Abstract
Recent cases of infections caused by glycopeptide-resistant enterococci (GRE) have highlighted the emergence of these organisms in the Republic of South Africa. During May 1998 we conducted a prevalence study in four hospitals in Johannesburg and obtained 184 rectal swabs from patients identified as being at high risk for GRE colonization. Twenty enterococcal isolates showing various glycopeptide resistance genotypes were recovered: 3 Enterococcus faecium vanA isolates, 10 E. faecium vanB isolates, 6 E. gallinarum vanC1 isolates, and 1 E. avium vanA isolate. Macrorestriction analysis was used to demonstrate the clonal spread of GRE strains within hospitals. Evidence also demonstrated the likely persistence of the original E. faecium vanA isolate associated with the first confirmed death contributed to by GRE infection in South Africa in March 1997.
Glycopeptide-resistant enterococci (GRE) have emerged over the last decade as important nosocomial pathogens (17, 27). The epidemiology of GRE colonization, infection, and rapid dissemination in the United States and in western Europe is well described, and prevalence rates vary among different centers (11). Control of these infections involves stringent infection control measures, prudent antibiotic use, and an understanding of the epidemiology of infections through improved surveillance (14). Risk factors for colonization with GRE have been documented and are similar for various centers (2, 13, 14, 19, 21).
Once GRE infections are detected in a country, a rapid increase in the number of cases is generally demonstrated (1, 14, 18). The preponderance of the vanA genotype has been shown in some countries (14, 18) but may become evident only after several years of GRE endemicity (16). In a study from Australia the predominant genotype of clinical Enterococcus faecium isolates was vanB (1), suggesting a different epidemiology for these organisms.
The first two documented GRE infections in the Republic of South Africa corresponded phenotypically with VanA (3). In the present study we characterized the strains genotypically and compared them with those colonizing high-risk patients. Subsequent to this prevalence study, an additional five patients in Johannesburg were confirmed to be colonized with GRE, and details about them are included in this report.
Patients were screened for GRE in high-risk wards in four Johannesburg hospitals. These included intensive care units (ICU), oncology wards, renal dialysis units, and a burn unit. Criteria for screening included admission to one of these units for ≥5 days or regular outpatient attendance at oncology or renal dialysis units. After written informed consent was obtained, demographic patient data and possible risk factors were recorded and a rectal swab was taken. Data collected included age, sex, ward and hospital, number of days in the hospital, transfer from another hospital, primary diagnosis on admission, underlying risk factors, history of diarrhea, history of surgical procedures and other invasive medical procedures, and current and previous antibiotics (up to 4 weeks prior) used. Of 184 rectal swabs collected, 78 (42.4%) came from state hospital S1, a tertiary-care academic hospital with 1,165 beds. In S1 a total of 122 patients were considered high-risk inpatients or were on chronic hemodialysis: 6% did not fulfill inclusion criteria, and others did not give consent or were not approached due to time constraints. In state hospital S2, a tertiary-care academic hospital with 2,500 beds, rectal swabs were collected from 55 patients, constituting a convenience sample of 29.9% of a total of 172 high-risk inpatients or chronic dialysis patients. No patients from this hospital were excluded on the basis of inclusion criteria. From P1, a 408-bed private hospital, with 11 patients considered eligible in ICU and 50 patients on chronic dialysis, 22 rectal swabs (12% of all swabs) were collected. Of these, 8 came from ICU patients (informed consent was not obtained from other patients) and 14 came from renal patients (representing a convenience sample). Private hospital P2, with 321 beds, had 58 ICU patients, and swabs were taken from 29 patients (15.8% of total) who either satisfied inclusion criteria or gave informed consent.
Four media were used for screening for vancomycin-resistant enterococci: bile esculin azide agar (Enterococcosel agar; Difco Laboratories, Detroit, Mich.) (15), bile esculin (Difco Laboratories) plus staphylococcus or streptococcus supplement (Oxoid, Basingstoke, United Kingdom), colistin-nalidixic acid agar (Difco Laboratories) plus 2.5 μg of amphotericin B (Squibb Laboratories, SA) per ml (26), and bile esculin azide broth (Enterococcosel broth; Difco Laboratories). Each medium contained 10 μg of vancomycin (Eli Lilly, SA) per ml.
Cotton wool-tipped swabs with Stuart's transport medium were distributed to the investigators at each of the hospitals. Rectal or perianal swabs were taken by the investigators or nursing staff or by the patients themselves after adequate instruction. Swabs were plated onto media within 24 h of collection in our central laboratory. Care was taken to systematically alternate the sequence of inoculation of all the laboratory media. No broth enrichment step was used.
Media were incubated at 35°C in ambient air, and results were read at 24 and 48 h. Esculin hydrolysis in the broth medium was considered an indicator of the possible presence of GRE. Broths exhibiting esculin hydrolysis were subcultured onto blood agar plates, which were then incubated for 24 to 48 h.
The following strains were used as controls: E. casseliflavus ATCC 25788 (vanC2; vancomycin MIC, 16 μg/ml; teicoplanin MIC, 0.5 μg/ml); E. faecalis ATCC 51575 (vanB; vancomycin MIC, >64 μg/ml; teicoplanin MIC, <0.12 μg/ml) and ATCC 51299 (vanB; vancomycin MIC, 2 μg/ml; teicoplanin MIC, <0.12 μg/ml); and E. faecium ATCC 19434 (vancomycin MIC, 0.25 μg/ml; teicoplanin MIC, <0.12 μg/ml) and ATCC 51559 (vanA; vancomycin MIC, 64 μg/ml; teicoplanin MIC, 64 μg/ml). Additional isolates included two strains isolated from patients with GRE infection in 1997 from two of the study hospitals: E. faecium vanA from S1 and E. faecalis vanA from S2 (3). Five subsequent GRE isolates from patients in Johannesburg were also included. One was from a tracheal aspirate (for this patient, GRE were also found in urine, a rectal swab, and a catheter tip), two were from catheter tips, one was from fluid from an abdominal drain, and one was from a tissue biopsy in a burn patient.
All isolates were identified to the species level (4, 10). Isolates were tested for motility, determined on semisolid agar stabs, and for pigmentation. Biotyping was correlated with species identification using multiplex PCR (9). MICs were determined by the microdilution method according to the National Committee for Clinical Laboratory Standards criteria. Isolates were tested for β-lactamase production with the chromogenic cephalosporin nitrocefin (23).
Multiplex PCR based on the method developed by Dutka-Malen et al. (9) was performed on all enterococcal isolates. Macrorestriction analysis was performed using the methods described by Murray et al. (22). EpiInfo (version 6.01) was used for data analysis. For categorical variables Fisher's exact two-tailed test or Yates' corrected chi-square test was used, and the Mann-Whitney U test was used for continuous variables.
Twenty of the 184 patients (10.9%) who were included in the study were colonized with GRE: 3 with E. faecium vanA, 10 with E. faecium vanB, 6 with E. gallinarum vanC1, and 1 with E. avium vanA. Details for those patients harboring GRE isolates are shown in Table 1. Of the 184 patients included in the study, 85 (46.2%) were female, and 99 (53.8%) were male; 177 (97.8%) were human immunodeficiency virus (HIV) seronegative, and 4 (2.2%) were HIV seropositive. The mean age of those patients above the age of 1 year was 36.3 years. Ten patients were less than 1 year of age. Of the patients selected, 94 (51.1%) had a history of recent surgery and 77 (42.1%) were on hemodialysis. Three patients (1.6%) were renal transplant recipients, 5 (2.7%) were premature infants, 34 (18.5%) had documented malignancies, and 13 (7.1%) were burn patients. History of previous or current antibiotic use was obtained for 183 patients, with 105 (57.3%) receiving antimicrobials.
TABLE 1.
Clinical features of patients colonized with GRE
| Hospital and patient/lane no.a | Sex | Age (yrs) | Risk factor(s) | Received antibiotics | Received vancomycin | No. of days in ward |
|---|---|---|---|---|---|---|
| Hospital S1 | ||||||
| 1 | Female | 51 | Renal transplant, pneumonia | Yes | No | 22 |
| 2 | Female | 8 | Nephroblastoma, nephrectomy | Yes | Yes | 42 |
| 3 | Female | 20c | Leukemia, sepsis | Yes | No | 8 |
| 4 | Male | 16 | Lymphoma, chemotherapy | No | No | None (outpatientd) |
| 5 | Female | 2 | Aplastic anemia, sepsis | Yes | No | 56 |
| NAb | Male | 13 | Osteogenic sarcoma | Yes | No | 5 |
| 6 | Female | 58 | Hemodialysis | Yes (oral) | No | None (outpatient) |
| NAb | Female | 26 | Hemodialysis | No | No | None (outpatient) |
| NAe | Male | 39 | Chronic renal failure | Yes | Yes | 6 |
| Hospital S2, NAb | Female | 22 | Burns | No | No | 6 |
| Hospital P1 | ||||||
| 7 | Female | 69 | Peritoneal dialysis | No | No | 14 |
| 8 | Female | 42 | Hemodialysis | Yes (oral) | No | 28 |
| 9 | Female | 51 | Hemodialysis | No | No | None (outpatient) |
| 10 | Male | 66 | Hemodialysis | No | No | None (outpatient) |
| 11 | Female | 42 | Hemodialysis | Yes | No | None (outpatient) |
| NAb | Female | 55 | Hemodialysis | No | No | None (outpatient) |
| Hospital P2 | ||||||
| NAb | Male | 22 | Motor vehicle accident | Yes | No | 21 |
| 12 | Female | 27 | Abdominal gunshot wound | Yes | No | 7 |
| 13 | Female | 72 | COPDf, heart failure | Yes | No | 13 |
| NA | Male | 13 | Brain tumor | Yes | Yes | 34 |
Lane in Fig. 1.
NA, not applicable; E. gallinarum isolate not run on PFGE.
Twenty months.
This patient had been seen regularly in the pediatric oncology outpatient department and therefore satisfied the inclusion criteria as an outpatient.
E. avium isolate not run on PFGE.
COPD, chronic obstructive pulmonary disease.
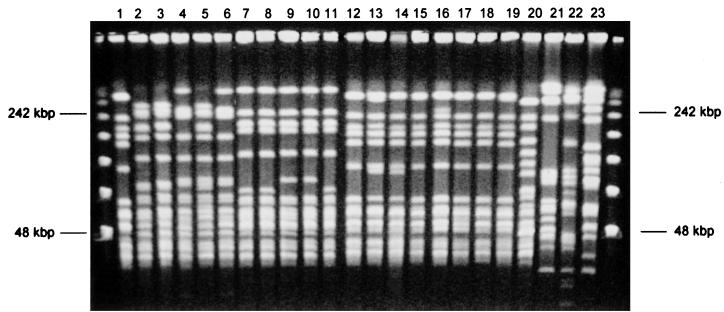
At hospital S1, swab samples were obtained from 78 patients from different wards; 9 (11.5%) patients were found to be colonized with GRE (Table 2). Five patients were colonized with E. faecium vanB (patients 2 to 6). Of these, patients 2 and 3 (Fig. 1, lanes 2 and 3) had identical strains, and these were very similar to the strain isolated from patient 5 (lane 5), with a two-band difference. Patients 4 and 6 (lanes 4 and 6) show identical fingerprinting patterns but had more than four band differences from the former strains. We therefore had three strains isolated from five patients, two strains being closely related and the third strain being more distantly related (26).
TABLE 2.
Results of GRE screening of 184 high-risk patients in four hospitals in Johannesburg
| Lane no.a | Isolate | Genotype | Restriction pattern on PFGE | MIC (μg/ml)
|
Hospital | Ward | ||
|---|---|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | Gentamicin | ||||||
| 1 | E. faecium | vanA | A | >64 | >64 | >64 | S1 | ICU |
| 2 | E. faecium | vanB | B | 64 | 2 | >64 | S1 | POb |
| 3 | E. faecium | vanB | B | >64 | 2 | >64 | S1 | PO |
| 4 | E. faecium | vanB | C | >64 | 2 | 4 | S1 | PO |
| 5 | E. faecium | vanB | B1 | 32 | 1 | 32 | S1 | PO |
| E. gallinarum | vanC1 | NAc | 16 | 1 | 2 | S1 | PO | |
| 6 | E. faecium | vanB | C | >64 | 2 | >64 | S1 | Renal OPDd |
| E. gallinarum | vanC1 | NA | 16 | 0.5 | 4 | S1 | Renal OPD | |
| E. avium | vanA | NA | >64 | >64 | >64 | S1 | Renal OPD | |
| E. gallinarum | vanC1 | NA | 1 | 0.25 | 1 | S2 | Burn unit | |
| 7 | E. faecium | vanB | D | 32 | 1 | 64 | P1 | ICU |
| 8 | E. faecium | vanB | D | 32 | 0.5 | 64 | P1 | ICU |
| 9 | E. faecium | vanB | D1 | 32 | 1 | 32 | P1 | Renal OPD |
| 10 | E. faecium | vanB | D1 | 32 | 2 | >64 | P1 | Renal OPD |
| 11 | E. faecium | vanB | D | 32 | 1 | 32 | P1 | Renal OPD |
| E. gallinarum | vanC1 | NA | 16 | 1 | 8 | P1 | Renal OPD | |
| E. gallinarum | vanC1 | NA | 16 | 1 | 32 | P2 | ICU | |
| 12 | E. faecium | vanA | A | >64 | >64 | >64 | P2 | ICU |
| 13 | E. faecium | vanA | A3 | >64 | >64 | >64 | P2 | ICU |
| E. gallinarum | vanC1 | NA | 16 | 0.25 | 4 | P2 | ICU | |
Lane in Fig. 1.
PO, pediatric oncology (inpatients and outpatients).
NA, not applicable; macrorestriction analysis of E. gallinarum and E. avium isolates was not performed.
Renal OPD, renal patients on hemodialysis.
FIG. 1.
Macrorestriction analysis of GRE isolates. Lanes 1 through 13, isolates from patients 1 through 13 as identified in Tables 1 and 2; lanes 14 to 19, E. faecium vanA (type A) isolated from S1 in 1997 and four different private hospitals in 1998; lane 20, E. faecium vanA (ATCC 51559); lanes 21 and 22, E. faecalis vanA isolates from S1 and S2; lane 23, E. faecalis vanA (ATCC 51299). The leftmost and rightmost lanes contain molecular size markers (PFGE marker 1 [λ ladder]; Boehringer GmbH, Mannheim, Germany).
Patient 1 was colonized with the same strain of E. faecium vanA as was isolated in March 1997 from a patient in S1 (Fig. 1, lane 14) and in hospital P3 (a privately run hospital not included in the surveillance study) before transfer (lane 15). Each of these strains has a one band variation, possibly as a consequence of repeated subculturing. This strain has subsequently been repeatedly isolated from a patient at a fourth private hospital, P4. The patient died 2 weeks after the first isolate was identified from a tracheal aspirate (lane 16). A second patient from the same hospital was also colonized with this strain a month later (lane 17). Two more private hospitals have yielded this strain from clinical specimens in patients thought only to be colonized (lanes 18 and 19). This strain was also isolated from most patients involved in an outbreak in hospital S1 (K. McCarthy, W. van Nierop, A. Dusé, W. Bezwoda, A. von Gottberg, M. Kassel, O. Perovic, and R. Smego, submitted for publication). One patient in hospital S1 was colonized with an E. avium strain with a vanA genotype.
Hospital S2 had only one patient (1.8%) colonized with GRE, an E. gallinarum strain. At hospital P1 a total of six patients (27%) were colonized with GRE. Five patients were colonized with E. faecium vanB; three of these patients carried an identical strain (patients 7, 8, and 11 [Fig. 1, lanes 7, 8, and 11]), while the remaining two patients (lanes 9 and 10) carried the same strain, which differed from the former by only two bands. Four patients (13.8%) in total were colonized with GRE in hospital P2. Two patients (lanes 12 and 13) were colonized with E. faecium vanA, and these two strains differed only by one band in pulse-field gel electrophoresis (PFGE).
Of the variables recorded, frequency of the isolation of GRE from different hospitals was statistically significant, the recovery of GRE from hospital P1 being the highest and the yield from S2 being the lowest (P = 0.011). The difference between state hospitals and private hospitals was also statistically significant: 10 of 133 (7.5%) patients in state hospitals were colonized, compared to 10 of 51 (19.6%) in private hospitals (P = 0.036). Females were more likely to be colonized with glycopeptide-resistant organisms than males (P = 0.04). History of diarrhea (3 of 11 patients with history of diarrhea versus 17 of 173 without [P = 0.10]), hemodialysis (7 of 77 versus 13 of 106 [P = 0.66]), and current or previous antibiotic use within 4 weeks of the rectal swab (14 of 105 versus 6 of 78 [P = 0.33]) did not show any statistical correlation with GRE carriage. The mean number of days in the ward for inpatients (96 patients) colonized with GRE was 19.3, and that for those not colonized was 27.2 (P = 0.386). Other variables, such as HIV status, renal transplantation, malignancy, history of surgery, and catheterization, showed no association with GRE colonization.
Discussion.
This study of GRE strains and their characterization with macrorestriction analysis has highlighted the colonization patterns of E. faecium of the vanB and vanA genotypes in our hospitals and their potential for causing disease outbreaks in high-risk patients. Although E. faecium vanA is more commonly reported in outbreaks, the importance of E. faecium vanB as a nosocomial pathogen has been previously described (2, 19, 20). We documented E. faecium vanB rectal colonization only in this study, with no clinically significant isolates yet identified in our hospitals.
We were able to demonstrate clonal spread of vanA and vanB strains within different hospitals, with possible interhospital spread and persistence of one E. faecium vanA strain (type A) within hospitals in the city, as has been described for other centers (8, 12, 22, 24). Spread of a strain from one hospital (P3) to a second hospital (S1) was documented when a patient who was colonized a vanA strain was transferred in 1997 to the ICU in hospital S1. This strain was isolated from a rectal swab from a patient in the same ward during the prevalence study and has been isolated from clinical specimens from three other hospitals in Johannesburg (Fig. 1, lanes 16 to 19). Subsequent to this surveillance study there was an outbreak in hospital S1 with the above-mentioned strain, isolated from clinical specimens from four patients in the affected ward (McCarthy et al., submitted).
The persistence of the type A strain over a period of more than 1 year, its widespread distribution (six different hospitals in Johannesburg during 1997 and 1998), and its predominance among all clinical isolates suggest the possibility of increased virulence and/or adaptation to spread. Similar instances of clonal spread of GRE have been described (2, 5, 12, 24), but such spread is poorly understood. Clonal spread is also seen once GRE colonization is well established in a particular center and may play a role in maintaining the endemicity of GRE (16).
No E. faecalis vanB strain has as yet been isolated in our diagnostic laboratories in Johannesburg; this may be due to the difficulty of identifying vanB isolates, especially if these have intermediate to low levels of resistance (6, 7, 25).
The inability to document significant clinical and epidemiological risk factors in this study, including no significant correlation with previous antibiotic use, may lie in the bias of our patient selection and the small numbers of patients who were colonized with GRE. Only high-risk patients in high-risk hospital settings were considered for rectal swabs to increase the possible yield and save costs, thus reducing the ability to differentiate risk factors that may have been associated with GRE colonization. The significantly higher prevalence of GRE in the two private hospitals suggests that such hospitals, which are less restricted financially and have fewer constraints on antibiotic usage, may be at greater risk. Rates of colonization may also be less representative due to limited sampling. Prevalence rates could also have been higher had we used the more sensitive broth enrichment technique. The prevalence of colonization documented in this study refers only to high-risk patients in tertiary-care hospitals, where patient profiles and antibiotic usage are very different from those in smaller or rural hospitals in South Africa.
Elsewhere in South Africa, four clinical isolates of E. faecalis vanB and one strain of E. gallinarum were isolated in a hospital in Bloemfontein, Free State, in 1995 (7). In a hospital in Cape Town, an E. faecium vanA strain was isolated in 1993 from a clinical specimen (7). Another large university hospital in Cape Town yielded only two E. gallinarum isolates when 230 consecutive clinical isolates from 1997 were screened by disk susceptibility testing, E-test quantitative MIC testing, and PCR (7).
Our findings confirm the potential for interhospital spread of GRE and highlight the importance of developing appropriate infection control protocols for early implementation (14). The media for screening for vancomycin-resistant enterococci may be successfully utilized for routine surveillance, even in smaller hospital laboratories, without much additional cost. Measures such as prudent antibiotic use, contact isolation, adequate handwashing by health-care workers, and decontamination of environmental surfaces and contaminated items should be adopted to prevent escalation of this nosocomial problem. Such measures have been successful in some outbreak interventions (2), although other authors have documented no decrease in endemic colonization or infections despite their implementation (21).
In conclusion, although data are limited, South Africa is presently experiencing an early stage in the epidemiology of GRE, with patients in certain hospitals colonized with single clones of resistant enterococci, while other large academic centers have yet to describe GRE infections (7). The rapid evolution of this epidemic is predicted, with one strain, E. faecium vanA type A, already exhibiting interhospital spread.
Acknowledgments
We thank Nancye Clark from the Centers for Disease Control and Prevention and Daniel Monget from Biomerieux S.A., Marcy L'Etoile, France, for assistance in the identification of organism 73 as E. avium and in the confirmation of the presence of the genotype vanA; thanks are also due to Heidi Toxopeus, Thora Capper, Marshagne Smith, and Lesley McGee for technical assistance and to the staff and patients of the participating hospitals.
REFERENCES
- 1.Bell J M, Paton J C, Turnidge J. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J Clin Microbiol. 1998;36:2187–2190. doi: 10.1128/jcm.36.8.2187-2190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J M, Opal S M, Chow J W, Zervos M J, Potter-Bynoe G, Sherman C B, Romulo R L C, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budavari S M, Saunders G L, Liebowitz L D, Khoosal M, Crewe-Brown H H. Emergence of vancomycin-resistant enterococci in South Africa. S Afr Med J. 1997;87:1557. [PubMed] [Google Scholar]
- 4.Carvalho M D G S, Teixeira L M, Facklam R R. Use of tests for acidification of methyl-α-d-glucopyranoside and susceptibility to efrotomycin for differentiation of strains of Enterococcus and some related genera. J Clin Microbiol. 1998;36:1584–1587. doi: 10.1128/jcm.36.6.1584-1587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow J W, Kuritza A, Shaes D M, Green M, Sahm D F, Zervos M J. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–1611. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cookson S T, Lopardo H, Marin M, Arduino R, Rial M J, Altschuler M, Galanternik L, Swenson J M, Tokars J I, Jarvis W R. Study to determine the ability of clinical laboratories to detect antimicrobial-resistant Enterococcus spp. in Buenos Aires, Argentina. Diagn Microbiol Infect Dis. 1997;29:107–109. doi: 10.1016/s0732-8893(97)00099-0. [DOI] [PubMed] [Google Scholar]
- 7.Derby P, Allan B, Lambrick M, Gay Elisha B. Detection of glycopeptide-resistant enterococci using susceptibility testing and PCR. S Afr J Epidemiol Infect. 1998;13:66–69. [Google Scholar]
- 8.Dunne W M, Wang W. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J Clin Microbiol. 1997;35:388–392. doi: 10.1128/jcm.35.2.388-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French G L. Enterococci and vancomycin resistance. Clin Infect Dis. 1998;27(Suppl. 1):S75–S83. doi: 10.1086/514910. [DOI] [PubMed] [Google Scholar]
- 12.Fridkin S K, Yokoe D S, Whitney C G, Onderdonk A, Hooper D C. Epidemiology of a dominant clonal strain of vancomycin-resistant Enterococcus faecium at separate hospitals in Boston, Massachusetts. J Clin Microbiol. 1998;36:965–970. doi: 10.1128/jcm.36.4.965-970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordts B, Van Landuyt H, Ieven M, Vandamme P, Goossens H. Vancomycin-resistant enterococci colonizing the intestinal tracts of hospitalized patients. J Clin Microbiol. 1995;33:2842–2846. doi: 10.1128/jcm.33.11.2842-2846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen B J. Screening specimens for vancomycin-resistant enterococcus. Lab Med. 1996;27:53–55. [Google Scholar]
- 16.Kim W, Weinstein R A, Hayden M K. The changing molecular epidemiology and establishment of endemicity of vancomycin resistance in enterococci at one hospital over a 6-year period. J Infect Dis. 1999;179:163–171. doi: 10.1086/314564. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 18.Marcus N, Peled N, Yagupsky P. Rapid increase in the prevalence of antimicrobial drug resistance among enterococcal blood isolates in southern Israel. Eur J Clin Microbiol Infect Dis. 1997;16:913–915. doi: 10.1007/BF01700558. [DOI] [PubMed] [Google Scholar]
- 19.McNeeley D F, Brown A E, Noel G J, Chung M, de Lancastre H. An investigation of vancomycin-resistant Enterococcus faecium within the pediatric service of a large urban medical center. Pediatr Infect Dis J. 1998;17:184–188. doi: 10.1097/00006454-199803000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Moreno F, Grota P, Crisp C, Magnon K, Melcher G P, Jorgensen J H, Patterson J E. Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in southern Texas. Clin Infect Dis. 1995;21:1234–1237. doi: 10.1093/clinids/21.5.1234. [DOI] [PubMed] [Google Scholar]
- 21.Morris J G, Jr, Shay D K, Hebden J N, McCarter R J, Jr, Perdue B E, Jarvis W, Johnson J A, Dowling T C, Polish L B, Schwalbe R S. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Establishment of endemicity in a university medical center. Ann Intern Med. 1995;123:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 22.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Callaghan C H, Morris A, Kirby S M, Shingler A H. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pegues D A, Pegues C F, Hibberd P L, Ford D S, Hooper D C. Emergence and dissemination of a highly vancomycin-resistant vanA strain of Enterococcus faecium at a large teaching hospital. J Clin Microbiol. 1997;35:1565–1570. doi: 10.1128/jcm.35.6.1565-1570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover F C. Laboratory methods for surveillance of vancomycin-resistant enterococci. Clin Microbiol Newsl. 1998;20:1–5. [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B A, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uttley A H C, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]