Abstract
Flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. utilis Tsen et Lee) is an important and extensively cultivated vegetable in south China, and its stalk development is mainly regulated by gibberellin (GA). DELLA proteins negatively regulate GA signal transduction and may play an important role in determining bolting and flowering. Nevertheless, no systematic study of the DELLA gene family has been undertaken in flowering Chinese cabbage. In the present study, we found that the two-true-leaf spraying of gibberellin A3 (GA3) did not promote bolting but did promote flowering, whereas the three-true-leaf spraying of GA3 promoted both bolting and flowering. In addition, we identified five DELLA genes in flowering Chinese cabbage. All five proteins contained DELLA, VHYNP, VHIID, and SAW conserved domains. Protein-protein interaction results showed that in the presence of GA3, all five DELLA proteins interacted with BcGID1b (GA-INSENSITIVE DWARF 1b) but not with BcGID1a (GA-INSENSITIVE DWARF 1a) or BcGID1c (GA-INSENSITIVE DWARF 1c). Their expression analysis showed that the DELLA genes exhibited tissue-specific expression, and their reversible expression profiles responded to exogenous GA3 depending on the treatment stage. We also found that the DELLA genes showed distinct expression patterns in the two varieties of flowering Chinese cabbage. BcRGL1 may play a major role in the early bud differentiation process of different varieties, affecting bolting and flowering. Taken together, these results provide a theoretical basis for further dissecting the DELLA regulatory mechanism in the bolting and flowering of flowering Chinese cabbage.
Keywords: flowering Chinese cabbage, DELLA, gibberellin signaling, bolting and flowering, flower bud differentiation
1. Introduction
Flowering Chinese cabbage is a subspecies of Chinese cabbage that originated in south China. It is now planted throughout the country owing to the increasing consumer demand [1]. The major food product of flowering Chinese cabbage is the stalk, the development of which is directly related to plant quality and yield [2]. Stem elongation, thickening, and flowering are key characteristics of flowering Chinese cabbage stem development. Factors that affect the timing of the bolting and flowering of flowering Chinese cabbage include temperature and plant hormones [3,4,5]. Low temperature and exogenous gibberellin A3 (GA3) treatment advance bolting time, stem elongation, and flowering time. Moreover, they display positive additive effects. Low temperature accelerates bolting and flowering by inducing gibberellin (GA) content in the shoot apices; therefore, GA is the main regulator of flowering and bolting in flowering Chinese cabbage.
The regulatory function of GA in plant growth and development is mainly achieved through the GA signaling pathway. DELLA proteins act as the negative regulators of GA signal transduction [6]. GA-induced DELLA degradation is a central regulatory system in the GA signaling pathway [7,8,9]. DELLA proteins contain highly conserved DELLA and TVHYNP domains at the N-terminus, which are important for GA signal perception domains [10,11]. The middle region harbors a nuclear localization signal domain, with a conserved amino acid domain VHIID, leucine repeats LZ, and ploy S/T/V (serine, threonine, and valine). The C-terminus has a conserved GRAS domain, which is a functional structural domain of DELLA proteins and plays a role in regulating DELLA protein activity. VHIID and SAW in the GRAS domain are blocking structural domains.
The Arabidopsis genome has five DELLA subfamily members: GA insensitive (GAI), repressor of gal-3 (RGA), RGA-like 1 (RGL1), RGL2, and RGL3 [12,13]. These DELLA proteins belong to the plant-specific GRAS regulatory protein gene family [6,11]. GAI and RGA are crucial for the regulation of plant stem elongation growth in response to GA [13,14,15] and have a functional overlap in inhibiting plant elongation [16,17]. RGL1 and RGL2 play important roles in controlling flower bud differentiation and flower development, respectively [16,17,18,19]. RGL3 acts as a positive regulator in the defense response but performs a minor function in plant development processes [20,21]. These five DELLA proteins in Arabidopsis have both redundant and specific functions [21,22]. There are many studies on DELLA proteins in Arabidopsis; however, research on flowering Chinese cabbage is still lacking. Advances in research on GA signal transduction pathways in model plants, such as the molecular mechanism of DELLA proteins blocking plant growth and development and the model of GA depression, provide an important basis for studying the mechanism of DELLA proteins in flowering Chinese cabbage.
To understand the role of DELLA genes in the bolting and flowering of flowering Chinese cabbage, we isolated five DELLA family genes from flowering Chinese cabbage. We then analyzed their expression patterns in different flowering Chinese cabbage tissues and their response to GA3 and cold treatment as well as their expression levels in two different cultivars of flowering Chinese cabbage. Finally, we investigated the interaction between DELLA proteins and the GA receptor BcGID1a/b/c using yeast two-hybrid. These data laid the foundation for the further study of DELLA protein function in flowering Chinese cabbage.
2. Results
2.1. Identification of Key Stage for GA3 Sensitivity in Bolting and Flowering of Flowering Chinese Cabbage ‘youlv501’
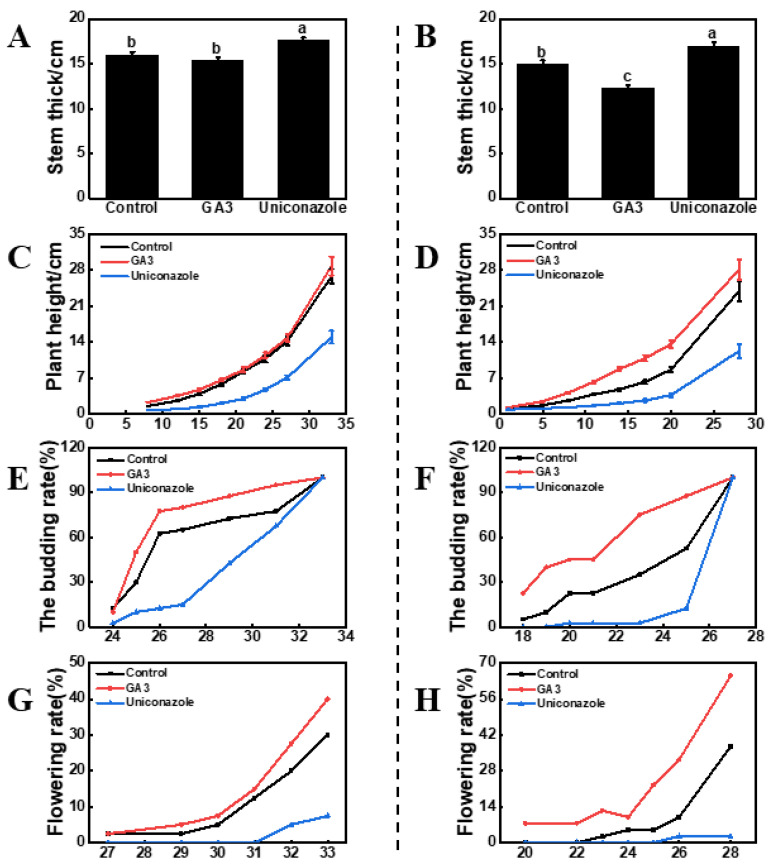
To identify the key stage of stalk development in flowering Chinese cabbage in response to GA3 treatments, we treated the plants at the two-true-leaf and three-true-leaf stages with GA3 and its inhibitor (uniconazole) separately. To check the effect of these treatments on plant bolting and flowering, we calculated stem diameter, plant height, budding rate, and flowering rate. As shown in Figure 1, there was a different effect on the stem diameters between the treated stages, and the three-true-leaf stage appeared to be more sensitive to GA3 treatment. After the two-true-leaf stage treatment, there was no significant difference in stem diameter between the GA3-treated and non-treated plants, whereas for the three-true leaf stage, the stem diameter of the GA3-treated plants was thinner than that of the control. Similar results were observed for plant height. After the two-true-leaf stage treatment, there was no significant difference in plant height between the GA3-treated and non-treated plants, whereas after the three-true-leaf stage treatment, the plant height of the GA3-treated plants was higher than that of the control. GA3 treatment had the same effect on the bolting and flowering of flowering Chinese cabbage, accelerating both bolting and flowering time regardless of the treatment stage (Figure 1). The three-true-leaf stage promoted bolting and flowering more significantly than the two-true-leaf stage. Regardless of the stage, the stem diameter was significantly increased, the plant height was obviously shorter, and the bolting and flowering rates were significantly delayed after uniconazole treatment. Overall, the three-true-leaf stage may be the key stage for stalk development.
Figure 1.
Effect of gibberellin A3 (GA3) and uniconazole spraying on the growth of flowering Chinese cabbage. (A,B) are at 33 and 28 days after treatment, respectively, which is also the harvesting period, and we measured the stem thickness. (A,C,E,G): two-true-leaf treatment, (B,D,F,H): three-true-leaf treatment. The units on the lower axis are the number of days after treatment in (C–F). The data represent an average of three replicates ± standard error. Values followed by the same letter are not significantly different using Duncan’s test at p < 0.05.
2.2. Identification and Expression of DELLA Genes in Flowering Chinese Cabbage ‘youlv501’
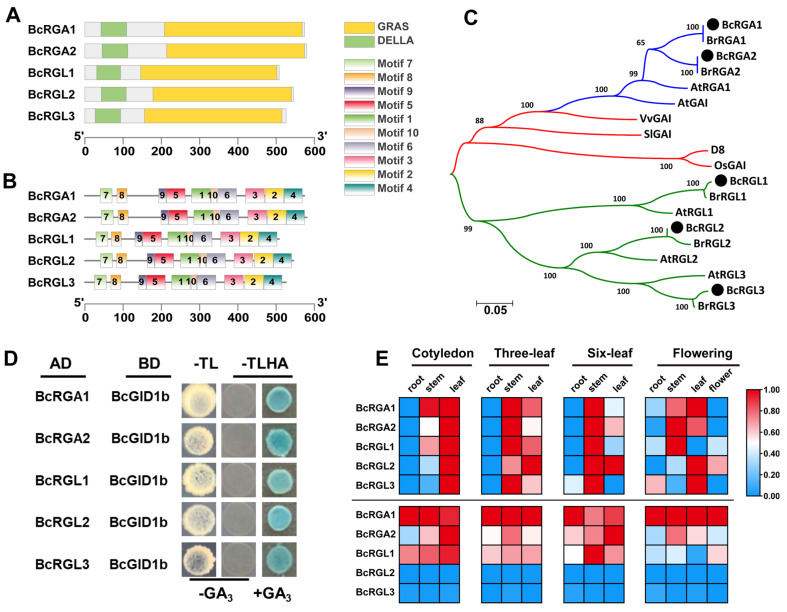
As GA regulates plant growth and development mainly through the GA signaling pathway, in which DELLA proteins act as key regulators, we characterized the DELLA proteins of flowering Chinese cabbage. An in silico search was performed in the Brassica Database (http://brassicadb.cn/#/BLAST/, accessed on 20 August 2021) using Arabidopsis DELLA protein RGA sequences as queries for BLAST searches. Five genes were predicted to encode the putative DELLA proteins. The full-length cDNA of the five DELLA genes was further confirmed by reverse transcription polymerase chain reaction (RT-PCR) amplification, indicating that the corresponding coding sequences ranged from 1524 to 1740 bp (Table 1, Figure S1 and Table S1). We used Expasy and Wolf Psort online software to analyze the physicochemical properties of the predicted proteins based on the sequence results. The physicochemical properties are shown in Table 1. All DELLA contained conserved DELLA and GRAS (Figure 2A). Multiple sequence alignment revealed that they also contained the DELLA, VHYNP, VHIID, and SAW domains (Figure S2). Motif searches in the MEME online program indicated that all DELLA contained the same conserved motif numbers (Table S2). Motif 7 and 8 represent the DELLA domain, whereas motifs 1, 2, 3, 4, 5, 6, 9, and 10 are in the GRAS domain (Figure 2B).
Table 1.
Physicochemical properties of DELLA genes in flowering Chinese cabbage.
| Gene | CDS (bp) |
AA | pI | MW (KD) |
Subcellular Localization |
|---|---|---|---|---|---|
| BcRGA1 | 1722 | 573 | 4.94 | 142.7 | chlo: 6, nucl: 6, cyto: 1 |
| BcRGA2 | 1740 | 579 | 4.93 | 143.4 | nucl: 6.5, nucl_plas: 4, chlo: 3, cyto: 3 |
| BcRGL1 | 1524 | 507 | 4.99 | 126.0 | nucl: 6, chlo: 3, cyto: 3, cysk: 2 |
| BcRGL2 | 1635 | 544 | 4.94 | 133.4 | chlo: 6, nucl: 3, cyto: 2, mito: 2 |
| BcRGL3 | 1578 | 525 | 4.95 | 129.4 | mito: 6, nucl: 3.5, nucl_plas: 3.5, plas: 2.5, chlo: 2 |
CDS: coding sequence, AA: amino acid, MW: molecular weight.
Figure 2.
Characterization of DELLA proteins in flowering Chinese cabbage. (A) Conserved domain of the DELLA genes. Green and yellow represent the DELLA and GRAS domain, respectively. (B) Conserved motifs of the DELLA genes. Different motifs are represented by different colored boxes with numbers 1−10. (C) Phylogenetic tree of DELLA proteins in flowering Chinese cabbage and other species, as determined by the neighbor-joining method. The protein IDs used in the phylogenetic tree are AtRGA1 (NP_178266.1), AtGAI (NP_172945.1), AtRGL1 (NP_176809.1), AtRGL2 (NP_186995.1), AtRGL3 (NP_197251.1), BrRGA1 (XP_009101333.1), BrRGA2 (XP_009114228.2), BrRGL1 (XP_009127484.1), BrRGL2 (XP_009130367.1), BrRGL3 (XP_009107522.1), SlGAI (NP_001234365.1), D8 (NP_001354393.1), OsGAI (XP_015631543.1), and VvGAI (XP_002284648.1). (D) Detection of interactions among DELLA proteins and BcGID1b using Y2H. Yeast cells with different construct combinations were grown on selective media without Trp and Leu (-TL) and were tested for interactions on selective media without Trp, Leu, His, and Ade (−TLHA). (E) Expression profiles of flowering Chinese cabbage DELLA genes in various organs. Above the line is a horizontal comparison, with each column exhibiting normalization independently, and below the line is a vertical comparison, with each row displaying normalization independently. GA3, gibberellin A3.
To adopt a nomenclature consistent with that of Arabidopsis DELLA proteins, we conducted phylogenetic analyses on different DELLA proteins from different plant sequenced genomes comprising Chinese cabbage, Arabidopsis thaliana, tomato, grape, rice, and maize sequences. The phylogenetic trees showed that the DELLA proteins in flowering Chinese cabbage had the closest relationship with Chinese cabbage, followed by Arabidopsis. The DELLA proteins in flowering Chinese cabbage were named according to their phylogenetic trees (Figure 2C). The evolutionary tree also showed that BcRGA1 and BcRGA2 are more closely related, whereas BcRGL1, BcRGL2, and BcRGL3 belong to one branch and are more closely related. Using the Arabidopsis protein database as a reference, online software STRING was used to predict the potential interaction proteins of DELLA family members. The results showed that BcRGA1, BcRGA2, BcRGL1, BcRGL2, and BcRGL3 interacted with the GID1A, GID1B, GID1C, PIF3, and SLY1 proteins (Figure S1B). To verify the interaction between the DELLA proteins and GA receptor GID1 in flowering Chinese cabbage, we performed yeast two-hybrid experiments in the presence or absence of GA3. The yeast double-hybrid result showed that the five DELLA proteins could only interact with BcGID1b in the presence of GA3, and none of the five DELLA proteins interacted with the GA receptors, BcGID1a and BcGID1c, either with or without GA3 (Figure 2D and Figure S1C,D). The expression patterns of the BcRGA1, BcRGA2, BcRGL1, BcRGL2, and BcRGL3 genes were examined in different tissues of flowering Chinese cabbage, including roots, stems, leaves, and flowers. The relative expression was calculated using the 2−ΔΔCt method. The results showed that all the DELLA genes were expressed in all the evaluated tissues. The expression profiles of these genes in the cotyledon, three-true-leaf, six-true-leaf, and flowering stages were similar, with high expression levels in the stems and leaves and low expression levels in the roots and flowers (Figure 2E and Figure S3). The expression levels of BcRGA1, BcRGA2, and BcRGL1 were higher than those of the other three genes (Figure 2E and Figure S3).
2.3. DELLA Genes Responded Differentially to GA3 between Two-True-Leaf Stage and Three-True-Leaf Stage Treatments
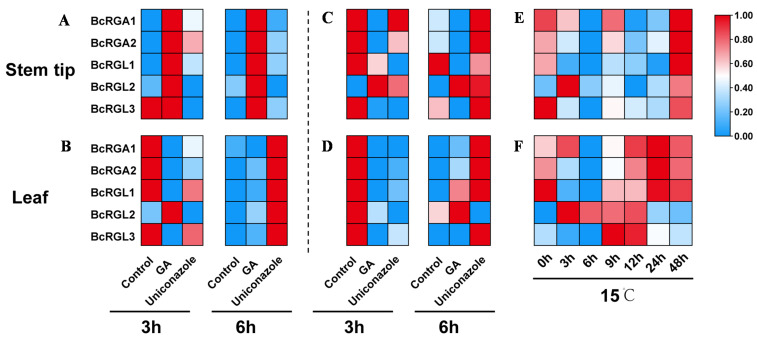
To determine whether the DELLA proteins respond to GA differentially between the two- and three-true-leaf stages, we examined the expression level of DELLA genes at these two stages after GA3 and uniconazole (GA inhibitor) treatment using RT-PCR-based expression analysis. The relative expression was calculated using the expression levels of the housekeeping gene Actin2/GAPDH and the 2−ΔΔCt method. The trend of DELLA genes expression in leaves was the same for both the two- and three-true-leaf treatments. The expression of DELLA genes was downregulated after 3 h of GA3 treatment and upregulated after 6 h of uniconazole treatment. In the stem tip, the expression pattern of the DELLA genes was different between the two- and three-true-leaf stage treatments. After two-true-leaf GA3 treatment, the expression of DELLA genes was upregulated, whereas the expression of the other DELLA genes, except BcRGL2, was downregulated after the three-true-leaf GA3 treatment (Figure 3, Figures S4 and S5).
Figure 3.
Expression profiles of flowering Chinese cabbage DELLA genes under gibberellin A3 (GA3), uniconazole, and 15 °C treatments. (A,B): two-true-leaf treatment, (C–F): three-true-leaf treatment. Each column displays normalization independently.
Low temperature promotes bolting and flowering of flowering Chinese cabbage ‘youlv501’ by inducing GA content. To evaluate whether temperature affects the DELLA genes, we treated flowering Chinese cabbage at 15 °C in the three-true-leaf stage. Most DELLA genes exhibited inhibition after 3 h of treatment, and the most significant downregulation in the expression was observed after 6 h of treatment. However, the expression of BcRGL2 was upregulated by the treatment (Figure 3E,F and Figure S6). Low temperature downregulated the expression of most of the DELLA genes, which was consistent with the spraying of GA3 in the three-true-leaf stage; this indicated that the three-true-leaf stage is the key stage for bolting and flowering, and DELLA genes play an important role in this process.
2.4. Bolting and Flowering-Related Genes Responded Differentially to GA between Two- and Three-True-Leaf Stage Treatments
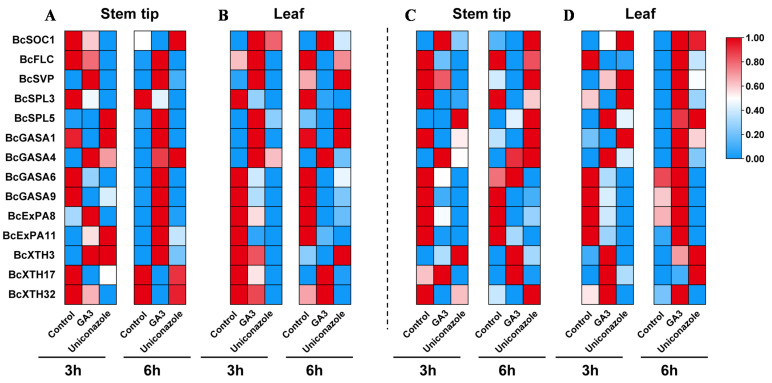
In combination with a previous study [2], we selected five flowering-related genes, SOC1, FLC, SVP, SPL3, and SPL5; five cell elongation-related genes, EXP8, EXP11, XTH3, XTH17, and XTH32; and four GA-regulated proteins, GASA1, -4, -6, and -9, to analyze the expression characteristics of these genes after GA3 treatment (Figure 4, Figures S7 and S8). SOC1 is a flowering promoter, and FLC and SVP are flowering repressors [23,24,25,26]. The expression of SOC1 was increased in the leaves and stem tips after GA3 treatment, except in the stem tips of the two-true-leaf stage. SPL is an interactive DELLA protein [27]. The expression of SPL3 and SPL5 was significantly different. After GA3 treatment, the expression of SPL3 was downregulated, whereas the expression of SPL5 was either upregulated or unchanged. The expression of GA-regulated proteins also differed, with GA3 treatment upregulating the expression of GASA4 and downregulating the expression of GASA1, -6, and -9. Expansin proteins (ExPA) and xyloglucan endotransferases (XTH) are key factors in cell elongation. The expression of XTH3 and XTH17 was increased by GA3 treatment, especially in the three-true-leaf stage.
Figure 4.
Expression profiles of flowering and elongation genes in flowering Chinese cabbage. (A,B): two-true-leaf treatment, (C,D): three-true-leaf treatment. Each column displays normalization independently. GA3, gibberellin A3.
Concomitantly, we analyzed the correlation of these genes with the DELLA genes in leaves and stem tips during different treatments with GA3 (Figure S9). The correlations between genes varied at different stages due to changes in gene expression. The correlation between genes was not significant in the two-true-leaf stage stem tips. In the three-true-leaf stage stem tips, BcRGA1 was significantly and negatively correlated with BcXTH17. In the two-true-leaf stage leaves, all the DELLA genes were positively correlated with BcSPL5 and BcXTH3. In the three-true-leaf stage leaves, BcRGL2 was significantly and positively correlated with BcGASA6, -9, BcExPA8, -11, and BcXTH32.
2.5. Five DELLA Genes Showed Distinct Expression Patterns in Two Varieties of Flowering Chinese Cabbage
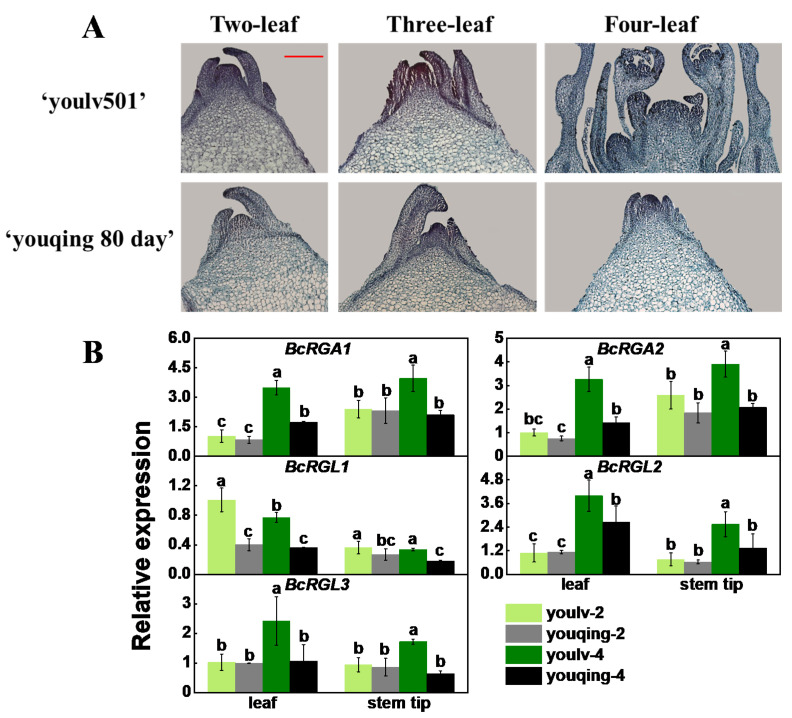
An inappropriate sowing season may cause abnormal bolting and flowering of flowering Chinese cabbage; therefore, various varieties suited to different seasons have been developed. ‘youlv501’ and ‘youqing 80 day’ are two varieties of flowering Chinese cabbage, which are an early-maturing variety and a late-maturing variety, respectively. Using paraffin sections, we observed the stem tip structure of two varieties of flowering Chinese cabbage. From the horizontal section of the stem tips, we found that ‘youlv501’ has larger pith cells than ‘youqing 80 day’ at the three- and four-true-leaf stages (Figure S10). As illustrated in the longitudinal section of the stem tips, ‘youlv501′ developed faster than ‘youqing 80 day’ at all three stages, especially during the four-true-leaf stage. In the two-true-leaf stage, both varieties did not complete the flower bud differentiation; however, in the four-true-leaf stage, the flower bud differentiation of ‘youlv501’ was completed (Figure 5A). The expression levels of all the DELLA genes were significantly different at the four-true-leaf stage, whereas only the expression level of BcRGL1 was significantly different at the two-true-leaf stage (Figure 5B), indicating its potential role in the bolting and flowering of flowering Chinese cabbage in these cultivar variations.
Figure 5.
Expression analysis of DELLA genes in the ‘youlv501’ and ‘youqing 80 day’ varieties of flowering Chinese cabbage. (A) Stem tip longitudinal structures of ‘youlv501’ and ‘youqing 80 day’ flowering Chinese Cabbage. All the pictures were taken under a 10 × microscope, and the scale was unified at 200 μm. (B) Among them, youlv-2 and youqing-2 represent the two-true-leaf stage of ‘youlv501’ and ‘youqing 80 day’ respectively. youlv-4, and youqing-4 represent the four-true-leaf stage of ‘youlv501’ and ‘youqing 80 day’, respectively. The data represents an average of three replicates ± standard error. Values followed by the same letter are not significantly different using Duncan’s test at p < 0.05.
3. Discussion
We identified five DELLA genes (BcRGA1, BcRGA2, BcRGL1, BcRGL2, and BcRGL3) in flowering Chinese cabbage, which are members of the plant-specific GRAS family. All five DELLA contain the DELLA, TVHYNP, VHIID, and SAW domains [10,11,19]. These domains have been implicated in the perception of GA and the subsequent destabilization of DELLA proteins [16,28,29,30]. Mutations to either of these domains can result in a semi-dominant, GA-insensitive, and dwarfed phenotype in various plant species [15,31,32]. Arabidopsis and Chinese cabbage have five members of the DELLA subfamily, whereas monocots, such as rice and barley, have only one [15,19,33]. The names of the individual DELLA introduced here are intended to imply functional homology to specific Arabidopsis DELLA proteins.
The interaction between GID1 and DELLA proteins in rice and plum is dependent on GA [34,35]. In addition, GID1 interacts with DELLA proteins even in the absence of GA, and the presence of GA enhances the interaction between GID1 and DELLA proteins [36]. Therefore, GA-dependent and -independent pathways are present in plants [37]. In our study, the DELLA proteins only interacted with the GA receptor BcGID1b in the presence of GA3, probably because BcGID1b has a greater binding ability to GA than BcGID1a and BcGID1c [38]. Gene knockouts, overexpression of individual DELLA genes, and further characterization of the DELLA proteins will help elucidate the role of these proteins in the development of flowering Chinese cabbage.
The transcript levels of four DELLA genes in cucumber have been analyzed in different tissues and organs, including roots, stems, leaves, male flower buds, female flower buds, and fruits, and all four genes are expressed in the examined tissues, with the highest expression in stems and male flower buds [39]. In Arabidopsis, five DELLA genes are expressed in different tissues, and the expression of RGA1, RGA2, and RGL1 is higher than that of RGL2 and RGL3, similar to the expression profiles of the DELLA genes in flowering Chinese cabbage [40]. We analyzed the expression of five DELLA genes in the roots, stems, leaves, and flowers of flowering Chinese cabbage found that the DELLA genes were mainly expressed in the stem tips and leaves, and the highly expressed genes were consistent with those in Arabidopsis. It is noteworthy that during the cotyledon stage, expression was mainly present in the leaves, whereas during the three- and six-true-leaf stages, the expression was higher in the stems than in the leaves. It is possible that DELLA genes act mainly in the stem tip as flowering Chinese cabbage enters the bud differentiation and rapid growth phase.
GA promotes bolting and flowering in mustard, radish, and Chinese cabbage, but the results vary among varieties, different stages, and different environmental conditions [41,42,43,44]. Uniconazole (a GA biosynthesis inhibitor) can inhibit bolting and flowering and improve yield in purple kale and cauliflower [41]. In flowering Chinese cabbage, two-true-leaf spraying of GA3 did not promote bolting but did promote flowering, whereas three-true-leaf spraying of GA3 promoted bolting and flowering. Uniconazole significantly inhibited bolting and flowering in the two- and three-true-leaf treatments. This indicates that GA3 can affect bolting and flowering, but the effect of GA3 depends on the treatment stage. The experiment was conducted with an early maturing variety, ‘youlv501′, which may have initiated floral bud differentiation earlier. In the two-true-leaf stage, flowering Chinese cabbage mainly undergoes floral bud differentiation, and the main effect of GA may affect floral bud differentiation, thus affecting plant flowering. In contrast, during the three-true-leaf stage, in addition to floral bud differentiation, the bolting signal is initiated; thus, GA may also affect stem elongation and flowering. At the three-true-leaf stage, the expression of the DELLA genes, except BcRGL2, was downregulated after 3 h of GA treatment. This indicates that BcRGA1, BcRGA2, BcRGL1, and BcRGL3 may inhibit stem growth and development, whereas GA3 treatment, which degrades them, initiates stem elongation and development. These findings are consistent with the current understanding of DELLA proteins as GA-responsive repressors of plant growth [17]. RGL2 is a negative regulator of GA responses that acts specifically to control seed germination rather than stem elongation [19]. We found that BcRGL2 responded differently to GA than the other four genes, suggesting that BcRGL2 may have a different function in flowering Chinese cabbage.
GA affects flowering Chinese cabbage bolting and flowering through DELLA proteins, but the exact molecular mechanism is not clear. We searched for the relationship between DELLA genes, cell elongation, and flower-related genes by detecting their expression and correlation. In the three-true-leaf stage stem tips, GA3 treatment caused the downregulation of the expression of DELLA genes, increased the expression of flowering-promoting factor BcSOC1, and decreased the expression of flowering-inhibiting factors BcFLC and BcSVP. Furthermore, GA3 treatment upregulated the expression of BcGASA4, BcXTH3, and BcXTH17. Previous studies have shown that DELLA proteins interact with FLC and SPL to repress the flowering transition [23,27]. GA induces the expression of EXPA and XTH genes through signal transduction pathways to promote cell wall relaxation and thus cell elongation [45,46]. Overexpression of AtGASA6 promotes cell elongation [47]. Therefore, we hypothesized that GA3 spraying causes the degradation of DELLA, BcFLC, and BcSPL3 and upregulates BcSOC1 expression, thus promoting flowering in flowering Chinese cabbage. Furthermore, GA3 spraying upregulated BcGASA4 expression and promoted BcXTH3 and BcXTH17 gene expression, which promotes cell wall relaxation and ultimately the elongation of flowering Chinese cabbage stems. Based on the Pearson correlation, BcRGA1 was significantly and negatively correlated with BcXTH17. It is possible that BcRGA1 and BcXTH17 play a key role in the stem growth of flowering Chinese cabbage, and follow-up studies should be conducted to verify this role.
In general, flower bud differentiation starts in the two- and three-true-leaf stages, and the flower bud differentiation stage varies among different varieties [48]. The early and late development of flower buds is different for the three varieties with different maturities, and they all start flower bud differentiation in the two- or three-true-leaf stages, but the development speed is generally early maturity < medium maturity < late maturity. ‘youlv501’ and ‘youqing 80 day’ are the two varieties of flowering Chinese cabbage, representing an early-maturing variety and a late-maturing variety, respectively. To investigate the relationship between DELLA genes and bud differentiation, we used the early-maturing variety ‘youlv501’ and the late-maturing variety ‘youqing 80 day’ for the experiment. Paraffin section results showed that ‘youlv501’ developed faster than ‘youqing 80 day’ at all stages. The quantitative results showed that BcRGL1 was significantly expressed at the two-true-leaf stage. RGL1 appears to be a more diverse negative regulator in Arabidopsis and is associated with seed germination, stem elongation, leaf expansion, and flower development [49]. Nevertheless, BcRGL1 may play a major role in the early bud differentiation process of different varieties, affecting plant bolting and flowering in flowering Chinese cabbage.
4. Materials and Methods
4.1. Cloning of DELLA Genes in Flowering Chinese Cabbage
As Arabidopsis DELLA genes have been reported previously [12], all the protein sequences from these two gene families were extracted from the Arabidopsis Information Resource database (https://www.arabidopsis.org/, accessed on 10 August 2021) and used as queries for a BLASTP search, with an e-value threshold of <1 in the Brassica Database (http://brassicadb.cn/#/BLAST/, accessed on 10 August 2021). Five DELLA genes were found in Brassica cabbage, and primers were designed based on their sequences (Table S3).
4.2. Phylogenetic and Sequence Analysis of DELLA Genes in Flowering Chinese Cabbage
The DELLA sequences of some other species and flowering Chinese cabbage were subjected to multiple sequence alignment. Multiple sequence alignment was conducted using Muscle with default parameters, and the results were used to construct a neighbor-joining phylogenetic tree in MEGA7.0, with the bootstrap value set to 1000 [50].
Conserved motifs were analyzed using the Multiple Em for Motif Elicitation (MEME) online program (http://meme-suite.org, accessed on 15 August 2021) with the following parameters: the number of repetitions was set to zero or one, and the maximum number of motifs was 10 [51]. The conserved domain of DELLA was analyzed using the NCBI Batch CDD online program (https://www.ncbi.nlm.nih.gov/Structrue/bwrpsb/bwrpsb.cgi, accessed on 15 August 2021). Conserved motifs and gene domains were visualized using TBtools software [52]. pI/MW was computed using the Expasy online program (https://web.expasy.org/compute_pi/, accessed on 15 August 2021) [53]. Protein subcellular localization was predicted using Wolf Psort (https://wolfpsort.hgc.jp/, accessed on 15 August 2021) [54].
4.3. Interaction Analysis of DELLA Proteins
Protein interaction analysis prediction for DELLA proteins was completed using the website STRING 11.5 (https://string-db.org/, accessed on 20 August 2021) [55]. PPI maps were visualized using Cytoscape 3.8.2 software [56]. The full-length coding sequences of five DELLA and BcGID1s genes were amplified from a mixture of stem and leaf cDNA. After amplification (95 °C for 2 min, followed by 35 cycles at 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min), the DELLA and BcGID1s genes were cloned into the pGAD and pDBD vectors (Clontech, Takara Bio Inc., Shiga, Japan), respectively. The yeast strain AH109 (Clontech) was used for transformation. Diploids were selected on a medium lacking Trp and Leu (TL), and interactions were validated using HIS3 and ADE2 reporter genes in a medium lacking Trp, Leu, His, and Ade (TLHA). Manipulation and analysis of Y2H were performed according to the manufacturer’s instructions in the Clontech Yeast Protocols Handbook, and all experiments were repeated three times independently.
4.4. Plant Growth Conditions, GA3, and Cold Treatment
Flowering Chinese cabbage plants were planted in pots with perlite and cultivated in the glasshouse of college horticulture at the South China Agriculture University. The growth conditions were as follows: 14 h-day/10 h-night cycles, 25/20 °C day/night temperature, 70–80% relative humidity, and 300 mol m−2 s−1 intense light.
The ‘youlv501′ flowering Chinese cabbage was used as the experimental material in the treatments with GA and its inhibitor (uniconazole). When the young seedlings were at the two- and three-true-leaf stages, plants were sprayed with GA3 (200 mg/L) and uniconazole (10 mg/L). Shoots (including the stem tip and leaf tissue) were collected from both the treated and control plants at 0, 3, and 6 h. In the low-temperature treatment, when the young seedlings were at the three-true-leaf stage, plants were treated at 15 °C, and shoots (including the stem tip and leaf tissue) were collected from both the treated and control plants at 0, 3, 6, 9, 12, 24, and 48 h. ‘Youlv501’ and ‘youqing 80 day’ were used as the experimental material to evaluate different varieties. Shoots (including the stem tip and leaf tissue) were collected when the young seedlings were at the two- and three-true-leaf stages. Three biological replicates were used for each condition, and ten seedlings were used for each biological replicate. All samples were subsequently frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
4.5. Histological Analysis
The stem tips (5 mm) of different varieties (‘youlv501’ and ‘youqing 80 day’) were collected and immersed in formaldehyde alcohol acetic acid solution (70% alcohol: acetic acid: formaldehyde = 90:5:5), placed under a vacuum for 20 min, and incubated for 48 h at 4 °C. The samples were then dehydrated in a gradient of ethanol and embedded in paraffin. Subsequently, 8 mm thick sections were stained with reddish-green stain and observed under a microscope.
4.6. RNA Isolation and RT-PCR Analysis
Total RNA was isolated from three replicates of each sample using the Promega Plant RNA Kit, which contained a genomic DNA elimination step. Total RNA was quantified spectrophotometrically by measuring the absorbance at 260 nm and the absorbance ratio of 260/280 nm using a Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA) and 2% w/v agarose gel. First-strand cDNA was reverse transcribed using an Eastep® RT Master Mix kit (Promega Corporation, Madison, WI, USA). Gene-specific primers were designed, and their sequences are listed in Table S3.
To determine the expression levels of the genes, fluorescent quantitative PCR was performed using a LightCycler 480 real-time PCR system (Roche, Basel, Switzerland) with SYBR Premix Ex Taq (Takara Bio Inc., Shiga, Japan), and the relative gene expression was calculated using the expression levels of the housekeeping gene Actin2/GAPDH and the 2−ΔΔCt method [57]. Gene expression profiles were visualized as a heatmap using TBtools software [52].
5. Conclusions
In this study, we isolated five DELLA genes from flowering Chinese cabbage, all of which interacted with the GA receptor BcGID1b in the presence of GA3. Based on the different morphologies of the two- and three-true-leaf stage GA3 treatments on the stem diameters and the reversible expression trend of DELLA genes, we believe that the three-true-leaf stage is likely the key stage for bolting regulation. Moreover, BcRGL1, which displayed variable expression between the early-maturing variety ‘youlv501’ and the late-maturing variety ‘youqing 80 day’ at the two-true-leaf stage, may be the key factor regulating bolting of flowering Chinese cabbage.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222212092/s1.
Author Contributions
H.G., X.H., Y.Z. and B.X. performed the experiments; Y.H., X.H. and R.C. participated in the design of the study; H.G. and Y.H. analyzed the data; H.G. wrote the manuscript; B.X., R.C., S.S., H.L. and Y.H. assisted in revising the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (32072656, 31972481), Key-Area Research and Development Program of Guangdong Province, China (2020B0202010006), and China Agriculture Research System of MOF and MARA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available in a publicly accessible repository.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lin L.-Z., Harnly J.M. Phenolic component profiles of mustard greens, Yu Choy, and 15 other Brassica vegetables. J. Agric. Food Chem. 2010;58:6850–6857. doi: 10.1021/jf1004786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang X., Lei Y., Guan H., Hao Y., Liu H., Sun G., Chen R., Song S. Transcriptomic analysis of the regulation of stalk development in flowering Chinese cabbage (Brassica campestris) by RNA sequencing. Sci. Rep. 2017;7:15517. doi: 10.1038/s41598-017-15699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song S., Lei Y., Huang X., Su W., Chen R., Hao Y. Crosstalk of cold and gibberellin effects on bolting and flowering in flowering Chinese cabbage. J. Integr. Agric. 2019;18:992–1000. doi: 10.1016/S2095-3119(18)62063-5. [DOI] [Google Scholar]
- 4.Dahanayake R., Galwey W. Effects of interactions between low-temperature treatments, gibberellin (GA3) and photostage on flowering and stem height of spring rape (Brassica napus var . annua) Ann. Bot. 1999;84:321–327. doi: 10.1006/anbo.1999.0920. [DOI] [Google Scholar]
- 5.Kou E., Huang X., Zhu Y., Su W., Liu H., Sun G., Chen R., Hao Y., Song S. Crosstalk between auxin and gibberellin during stalk elongation in flowering Chinese cabbage. Sci. Rep. 2021;11:3976. doi: 10.1038/s41598-021-83519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards D.E., King K.E., Ait-ali T., Harberd N.P. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- 7.Fleet C.M., Sun T.-P. A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Harberd N.P., Belfield E., Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an "inhibitor of an inhibitor" enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun T.-P. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154:567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas S.G., Sun T.-P. Update on gibberellin signaling. A tale of the tall and the short. Plant Physiol. 2004;135:668–676. doi: 10.1104/pp.104.040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Fernández R., Ardiles-Díaz W., Montagu M.V., Inzé D., May M.J. Cloning of a novel Arabidopsis thaliana RGA-like gene, a putative member of the VHIID-domain transcription factor family. J. Exp. Bot. 1998;49:1609–1610. doi: 10.1093/jexbot/49.326.1609. [DOI] [Google Scholar]
- 13.Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandler P.M., Marion-Poll A., Ellis M., Gubler F. Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 2002;129:181–190. doi: 10.1104/pp.010917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dill A., Sun T.-P. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King K.E., Moritz T., Harberd N.P. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004;131:1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- 19.Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16:646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wild M., Achard P. The DELLA protein RGL3 positively contributes to jasmonate/ethylene defense responses. Plant Signal. Behav. 2014;8:e23891. doi: 10.4161/psb.23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wild M., Davière J., Cheminant S., Regnault T., Baumberger N., Heintz D., Baltz R., Genschik P., Achard P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell. 2012;24:3307–3319. doi: 10.1105/tpc.112.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davière J., Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 23.Li M., An F., Li W., Ma M., Feng Y., Zhang X., Guo H. DELLA proteins interact with FLC to repress flowering transition. J. Integr. Plant Biol. 2016;58:642–655. doi: 10.1111/jipb.12451. [DOI] [PubMed] [Google Scholar]
- 24.Michaels S.D., Amasino R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon J., Suh S., Lee H., Choi K., Hong C.B., Paek N., Kim S., Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003;35:613–623. doi: 10.1046/j.1365-313X.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- 27.Yu S., Galvão V.C., Zhang Y.-C., Horrer D., Zhang T.-Q., Hao Y.-H., Feng Y.-Q., Wang S., Schmid M., Wang J.-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gubler F., Chandler P.M., White R.G., Llewellyn D.J., Jacobsen J.V. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 2002;129:191–200. doi: 10.1104/pp.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.-P. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boss P.K., Thomas M.R. Association of dwarfism and floral induction with a grape ’green revolution’ mutation. Nature. 2002;416:847–850. doi: 10.1038/416847a. [DOI] [PubMed] [Google Scholar]
- 32.Hynes L.W., Peng J., Richards D.E., Harberd N.P. Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgenic Res. 2003;12:707–714. doi: 10.1023/B:TRAG.0000005145.68017.6e. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa M., Kusano T., Katsumi M., Sano H. Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene. 2000;245:21–29. doi: 10.1016/S0378-1119(00)00018-4. [DOI] [PubMed] [Google Scholar]
- 34.El-Sharkawy I., Sherif S., El Kayal W., Mahboob A., Abubaker K., Ravindran P., Jyothi-Prakash P.A., Kumar P.P., Jayasankar S. Characterization of gibberellin-signalling elements during plum fruit ontogeny defines the essentiality of gibberellin in fruit development. Plant Mol. Biol. 2014;84:399–413. doi: 10.1007/s11103-013-0139-8. [DOI] [PubMed] [Google Scholar]
- 35.Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T., Hsing Y.C., Kitano H., Yamaguchi I., et al. Gibberellin insensitive DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.-P., et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao D., Cheng H., Wu W., Soo H.M., Peng J. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 2006;142:509–525. doi: 10.1104/pp.106.082289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima M., Shimada A., Takashi Y.-C., Kim Y., Park S.-H., Ueguchi-Tanaka M., Suzuki H., Katoh E., Iuchi S., Kobayashi M., et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Liu B., Yang S., An J., Chen C., Zhang X., Ren H. A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of B class floral homeotic genes. PLoS ONE. 2014;9:e91804. doi: 10.1371/journal.pone.0091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.-P. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang Y., Huang A., Yang X., Liu X. Effects of ABA and uniconazole on bolting characteristic and growth of flowering Chinese cabbage. Agric. Sci. Guangdong. 2010;12:49–51. (In Chinese) [Google Scholar]
- 42.Hu C., Chen Y., Liu S., Peng J. Regulation of flowering time of Chinese cabbage by paclobutrazol and gibberellin. J. Mt. Agric. Biol. 2016;35:73–75. (In Chinese) [Google Scholar]
- 43.Tang Q., Hu Y., Song M., Wang X., Wang Z. Effects of mustard bolting induced with gibberellin and abscisic acid. China Veg. 2008;12:18–20. (In Chinese) [Google Scholar]
- 44.Suge H., Rappaport L. Role of gibberellins in stem elongation and flowering in radish. Plant Physiol. 1968;43:1208–1214. doi: 10.1104/pp.43.8.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park J., Nguyen K.T., Park E., Jeon J.S., Choi G. DELLA proteins and their interacting RING finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell. 2013;25:927–943. doi: 10.1105/tpc.112.108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribeiro D.M., Araujo W.L., Fernie A.R., Schippers J.H.M., Mueller-Roeber B. Translatome and metabolome effects triggered by gibberellins during rosette growth in Arabidopsis. J. Exp. Bot. 2012;63:2769–2786. doi: 10.1093/jxb/err463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong C., Xu H., Ye S., Wang S., Li L., Zhang S., Wang X. Gibberellic acid-stimulated arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 2015;169:2288–2303. doi: 10.1104/pp.15.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang C., Guan P. Flower bud morphogenesis of Brassica parachinesis Bailey. Acta Hortic. Sin. 1983;3:183–186. (In Chinese) [Google Scholar]
- 49.Wen C., Chang C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell. 2002;14:87–100. doi: 10.1105/tpc.010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 54.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available in a publicly accessible repository.