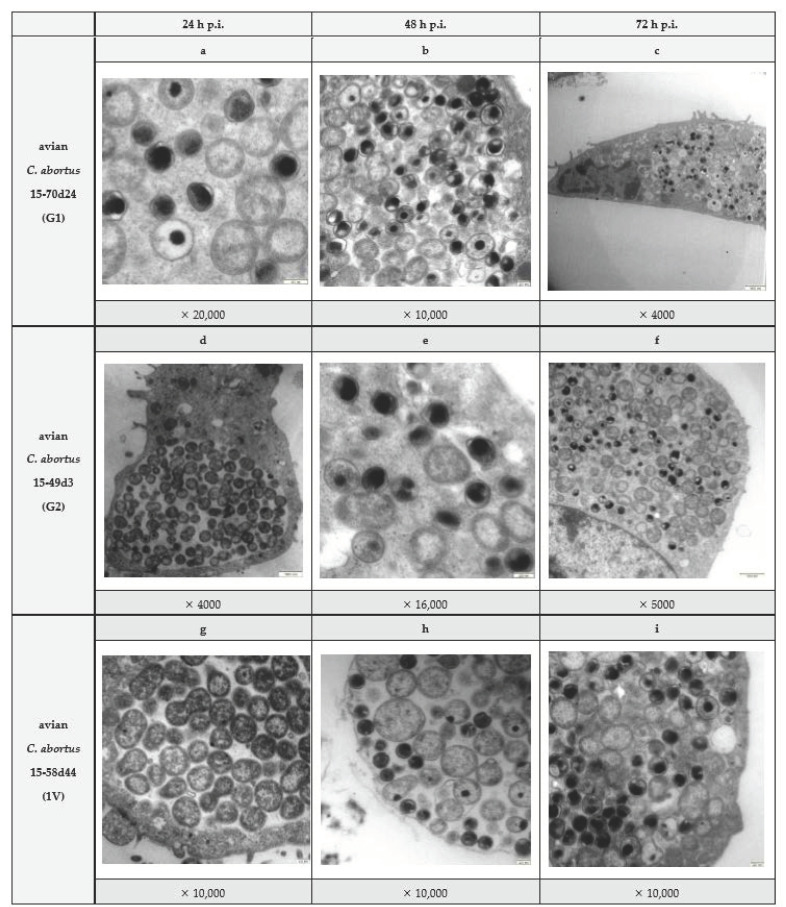
Strains were isolated and propagated according to routine established standard procedures in Buffalo Green Monkey (BGM) cells [19,22,23]. By transmission electron microscopy (TEM), the typical biphasic developmental cycle was observed as seen for all chlamydiae. Figure 1 shows inclusions in infected BGM cells including elementary bodies (EBs), reticulate bodies (RBs), dividing reticulate bodies (dividing RBs) and intermediate bodies (IBs). In all three isolates, EBs are round, electron dense and 240–400 nm in diameter. RBs are larger (400–1100 nm), round to oval and more electron lucent. Binary fission of RBs was observed at different stages of division. Occasionally, IBs with characteristic electron dense centres and a more lucent periphery were found. Immunofluorescence testing of all strains displayed positive reaction with a monoclonal antibody to chlamydial LPS (IMAGEN Chlamydia Kit, Oxoid, Wesel, Germany) (not shown).
Figure 1.
Transmission electron microscopy images of Buffalo Green Monkey (BGM) cell culture infected with avian C. abortus strains. Top row (images (a–c)) presents BGM cell culture infected with avian C. abortus 15-70d24 after 24, 48 and 72 h, respectively. Second row (images (d–f)) shows BGM cell culture infected with avian C. abortus 15-49d3 after 24, 48 and 72 h, respectively. Bottom row (images (g–i)) illustrates BGM cell culture infected with avian C. abortus 15-58d44 after 24, 48 and 72 h, respectively. The magnification is given below images.
2.3. Genome Analysis
2.3.1. 16S rRNA and 23S rRNA Phylogenetic Analyses
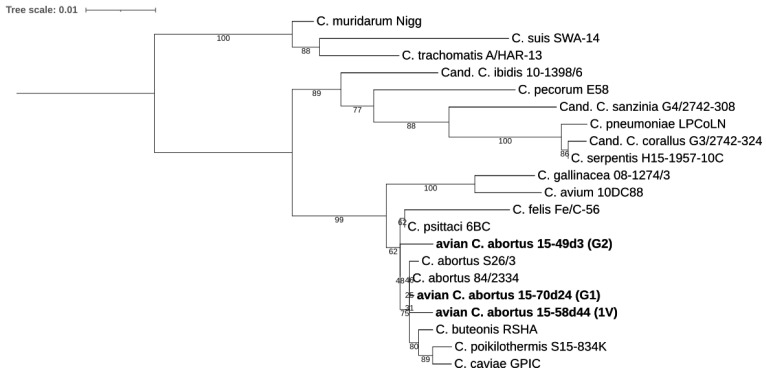
The taxonomic position of the avian C. abortus strains within the genus Chlamydia was evaluated based on pairwise sequence identities, with values greater than 92.5% and 91% for 16S rRNA (Figure 2) and 23S rRNA genes (Figure 3), respectively, identified [24]. Sequence homologies showed their closest relatedness was to C. abortus, C. psittaci and C. buteonis. Pairwise sequence identity values of representative strains for Chlamydiaceae and the three avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 are presented in Supplementary Table S2.
Figure 2.
Analysis of the complete 16S rRNA sequences of the avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 (G1, G2, 1V, respectively) and representative strains of Chlamydia spp. Phylogeny based on 1567 bp consensus alignment was constructed by Maximum Likelihood method with best-fit model according to Bayesian Information Criterion: TVM+F+I+G4. Bootstrap values are presented as percentages.
Figure 3.
Analysis of the complete 23S rRNA sequences of the strains 15-70d24, 15-49d3 and 15-58d44, representing genotypes G1, G2 and 1V, respectively, and members of Chlamydiaceae. Phylogeny based on 2943 bp consensus alignment was constructed by Maximum Likelihood method with best-fit model according to Bayesian Information Criterion: TPM3u+F+I+G4. Bootstrap values are presented as percentages.
2.3.2. Molecular Typing of Avian C. abortus Strains
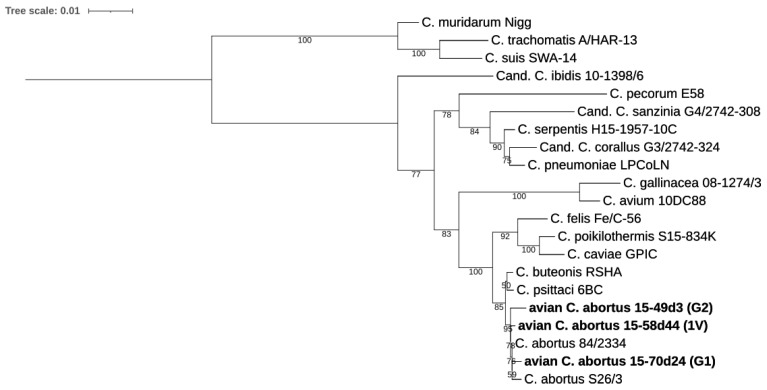
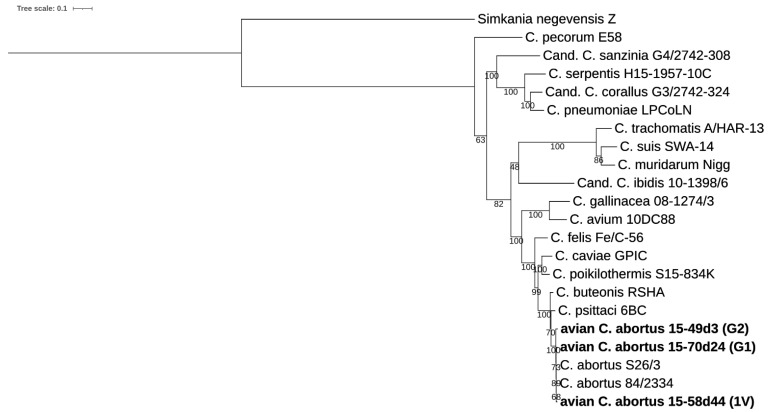
The identity values obtained for the strains 15-70d24, 15-49d3, 15-58d44 representing genotypes G1, G2 and 1V, respectively, are above the accepted cut-off for a new species assignment, so the isolates can be assigned to the Chlamydia genus (DnaA ≥ 70%, Fabl ≥ 78%, Hyp325 ≥ 57% and SucA ≥ 64%) and strains of C. abortus (RpoN > 96%, PepF > 96%, Adk > 95%, FtsK > 98%, HemL > 95%) [24]. Detailed results are shown in Figure 4 and Supplementary Tables S3–S5.
Figure 4.
Analysis of the nine phylogenetically informative markers. The dendrogram was constructed by Maximum Likelihood method with best-fit model according to Bayesian Information Criterion: JTT+F+G4 model based on the concatenated alignment of 9 individual genes (DnaA, SucA, Hyp325, Fabl, RpoN, FtsK, PepF, Adk, HemL), and includes avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 (G1, G2, 1V, respectively), and representative strains of the genus Chlamydia. Bootstrap values are presented as percentages. The tree scale bar indicates the number of amino acid substitutions per site.
2.3.3. ANIb and Tetra-Nucleotide Signatures
Average nucleotide identity by BLAST (ANIb) and tetranucleotide regressions were calculated based on avian C. abortus strains (15-70d24, 15-49d3 and 15-58d44, representing genotypes G1, G2 and 1V, respectively) in comparison to genomic sequences of Chlamydiaceae representatives. The results were presented in Supplementary Table S6. Highest values of 0.99881, 0.99783, 0.9989 (tetranucleotide regression) and 97.93, 97.47, 98.36 (ANIb) were noted between 15-70d24, 15-49d3, 15-58d44, respectively, and UK reference strain C. abortus S26/3.
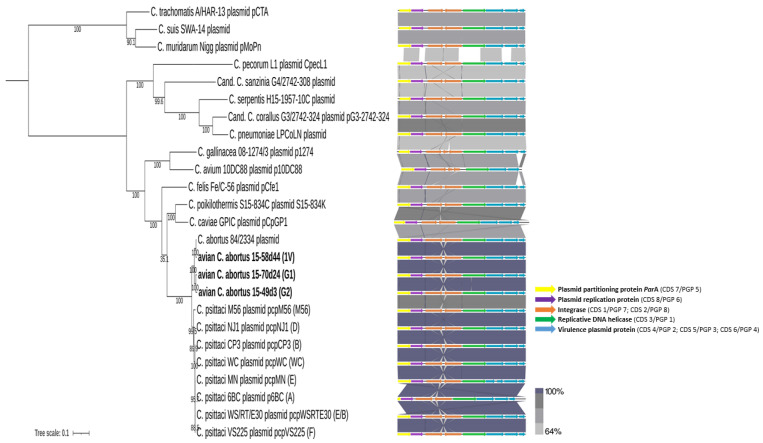
2.3.4. Plasmid Comparisons
Plasmids were detected in all three avian C. abortus genotypes. They are organised into eight open reading frames (ORFs) (Figure 5), as observed for all other chlamydial plasmids. Avian C. abortus 15-70d24, 15-49d3, 15-58d44 and C. abortus 84/2334 show the greatest sequence similarity: 97.63%, 97.33% and 99.54%, respectively. C. psittaci plasmids have significant sequence similarity, ranging from 94.48% to 95.75% nucleotide identity depending on genotype (Figure 5; Supplementary Table S7). C. psittaci 6BC (genotype A) shows the lowest nucleotide identity compared to 15-70d24, 15-49d3, 15-58d44 plasmids (94.61%, 94.48%, 94.68%, respectively). The highest level of nucleotide identity among C. psittaci is observed between avian C. abortus 15-70d24 and C. psittaci CP3 and C. psittaci NJ1, representing genotypes B and D, respectively (95.71%); avian C. abortus 15-49d3 and C. psittaci WS/RT/E30 and C. psittaci VS225 representing genotypes E/B and F, respectively (95.47%); and avian C. abortus 15-58d44 and C. psittaci CP3 being representative of genotype B (95.75%).
Figure 5.
Phylogenetic analysis of avian Chlamydia abortus strains corresponding to genotypes G1, G2 and 1V (strains 15-70d24, 15-49d3 and 15-58d44, respectively) and Chlamydia spp. based on plasmid sequences comparison and chlamydial plasmids arrangement. Support values are presented on the branches. Figure was constructed using EasyFig. Grey shading represents tBLASTx matches, while coloured arrows represent coding regions.
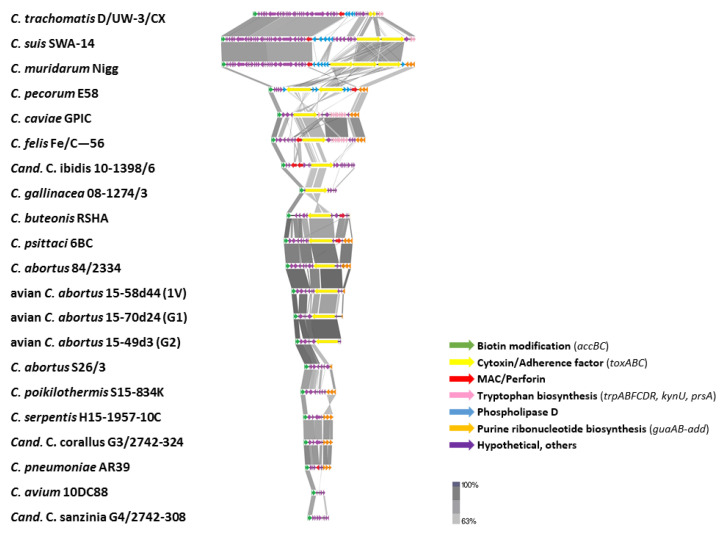
2.3.5. Plasticity Zone
The plasticity zone (PZ) region (19,443-22,657 bp) of avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 (G1, G2, 1V, respectively) is composed of genes required for several biochemical pathways such as acetyl-CoA-carboxylase (accBC) and partial purine and pyrimidine biosynthesis genes (guaAB-add). When compared with other chlamydial species, PZs of avian C. abortus isolates are most related to the C. abortus 84/2334 (51.50–77.67%), C. buteonis RSHA (54.47–70.85%), C. psittaci 6BC (49.12–64.19%), Cand. C. ibidis 10-1398/6 (39.67–43.08%), C. abortus S26/2 (25.73–38.38%) and C. gallinacea 08-1274/3 (25.61–26.24%) (Figure 6, Supplementary Table S8). In contrast to classical C. abortus S26/3 strain, the avian C. abortus 15-70d24, 15-49d3 and 15-58d44 strains contain the cytotoxin virulence factor in their genomes, which is characteristic for C. abortus 84/2334, C. psittaci 6BC and C. buteonis RSHA strains. On the other hand, C. psittaci 6BC and C. buteonis RSHA strains have MAC/perforin genes which are absent in avian Chlamydia abortus genomes and classical C. abortus strains. Additionally, avian and typical C. abortus representatives have a smaller PZ than most of chlamydial species. No tryptophan operon (trpABFCDR, kynU, prsA) and phospholipase D were detected in the plasticity zone of avian C. abortus strains.
Figure 6.
Graphical presentation of plasticity zones of avian Chlamydia abortus strains 15-70d24, 15-49d3, 15-58d44 representing genotypes G1, G2, 1V, respectively, and representatives of Chlamydia spp. and Candidatus to this species via tBLASTx analysis and their arrangement plotted in EasyFig. The coloured arrows represent genes of plasticity zones according to their function, while grey shading represents sequence homology (as listed in the legend).
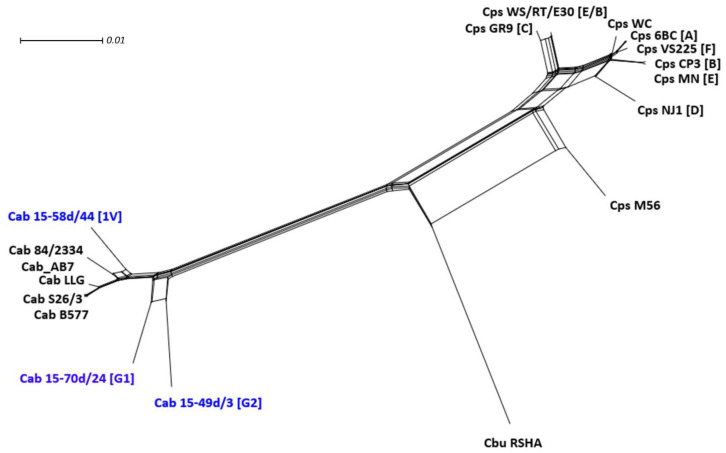
2.3.6. Phylogenetic Network Analysis
The results show that the avian C. abortus G1, G2 and 1V strains are placed together with C. abortus representatives. C. psittaci strains create a separate clade, while C. buteonis is positioned between C. abortus and C. psittaci representatives (Figure 7).
Figure 7.
NeighborNet tree of C. abortus, C. psittaci (genotypes A, B, C, D, E, E/B, F, M56 and WC), C. buteonis and avian C. abortus (genotypes G1, G2 and 1V) representatives. The scale bar indicates the number of substitutions per site.
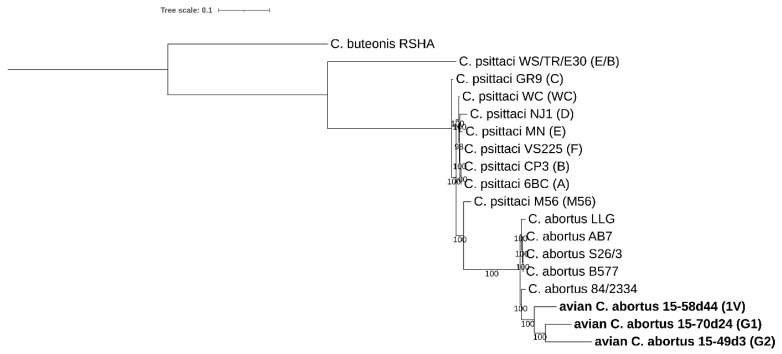
2.3.7. SNP Analysis
Whole-genome single-nucleotide polymorphism (SNP) analysis clearly clustered all strains into three groups: C. buteonis, C. psittaci and C. abortus (Figure 8). Avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 are grouped together with C. abortus strains but create a subclade. The number of SNP differences for 15-70d24, 15-49d3, 15-58d44 within subclade is ranging from 13,071 to 16,891, while within C. abortus clade is ranging from 8872 to 16,730. Interestingly, the number of SNP differences between 15-58d44 and classical C. abortus strains and C. abortus 84/2334 is remarkably lower (8872–9554) than for 15-70d24 (11,957–12,637) or 15-49d3 (16,064–16,730). Significantly greater number of SNP differences is observed between avian C. abortus strains and C. psittaci representatives (21,854–69,925) and C. buteonis (72,206–74,315) (Supplementary Table S9).
Figure 8.
Single-nucleotide polymorphism-based phylogeny of C. abortus, C. psittaci (genotypes A, B, C, D, E, E/B, F, M56 and WC), C. buteonis and avian C. abortus (genotypes G1, G2 and 1V) strains. The dendrogram was constructed by Maximum Likelihood method with best-fit model according to Bayesian Information Criterion: K3P. Bootstrap values are presented as percentages. The tree was rooted with C. buteonis as an outgroup. The scale bar indicates the number of substitutions per site.
3. Discussion
Molecular analysis of avian C. abortus performed in our previous study [19] did not lead to final determination of their taxonomic position because the isolates demonstrated intermediate features of both C. abortus and C. psittaci. Previous research has suggested that these three strains should be classified as avian C. abortus strains based on preliminary analysis. Although, MLST analysis indicated they should be classified as C. abortus, it was not clear if they should be considered as a part of the established classical C. abortus species or be designated a completely new species. Therefore, complete information based on WGS analysis was required. In this study, a more detailed molecular analysis of the genome sequences was performed to fully characterize these strains. In the last decade, whole-genome sequence analysis has enabled identification and characterization of new taxa: C. avium, C. gallinacea [18,25], C. buteonis [5], C. poikilothermis [3], C. serpentis [3], Cand. C. ibidis [6], Cand. C. sanzinia [7], Cand. C. corallus [8] and Cand. C. testudinis [9]. Genome sequencing based on short-read technologies is relatively inexpensive and widely accessible, but due to repetitive regions and/or high level of %G+C content it can be challenging to close the genome. Long-read sequencing however can span problematic repetitive regions thus improving assembly success. On the other hand, long-read sequencing has several drawbacks including higher error rates affecting both insertions and deletions [26] but it should be noted that error rates have significantly come down to nearer 5% using the latest Nanopore sequencing technologies. In this study, a hybrid sequencing approach, combining the high-throughput (Oxford Nanopore Technologies) and high-accuracy short read data (Illumina), was utilized to produce single contiguous genome sequences of avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 corresponding to genotypes G1, G2 and 1V, respectively [22,23]. This approach provides reliable contigs and scaffolds by firstly assembling short-read data to produce an assembly graph and then using long-reads to create bridges between contigs and scaffolds. High-quality hybrid assembly can help resolve difficulties associated with repeat regions and with the identification of SNPs. Therefore, hybrid sequencing seems to be the gold-standard in genomic analysis [27].
Analysis of the three strains, 15-70d24, 15-49d3 and 15-58d44, originating from Anas crecca, Anas platyrhynchos and Pica pica, respectively, revealed that they belong to the order Chlamydiales (identity values of ≥80% for 16S rRNA and 23S rRNA) and family Chlamydiaceae (identity values of ≥92.5% for 16S rRNA, ≥91% for 23S rRNA) based on the classification scheme proposed by Pillonel et al [24]. According to the comparison of nine informative marker proteins with 15 members of the Chlamydia genus and three Candidatus species, the new isolates could be assigned to species C. abortus (RpoN > 96%, PepF > 96%, Adk > 95%, FtsK > 98%, HemL > 95%) of the Chlamydia genus (DnaA ≥ 70%, Fabl ≥ 78%, Hyp325 ≥ 57% and SucA ≥ 64%) [24]. G+C content of G1, G2 and 1V (39.9%, 39.6%, 39.8%) was mostly similar to C. abortus 84/2334 (39.9%) and classical C. abortus S26/3 strain (39.9%). The ANIb and tetra nucleotide regression values calculated for 15-70d24, 15-49d3 and 15-58d44 also indicated they should be classified as C. abortus species.
Interestingly, comparative genome analysis revealed that the G1, G2 and 1V strains significantly differ from classical C. abortus strains. Comparative genomics of Chlamydia spp. is mainly focused on the plasticity zone (also known as the replication termination region–RTR) as a key region of chlamydial genomes associated with pathogenesis [28,29,30]. The size of plasticity zones in chlamydial genomes ranges usually from 6 kb to 83 kb [3,5,6,7,8,9,28,29,30] dependent on species and is bounded by acetyl-CoA carboxylase (accB) and einosine-5′-monophosphate dehydrogenase (guaB) [28,29]. The lack or truncation of the guaAB-add operon is common for some species including C. trachomatis, C. suis, C. gallinacea, C. avium, C. buteonis and avian C. abortus. This highly variable region contains genes encoding proteins responsible for regulation of fatty acid synthesis and degradation (accBC), purine biosynthesis (guaAB-add) and tryptophan biosynthesis (trpABFCDR, kynU, prsA). Moreover, it comprises several virulence factors including cytotoxin genes/adherence factor, membrane attack complex/perforin (MAC/Perforin) and phospholipase D (PLD) enzymes as well as a number of hypothetical proteins of unknown function [28,29]. Avian C. abortus strains have a more extensive PZ when compared to the classical C. abortus S26/3 representative. Considering the size, PZs of avian C. abortus 15-70d24 (20,909 bp), 15-49d3 (19,443 bp) and 15-58d44 (22,657 bp) are most similar to C. abortus 84/2334 (27,678 bp), C. psittaci 6BC (29,144 bp) and C. buteonis RSHA (26,735 bp), while PZ of C. abortus S26/3 is about half the size (11,776 bp). Analysis of nucleotide identity of avian C. abortus strains showed their structure is most similar to: C. abortus 84/2334 (51.50–77.67%), C. buteonis RSHA (54.47–70.85%) and C. psittaci 6BC (49.12–64.19%). Moreover, the avian C. abortus strains in this study carry the cytotoxin gene which is absent in classical C. abortus S26/3 but present in C. abortus 84/2334, which is also of avian origin [21]. However, in contrast to C. psittaci 6BC and C. buteonis RSHA, the MAC/Perforin gene is absent from the avian C. abortus PZ, as previously observed for C. abortus S26/3 and C. abortus 84/2334 [21,31]. It has been suggested that MAC/Perforin might be involved in assisting PLD proteins in lipid acquisition and processing [29] but no PLDs were detected in avian C. abortus strains neither were they detected in C. abortus S26/3, C. psittaci 6BC or C. buteonis RSHA, or observed in the recently published avian C. abortus 84/2334 strain [21]. The complete trp operon is absent in avian C. abortus which is a characteristic feature of the majority of Chlamydia spp. excluding C. caviae and C. felis. It probably suggests a different pathway for synthesising tryptophan.
Most chlamydial species, but not all strains, carry an endogenous 7.5 kbp circular plasmid which has been identified in C. avium [18], C. caviae [32], C. felis [33], C. gallinacea [18], C. muridarum [34], C. pecorum [35], C. pneumoniae [36], C. poikilothermis [3], C. psittaci [37,38], C. serpentis [3], C. suis [39], C. trachomatis [2], C. buteonis [5], Cand. C. sanzinia [7], Cand. C. corallus [8] and Cand. C. testudinis [9]. It is assumed that the presence of plasmid is associated with the virulence of the strain. Plasmids are highly conserved and not integrated into the genome [36,40,41]. They usually contain eight coding sequences (CDS) encoding for genes involved in plasmid maintenance and glycogen synthesis [42,43] and non-coding RNA with unknown functionality [41]. Microbial plasmids can carry antibiotic resistance genes, but there is no proof of antibiotic resistance located on chlamydial plasmids [44]. Interestingly, all of the avian C. abortus strains reported here possess plasmids, similar to that found in the avian C. abortus strain 84/2334, while to date no plasmids have been identified in classical C. abortus strains [21]. The size of the avian C. abortus plasmids ranging from 7553 to 7556 bp and their sequences are most closely related to C. abortus 84/2334 (97.33–99.54%) and C. psittaci strains ranging from 94.48% to 95.75%. It should be highlighted that avian C. abortus (strains 15-70d24, 15-49d3 and 15-58d44) plasmids share the most similar structure with the plasmids of C. abortus 84/2334, isolated from a parrot, and genotypes B, D, E/B and F of C. psittaci, which were isolated from pigeon, turkey, mallard and parakeet, respectively, whereas C. psittaci genotype A strain associated with parrot shares the lowest nucleotide identity with plasmids of avian C. abortus. Plasmid proteins are commonly used for diagnostic targets and vaccine candidates, therefore future research on avian C. abortus plasmids might lead to the development of new vaccines against avian C. abortus and/or C. psittaci strains [45].
Whole-genome SNP and NeighborNet analyses clearly show that the avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 are phylogenetically positioned as C. abortus species confirming that they should be classified as C. abortus. Interestingly, the number of SNP differences indicate that the strain 15-58d44 is phylogenetically more closely related to classical C. abortus strains and C. abortus 84/2334 (8872–9554) than to the 15-70d24 (13,071) and 15-49d3 (16,891) strains.
Avian C. abortus strains can be found in birds, mainly in waterfowl (genotype G1 and G2) and in corvids (genotype 1V). The microorganism can be isolated from cloaca and oropharynx. The route of transmission has not yet been determined but by analogy with other Chlamydiae, infection may take place through airborne aerosols containing faecal and dust particles or direct contact with fresh faeces. Zoonotic potential of these strains remains unknown but may become clearer in time as more strains are identified in human populations. Due to the fact that more than 50% of avian C. abortus DNA extracts are non-typable by the available PCR assays [19], our team is currently working on development of a specific real-time PCR assay.
Taking into consideration the above, the plasmid and chromosomal genome characteristics and WGS comparison indicated that avian C. abortus show features of both C. abortus and C. psittaci species, in agreement with analyses recently published on avian C. abortus strain 84/2334 [21]. However, based on species classification rules established by the International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of Chlamydiae, they should be classified as C. abortus. Furthermore, as previously suggested, current definition of C. abortus is outdated and should be expanded to include not only the classical mammalian isolates, but also strains isolated from birds 15-70d24, 15-49d3, 15-58d44 (G1, G2, 1V, respectively), Prk/Daruma and 84/2334 [4,21,46,47,48].
4. Materials and Methods
4.1. Bacterial Strains
During the previously published scientific work [19,22,23], cloacal/faecal swabs were taken from birds transiently living in bird rehabilitation centres and free-living birds caught randomly by authorised veterinarians or ornithologists during clinical studies. According to the Local Ethical Committee on Animal Testing at the University of Life Sciences in Lublin (Poland), formal ethical approval is not required for this kind of study [19]. The individual health status of a bird was not recorded. Avian C. abortus strains representing genotype G1 (15-70d24) from Eurasian teal (Anas crecca), genotype G2 (15-49d3) from mallard (Anas platyrhynchos) and 1V (15-58d44) from magpie (Pica pica) were successfully isolated in BGM cell culture and propagated in 25 cm2 tissue culture flasks for 72 h based on procedures published previously [19,22,23]. To confirm proper multiplication of the strains, a Chlamydiaceae-specific real-time PCR targeting the 23S rRNA gene fragment [49] and immunofluorescence staining using IMAGEN Chlamydia kit (Oxoid, Wesel, Germany), according to manufacturer’s instructions, were performed. Deep molecular analysis of ompA, 16S rRNA, IGS, partial 23S rRNA sequences and MLST as well as announcements of the draft genomes were described in our previous studies [19,22,23]. Basic data of the cultured isolates presented in this work are specified in Supplementary Table S10. Strains used in this study and their GenBank accession numbers are listed in Supplementary Table S11.
4.2. Electron Microscopy
BGM cells were infected with avian C. abortus strains corresponding to genotypes G1, G2 and 1V (strains 15-70d24, 15-49d3 and 15-58d44, respectively). At 24, 48 and 72 h post-infection, infected cells plus medium were harvested by scraping from the flasks and transferred to separate micro-centrifuge tubes before adding 4% glutaraldehyde solution to each tube at a 1:1 ratio. Tubes were centrifuged at 1000× g for 10 min and pellets were used for preparing microscope slides. Chlamydia-infected cells were fixed in 2% paraformaldehyde/2.5% glutaraldehyde (Roth, Karlsruhe, Germany) in 100 mM phosphate buffer, pH 7.2, for 1 h at room temperature. Cells were washed in phosphate buffer and post-fixed in 1% osmium tetroxide (Roth, Karlsruhe, Germany) for 1 h. After several rinses in distilled water, samples were dehydrated in a graded series of ethanol and embedded in Epon 812 resin (Sigma Aldrich, Saint Louis, MO, USA). Sections of 70–80 nm were cut, stained with uranyl acetate and lead citrate, put on grids and viewed on a Zeiss Libra 120 transmission electron microscope (Carl Zeiss, Oberkochen, Germany). Evaluation of phenotypic features was performed including diameter of elementary/reticulate bodies and confirmation of biphasic developmental cycle.
4.3. Genomic DNA Preparation
4.3.1. Illumina Sequencing
Genomic DNA was extracted after several passages from BGM cell culture infected with avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 by DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Removal of methylated host DNA from obtained genomic DNA samples was performed according to the manufacturer’s protocol of NebNext Microbiome DNA Enrichment Kit (BioLabs, Ipswich, UK) [22,23].
4.3.2. Nanopore (MinION) Sequencing
Infected cells from ten 225 cm2 infected tissue culture flasks were harvested using sterile glass beads and centrifuged at 153× g for 10 min at 4 °C to remove gross cellular debris. Supernatants were then centrifuged at 22,100× g for 30 min at 4 °C and pellets were washed with ice-cold PBS and centrifuged as previously. Each pellet was resuspended in 20 mM Tris-HCl, pH 7.5/150 mM KCl/1% sarkosyl, lightly homogenised using a ground glass homogenizer and carefully layered onto 3 mL cushions of 15% sucrose in 20 mM Tris-HCl, pH 7.5/150 mM KCl/1% sarkosyl before centrifuging at 70,000× g for 45 min at 4 °C. Genomic DNA was extracted from pellets using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) as per manufacturer’s instructions.
4.3.3. Quality Assessment and Quantification
To confirm the quality and quantity of extracted DNA, spectrophotometric and fluorometric methods were used. DNA extracts were checked qualitatively and quantitatively by means of Nanodrop spectrophotometer (DeNovix, Wilmington, NC, USA) and Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
4.4. Genome Sequencing, Assembly and Draft Annotation
Illumina and Nanopore Sequencing
Genomic DNA libraries were prepared using the Nextera XT DNA library preparation kit and Nextera XT index kit (Illumina, San Diego, CA, USA). Sequencing was carried out on MiSeq sequencer (Illumina, San Diego, CA, USA) with the 2 × 300-bp paired-end protocol [22,23]. Long-read genomic DNA libraries were prepared using a rapid sequencing kit (SQK-RAD004; Oxford Nanopore Technologies [ONT], Oxford, UK). Sequencing was conducted in a MinION using a FLO-MIN106D (R9) flow cell according to a standard protocol (ONT, Oxford, UK) and basecalled with Guppy v.3.0.7. Illumina raw sequencing data were processed using fastp v.0.20.1 [50] for the purpose of trimming adapters and low-quality data. Long reads were adapter trimmed with Porechop v.0.2.4. BBDuk v.38.34 [51] was applied to remove non-chlamydial reads pertaining to the host DNA (from BGM cells) based on standard operating procedures. De novo genome assembly based on both the Illumina and Nanopore (MinION) data was performed using the hybrid assembly mode of Unicycler version 0.4.1 [52]. Each genome was assembled into a single chromosome and plasmid contig. The genome sequences were annotated using the Rapid Annotations via Prokka—Prokaryotic genome annotation (Galaxy Version 1.14.6) [53,54,55]. Default parameters were used for all software unless otherwise specified.
4.5. Genome Analysis
4.5.1. 16S rRNA and 23S rRNA Phylogenetic Analyses
The 16S and 23S rRNA gene sequences were extracted from avian C. abortus 15-70d24, 15-49d3, 15-58d44 genomes and representatives of family Chlamydiaceae, then aligned using MAFFTv7.013 by Geneious Pro 8.0 software (Biomatters, Auckland, New Zealand). Dendrograms were constructed by IQ-TREE v1.6.12 [56], with the best-fit model according to Bayesian Information Criterion (BIC) model and 1000 bootstrap replicates. The models were calculated by Model Finder of IQ-TREE v1.6.12 [57,58,59] and MEGA X [60,61]. The phylogeny was visualized by ITOL [62].
4.5.2. Molecular Typing of Avian C. abortus Strains
The genetic relationship of avian C. abortus strains described in this study and other chlamydial species was assessed using the classification system published by Pillonel et al. [24] and recommended by the International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of Chlamydiae [4,46]. Nine phylogenetically informative markers were identified and extracted from 15 reference strains of Chlamydia spp., three Candidatus Chlamydia spp. (available in NCBI database) and one Simaniaceae representative. Amino acid sequences were aligned with MAFFTv7.013 by Geneious Pro 8.0 software (Biomatters, Auckland, New Zealand). Alignments were concatenated to build a reference phylogeny using IQ-TREE v1.6.12 [56], with the best-fit model according to BIC and 1000 bootstrap replicates. The model was calculated by Model Finder of IQ-TREE v1.6.12 [57,58,59] and MEGA X [60,61]. The phylogeny was visualized by ITOL [62]. Pairwise amino acid sequence identities were determined based on the MAFFT alignment by Geneious Pro 8.0 software (Biomatters, Auckland, New Zealand).
4.5.3. ANIb and Tetra-Nucleotide Signatures
The Average Nucleotide Identity (ANIb) and correlation indexes of their Tetra-nucleotide signatures were calculated based on the program JSpecies v3.7.2 with default parameters for avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 and selected chlamydial representatives [63].
4.5.4. Plasmid Comparisons
Phylogenetic analysis based on plasmid sequences was performed on the 15-70d24, 15-49d3 and 15-58d44 strains and other Chlamydiaceae members (available in NCBI database), including C. abortus 84/2334 and C. psittaci genotypes A, B, D, E, E/B, F, M56 and WC. Nucleotide sequences were aligned using MAFFT in Geneious Pro 8.0 software (Biomatters, Auckland, New Zealand) and a phylogenetic tree was constructed using IQ-TREE v1.6.12 [56], with the best-fit model according to BIC and 1000 bootstrap replicates. The model was calculated by Model Finder of IQ-TREE v1.6.12 [57,58,59] and MEGA X [60,61]. The generated tree was visualized using ITOL [62]. Chlamydial plasmids arrangement was constructed using EasyFig [64].
4.5.5. Plasticity Zone
In order to further characterise the genomes of avian Chlamydia abortus, the plasticity zone regions were analysed in comparison to other chlamydial species. PZs were extracted from Chlamydia spp. reference strains and avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 (G1, G2, 1V, respectively) by Geneious Pro 8.0 software (Biomatters, Auckland, New Zealand). The visualisation of the BLAST comparisons were generated using Easyfig [64].
4.5.6. Phylogenetic Network Analysis
Whole-genome sequences of C. abortus, C. psittaci (genotypes A, B, C, D, E, E/B, F, M56 and WC), C. buteonis and avian C. abortus G1, G2, 1V representative strains were aligned using MAFFT. The generated alignment in fasta format was imported into SplitsTree (v4.16.2) to generate a NeighborNet tree [65].
4.5.7. SNP Analysis
Analysis included representatives of C. abortus, C. psittaci (genotypes A, B, C, D, E, E/B, F, M56 and WC), C. buteonis and avian C. abortus strains 15-70d24, 15-49d3, 15-58d44 (G1, G2, 1V, respectively). A representative batch of sequenced reads for each genome were taken from the Sequence Read Archive. All available next generation sequencing reads were quality checked and aligned against the reference genome (C. abortus 84/2334) using Burrows-Wheeler Aligner (BWA-MEM) (v. 0.7.17) [66]. Variant calling was performed using FreeBayes (v. 1.3.2) [67], while called variants were filtered by VcfFilter, part of the VCFlib (v. 1.0.2) [68]. SNP detection on downloaded assemblies were conducted using NUCmer (v.3.1) and Show-SNPs, in the MUMmer package [69]. Bcftools (v. 1.9) [70] was applied to combined the vcf files containing SNPs for each strain together. VCF-kit (v.0.2.9) was utilized to generate a single file in fasta format to be used for the construction of a phylogenetic tree using IQ-TREE v1.6.12 [56], with the best-fit model according to BIC and 1000 bootstrap replicates. The model was calculated by Model Finder of IQ-TREE v1.6.12 [57,58,59] and MEGA X [60,61]. The generated tree was visualized using ITOL [62]. As C. abortus 84/2334 is closely related to the sequenced samples it was used as a reference genome.
Acknowledgments
The authors would like to thank Arkadiusz Bomba and co-workers (Department of Omics Analyses, NVRI, Puławy, Poland) for excellent technical assistance and unparalleled support. Małgorzata Mazur (Department of Hygiene of Animal Feedingstuffs, NVRI, Puławy, Poland) is acknowledged for valuable advices and support. Teresa Karpińska and Magdalena Wasiak (Department of Pathology, NVRI, Puławy, Poland) are acknowledged for assistance in preparing TEM images. The authors are grateful to Christiane Schnee and Sabine Scharf (FLI, Jena, Germany) for help with isolation of the strains.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10111405/s1, Table S1. Basic genomic parameters of avian C. abortus strains and selected representatives of Chlamydia spp. Table S2. Lower triangle represents pairwise sequence identity values for 16S rRNA, while upper triangle shows pairwise sequence identity values for 23S rRNA genes extracted from selected representatives of Chlamydia spp., Candidatus and avian C. abortus strains. Tables S3–5. Nine taxonomically conserved protein sequences extracted from the avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 (G1, G2 and 1V, respectively) compared to strains belonging to Chlamydia spp., Candidatus to Chlamydia species and Simkania negevensis Z. Red cells present identity values higher than the defined threshold values, whereas green cells show identity values below the threshold values. Table S6. Pairwise comparison of selected representatives of Chlamydia spp. and avian C. abortus strains. Lower triangle represents pairwise sequence identity values for tetranucleotide signature correlation index, while upper triangle shows pairwise sequence identity values for average nucleotide identity (ANIb). Table S7. Nucleotide identity values (%) of plasmid sequences of selected Chlamydia spp. Distance matrices based on multiple sequence alignment were used for calculation of nucleotide sequence identity values (Geneious Pro 8.0 software; Biomatters, Auckland, New Zealand). Table S8. Nucleotide identity values (%) of plasticity zone sequences of selected representative strains of Chlamydia spp. Distance matrices based on multiple sequence alignment were used for calculation of nucleotide sequence identity values (Geneious Pro 8.0 software; Biomatters, Auckland, New Zealand). Table S9. The number of SNP differences of selected representatives of Chlamydia spp. Table S10. Characterization of avian C. abortus strains. Table S11. Strains used in this study and their GenBank accession numbers.
Author Contributions
Conceptualization, M.S.-C. and K.N.; Data curation, K.Z.-M.; Formal analysis, K.Z.-M., M.S.-C., M.L. and D.L.; Funding acquisition, K.N. and D.L.; Investigation, K.Z.-M., M.L. and D.L.; Methodology, K.Z.-M.; Project administration, M.S.-C.; Resources, M.S.-C. and K.N.; Supervision, M.S.-C. and K.N.; Visualization, K.Z.-M. and D.L.; Writing—original draft, K.Z.-M. and M.S.-C.; Writing—review and editing, K.Z.-M., M.S.-C., M.L., D.L. and K.N. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by KNOW (Leading National Research Centre) Scientific Consortium “Healthy Animal–Safe Food”, decision of Ministry of Science and Higher Education number 05-1/KNOW2/2015. DL and ML are funded by the Scottish Government Rural and Environment Science and Analytical Services (RESAS) division.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequences of strains: 15-70d24, 15-49d3 and 15-58d44 obtained in this study have been deposited at ENA/GenBank/DDBJ under the accession numbers: LS450958.2, LS450956.2, OU508367.1 (chromosomes); LS450959.2, LS450957.2, OU508368.1 (plasmids), respectively. The Nanopore raw reads have been deposited under accession numbers: ERR6415086, ERR6415087, ERR6415088, respectively. All data related to WGS of avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 are included as a part of BioProject PRJEB26715.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vorimore F., Hölzer M., Liebler-Tenorio E.M., Barf L.M., Delannoy S., Vittecoq M., Wedlarski R., Lécu A., Scharf S., Blanchard Y., et al. Evidence for the existence of a new genus Chlamydiifrater gen. nov. inside the family Chlamydiaceae with two new species isolated from flamingo (Phoenicopterus roseus): Chlamydiifrater phoenicopteri sp. nov. and Chlamydiifrater volucris sp. nov. Syst. Appl. Microbiol. 2021;44:126200. doi: 10.1016/j.syapm.2021.126200. [DOI] [PubMed] [Google Scholar]
- 2.Sachse K., Bavoil P.M., Kaltenboeck B., Stephens R.S., Kuo C.C., Rosselló-Móra R., Horn M. Emendation of the family Chlamydiaceae: Proposal of a single genus, Chlamydia, to include all currently recognized species. Syst. Appl. Microbiol. 2015;38:99–103. doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Staub E., Marti H., Biondi R., Levi A., Donati M., Leonard C.A., Ley S.D., Pillonel T., Greub G., Seth-Smith H.M.B., et al. Novel Chlamydia species isolated from snakes are temperature-sensitive and exhibit decreased susceptibility to azithromycin. Sci. Rep. 2018;8:5660. doi: 10.1038/s41598-018-23897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borel N., Greub G. International committee on systematics of prokaryotes (ICSP) subcommittee on the taxonomy of chlamydiae. Minutes of the closed meeting, 5 July 2018 Woudschoten, Zeist, The Netherlands. Int. J. Syst. Evol. Microbiol. 2019 July 5;69:2606–2608. doi: 10.1099/ijsem.0.003418. [DOI] [PubMed] [Google Scholar]
- 5.Laroucau K., Vorimore F., Aaziz R., Solmonson L., Hsia R.C., Bavoil P.M., Fach P., Hölzer M., Wuenschmann A., Sachse K. Chlamydia buteonis, a new Chlamydia species isolated from a red-shouldered hawk. Syst. Appl. Microbiol. 2019;42:125997. doi: 10.1016/j.syapm.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Vorimore F., Hsia R.C., Huot-Creasy H., Bastian S., Deruyter L., Passet A., Sachse K., Bavoil P.M., Myers G., Laroucau K. Isolation of a New Chlamydia species from the Feral Sacred Ibis Threskiornis aethiopicus Chlamydia ibidis. PLoS ONE. 2013;8:e74823. doi: 10.1371/journal.pone.0074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor-Brown A., Bachmann N.L., Borel N., Polkinghorne A. Culture-independent genomic characterisation of Candidatus Chlamydia sanzinia, a novel uncultivated bacterium infecting snakes. BMC Genom. 2016;17:10. doi: 10.1186/s12864-016-3055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor-Brown A., Spang L., Borel N., Polkinghorne A. Culture-independent metagenomics supports discovery of uncultivable bacteria within the genus Chlamydia. Sci. Rep. 2017;7:10661. doi: 10.1038/s41598-017-10757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laroucau K., Ortega N., Vorimore F., Aaziz R., Mitura A., Szymańska-Czerwińska M., Cicerol M., Salinas J., Sachse K., Caro M.R. Detection of a novel Chlamydia species in captive spur-thighed tortoises Testudo graeca in southeastern Spain and proposal of Candidatus Chlamydia testudinis. Syst. Appl. Microbiol. 2020;43:126071. doi: 10.1016/j.syapm.2020.126071. [DOI] [PubMed] [Google Scholar]
- 10.Borel N., Polkinghorne A., Pospischil A. A Review on Chlamydial Diseases in Animals: Still a Challenge for Pathologists? Vet. Pathol. 2018;55:374–390. doi: 10.1177/0300985817751218. [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Liu P., Cao X., Lou Z., Zaręba-Marchewka K., Szymańska-Czerwińska M., Niemczuk K., Hu B., Bai X., Zhou J. First report of Chlamydia abortus in farmed fur animals. Biomed Res. Int. 2018;2018:4289648. doi: 10.1155/2018/4289648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knittler M.R., Sachse K. Chlamydia psittaci: Update on an underestimated zoonotic agent. Pathog. Dis. 2015;73:1–15. doi: 10.1093/femspd/ftu007. [DOI] [PubMed] [Google Scholar]
- 13.Sachse K., Laroucau K., Vanrompay D. Avian Chlamydiosis. Curr. Clin. Microbiol. Rep. 2015;2:10–21. doi: 10.1007/s40588-014-0010-y. [DOI] [Google Scholar]
- 14.Szymańska-Czerwińska M., Niemczuk K. Avian Chlamydiosis Zoonotic Disease. Vector-Borne Zoonotic Dis. 2016;16:1–3. doi: 10.1089/vbz.2015.1839. [DOI] [PubMed] [Google Scholar]
- 15.Pantchev A., Sting R., Bauerfeind R., Tyczka J., Sachse K. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet. J. 2009;181:145–150. doi: 10.1016/j.tvjl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 16.Guo W., Li J., Kaltenboeck B., Gong J., Fan W., Wang C. Chlamydia gallinacea not C. psittaci is the endemic chlamydial species in chicken Gallus gallus. Sci. Rep. 2016;6:19638. doi: 10.1038/srep19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szymańska-Czerwińska M., Mitura A., Zaręba K., Schnee C., Koncicki A., Niemczuk K. Poultry in Poland as Chlamydiaceae carrier. J. Vet. Res. 2017;61:411–419. doi: 10.1515/jvetres-2017-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachse K., Laroucau K., Riege K., Wehner S., Dilcher M., Huot C.H., Weidmann M., Myers G., Vorimore F., Vicari N., et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst. Appl. Microbiol. 2014;37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Szymańska-Czerwińska M., Mitura A., Niemczuk K., Zaręba K., Jodełko A., Pluta A., Scharf S., Vitek B., Aaziz R., Vorimore F., et al. Dissemination and genetic diversity of chlamydial agents in Polish wildfowl: Isolation and molecular characterisation of avian Chlamydia abortus strains. PLoS ONE. 2017;12:e0174599. doi: 10.1371/journal.pone.0174599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaręba-Marchewka K., Szymańska-Czerwińska M., Niemczuk K. Chlamydiae-What’s New? J. Vet. Res. 2020;64:461–467. doi: 10.2478/jvetres-2020-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longbottom D., Livingstone M., Ribeca P., Beeckman D.S.A., van der Ende A., Pannekoek Y., Vanrompay D. Whole genome de novo sequencing and comparative genomic analyses suggests that Chlamydia psittaci strain 84/2334 should be reclassified as Chlamydia abortus species. BMC Genom. 2021;22:159. doi: 10.1186/s12864-021-07477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaręba-Marchewka K., Szymańska-Czerwińska M., Mitura A., Niemczuk K. Draft genome sequence of avian Chlamydia abortus genotype G1 strain 15-70d24, isolated from Eurasian teal in Poland. Microbiol. Resour. Announc. 2019;8:e00658-19. doi: 10.1128/MRA.00658-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaręba-Marchewka K., Szymańska-Czerwińska M., Niemczuk K. Draft Genome Sequences of Avian Chlamydia abortus Genotype G2 Strain 15-49d3, Isolated from Mallard, and Genotype 1V Strain 15-58d44, Isolated from Magpie in Poland. Microbiol. Resour. Announc. 2021;4:e01203-19. doi: 10.1128/MRA.01203-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillonel T., Bertelli C., Salamin N., Greub G. Taxogenomics of the order Chlamydiales. Int. J. Syst. Evol. Microbiol. 2015;65:1381–1393. doi: 10.1099/ijs.0.000090. [DOI] [PubMed] [Google Scholar]
- 25.Holzer M., Laroucau K., Creasy H.H., Ott S., Vorimore F., Bavoil P.M., Sachse. K. Whole-genome sequence of Chlamydia gallinacea type strain 08-1274/3. Genome Announc. 2016;4:e00708-16. doi: 10.1128/genomeA.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rang F.J., Kloosterman W.P., De Ridder J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein S., Beka L., Graf J., Klassen J.L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genom. 2019;20:23. doi: 10.1186/s12864-018-5381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachmann N.L., Polkinghorne A., Timms P. Chlamydia genomics: Providing novel insights into chlamydial biology. Trends Microbiol. 2014;22:464–472. doi: 10.1016/j.tim.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 29.Nunes A., Gomes J.P. Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infect. Genet. Evol. 2014;23:49–64. doi: 10.1016/j.meegid.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 30.Read T.D., Brunham R.C., Shen C., Gill S.R., Heidelberg J.F., White O., Hickey E.K., Peterson J., Utterback T., Berry K., et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson N.R., Yeats C., Bell K., Holden M.T., Bentley S.D., Livingstone M., Cerdeño-Tárraga A.M., Harris B., Doggett J., Ormond D., et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15:629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Read T.D., Myers G.S., Brunham R.C., Nelson W.C., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): Examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Lent S., Piet J.R., Beeckman D., van der Ende A., Van Nieuwerburgh F., Bavoil P., Myers G., Vanrompay D., Pannekoek Y. Full genome sequences of all nine Chlamydia psittaci genotype reference strains. J. Bacteriol. 2012;194:6930–6931. doi: 10.1128/JB.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Chen C., Gong S., Hou S., Qi M., Liu Q., Baseman J., Zhong G. Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J. Bacteriol. 2014;196:989–998. doi: 10.1128/JB.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelocnik M., Bachmann N.L., Kaltenboeck B., Waugh C., Woolford L., Speight K.N., Gillett A., Higgins D.P., Flanagan C., Myers G.S., et al. Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genom. 2015;16:893. doi: 10.1186/s12864-015-2053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas N.S., Lusher M., Storey C.C., Clarke I.N. Plasmid diversity in Chlamydia. Microbiol.-Sgm. 1997;143:1847–1854. doi: 10.1099/00221287-143-6-1847. [DOI] [PubMed] [Google Scholar]
- 37.Grinblat-Huse V., Drabek E.F., Creasy H.H., Daugherty S.C., Jones K.M., Santana-Cruz I., Tallon L.J., Read T.D., Hatch T.P., Bavoil P., et al. Genome sequences of the zoonotic pathogens Chlamydia psittaci 6BC and Cal10. J. Bacteriol. 2011;193:4039–4040. doi: 10.1128/JB.05277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seth-Smith H.M., Harris S.R., Rance R., West A.P., Severin J.A., Ossewaarde J.M., Cutcliffe L.T., Skilton R.J., Marsh P., Parkhill J., et al. Genome sequence of the zoonotic pathogen Chlamydophila psittaci. J. Bacteriol. 2011;193:1282–1283. doi: 10.1128/JB.01435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donati M., Huot-Creasy H., Humphrys M., Di Paolo M., Di Francesco A., Myers G.S. Genome Sequence of Chlamydia suis MD56, Isolated from the Conjunctiva of a Weaned Piglet. Genome Announc. 2014;2:e00425-14. doi: 10.1128/genomeA.00425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hugall A., Timms P., Girjes A.A., Lavin M.F. Conserved DNA sequences in Chlamydial plasmids. Plasmid. 1989;22:91–98. doi: 10.1016/0147-619X(89)90018-8. [DOI] [PubMed] [Google Scholar]
- 41.Pawlikowska-Warych M., Śliwa-Dominiak J., Deptuła W. Chlamydial plasmids and bacteriophages. Acta Biochim. Pol. 2015;62:1–6. doi: 10.18388/abp.2014_764. [DOI] [PubMed] [Google Scholar]
- 42.Song L., Carlson J.H., Whitmire W.M., Kari L., Virtaneva K., Sturdevant D.E., Watkins H., Zhou B., Sturdevant G.L., Porcella S.F., et al. Chlamydia trachomatis Plasmid-Encoded Pgp4 Is a Transcriptional Regulator of Virulence-Associated Genes. Infect. Immun. 2013;81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joseph S.J., Li B., Ghonasgi T., Haase C.P., Qin Z.S., Dean D., Read T.D. Direct amplification, sequencing and profiling of Chlamydia trachomatis strains in single and mixed infection clinical samples. PLoS ONE. 2014;9:e99290. doi: 10.1371/journal.pone.0099290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo K.V., O’Neill C.E., Clarke I.N. Diversity in Chlamydial plasmids. PLoS ONE. 2020;15:e0233298. doi: 10.1371/journal.pone.0233298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips S., Quigley B.L., Timms P. Seventy Years of Chlamydia Vaccine Research-Limitations of the Past and Directions for the Future. Front. Microbiol. 2019;10:70. doi: 10.3389/fmicb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greub G., Bavoil P. International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of Chlamydiae. Minutes of the closed meeting, 7 September 2016, Oxford, UK. Int. J. Syst. Evol. Microbiol. 2018;68:3683–3684. doi: 10.1099/ijsem.0.003049. [DOI] [PubMed] [Google Scholar]
- 47.Van Loock M., Vanrompay D., Herrmann B., Vander Stappen J., Volckaert G., Goddeeris B.M., Everett K.D.E. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int. J. Syst. Evol. Microbiol. 2003;53:761–770. doi: 10.1099/ijs.0.02329-0. [DOI] [PubMed] [Google Scholar]
- 48.Pannekoek Y., Dickx V., Beeckman D.S., Jolley K.A., Keijzers W.C., Vretou E., Maiden M.C., Vanrompay D., van der Ende A. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS ONE. 2010;5:e14179. doi: 10.1371/journal.pone.0014179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehricht R., Slickers P., Goellner S., Hotzel H., Sachse K. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell. Probe. 2006;20:60–63. doi: 10.1016/j.mcp.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Chen S., Zhou Y., Chen Y., Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bushnell B. BBTools Software Package. [(accessed on 17 August 2021)];2014 Available online: http://bbtools.jgi.doe.gov.
- 52.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 54.Cuccuru G., Orsini M., Pinna A., Sbardellati A., Soranzo N., Travaglione A., Uva P., Zanetti G., Fotia G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics. 2014;30:1928–1929. doi: 10.1093/bioinformatics/btu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:537–544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;8:232–235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection foraccurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York, NY, USA: 2000. [Google Scholar]
- 61.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 66.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 20121207.3907 [Google Scholar]
- 68.Garrison E., Kronenberg Z.N., Dawson E.T., Pedersen B.S., Prins P. Vcflib and tools for processing the VCF variant call format. BioRxiv. 2021 doi: 10.1101/2021.05.21.445151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P., McCarthy S.A. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics. 2017;33:2037–2039. doi: 10.1093/bioinformatics/btx100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of strains: 15-70d24, 15-49d3 and 15-58d44 obtained in this study have been deposited at ENA/GenBank/DDBJ under the accession numbers: LS450958.2, LS450956.2, OU508367.1 (chromosomes); LS450959.2, LS450957.2, OU508368.1 (plasmids), respectively. The Nanopore raw reads have been deposited under accession numbers: ERR6415086, ERR6415087, ERR6415088, respectively. All data related to WGS of avian C. abortus strains 15-70d24, 15-49d3 and 15-58d44 are included as a part of BioProject PRJEB26715.