Abstract
Alocasia longiloba is a popular ornamental plant in China, however pests and diseases associated with A. longiloba reduce the ornamental value of this plant. From 2016 to 2021, stem and root rot has been observed on A. longiloba in Guangdong Province, China. Once the disease became severe, plants wilted and died. A fungus was isolated from the diseased stem and identified as Fusarium elaeidis using both morphological characteristics and molecular analysis of DNA-directed RNA polymerase II subunit (rpb2), translation elongation factor-1α (tef1) gene and β-tubulin (tub2) sequence data. The pathogenicity test showed the fungus was able to produce typical symptoms on A. longiloba similar to those observed in the field. The original pathogen was reisolated from inoculated plants fulfilling Koch’s postulates. This is the first report of Fusarium elaeidis causing stem rot on A. longiloba. These results will provide a baseline to identify and control diseases associated with A. longiloba.
Keywords: Fusarium rot, first report, ornamental plants, pathogenicity, Nectriaceae
1. Introduction
The ornamental plant industry has become a high demanding industry with the increased interest in modern architecture. The values of ornamental plants are based on the appearance of the plant. However, they are susceptible to several diseases including wilts, rots and leaf spots which reduce the economical values of the plants [1]. In addition, the ornamental plant industry is the main global source of planting material exchanges. Thus, it is possible to introduce a new pathogen to a new locality via these planting materials which can affect already established plants as well [2] Therefore, the identification and characterisation of phytopathogenic genera on ornamental plants are crucial.
Four Alocasia species are grown in China: A. cucullata, A. hainanica, A. longiloba and A. macrorrhiza. Alocasia longiloba is a perennial herb belonging to the Araceae (L). It is native to tropical Asia and in China this plant is mainly distributed in Hainan, Yunnan and Guangdong provinces. Among these A. longiloba is widely cultivated in Guangdong Province as an ornamental plant due to its distinctive ‘elephant ear’ foliage. There are several diseases and pests associated with Alocasia spp., including fungal diseases [3,4]. Alternaria alocasiae and Ceratocystis fimbriata have been reported to cause leaf spots [4] and leaf blight [5], respectively, on A. macrorrhiza in China [4]. In addition, anthracnose of A. macrorrhiza caused by Colletotrichum karstii [3], root rot on Alocasia sp. caused by Pythium sp. and Rhizoctonia solani in Florida, USA [6] are among reported diseases. Most of these studies were based on both morphological and phylogenetic aspects [3]. However, so far there are no records of fungal diseases on A. longiloba [7].
Fusarium is one of the well-known phytopathogenic genera that belong to the Nectriaceae [8,9]. In addition, they are also saprobes, endophytes, soil-borne or can be isolated from water [10]. Fusarium species are cosmopolitan, and the traditional classification of these fungi was based on morphology which has led to controversial arguments for many years. Species belonging to F. solani species complex were transferred to the Neocosmospora [11,12], however, this was not accepted by O’Donnell et al. 2020 [13] who suggested that the F. solani species complex should remain as a species complex within the Fusarium [13]. Species delineation in Fusarium is based on morphological characters and molecular phylogeny [14]. In the Fusarium taxonomy and molecular phylogeny, the rDNA fragments; the nuclear ribosomal internal transcribed spacer (ITS) and large subunit (LSU) are uninformative in species-level identification [15,16]. However, the tef1 is highly informative at the species level [16]. In addition, DNA-directed RNA polymerase II subunit RPB1 (rpb1) and rpb2 are also informative for species identification in Fusarium [14,17].
During 2016–2021, a new stem and rot rot disease was observed on A. longiloba in Guangzhou City (Guangdong Province, China). This study aims to identify the causal organism of this disease. Based on both morphological and molecular approaches, the pathogen was identified as Fusarium elaeidis. The pathogenicity of this species was confirmed on potted A. longiloba plants.
2. Results
2.1. Field Symptoms
In the field, a one pot usually contains three to four plants. In these pots one or two plants showed wilt symptoms. The disease begins with a progressive leaf yellowing, followed by wilting, until the whole plant collapsed at the end (Figure 1a). Roots of diseased plants became black and rotted (Figure 1b). When diseased plants were cut open longitudinally the inside of the stems appeared brown (Figure 1c). Disease incidence was as high as 10% in some nurseries in Guangzhou City.
Figure 1.
Field symptoms of stem and root rot on Alocasia longiloba. (a–c) Field symptoms of stem and root rot on A. longiloba; (d–f) Healthy plants of A. longiloba in the field.
2.2. Phylogenetic Analyses and Species Identification
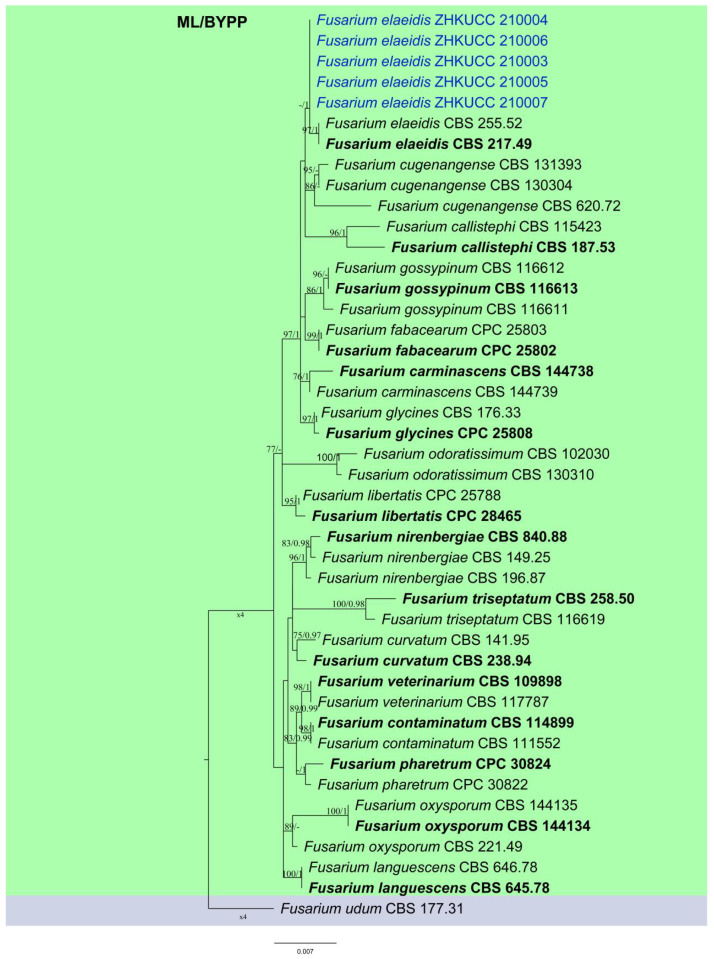
The combined sequence data set of rpb2, tef1, and tub2, comprised five Fusarium isolates from this study and 38 reference sequences of Fusarium from Lomboard et al. [18]. The tree was rooted with Fusarium udum (CBS 177.31). The tree topology of the maximum likelihood (ML) analysis was similar to the Bayesian posterior probability analysis (BYPP). The best scoring RAxML tree with a final likelihood value of −4431.098940 is presented (Figure 2). The matrix had 150 distinct alignment patterns, with 0.42% undetermined characters or gaps.
Figure 2.
Phylogenetic tree of the Fusarium based on maximum likelihood and MrBayes of rpb2, tef1 and tub2 gene sequences. The ML bootstrap support values above 75% and BYPP higher than 0.90 are shown as ML/BYPP at each node. Some branches were shortened to fit them to the page. Ex-type isolates are bold, the isolates in the study are marked in blue.
Estimated base frequencies were as follows: A = 0.257323, C = 0.268459, G = 0.237268, T = 0.236950; substitution rates AC = 1.805852, AG = 3.992536, AT = 0.764421, CG = 1.200643, CT = 8.476617, GT = 1.000000; gamma distribution shape parameter α = 1.082532. In the phylogenetic tree, isolates from A. longiloba were grouped with F. elaeidis strains with 97% ML bootstrap support and 1.00 BYPP.
2.3. Taxonomy
Fusarium elaeidis L. Lombard & Crous, Persoonia 41: 23 (2018) (Figure 3).
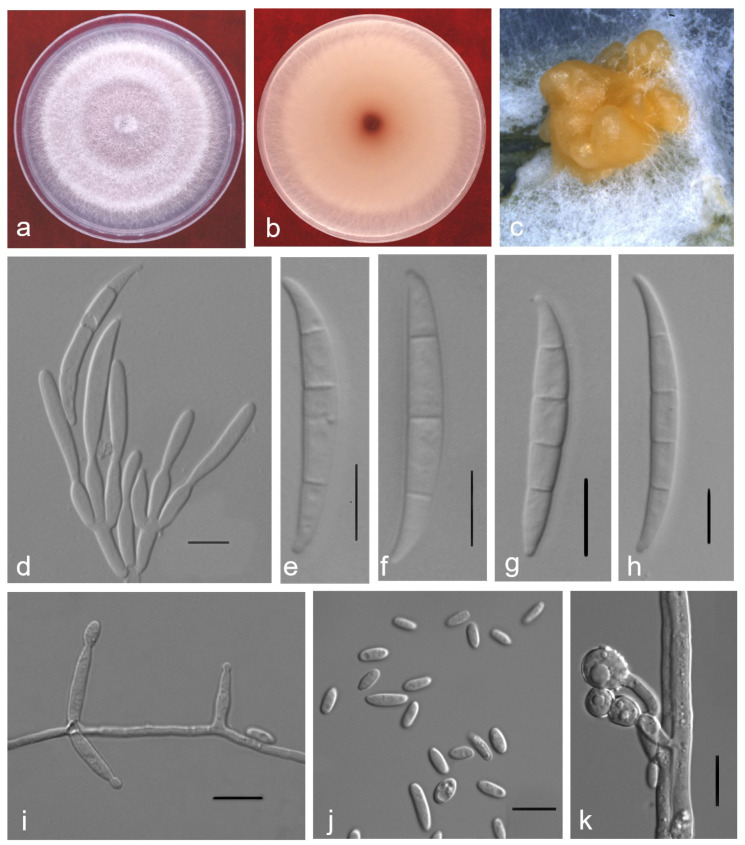
Figure 3.
Fusarium elaeidis (ZHKUCC 21-0006). (a) surface of colony on PDA at 25 °C after three days; (b) reverse of colony on CLA at 25 °C after seven days; (c) sporodochia on carnation leaves; (d) sporodochial conidiophores and phialides; (e–h) macroconidia; (i) Aerial conidiophores phialides; (j) microconidia; (k) chlamydospores. Scale bars: d–k = 10 μm.
IF 826838,
MycoBank MB826838
Pathogenic on Alocasia longiloba Miq. Sexual morph: Not observed. Asexual morph: The colour of sporodochia was orange on the surface of carnation leaves. Conidiophores 8–27 × 2–5 μm ( 13 × 4 μm, n = 50) irregular, branched; sporodochial phialides doliiform to ampulliform. Macroconidia (1–) 3–4 (–5)-septa, 3-septate 30–50 × 3–7 μm ( 39 × 5 μm, n = 50), 4-septate 35–55 × 3–6 μm ( 46 × 4 μm, n = 50), hyaline, straight to slightly curved. Apical cell tapered and curved. Basal cell was notched or foot-shaped. Conidiophore 7–26 × 2–5 μm ( 13 × 3 μm, n = 50) phialides, on the aerial mycelium cylindrical, monophialidic. Microconidia 6–10 × 2–4 μm ( 8 × 3 μm, n = 50) produced in false heads on short phialides, hyaline with zero to one septum, ellipsoidal, reniform, or oval. Chlamydospores 6–15 × 4–11 μm ( 9 × 8 μm, n = 50), formed solitary, in pairs or chains, either terminal or intercalary in hyphae. Culture characteristics: Colonial growth rate was 6–7 mm on PDA at 25 °C per day. The colony surface was white, flat, and floccose with a regular margin. In reverse colony white at first and turned pink in the centre at the end. Sporodochia produced on carnation leaf agar (CLA) after 7 days.
Material examined: CHINA, Guangdong Province, Guangzhou, isolated from diseased stems of A. longiloba Miq., from November 2016 to April 2021, YX Zhang and CP You, dried cultures ZHKU 21-0003–21-0007; living culture ZHKUCC 21-0003–21-0007.
Notes: Isolates obtained from this study formed a well-supported clade with the representative strains of Fusarium elaeidis (CBS 217.49; Ex-type strain and CBS 255.52). Most of the morphological characteristics in this study were similar to the original description of Fusarium elaeidis by Lombard et al. [18]. However, the isolates obtained in this study differed by 3-septate and 4-septate of macroconidia. Moreover, in this study isolates produce larger macroconidia than those of the original description, and no polyphialides were observed in this study. The base pairs of both rpb2 and tub2 in this study are same as that of F. elaeidis (strain no?). Only three base pairs of tef1 (totally 616 bp) were different with that of F. elaeidis (?) in Lombord et al. study [18]. Therefore, we identified our strains as F. elaeidis based on both morphology and molecular phylogeny.
2.4. Pathogenicity Test
The pathogenicity of isolates ZHKU 21-0005 and ZHKU 21-0006 was determined by inoculating healthy A. longiloba with mycelium suspension. Leaves of inoculated plants turned yellow after ten days (Figure 4a). Root and stem showed black and started to rot after fifteen days of inoculation (Figure 4b,c). The root density of inoculated plants was lower compared to healthy plants. Symptoms on the inoculated plants were similar to those on diseased plants in the field. None of the control plants developed symptoms (Figure 4d–f). From the diseased plants, the pathogen was reisolated. The colonies and morphological characteristics of isolates from inoculated plants were the same as those from the field.
Figure 4.
Inoculation assays of stem and root rot on Alocasia longiloba (inoculated with ZHKUCC 21-0005). (a–c) Symptom of stem and root rot after 15 days of inoculation; (d–f) Control plants of A. longilob after 15 days of inoculation.
3. Discussion
Fusarium is an important genus including large numbers of phytopathogenic species. Therefore, correct species identification is important for disease diagnosing and control. Twenty monophyletic species complexes have been reported in Fusarium [18]. The morphological species of F. oxysporum was recognized as a species complex because of its high level of phylogenetic diversity [18,19,20,21]. Laurence et al. [21] divided 17 independent evolutionary lineages into two phylogenetic species using genealogical concordance phylogenetic species recognition (GCPSR) criteria. Moreover, Lombard et al. [18] identified fifteen cryptic taxa using five gene regions in the Fusarium. Multi-genes analysis, such as ITS, IGS rDNA (the nuclear ribosomal intergenic spacer region), mating-type genes (mat1-1-1α-box), tef1 and tub2, are used to delineate species in Fusarium and beyond species level within the F. oxysporum species complex [2,20,22,23,24]. In this study, we combined morphological characteristics with DNA sequence analysis of rpb2, tef1 and tub2 genes to ensure species identification accurately.
Fusarium oxysporum species complex is a well-established and diverse complex in this genus. It can infect around 150 plant species [10], including vegetables, fruits, and ornamental plants, and cause severe diseases. Among these F. oxysporum f. sp. cubense which is the causal agent of banana wilt is one of the well-know and destructive disease on banana worldwide [25]. Fusarium fabacearum infect Malay apple plants (Syzygium malaccensis) and caused tree death in Brazil in 2017 [25]. On ornamentals, the disease caused by F. oxysporum cause wilt, crown rot and root rot. Moreover, these hosts included several valuable ornamental plants, such as Cymbidium spp., Chrysanthemum spp., and Gladiolus spp. [1]. Fusarium wilt is a destructive disease in cyclamen production. This disease was the first outbreak in Germany and then quickly spread to all European production regions resulting in a huge effect on the cyclamen industry [2,26]. Fusarium wilt on Cymbidium spp. is a well-known disease that leads to stem rot and plant death [27,28]. Fusarium oxysporum can infect various cultivars of Cymbidium and hybrid Cymbidium. The disease incidence was up to 30% in some plantations in Guangdong, China [28]. Over 30 ornamental plant genera have been recorded as hosts of F. oxysporum [1]. In the present study, we isolated and identified F. elaeidis from A. longiloba causing stem and root rot.
Fusarium oxysporum f. sp. elaeidis was upgraded to the species level as Fusarium elaeidis by Lombard et al. [18]. In that study, they resolved 15 cryptic taxa which were named as forma specialis in Fusarium oxysporum [18]. Because of this, almost all the previous reports on this species are under Fusarium oxysporum f. sp. elaeidis. This species has been reported to cause Fusarium wilt in oil palm worldwide [29]. This pathogen is a soilborne fungus and difficult to control [30]. However, this is the first report of stem and root rot on A. longiloba caused by F. elaeidis. Therefore, further studies are necessary to understand the host range of this pathogen. Fusarium is a well-established genus with a larger number of forms and races. Given the same weight as other respective fields in mycology, identification of pathogenic species is the most critical step in plant pathology [31,32]. Moreover, in plant pathology, this identification is necessary to go beyond the species level. However, for most of the phytopathogenic fungal genera except like Fusarium, defining beyond species levels are not practised [32]. Even though lower-level ranking is well defined in Fusarium, in recent studies, some of these lower levels are upgraded into species level. Therefore, it is necessary to conclude how to delineate each species and define lower levels in Fusarium.
4. Materials and Methods
4.1. Sample Collection
A stem rot disease was observed on A. longiloba in a nursery in Guangzhou City (Guangdong Province, China) from 2016–2021. Diseased plants showed yellow leaves, rotted stems and wilting. Twenty plants from different greenhouses were collected in 2016, 2019 and 2021, respectively. Samples were placed in sterile, transparent plastic bags. Relevant photographs were taken on-site and samples were taken to the laboratory for further studies. In addition to that disease symptoms, sampling time and diseases severity were recorded at the time of sample collection.
4.2. Fungal Isolation and Purification
Infected stems from diseased plants were first washed with running tap water to remove debris. Then the samples were cut into 0.5 × 0.5 cm sections including both healthy and diseased tissues. Those cuttings were surface sterilized by immersion in 75% ethanol for 10 s, 2.5% NaOCl for 40 s, and rinsed in sterile water three times. After that samples were dried in sterilized tissue paper. Dried cuttings were placed onto potato dextrose agar (PDA) and incubated at 25 °C. Pure cultures were obtained after three times hyphal tip isolations. In total four isolates were obtained. The cultures were deposited in the Culture Collection of Zhongkai University of Agriculture and Engineering (ZHKUCC 21-0003, ZHKUCC 21-0004, ZHKUCC 21-0005, ZHKUCC 21-0006, ZHKUCC 21-0007).
4.3. DNA Extraction, PCR Amplification and Sequencing
The genomic DNA of four isolates was extracted using a DNA rapid Extraction Kit (Aidlab Biotechnologies Co., Ltd, Beijing, China). A portion of DNA-directed RNA polymerase II subunit (rpb2), the translation elongation factor-1α gene (tef1) and the β-tubulin genes (tub2) were amplified and sequenced. The tef1 region was amplified using primer pair EF-1H and EF-2T [33]. The tub2 region was amplified using primer pair T1 and CYLTUB1R [22,34]. The rpb2 region was amplified using primer pair 5f2 and 7cr [35,36]. PCR reactions were conducted in 25 μL volumes containing 12.5 μL of 2× Easy Taq PCR SuperMix (TransGen Biotech, Beijing, China), 1 μL DNA, each of 5μM premier (1 μL), ddH2O (9.5 μL). PCR amplification was performed with an initial denaturation step of 94 °C for 5 min, followed by 35 cycles of 94 °C for 40 s, annealing at 55 °C for 45 s, and extension at 72 °C for 45 s, and a final extension at 72 °C for ten min. The PCR products were sequenced by Guangzhou Tianyi Huiyuan Science and Technology Co. Ltd (Guangzhou, China). For sequencing in both directions with forward and reverse primers were used. The DNA sequence of rpb2, tef1 and tub2 were deposited in the GenBank (Table 1).
Table 1.
GenBank accession numbers of Fusarium strains used in phylogenetic analysis. The isolates obtained in this study are bold.
| Species | Culture Accession | GenBank Accession Numbers | ||
|---|---|---|---|---|
| rpb2 | tef1 | tub2 | ||
| Fusarium callistephi | CBS 187.53 T | MH484875 | MH484966 | MH485057 |
| CBS 115423 | MH484905 | MH484996 | MH485087 | |
| F. carminascens | CBS 144739 = CPC 25792 | MH484934 | MH485025 | MH485116 |
| CBS 144738 = CPC 25800 T | MH484937 | MH485028 | MH485119 | |
| F. contaminatum | CBS 111552 | MH484900 | MH484991 | MH485082 |
| CBS 114899 T | MH484901 | MH484992 | MH485083 | |
| F. cugenangense | CBS 620.72 = DSM 11271 = NRRL 36520 | MH484879 | MH484970 | MH485061 |
| CBS 130304 = BBA 69050 = NRRL 25433 | MH484921 | MH485012 | MH485103 | |
| CBS 131393 | MH484928 | MH485019 | MH485110 | |
| F. curvatum | CBS 238.94 = NRRL 26422 = PD 94/184 T | MH484893 | MH484984 | MH485075 |
| CBS 141.95 = NRRL 36251 = PD 94/1518 | MH484894 | MH484985 | MH485076 | |
| F. elaeidis | CBS 217.49 = NRRL 36358 | MH484870 | MH484961 | MH485052 |
| CBS 255.52 = NRRL 36386 | MH484874 | MH484965 | MH485056 | |
| ZHKUCC 21-0003 | MZ439841 | MZ325284 | MZ439836 | |
| ZHKUCC 21-0004 | MZ439842 | MZ325285 | MZ439837 | |
| ZHKUCC 21-0005 | MZ439843 | MZ325286 | MZ439838 | |
| ZHKUCC 21-0006 | MZ439844 | MZ325287 | MZ439839 | |
| ZHKUCC 21-0007 | MZ439845 | MZ325288 | MZ439840 | |
| F. fabacearum | CBS 144743 = CPC 25802 T | MH484939 | MH485030 | MH485121 |
| CBS 144744 = CPC 25803 | MH484940 | MH485031 | MH485122 | |
| F. glycines | CBS 176.33 = NRRL 36286 | MH484868 | MH484959 | MH485050 |
| CBS 144746 = CPC 25808 T | MH484942 | MH485033 | MH485124 | |
| F. gossypinum | CBS 116611 | MH484907 | MH484998 | MH485089 |
| CBS 116612 | MH484908 | MH484999 | MH485090 | |
| CBS 116613 T | MH484909 | MH485000 | MH485091 | |
| F. languescens | CBS 645.78 = NRRL 36531 T | MH484880 | MH484971 | MH485062 |
| CBS 646.78 = NRRL 36532 | MH484881 | MH484972 | MH485063 | |
| F. libertatis | CBS 144747 = CPC 25788 | MH484933 | MH485024 | MH485115 |
| CBS 144749 = CPC 28465 T | MH484944 | MH485035 | MH485126 | |
| F. nirenbergiae | CBS 840.88 T | MH484887 | MH484978 | MH485069 |
| CBS 149.25 = NRRL 36261 | MH484865 | MH484956 | MH485047 | |
| CBS 196.87 = NRRL 26219 | MH484886 | MH484977 | MH485068 | |
| F. odoratissimum | CBS 102030 | MH484898 | MH484989 | MH485080 |
| CBS 130310 = NRRL 25603 | MH484922 | MH485013 | MH485104 | |
| F. oxysporum | CBS 221.49 = IHEM 4508 = NRRL 22546 | MH484872 | MH484963 | MH485054 |
| CBS 144134 ET | MH484953 | MH485044 | MH485135 | |
| CBS 144135 | MH484954 | MH485045 | MH485136 | |
| F. pharetrum | CBS 144750 = CPC 30822 | MH484951 | MH485042 | MH485133 |
| CBS 144751 = CPC 30824 T | MH484952 | MH485043 | MH485134 | |
| F. triseptatum | CBS 258.50 = NRRL 36389 T | MH484873 | MH484964 | MH485055 |
| CBS 116619 | MH484910 | MH485001 | MH485092 | |
| F. veterinarium | CBS 109898 = NRRL 36153 T | MH484899 | MH484990 | MH485081 |
| CBS 117787 | MH484912 | MH485003 | MH485094 | |
CBS: Westerdijk Fungal Biodivesity Institute (WIFB), Utrecht, The Netherlands. ZHKUCC: Zhongkai University of Agriculture and Engineering culture collection. Sequences produced in this study are shown in bold. Type sequnces are given as T.
4.4. Phylogenetic Analyses
Consensus sequences were derived from forward and reverse primer sequences for rpb2, tef1 and tub2, and compared against the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 18 June 2021). Relevant sequence data of Fusarium species were obtained from the National Center for Biotechnology Information (NCBI) following Lomboard et al. [18]. Sequences were aligned using MAFFT version 7. Sequences were aligned manually using BioEdit where necessary. Concatenated dataset of rpb2, tef1, and tub2 were used for the phylogenetic analyses.
Phylogenetic analyses were conducted based on Maximum-likelihood in RAxML [37] and Bayesian analyses in MrBayes (v. 3.0b4) [38] The maximum likelihood analyses were conducted using RAxML-HPC2 on XSEDE (8.2.8) [39] in the CIPRES Science Gateway platform. The GTR+I+G evolution model was used with 1000 non-parametric bootstrapping iterations. Bayesian analysis was performed with six simultaneous Markov chains were run for 106 generations, sampling the trees at every 200th generation. From the 10,000 trees obtained, the first 2000 representing the burn-in phase were discarded. The remaining 8000 trees were used to calculate posterior probabilities (PPs) in a majority rule consensus tree. The final sequence alignment generated in this study was submitted to TreeBASE (https://treebase.org/treebase-web/home.html, accessed on 5 July 2021) under submission ID 28480.
4.5. Morphological Characterisation
Pure cultures incubated on PDA for 7 days at 25 °C under a 12 h light-dark cycle were used for colony characteristics and colonial growth rates. Cultures grown on carnation leaves agar (CLA) for 7–10 days at 25 °C under a 12 h light-dark cycle were used for microscopic characters observation. Morphological characters were photographed using an ECLIPSE 80i microscope (Nikon, Tokyo, Japan) and measurements were taken using NIS-Elements BR 3.2. Measurements of spore length and width of 50 spores were taken. The mean values were calculated with Microsoft Excel.
4.6. Pathogenicity Tests
Two representative isolates ZHKU21-0005 and ZHKU21-0006 were used to inoculate potted plants from the nursery. Representative isolates were incubated in PDA for three days to obtain mycelia. One gram of mycelium added into 100 mL sterilised water and blend to make into 3% mycelium suspension. Healthy plants were inoculated by pouring 50 mL of mycelium suspension into the substrate around the roots. Sterilised water (50 mL) was used as the control. For each treatment three replicates were used. All treated plants were maintained at 25 °C in a growth chamber. To fulfil Koch’s postulates, the fungus was re-isolated from inoculated plants showing typical disease symptoms and compared to the original isolate.
Author Contributions
Conceptualization, Y.Z. and I.S.M.; methodology, Y.Z.; software, C.C. (Cantian Chen); validation, C.C. (Chao Chen), J.L. and J.Z.; formal analysis, C.C. (Cantian Chen) and C.C. (Chao Chen); investigation, Y.Z. and C.Y.; resources, Y.Z. and C.Y.; data curation, C.C. (Chao Chen) and J.L.; writing—original draft preparation, Y.Z.; writing—review and editing, Y.Z., I.S.M., R.S.J.; visualization, Y.Z. and C.C. (Cantian Chen); supervision, M.X., I.S.M. and C.Y.; project administration, Y.Z.; funding acquisition, Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key Realm R&D Program of Guangdong Province, grant number 2018B020205003 and the Modern Agricultural Industry Technology System Flower Innovation Team of Guangdong Province, grant number 2021KJ121.It was also funded by National Natural Science Foundation of China, grant number 31600019.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are given in Table 1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lecomte C., Alabouvette C., Edel-Hermann V., Robert F., Steinberg C. Biological control of ornamental plant diseases caused by Fusarium oxysporum: A review. Biol. Control. 2016;101:17–30. doi: 10.1016/j.biocontrol.2016.06.004. [DOI] [Google Scholar]
- 2.Lecomte C., Edel-Hermann V., Cannesan M.A., Gautheron N., Langlois A., Alabouvette C., Robert F., Steinberg C. Fusarium oxysporum f. sp. cyclaminis: Underestimated genetic diversity. Eur. J. Plant Pathol. 2016;145:421–431. [Google Scholar]
- 3.He Y., Shu C., Chen J., Zhou E. First report of anthracnose of Alocasia macrorrhiza caused by Colletotrichum karstii in Guangdong, China. Plant Dis. 2014;98:696. doi: 10.1094/PDIS-10-13-1046-PDN. [DOI] [PubMed] [Google Scholar]
- 4.Li J., Yang J.Y., Xu K.C., Sun Y.X., Huang Q. First report of Ceratocystis fimbriata causing leaf blight on Alocasia macrorrhiza in China. Plant Dis. 2016;100:2172–2173. doi: 10.1094/PDIS-12-15-1464-PDN. [DOI] [Google Scholar]
- 5.Zhang T.Y., Zhang J.Z., Gao M.X. Taxonomic studies of Alternaria from China III. New species and new record on Anacardiaceae, Araceae and Araliaceae. Mycosystema. 1999;18:125–129. [Google Scholar]
- 6.Alfieri S.A., Jr., Langdon K.R., Wehlburg C., Kimbrough J.W. Index of Plant Diseases in Florida, Revised. Florida Dept. Agric. and Consumer Serv.; Tallahassee FL, USA: 1984. pp. 1–389. [Google Scholar]
- 7.Farr D.F., Rossman A.Y. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Retrieved 12 July 2021. [(accessed on 30 July 2021)]; Available online: https://nt.ars-grin.gov/fungaldatabases/
- 8.Hyde K.D., Norphanphoun C., Maharachchikumbura S.S.N., Bhat D.J., Jones E.B.G., Bundhun D., Chen Y.J., Bao D.F., Boonmee S., Calabon M.S., et al. Refined families of Sordariomycetes. Mycosphere. 2020;11:1060–1456. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 9.Rana A., Sahgal M., Johri B.N. Fusarium oxysporum: Genomics, diversity and Plant–Host interaction. In: Satyanarayana T., Deshmukh S., Johri B., editors. Developments in Fungal Biology and Applied Mycology. Springer; Singapore: 2017. pp. 159–199. [Google Scholar]
- 10.Lombard L., Merwe N.A.V.D., Groenewald J.Z., Crous P.W. Generic concepts in Nectriaceae. Stud Mycol. 2015;80:189–245. doi: 10.1016/j.simyco.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandoval-Denis M., Crous P.W. Removing chaos from confusion: Assigning names to common human and animal pathogens in Neocosmospora. Persoonia. 2018;41:109–129. doi: 10.3767/persoonia.2018.41.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnell K., Al-Hatmi A.M.S., Aoki T., Brankovics B., Cano-Lira J.F., Coleman J.J., Hoog G.S.D., Antonio Di Pietro A.D., Frandsen R.J.N., Geiser D.M., et al. No to Neocosmospora: Phylogenomic and Practical Reasons for Continued Inclusion of the Fusarium solani Species Complex in the Genus Fusarium. mSphere. 2020;5:00810–00820. doi: 10.1128/mSphere.00810-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyde K.D., Nilsson R.H., Alias S.A., Ariyawansa H.A., Blair J.E., Lei C., Cock A.W.A.M.D., Dissanayake A.J., Glockling S.L., Goonasekara I.D., et al. One stop shop: Backbones trees for important phytopathogenic genera: I (2014) Fungal Divers. 2014;67:21–125. doi: 10.1007/s13225-014-0298-1. [DOI] [Google Scholar]
- 14.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiser D.M., Jime´nez-Gasco M.D.M., Kang S., Makalowska I., Veeraraghavan N., Ward T.J., Zhang N., Kuldau G.A., O’donnell K. Fusarium-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004;110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 16.O’Donnell K., Rooney A.P., Proctor R.H., Brown D.W., McCormick S.P., Ward T.J., Frandsen R.J., Lysøe E., Rehner S.A., Aoki T., et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013;52:20–31. doi: 10.1016/j.fgb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Lombard L., Sandoval-Denis M., Lamprecht S.C., Crous P.W. Epitypification of Fusarium oxysporum–clearing the taxonomic chaos. Persoonia. 2019;43:1–47. doi: 10.3767/persoonia.2019.43.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon T.R., Martyn R.D. The evolutionary biology of Fusarium oxysporum. Ann. Rev. Phyto. 1997;35:111–128. doi: 10.1146/annurev.phyto.35.1.111. [DOI] [PubMed] [Google Scholar]
- 19.O’Donnell K., Gueidan C., Sink S., Johnston P.R., Crous P.W., Glenn A., Riley R., Zitomer N.C., Colyer P., Waalwijk C. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 2009;46:936–948. doi: 10.1016/j.fgb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Laurence M.H., Summerell B.A., Burgess L.W., Liew E.C.Y. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biol. 2014;118:374–384. doi: 10.1016/j.funbio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 21.O’Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 22.O’Donnell K., Sutton D.A., Rinaldi M.G., Magnon K.C., Cox P.A., Revankar S.G., Sanche S., Geiser D.M., Juba J.H., van Burik J.-A.H., et al. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: Evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 2004;42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell K., Ward T.J., Robert V.A.R.G., Crous P.W., Geiser D.M., Kang S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica. 2015;43:583–595. doi: 10.1007/s12600-015-0484-z. [DOI] [Google Scholar]
- 24.Farias O.R., Cruz J.M.F.L.J., Veloso J.S., Silva H.A.O., Oliveira M.D.M., Félix M.R.F., Varanda C.M.R., Materatski P., Podestá G.S., Nascimento L.C. First Report of Wilt of Syzygium malaccensis Caused by a Member of the Fusarium oxysporum Species Complex in Brazil. Plant Dis. 2021;105:1207. doi: 10.1094/PDIS-04-20-0708-PDN. [DOI] [Google Scholar]
- 25.Rouxel F., Grouet D. Première observation de la fusariose vasculaire du cyclamen en France. Ann. Phytopathol. 1974;6:475–478. [Google Scholar]
- 26.Lee B.D., Kim W.G., Cho W.D., Cho W.D., Sung J.M. Occurrence of Dry Rot on Cymbidium Orchids Caused by Fusarium spp. in Korea. Plant Pathol. J. 2002;18:156–160. [Google Scholar]
- 27.Zhang Y.X., Shi Z.R., Huang J.H., Xiang M.M. Identification and Control of Fusarium Wilt of Chinese Orchid. Guangdong Agric. Sci. 2011;38:21–22. [Google Scholar]
- 28.Adusei-Fosu K., Dickinson M. Detection of Fusarium oxysporum f. sp. elaeidis Causing Fusarium Wilt of Oil Palm Using Loop-Mediated Isothermal Amplification (LAMP) J. Plant Pathol. Microbiol. 2019;10:1–6. [Google Scholar]
- 29.Flood J. A review of Fusarium wilt of oil palm caused by Fusarium oxysporum f. sp. elaeidis. Phytopathology. 2006;96:660–662. doi: 10.1094/PHYTO-96-0660. [DOI] [PubMed] [Google Scholar]
- 30.Jayawardena R.S., Hyde K.D., de Farias A.R.G., Bhunjun C.S., Ferdinandez H.S. What is a species in fungal plant pathogens? Fungal Divers. 2021 doi: 10.1007/s13225-021-00484-8. in press. [DOI] [Google Scholar]
- 31.Manawasinghe I.S., Phillips A.J.L., Xu J., Balasuriya A., Hyde K.D., Tępień L., Harischandra D.L., Karunarathna A., Yan J., Weerasinghe J., et al. Defining a species in plant pathology: Beyond the species level. Fungal Divers. 2021 doi: 10.1007/s13225-021-00481-x. in press. [DOI] [Google Scholar]
- 32.O’Donnell K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia. 2000;92:919–938. doi: 10.2307/3761588. [DOI] [Google Scholar]
- 33.Crous P.W., Groenewald J.Z., Risede J., Simoneau P., Hywel-Jones N.L. Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Stud. Mycol. 2004;50:415–430. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y.J., Whelen S., Hall B.D. Phylogenetic Relationships among Ascomycetes: Evidence from an RNA Polymerse II Subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 35.Sung G.H., Sung J.M., Hywel-Jones N.L., Spatafora W.J. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Silvestro D., Michalak I. RaxmlGUI: A Graphical Front-End for RAxML. Org. Divers. Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 37.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 38.Miller M.A., Pfeiffer W.T., Schwartz T. Gateway Computing Environments Workshop (GCE) Institute of Electrical and Electronics, Engineers; New Orleans, LA, USA: 2010. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. [Google Scholar]
- 39.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are given in Table 1.