Abstract
We have comparatively evaluated Quantiplex version 3.0 and version 2.0 on 133 plasma samples and a repetitive dilution series. Version 3.0 yielded higher human immunodeficiency virus RNA values, and the ratio of version 3.0 results to version 2.0 results decreased from 3.47 below 1,000 copies/ml to 1.97 above 50,000 copies/ml [linear regression, log (version 3.0) = 0.915 + 0.871 × log (version 2.0); r2 = 0.952].
Viral load is strongly correlated with human immunodeficiency virus (HIV) disease progression (10, 11), and in patients receiving antiretroviral therapy, the degree of viral load reduction is the best prognostic parameter for clinical benefit and for sustainability of treatment efficacy (6, 13). Suppression of viral load to below 50 copies/ml is necessary to achieve durable response to treatment and to delay development of resistance to antiretroviral drugs (2, 7, 14, 15, 17). We therefore comparatively evaluated two branched-DNA assays with a low detection limit, which was reduced from 500 copies/ml in Quantiplex version 2.0 to 50 copies/ml in version 3.0 (3, 8).
Besides limitations of some assay types with certain subtypes of HIV type 1 (HIV-1) (12), the lack of approved quantitative HIV-1 standards constitutes a drawback for all HIV quantification systems. Investigations comparing systems of different manufacturers with clinical samples show significant differences (4, 9, 16) but have not yet been performed thoroughly with the new-generation systems that have lower detection limits of 40 to 80 copies/ml. Thus, if assays from different manufacturers or different generations of the same system are used, interpretation and direct comparison of results should be done with caution.
One hundred thirty-three samples were taken successively from HIV-1-infected patients from our outpatient department. One half of the samples were taken from patients with a viral load of <500 copies/ml and the other half were taken from patients with higher values in the most recent prior measurement (Quantiplex 2.0). Blood was collected with Vacutainer PPT tubes (Becton Dickinson, Heidelberg, Germany), centrifuged within 30 min at room temperature, and stored at −70°C in the same Vacutainer tubes until batch analysis. Plasma specimens as well as positive and negative controls were prepared in duplicate for version 2.0 and singly for version 3.0 according to the manufacturer's instructions. For dilution series, six additional stored plasma samples with a previously determined viral load of more than 400,000 copies/ml and fresh frozen plasma from the blood bank were used. Fresh frozen plasma was initially added to the 1.5- to 3-ml samples to a total volume of 13 ml each. An aliquot of 6.5 ml was then repeatedly diluted 1:2 for a total number of 12 samples (dilution factors 1 to 2,048). All dilution samples were aliquoted, frozen at −70°C for at least 1 h, and thawed immediately before testing. Two separate runs were performed on each dilution series. Computational statistics were performed with SAS system 6.12 (SAS Institute Inc., Cary, N.C.). Analysis of agreement was done according to the difference-against-mean method (1). Curves and regression lines were graphically fitted by Origin 5.0 (Microcal Software Inc., Northampton, Mass.).
Seventy-nine of 133 serum samples (59%) returned virus counts below the lower detection limit of 500 copies/ml with version 2.0. Fifteen (19%) of these samples yielded values above 500 copies/ml with the 3.0 assay (mean, 1,504; median, 1,259; range, 505 to 4,599). Of 54 samples with more than 500 copies/ml in the 2.0 assay, 3 (5.6%) had fewer than 500 copies/ml in the new assay. Of 69 samples that showed ≥500 copies/ml with at least one of the tests, 1 yielded equal values in both assays and only 4 samples yielded lower values with assay version 3.0 than with version 2.0. Regression analysis of 50 serum samples with all values within the valid range of 500 to 500,000 copies/ml on log-transformed values returned the following: [log10 (version 3.0)] = 0.915 + {0.871 × [log10 (version 2.0)]} (r2 = 0.952).
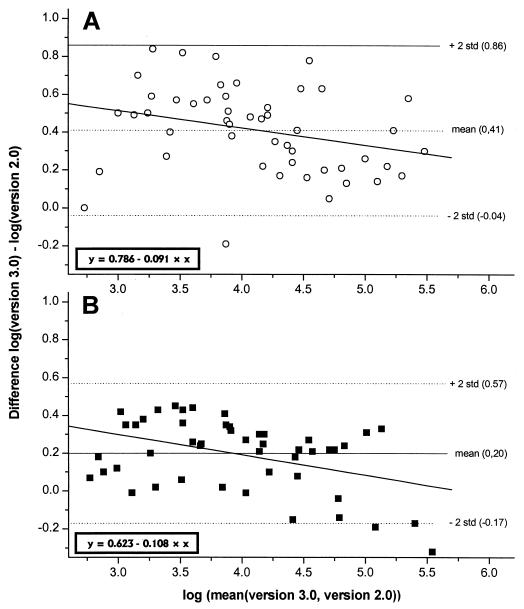
Analysis of agreement (Fig. 1) showed that the difference between both assays was normally distributed (P = 0.725) around the mean of the difference. The mean difference of log10 (version 3.0) − log10 (version 2.0) was 0.41 (standard deviation, 0.22). However, there was a clear trend of a decreasing difference with increasing viral load [log10 (version 3.0)] −[log10 (version 2.0)] = 0.786 − {0.091 × [log10 (version 2.0)]}; r2 = 0.079, P = 0.0485}. The same slope was observed with the dilution series ([log10 (version 3.0)] − [log10 (version 2.0)] = 0.623 − 0.108 × [log10 (version 2.0)]; r2 = 0.175, P = 0.003) although the mean of the difference was lower (0.20). There was a negative difference between version 3.0 and version 2.0 values in one of the six dilution series that mainly contributed to this lower mean of difference. Table 1 further illustrates the trend of a decreasing difference or ratio from lower to higher viral load ranges.
FIG. 1.
Analysis of agreement. The difference values are normally distributed around the mean but have a negative slope. The slopes for serum samples (A) and the dilution series (B) are similar, while the mean differences are different. std, standard deviation.
TABLE 1.
Ratio of assay version 3.0 and 2.0 resultsa
| VL range (version 2.0) (copies/ml) | Single plasma samples
|
Dilution series
|
|||
|---|---|---|---|---|---|
| Mean ratio (3.0/2.0) | CV (%) | n | Mean ratio (3.0/2.0) | CV (%) | |
| 500–1,000 | 3.47 | 54 | 8 | 1.99 | 30 |
| 1,001–10,000 | 3.45 | 40 | 19 | 1.91 | 31 |
| 10,001–50,000 | 2.50 | 56 | 14 | 1.57 | 25 |
| 50,001–500,000 | 1.97 | 41 | 9 | 1.09 | 64 |
VL, viral load.
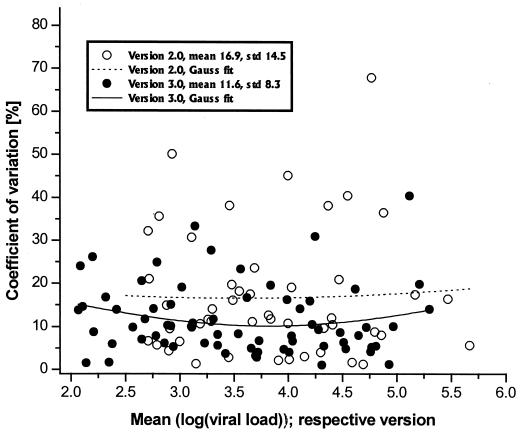
The variation coefficient (CV), which is only slightly lower in version 3.0 than in version 2.0 (mean CV over two runs, 11.6 versus 16.9%, respectively) does not explain this trend. The CV distribution over viral load and the Gauss fit curve (Fig. 2) illustrate only that there is, if anything, a slightly higher CV on either side of the viral load range in version 3.0.
FIG. 2.
Distribution of the CV over the measurement range. The CV is low and evenly distributed over the whole range, indicating a high level of precision of both assays. The CV of version 3.0 is lower than that of version 2.0, with a weak tendency of CV elevation at both measurement edges, demonstrated by the Gauss fit curves. std, standard deviation.
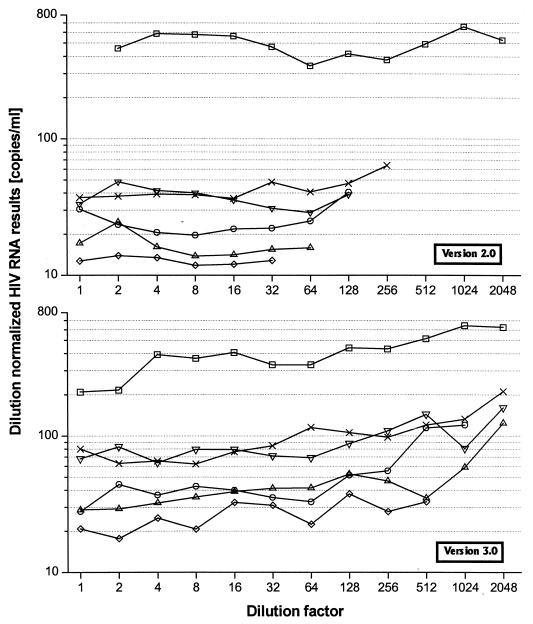
Figure 3 explains better the reason for the between-assay discrepancy, as it shows the “dilution normalized” RNA results of each individual dilution series that were calculated by dividing the RNA results by 2,048 (the highest dilution step) and multiplying them by the respective dilution degree of the individual test sample. In an ideal case, all points of one series should show the same value. However, there is an obvious tendency of increasing values with higher dilution in version 3.0, compared to version 2.0. Therefore, version 3.0 does relatively overestimate RNA values in the lower measurement range.
FIG. 3.
Test of linearity. Dilution-normalized results are calculated by dividing the RNA values of one dilution series by 2,048 (the highest dilution step) and multiplying by the respective dilution of the individual test sample. For version 3.0, the assay is not completely linear, with a trend of overestimation towards the lower concentration range.
In conclusion, both assay versions show high linearity, precision, and concordance. On a linear scale, values from version 3.0 are 3.5 times higher in the lower measurement range and 2 times higher in the upper range. From the clinical point of view, viral load changes of less than 0.5 log are not considered to be significant (5), but the between-assay difference should be taken into account if both test versions are used.
Acknowledgments
We thank S. Würden for his valuable technical and logistic assistance and J. Hemmer and C. Chiwakata for critically reviewing the manuscript.
The study was supported by Bayer Diagnostics (Emeryville, Calif.), the manufacturer of the Quantiplex kits that were evaluated.
REFERENCES
- 1.Bland J M, Altman D G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;i:307–310. [PubMed] [Google Scholar]
- 2.Cavert W, Notermans D W, Staskus K, Wietgrefe S W, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, Schuwirth C M, Goudsmit J, Danner S A, Haase A T. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. . (Erratum, 276:1321.) [DOI] [PubMed] [Google Scholar]
- 3.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coste J, Montes B, Reynes J, Peeters M, Segarra C, Vendrell J P, Delaporte E, Segondy M. Comparative evaluation of three assays for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J Med Virol. 1996;50:293–302. doi: 10.1002/(SICI)1096-9071(199612)50:4<293::AID-JMV3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Deeks S G, Coleman R L, White R, Pachl C, Schambelan M, Chernoff D N, Feinberg M B. Variance of plasma human immunodeficiency virus type 1 RNA levels measured by branched DNA within and between days. J Infect Dis. 1997;176:514–517. doi: 10.1086/517278. [DOI] [PubMed] [Google Scholar]
- 6.Hughes M D, Johnson V A, Hirsch M S, Bremer J W, Elbeik T, Erice A, Kuritzkes D R, Scott W A, Spector S A, Basgoz N, Fischl M A, D'Aquila R T. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. ACTG 241 Protocol Virology Substudy Team. Ann Intern Med. 1997;126:929–938. doi: 10.7326/0003-4819-126-12-199706150-00001. [DOI] [PubMed] [Google Scholar]
- 7.Kempf D J, Rode R A, Xu Y, Sun E, Heath-Chiozzi M E, Valdes J, Japour A J, Danner S, Boucher C, Molla A, Leonard J M. The duration of viral suppression during protease inhibitor therapy for HIV-1 infection is predicted by plasma HIV-1 RNA at the nadir. AIDS. 1998;12:F9–F14. doi: 10.1097/00002030-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H J, Pedneault L, Hollinger F B. Intra-assay performance characteristics of five assays for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:835–839. doi: 10.1128/jcm.36.3.835-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellors J W, Rinaldo C R J, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. . (Erratum, 275:14, 1997.) [DOI] [PubMed] [Google Scholar]
- 11.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R J. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 12.Nolte F S, Boysza J, Thurmond C, Clark W S, Lennox J L. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:716–720. doi: 10.1128/jcm.36.3.716-720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Brien W A, Hartigan P M, Daar E S, Simberkoff M S, Hamilton J D. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. VA Cooperative Study Group on AIDS. Ann Intern Med. 1997;126:939–945. doi: 10.7326/0003-4819-126-12-199706150-00002. [DOI] [PubMed] [Google Scholar]
- 14.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 15.Raboud J M, Montaner J S, Conway B, Rae S, Reiss P, Vella S, Cooper D, Lange J, Harris M, Wainberg M A, Robinson P, Myers M, Hall D. Suppression of plasma viral load below 20 copies/ml is required to achieve a long-term response to therapy. AIDS. 1998;12:1619–1624. doi: 10.1097/00002030-199813000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Vandamme A M, Schmit J C, Van Dooren S, Van Laethem K, Gobbers E, Kok W, Goubau P, Witvrouw M, Peetermans W, De Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the AMPLICOR HIV monitor test. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 17.Wong J K, Guenthard H F, Havlir D V, Zhang Z Q, Haase A T, Ignacio C C, Kwok S, Emini E, Richman D D. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc Natl Acad Sci USA. 1997;94:12574–12579. doi: 10.1073/pnas.94.23.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]