Abstract
Twenty-five clarithromycin-resistant Helicobacter pylori strains (selected by agar dilution) were studied to detect A2142G and A2143G mutations in the 23S rRNA gene by a PCR-restriction fragment length polymorphism method and an A2142C mutation by PCR using a 3′-mismatched specific primer. A 700-bp amplified fragment was obtained by the mismatched PCR only in strains without an A2142G or A2143G mutation, indicating that those strains had the A2142C mutation.
Helicobacter pylori is a gram-negative rod involved in digestive diseases such as peptic ulcer, gastritis, or mucosa-associated lymphoid tissue lymphoma or as a risk factor in the development of gastric cancer (5). Nowadays, the National Institutes of Health in the United States, the Maastricht Consensus in Europe, or the Canadian consensus (9) recommends treatment of some of these digestive diseases with antibiotics. Among the antibiotics used for the treatment of H. pylori infections, amoxicillin, tetracycline, metronidazole, and new macrolides (mainly clarithromycin) are the most commonly used. However, infection by a resistant strain is considered a factor in treatment failure (1, 14). The eradication rate when a clarithromycin-resistant strain is involved decreases from 62 to 20% with clarithromycin plus ranitidine (13) or from 92 to 50% with clarithromycin, amoxicillin, and omeprazole (21).
Monomethylation or dimethylation of Escherichia coli-equivalent base A-2058 in 23S rRNA by methyltransferases encoded by erm genes is the most common mechanism of macrolide resistance. However, transition mutations in the peptidyltransferase region of the 23S rRNA of Mycobacterium spp. or Propionibacterium spp. have been described previously (12). The mechanism of resistance to clarithromycin in H. pylori seems to be a decrease in binding of macrolides to the ribosome (10), associated with point mutations on the 23S rRNA gene (18), mainly at positions 2142 and 2143 (17). Point mutations in which a guanine or a cytosine residue replaces an adenine residue (A2142G, A2142C, and A2143G) have been described previously (2, 4, 7, 10, 14, 15, 17, 18). On initiation of these studies, mutations at position 2058 or 2059 were considered, based on the E. coli gene. Furthermore, positions 2143 and 2144 (19) or position 2514 or 2515 (2) was used, but after the paper published by Taylor et al. in 1997, positions 2142 and 2143 should be considered based on the structure of the gene in H. pylori (17).
A PCR-restriction fragment length polymorphism (RFLP) method has been widely used to detect A-to-G mutations (2, 7, 10, 16, 18), but it allows the detection of only A2142G and A2143G mutations. The A2142C mutation has also been described for clinical isolates, although less frequently.
Ge and Taylor reported that it is possible to distinguish mutant from wild-type sequences with a 3′-terminal nucleotide primer complementary to only the mutated nucleotide (6). Therefore, we decided to use a PCR method using a 3′-mismatched primer to detect the A2142C mutation in clinical isolates of H. pylori resistant to clarithromycin.
Twenty-five H. pylori strains were obtained from gastric biopsy samples transported to the Clinical Microbiology Department and cultured on selective and nonselective medium plates. Plates were incubated at 37°C in a microaerobic atmosphere for 7 to 10 days for primary culture and every 2 to 3 days for subculture. Strains were identified according to colony morphology, Gram stain, and positive reaction with urease, catalase, and oxidase and stored until use at −80°C in Trypticase soy broth containing 20% glycerol.
Clarithromycin resistance was determined by an agar dilution method. Briefly, isolates were grown for 48 h in brain heart infusion plus 10% fetal calf serum, and a suspension of 109 CFU/ml was applied to plates containing antibiotic by using a Steers replicator. Serial dilutions of the antibiotic ranging from 0.016 to 128 mg/liter were prepared in Mueller-Hinton agar supplemented with 10% horse blood. Plates were incubated for 3 to 5 days, and the MIC was recorded as the lowest concentration of the antibiotic inhibiting visible growth. Resistance was defined as the clarithromycin MIC being ≥4 mg/liter.
DNA was extracted by the cetyltrimethylammonium bromide reagent method and phenol chloroform extraction as previously described (20).
A2142G and A2143G mutations were determined by a PCR-RFLP method using primers described in Table 1 (18). The amplified fragment was digested with MboII (Amersham Pharmacia) or BsaI (New England Biolabs, Inc., Beverly, Mass.), which allows discrimination among the wild type, the A2142G mutant (MboII restriction site), and the A2143G mutant (BsaI restriction site). The restriction products were analyzed by electrophoresis on a 2% agarose gel.
TABLE 1.
Primers used in both PCRsa
| Primer | Sequence | Reference |
|---|---|---|
| CLA 18 | AGTCGGGACCTAAGGCGAG | 18 |
| CLA 21 | TTCCCGCTTAGATGCTTTCAG | 18 |
| CLA 3 | AGGTCCACGGGGTCTTG | This study |
Primers CLA 18 and CLA 21 were used in PCR-RFLP to obtain a 1.4-kbp amplified fragment. Primers CLA 18 and CLA 3 were used in 3′-mismatched PCR to obtain a 700-bp amplified fragment.
A 3′-mismatched PCR was developed using specific primers (described in Table 1). For PCR amplification, approximately 5 ng of genomic DNA of H. pylori was added to a 50-μl mixture containing buffer (1 mM MgCl2, 50 μM deoxynucleoside triphosphates, 1 μM [each] primer, and 1.25 U of Taq DNA polymerase [Promega]). Amplification was carried out in a DNA Thermal Cycler 2400 (Perkin-Elmer Corporation, Norwalk, Conn.). Thirty-five cycles, each for 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C, were performed after 10 min of denaturation at 94°C. A 700-bp fragment was detected with a 1.5% agarose gel only when the 23S rRNA gene had the A2142C mutation.
A 1.4-kbp fragment of the 23S rRNA gene was amplified in all the strains studied herein. Furthermore, MboII cut the fragment obtained in 10 strains and BsaI cut the other 10 amplified fragments. In the last five strains, neither MboII nor BsaI was able to digest the amplified fragment.
In all clarithromycin-resistant strains in which the mutation was neither A2142G nor A2143G, a 700-bp amplified fragment was obtained by A2142C-specifically primed 3′-mismatched PCR, indicating that the strains had the A2142C mutation. No amplification was obtained with clarithromycin-resistant strains with either the A2142G or A2143G mutation. The PCR was also used as nested PCR after amplification of 1.4 kbp of the 23S rRNA gene, and 700 bp was obtained when the A2142C mutation was present. In Table 2, the results obtained with the PCR-RFLP method and with the 3′-mismatched PCR method for the clinical isolates tested according to the clarithromycin MIC data are shown. The amplified fragments obtained by both methods are shown in Fig. 1 and 2. As indicated by other authors, the MIC for strains with the A2143G mutation was lower than that for strains with the A2142G or A2142C mutation.
TABLE 2.
Results obtained with the PCR-RFLP method and with the 3′-mismatched PCR method for the clinical isolates tested according to the clarithromycin MIC data
| Mutation | No. of strains | PCR-RFLP result with:
|
3′-mismatched PCR | MIC range (mg/liter) | |
|---|---|---|---|---|---|
| BsaI | MboII | ||||
| A2143G | 10 | Digestion | Negative | Negative | 4–8 |
| A2142G | 10 | Negative | Digestion | Negative | 16–64 |
| A2142C | 5 | Negative | Negative | Positive | 8–64 |
FIG. 1.
PCR-RFLP patterns obtained after digestion with BsaI or MboII. BsaI digested the 1.4-kbp fragment, producing 1,000- and 400-bp fragments in either susceptible or resistant strains. However, if the A2143G mutation was present, the 1,000-bp fragment was converted to 700- and 300-bp fragments. MboII digested the fragment, producing two fragments of 700 bp only when the A2142G mutation was present. Lanes: 1 and 2, strain 1 digested with BsaI and MboII, respectively; 3 and 4, strain 2 digested with BsaI and MboII, respectively; 5 and 6, strain 3 digested with BsaI and MboII, respectively; 7 and 8, strain 4 digested with BsaI and MboII, respectively; 9, DNA markers. BsaI digested the fragment in strains 1 and 2. BsaI or MboII did not digest strains 3 and 4.
FIG. 2.
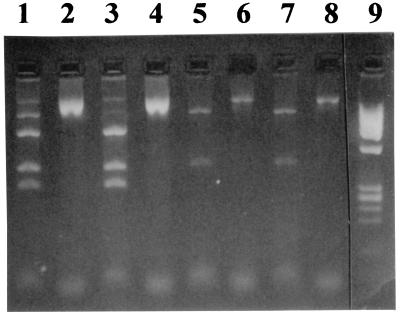
A 3′-mismatched PCR pattern after amplification of different strains. Lanes 1 to 5, 3′-mismatched positive PCRs; lanes 6 to 8, 3′-mismatched negative PCRs; lane 9, DNA markers.
Detection of resistance in H. pylori is generally performed by MIC determination. This method requires growth of bacteria and takes at least 10 days; thus, the applicability of the data in the clinical setting is sometimes controversial.
Although Hulten et al. (7) found two new mutations involved in clarithromycin resistance in H. pylori, the three mutations studied herein are the ones most commonly found in H. pylori clinical isolates (2, 18, 19). By site-directed mutagenesis, some authors found mutation A2142C, -G, or -T and A2143C, -G, or -T (3). However, they found that an apparent preferential mutation, A2142G or A2143G, exists, due to the fact that these mutants have the highest growth rates and more stable resistance as well as that the MICs for them are higher (3).
Several authors have used other methods to detect mutations related to H. pylori clarithromycin resistance. Stone et al. (15) proposed a PCR-oligonucleotide ligation assay for detection of clarithromycin resistance in H. pylori, and Pina et al. (11) used a test involving amplification and colorimetric hybridization in liquid phase. PCR-RFLP has been widely used due to its simplicity and may be performed in a few hours.
By our 3′-mismatched PCR method, obtaining a 700-bp amplified fragment indicates a mutation in the A2142C position. We found this fragment in 5 of the 25 strains tested. An amplified fragment was not obtained by 3′-mismatched PCR when the strains showed mutation A2142G or A2143G. This method seems to be useful in detecting the mutation in the A2142C position. After using PCR-RFLP to detect the A2142G or A2143G mutation, these 3′-mismatched PCRs can be applied to detect the A2142C mutation.
According to our results, similar to data previously reported by other authors, the A2143G mutation is related to the lowest clarithromycin MICs while the A2142G mutation was associated with higher MICs. For the strains showing A2142C, we found several MICs ranging from 8 to 64 mg/liter.
Resistance to clarithromycin is the main predictor of failure for eradication treatments using this compound, and the detection of resistance is becoming of major importance. This method provides a result within a few hours once the strain has been isolated, and no special technology apart from PCR is needed. The results could be more practical if the method were applied directly to biopsy specimens, which would provide a faster result.
Acknowledgments
We thank Brenda Ashley for English language assistance.
REFERENCES
- 1.Alarcón T, Domingo D, López-Brea M. Antibiotic resistance problems with Helicobacter pylori. Int J Antimicrob Agents. 1999;12:19–26. doi: 10.1016/s0924-8579(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 2.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J J, Vandenbroucke-Grauls C M J E. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 3.Debets-Ossenkopp Y J, Brinkman A B, Kuipers E J, Vandenbroucke-Grauls C M J R, Kusters J G. Explaining the bias in the 23S rRNA gene mutations associated with clarithromycin resistance in clinical isolates of Helicobacter pylori. Antimicrob Agents Chemother. 1998;42:2749–2751. doi: 10.1128/aac.42.10.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domingo D, Alarcon T, Sanz J C, Sánchez I, López-Brea M. High frequency of mutations at position 2144 of the 23S rRNA gene in clarithromycin resistant Helicobacter pylori strains isolated in Spain. J Antimicrob Chemother. 1998;41:573–574. doi: 10.1093/jac/41.5.573. [DOI] [PubMed] [Google Scholar]
- 5.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge Z, Taylor D E. Rapid polymerase chain reaction screening of Helicobacter pylori chromosomal point mutations. Helicobacter. 1997;2:1–5. doi: 10.1111/j.1523-5378.1997.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 7.Hulten K, Gibreel A, Skold O, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Megraud F. Resistance of Helicobacter pylori to antibiotics. Aliment Pharmacol Ther. 1997;11(Suppl. 1):43–53. doi: 10.1046/j.1365-2036.11.s1.11.x. [DOI] [PubMed] [Google Scholar]
- 9.NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in peptic ulcer disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- 10.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pina M, Occhialini A, Monteiro L, Doermann H P, Megraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross J I, Eady E A, Cove J H, Jones C E, Ratyal A H, Miller Y W, Vyakrnam S, Cunliffe W J. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob Agents Chemother. 1997;41:1162–1165. doi: 10.1128/aac.41.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schütze K, Hentschel E, Hirschl A M. Clarithromycin or amoxicillin plus high dose ranitidine in the treatment of Helicobacter pylori positive functional dyspepsia. Eur J Gastroenterol Hepatol. 1996;8:41–46. doi: 10.1097/00042737-199601000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Stone G G, Shortridge D, Flamm R K, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka S K. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;4:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 15.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Graham D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczebara F, Dhaenens L, Vincent P, Husson M O. Evaluation of rapid molecular methods for detection of clarithromycin resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1997;16:162–164. doi: 10.1007/BF01709478. [DOI] [PubMed] [Google Scholar]
- 17.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M G. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Versalovic J, Osato M S, Spakovsky K, Dore M P, Reddy R, Stone G G, Shortridge D, Flamm R K, Tanaka S K, Graham D Y. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 20.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R E, Kingston D D, Moore J G, Seidman J G, Smith J A, et al., editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1987. pp. 241–245. [Google Scholar]
- 21.Wurzer H, Rodrigo L, Stamler D, et al. Short course therapy with amoxicillin-clarithromycin triple for 10 days (ACT-10) eradicates H. pylori and heals duodenal ulcer. Aliment Pharmacol Ther. 1998;11:943–952. doi: 10.1046/j.1365-2036.1997.00223.x. [DOI] [PubMed] [Google Scholar]