Abstract
The NAC (NAM, ATAF and CUC) gene family plays a crucial role in the transcriptional regulation of various biological processes and has been identified and characterized in multiple plant species. However, genome-wide identification of this gene family has not been implemented in Juglans mandshurica, and specific functions of these genes in the development of fruits remain unknown. In this study, we performed genome-wide identification and functional analysis of the NAC gene family during fruit development and identified a total of 114 JmNAC genes in the J. mandshurica genome. Chromosomal location analysis revealed that JmNAC genes were unevenly distributed in 16 chromosomes; the highest numbers were found in chromosomes 2 and 4. Furthermore, according to the homologues of JmNAC genes in Arabidopsis thaliana, a phylogenetic tree was constructed, and the results demonstrated 114 JmNAC genes, which were divided into eight subgroups. Four JmNAC gene pairs were identified as the result of tandem duplicates. Tissue-specific analysis of JmNAC genes during different developmental stages revealed that 39 and 25 JmNAC genes exhibited upregulation during the mature stage in walnut exocarp and embryos, indicating that they may serve key functions in fruit development. Furthermore, 12 upregulated JmNAC genes were common in fruit ripening stage in walnut exocarp and embryos, which demonstrated that these genes were positively correlated with fruit development in J. mandshurica. This study provides new insights into the regulatory functions of JmNAC genes during fruit development in J. mandshurica, thereby improving the understanding of characteristics and evolution of the JmNAC gene family.
Keywords: Juglans mandshurica, NAC family, genome-wide analysis, walnut exocarp, embryos, expression pattern
1. Introduction
Transcription factors (TFs) are proteins that specifically bind to cis-acting elements in the promoter regions and play a vital regulatory role in abiotic and biotic stress responses in many plants [1,2]. They can bind to specific nucleotide sequences upstream to regulate the transcription or gene expression of functional genes, which can significantly enhance stress resistance or tolerance to various factors and regulate plant growth and development [3,4]. The NAC protein family is one of the largest plant-specific TF families [5]. The family name is derived from the common proteins of no apical meristem (NAM) from Petunia hybrida, ATAF1–2 and cup-shaped cotyledon (CUC) from Arabidopsis thaliana, which prompted the formation of a conserved NAC domain [6,7]. Furthermore, the N-terminal of encoded NAC proteins possesses a highly conserved DNA-binding domain with approximately 150 amino acids and consists of five typical subdomains that are divided into A–E, wherein the A, C, and D subdomains are relatively conserved [8,9]. The C-terminal contains highly varying transcriptional regulatory regions and is specifically involved in DNA binding [10,11].
Owing to their crucial function in the formation of the embryos and flowers of petunias, NAC TFs were widely studied, and multiple reports identified its key role in plant growth and development [12,13]. A large number of NAC-encoding genes have been identified and confirmed in various plants for their role in regulating wood formation [14], leaf senescence [15], fruit development [16,17], and flower morphogenesis [18,19]. Furthermore, NAC TFs also are involved in various biotic and abiotic stress responses in many plants such as Populus tomentosa [20], Arabidopsis thaliana [21], Oryza sativa [22,23], Glycine max [24], Arachis hypogaea [25], and Xanthoceras sorbifolia [26]. The overexpression of many NAC genes increases stress tolerance in plants. In a study on A. thaliana, the overexpression of an NAC gene from Malus baccata indicated that MbNAC25 significantly increased cold and salinity tolerance [27]. The overexpression of ChNAC1 in Cerasus humilis exhibited high drought stress tolerance since it positively regulated the expression of abscisic acid (ABA)-responsive genes [27,28]. Furthermore, in Nicotiana tabacum, the overexpression of LpNAC13 from Lilium pumilum caused significant negative regulation to drought stress tolerance but positively regulated the salt stress response [29]. The NAC family plays a crucial role in secondary cell wall biosynthesis. The overexpression of BpNAC012 in Betula platyphylla positively regulated ectopic secondary cell wall deposition [30]. In O. sativa, OsSND2, an NAC TF, was involved in regulating secondary cell wall biosynthesis [31]. NAC TFs can not only regulate the expression of downstream genes but also cooperate with other TFs to regulate the target genes. Previous studies have demonstrated that MaNAC5 cooperates with MaWRKY1 and MaWRKY2 for regulating pathogenesis-related gene response to diseases [32]. In addition, a study on Glycine max indicated that GmWRKY27 interacted with GmMYB174 and suppressed GmWRKY29, which significantly enhances drought stress tolerance [33].
Fruits are mainly involved in reproduction and gene transmission of plants, and they also provide many essential nutrients when consumed [34]. Fruit development and ripening are complex processes regulated by multiple factors such as gene regulation, hormone induction, temperature, and light. In particular, it was regulated by many transcription factors, which affected the expression of downstream target genes at the transcriptional level [35]. Previous studies have confirmed that NAC transcription factors participate in fruit development during ethylene biosynthesis and signal transduction [36]. Furthermore, some genes involved in plant hormone biosynthesis and signal transduction also are directly regulated by NAC transcription factors during fruit development and ripening process [37]. A recent study suggested that NAC TFs serve specific functions in fruit development and ripening in model species and crops of commercial interest and are involved in several cellular processes such as pigment accumulation, tissue softening, and organic acid biosynthesis [38]. In Picea abies, the overexpression of PaNAC03 influences flavanol biosynthesis and embryo development [39]. Furthermore, the seed and fruit development of Vitis vinifera is influenced by the interaction of VvNAC26 and VvMADS9 [40]. Owing to their specific expression in biological processes, NAC genes are important for conferring specificity to plant growth, development and adaptability.
To date, the NAC gene family has been widely identified and studied in many species, such as Musa acuminata [41], Medicago sativa [42], Panax ginseng [43], Dimocarpus longan [44], and Actinidia chinensis [45]. However, no comprehensive study has been conducted on the NAC gene family of J. mandshurica. The recent completion of J. mandshurica genome sequencing (https://ngdc.cncb.ac.cn/gwh/Assembly/22547/show) (accessed on 12 October 2021) allowed us to investigate the NAC gene family of J. mandshurica at the genome level. In the present study, we systematically performed genome-wide identification and analysis of the NAC gene family, which bridges the research gap for gene family studies in J. mandshurica. The analyses of physical and chemical characteristics, chromosomal location, phylogeny and evolutionary relationships, conserved motifs, regulatory network, and expression profiling of JmNAC genes were comprehensively performed. This study will improve the understanding of the functional characteristics of JmNAC genes to identify their role in the development of walnut exocarp and embryos in J. mandshurica and will also lay a foundation for further study of molecular mechanisms of fruits.
2. Results
2.1. Identification and Chromosomal Location of JmNAC Genes
To identify the NAC gene family members of J. mandshurica, sequence alignment of NAC genes in A. thaliana and J. mandshurica was performed using two BLASTP (Basic Local Alignment Search Tool for Protein) searches. After redundancy removal, a total of 114 highly confident NAC genes were screened and identified in the J. mandshurica genome. Furthermore, the names of JmNAC genes were encoded from JmNAC1 to JmNAC114 according to the gene annotation information (Table S1). The basic information of these genes was analyzed and summarized in detail, including their MW, pI, CDS length, and subcellular localization. Of the identified 114 JmNAC genes, the largest protein was encoded by JmNAC046 with 1160 amino acids and CDS length of 3483 bp, whereas the smallest protein was encoded by JmNAC089 with 77 amino acids and CDS length of 234 bp. The calculated theoretical MW of the proteins ranged from 8773.86 kDa (JmNAC089) to 132,712.8 kDa (JmNAC046), and pI varied from 4.21 (JmNAC089) to 9.83 (JmNAC081). With regard to subcellular localization, all JmNAC proteins were predicted, and the results showed that they may located in the nucleus (Table S1).
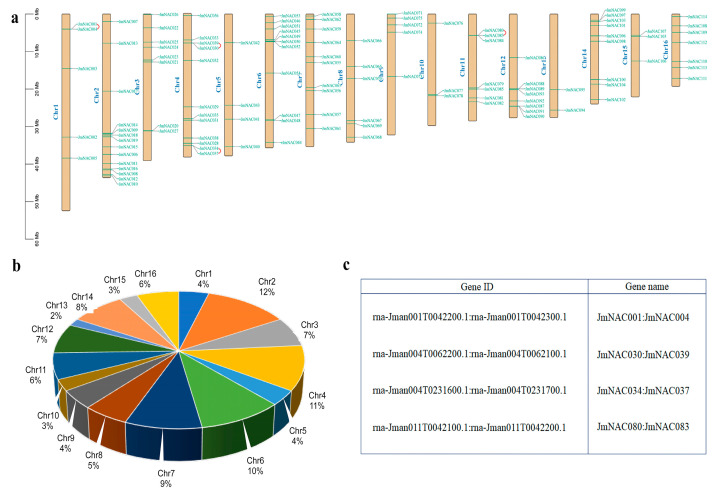
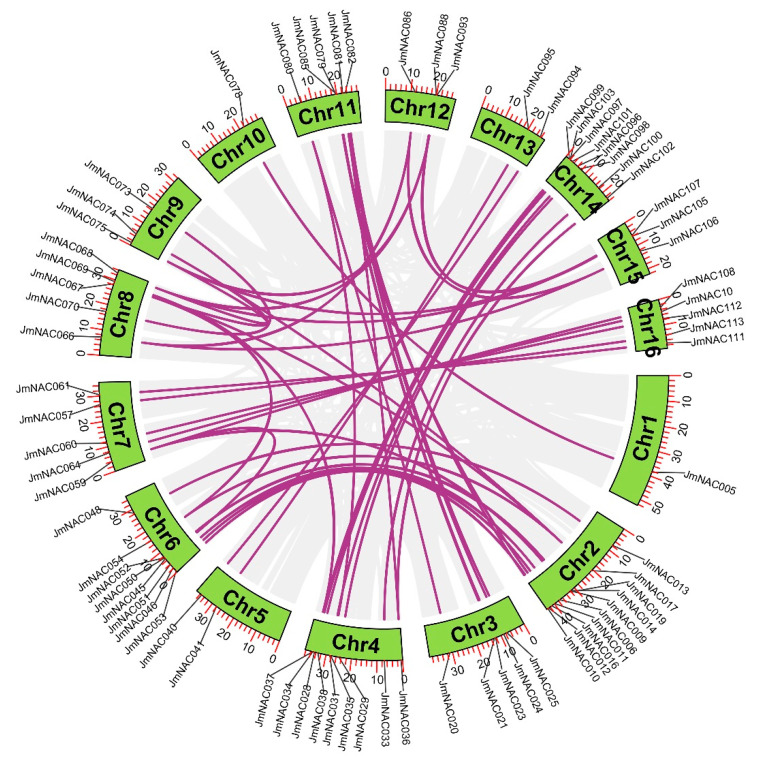
The identified 114 JmNAC proteins were unevenly distributed on 16 chromosomes of J. mandshurica (Figure 1a), indicating the diversification and complexity of the NAC family. Chromosome 2 contained the largest number of JmNAC genes (14 genes, ~12%), followed by chromosome 4 (12 genes, ~11%), and chromosome 13 contained the smallest number of JmNAC genes (2 genes, ~2%) (Figure 1b). In addition, a total of 5 (~4%), 8 (~7%), 4 (~4%), 11 (~10%), 10 (~9%), 6 (~5%), 5 (~4%), 3 (~3%), 7 (~6%), 8 (~7%), 9 (~8%), 3 (~3%) and 7 (~6%) genes were located on chromosomes 1, 3, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, and 16, respectively. Furthermore, JmNAC genes on chromosomes 2, 7, 10, 13, and 14 were mainly located at one end of the chromosome. Duplication events are of great importance for studying plant evolutionary patterns. According to the chromosomal location and genome annotation information, a total of four tandemly duplicated gene pairs were obtained, which were mainly distributed in chromosomes 1 and 4, with two gene pairs each (Figure 1c).
Figure 1.
Chromosomal distribution of JmNAC genes on eight J. mandshurica chromosomes. (a) Distribution of JmNAC genes in 16 chromosomes. Tandemly duplicated genes are marked with red. (b) The pie chart represents the distribution of JmNAC genes on each chromosome. (c) Tandemly duplicated JmNAC gene pairs in J. mandshurica.
2.2. Phylogenetic Analysis of JmNAC Genes
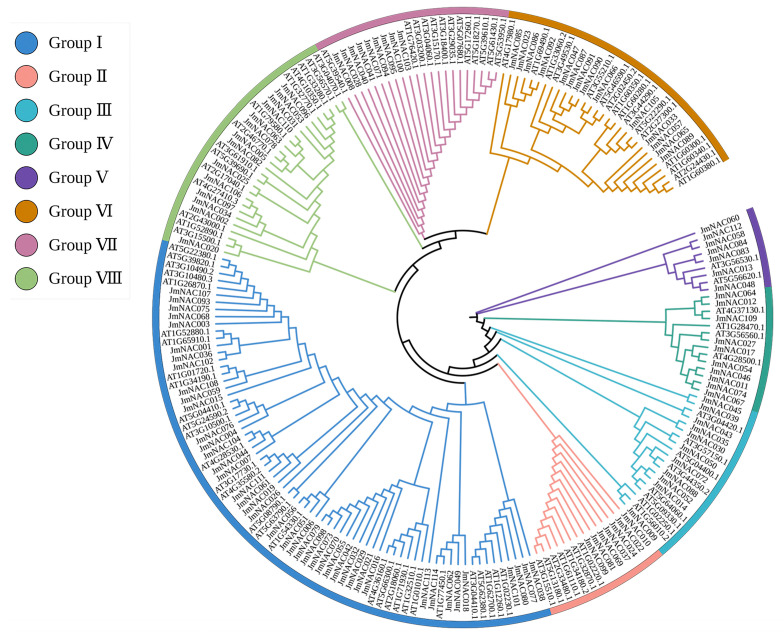
To understand the phylogenetic relationship and potential molecular function of the JmNAC gene family, phylogenetic analysis was performed using all NAC full-length protein sequences from J. mandshurica (114 genes) and A. thaliana (96 genes). Based on the homology of NAC proteins, 114 JmNAC genes in the constructed phylogenetic tree were classified into eight subgroups, namely, Group I, Group II, Group III, Group IV, Group V, Group VI, Group VII and Group VIII (Figure 2). Group I consisted of the largest number of JmNAC genes, that is, 42 genes, followed by Group VI and VIII with 14 genes each. Group III and IV included 11 and 10 JmNAC genes, respectively, whereas Group II, VI and VII contained less than 10 JmNAC genes. Furthermore, JmNAC060, JmNAC112, JmNAC058, JmNAC084, JmNAC083, JmNAC013, and JmNAC048 exhibited relatively low similarity with genes in other subgroups; therefore, they were included in Group V, representing a single clade.
Figure 2.
Phylogenetic tree representing the relationship between NAC genes of J. mandshurica and A. thaliana. Different colors represent the subgroups of the NAC gene family in J. mandshurica and A. thaliana.
2.3. Gene Structure, Motif Composition and Protein Interaction Analysis of JmNAC Genes
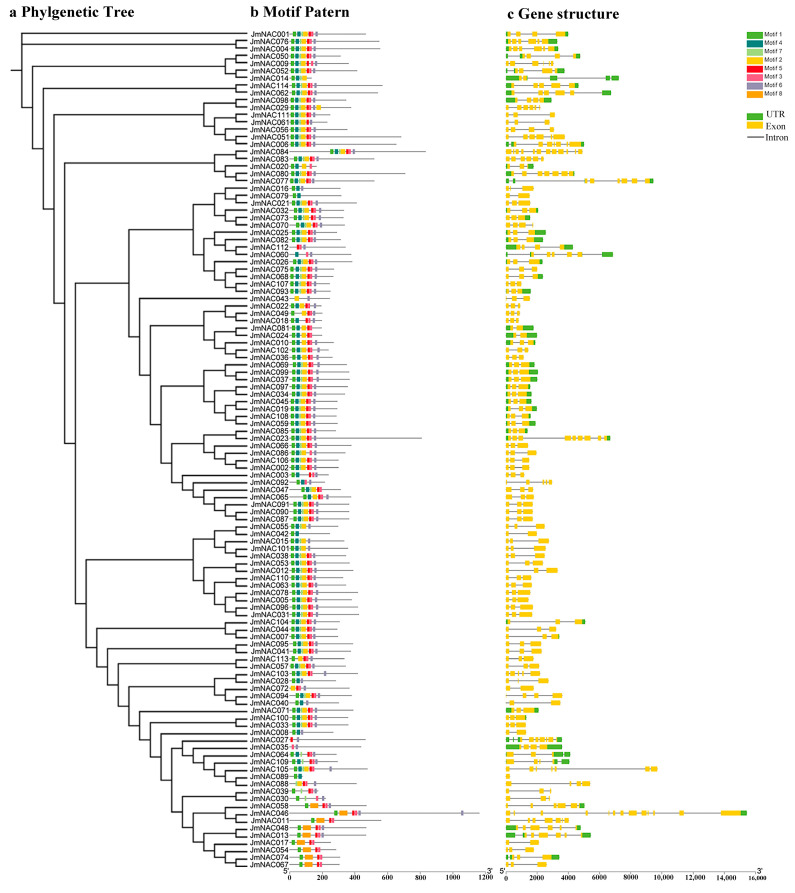
Structural differences in genes are the basis of gene family evolution, which contribute to understanding the diversity of genes and environmental adaptability in plants. To investigate the diversity of gene structure, the structure of JmNAC genes, including untranslated regions (UTR), exons, and introns was identified using TBtools. Genes grouped in the same class had a similar gene structure. We found that the number of exons in all JmNAC genes ranged from 3 to 13, and the number of introns varied from 1 to 12, exhibiting significant variation. JmNAC046 contained the largest number of exons and introns, followed by JmNAC084, which consisted of 12 exons and 11 introns. Some JmNAC genes such as JmNAC042, JmNAC055, JmNAC087, JmNAC090, and JmNAC091 contained exons and introns without the UTR region, indicating a specific evolutionary process of these genes.
Motifs are conserved sequences with biological functions, and different motifs usually have specific functional domains. To further analyze the functional regions of JmNAC proteins, the amino acid motifs of 114 JmNAC proteins were investigated using the MEME online tool (Figure 3). After MEME analysis, eight conserved motifs were predicted in JmNAC genes. A majority of JmNAC genes clustered into the same clade shared similar motif composition with the same position, indicating their similar biological functions. Motifs 1, 2, 4, 5, and 7 were found in most JmNAC proteins. Furthermore, most JmNAC proteins contained at least 5–6 conserved motifs, except for several specific proteins. A unique motif (motif 8) was identified in nine JmNAC proteins, which indicated that this specific motif was related to specific functions in different clades.
Figure 3.
Phylogenetic relationships, gene structure and protein motifs in JmNAC genes. (a) The phylogenetic tree of all NAC proteins in J. mandshurica was constructed using the neighbour-joining method with 1000 replicates in the MEGA 7.0 software (Mega Limited, Auckland, New Zealand). (b) The motif composition of J. mandshurica NAC proteins. Motifs 1–10 are displayed in different coloured boxes. (c) CDS–UTR structure of JmNAC genes. Yellow boxes represent CDS; black lines indicate introns; green boxes represent the untranslated 5′ and 3′ regions.
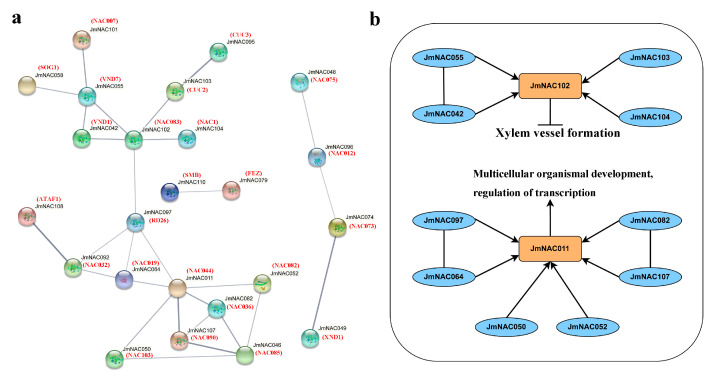
To further analyze the potential function of JmNAC genes, an interaction network of 114 JmNAC genes was constructed using the STRING website tools according to the homologous proteins of A. thaliana (Figure 4). The results showed that five JmNAC genes including JmNAC085, JmNAC011, JmNAC097, JmNAC102, and JmNAC055 may be the key nodes in this network, which interacted with other TFs to form a complicated regulatory network. A relatively high level of homology was found between JmNAC102 and AtNAC083, which is a crucial transcriptional activator related to xylem vessel formation with a positive regulatory function. Furthermore, high homology was also found between JmNAC011 and AtNAC044, which is a transcriptional regulator that positively regulates the expression of genes involved in the development of multicellular organisms. JmNAC011 was closely related to JmNAC097, JmNAC064, JmNAC050, JmNAC052, JmNAC107, and JmNAC082, which may contribute to the formation of a strong interaction network.
Figure 4.
Functional network of JmNAC genes. (a) Network of JmNAC genes in J. mandshurica based on the orthologues in A. thaliana. (b) A schematic representation of a regulatory network among JmNAC genes.
2.4. Chromosomal Distribution and Synteny Analysis of JmNAC Genes
Gene duplication events are common and widely occur in plant gene family formation, which is important for understanding the adaptive evolution of species. To understand the duplication events of all JmNAC genes, synteny analysis was performed using MCscanX and the Advanced Circos software in TBtools (Figure 5). The results revealed that a total of 50 orthologous gene pairs were identified in 114 JmNAC genes, indicating a close relationship between them. Furthermore, syntenic genes distributed on chromosome 2 (ten gene pairs) were the most common, followed by chromosome 4 (nine gene pairs) and chromosome 14 (eight gene pairs), whereas only one duplication gene pair was found on chromosomes 1 and 10. In addition, some JmNAC genes corresponded to more than one duplication gene pair on other chromosomes (Figure 5).
Figure 5.
Genomic localization and gene duplication of JmNAC genes on J. mandshurica chromosomes. Grey lines indicate all syntenic blocks in the J. mandshurica genome; purple lines indicate the inter-chromosomal relationships of JmNAC genes.
2.5. Syntenic Relationships of JmNAC Genes and NAC Genes of Several Different Species
To further analyze the evolutionary relationship between NAC genes of J. mandshurica and other different species including Juglans regia, Juglans sigillata, Populus trichocarpa, A. thaliana and Vitis vinifera, we performed comparative synteny analysis (Figure S1). J. mandshurica, J. regia and J. sigillata exhibited the highest level of homology. Furthermore, orthologous gene pairs in J. mandshurica and J. regia were mainly distributed on chromosomes 2 and 3, respectively. J. mandshurica and J. sigillata contained the largest number of orthologous gene pairs of chromosomes 6 and 3, respectively. J. mandshurica (orthologous gene pairs were mainly distributed on chromosome 2) and P. trichocarpa (orthologous gene pairs were mainly distributed on chromosome 1) also exhibited higher homology, whereas J. mandshurica (orthologous gene pairs were mainly distributed on chromosome 2) and A. thaliana (orthologous gene pairs were mainly distributed on chromosome 1) exhibited relatively low homology. Homology among NAC genes in J. mandshurica and other two genus Juglans plants (J. regia and J. sigillata) was higher than that in J. mandshurica and other several species, which may be related to species evolution and genetic relationships. Furthermore, we found that syntenic gene pairs on chromosome 2 were most abundant and diverse according to the intra- and inter-species collinearity analysis of 114 JmNAC genes. These results suggested that the NAC genes were relatively conserved in J. mandshurica and other plants.
2.6. Differential Expression of JmNAC Genes during Fruit Development in J. mandshurica
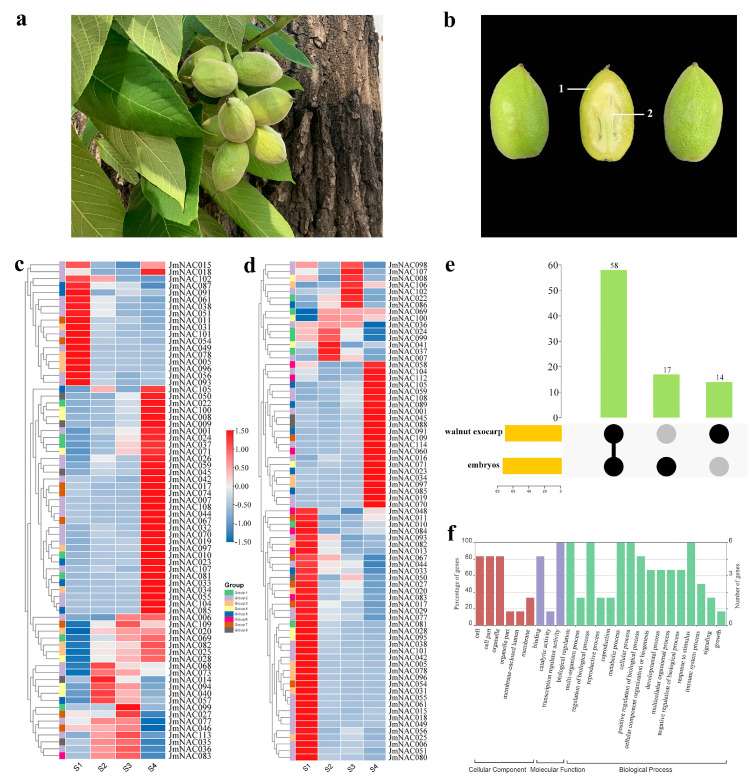
Extensive studies have revealed that the NAC gene family plays a crucial role during fruit development and ripening stages. The expression pattern of all identified 114 JmNAC genes was further analyzed in walnut fruit (including exocarp and embryos) at different developmental stages (from S1 to S4 stages) combining the transcriptome data and RT-qPCR analysis (Figure 6). The expression levels of 114 JmNAC genes are shown in Figure 6. Our results revealed that most JmNAC genes exhibited different expression patterns in the two tissues (walnut exocarp and embryos) during fruit development. Among the identified genes, a total of 72 and 75 JmNAC genes were specifically expressed in walnut exocarp and embryos, respectively (Figure 6c,d), whereas 58 JmNAC genes were commonly expressed in the two tissues. A total of 17 and 14 JmNAC genes were specifically expressed in walnut exocarp and embryos, respectively (Figure 6e). To analyze the biological function of 58 genes expressed in all fruit samples, we performed function annotation analysis (Figure 6f). The GO analysis demonstrated that these genes were mainly divided into 25 functional categories, including 16 biological process (BP) categories, 6 cellular component (CC) categories and 3 molecular function (MF) categories. In the BP category, the most abundant terms were biological regulation (GO:0065007), regulation of biological process (GO:0050789) and metabolic process (GO:0008152). In the CC category, the most abundant term was cell and cell part. In the MF category, the most abundant term was transcription regulator activity (GO:0140110).
Figure 6.
Expression profiles and functional analysis of JmNAC genes in different stages of fruit development and ripening. (a) Photographs of J. mandshurica fruits. (b) The tissue structure of J. mandshurica fruit. “1” indicates the walnut exocarp. “2” indicates the walnut embryos. (c) Heatmap representing 75 JmNAC gens in walnut exocarp at 30 days (S1), 50 days (S2), 70 days (S3) and 90 days (S4) after natural pollination. (d) Heatmap representing 75 JmNAC gens in walnut embryos at 30 days (S1), 50 days (S2), 70 days (S3) and 90 days (S4) after natural pollination. Group I–VIII correspond to the subgroups in the phylogenetic tree in Figure 2. (e) Upset plot of JmNAC genes expressed in walnut exocarp and embryos. (f) GO annotation analyses of the common JmNAC genes expressed in walnut exocarp and embryos.
In walnut exocarp, a total of 39 JmNAC genes were significantly upregulated in the S4 stage, whereas 17 JmNAC genes exhibited higher expression levels in the S1 stage. A total of eight JmNAC genes (JmNAC109, JmNAC020, JmNAC069, JmNAC082, JmNAC025, JmNAC028, JmNAC068, and JmNAC073) exhibited higher expression levels in S2-S4 stages than that in the S1 stage during development, indicating their positive roles in fruit development. Among the identified subgroups, Group II contained the largest number of JmNAC genes that were upregulated or downregulated during the development of walnut exocarp. We found that 25 JmNAC genes exhibited high expression during the development of walnut embryos, especially in the S4 stage. In addition, a total of 35 JmNAC genes were significantly upregulated in the S1 stage. Furthermore, the expression of five JmNAC genes (JmNAC024, JmNAC099, JmNAC041, JmNAC037, and JmNAC007) and seven JmNAC genes (JmNAC098, JmNAC107, JmNAC008, JmNAC106, JmNAC102, JmNAC022, and JmNAC086) was relatively high in the S2 and S3 stages, respectively. Genes with high expression in the S4 stage were mainly derived from Group II. A total of 12 JmNAC genes with high expression were commonly expressed in the S4 stage in walnut exocarp and embryos.
2.7. Validation of Expression Pattern of RNA-seq Data Using RT-qPCR
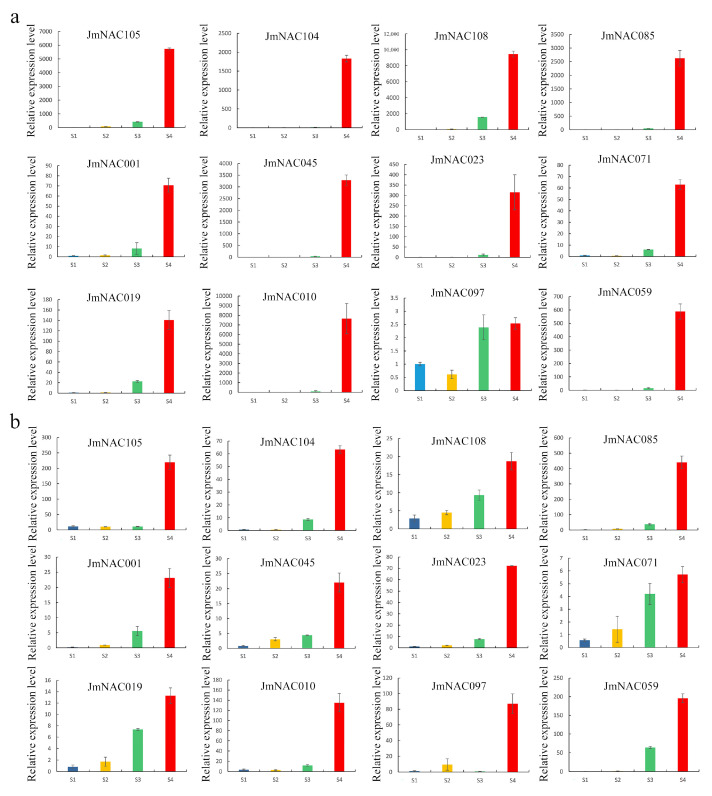
To better understand the expression pattern of JmNAC genes during the development and ripening of fruits, 12 potential JmNAC genes from four developmental stages of walnut exocarp and embryos were selected and used for RT-qPCR analysis with specific primers (Table S2). As demonstrated in Figure 7, all selected JmNAC genes exhibited relatively high expression in the S4 stage in the two different tissues, which is consistent with the results obtained from RNA-seq analysis. Furthermore, the expression level of six JmNAC genes, including JmNAC105, JmNAC104, JmNAC085, JmNAC045, JmNAC010, and JmNAC023, in the S4 stage was significantly higher than that in the S1 stage, which indicated a positive role of JmNAC genes during the development of fruits. Therefore, RT-qPCR results supported the credibility and accuracy of RNA-seq data in this study.
Figure 7.
Expression profiles of 12 JmNAC genes during the development and ripening of fruits. (a) Relative expression of 12 JmNAC genes in walnut exocarp using RT-qPCR. (b) Relative expression of 12 JmNAC genes in walnut embryos using RT-qPCR. The y-axis shows the relative gene expression levels (2−ΔΔCt) analyzed by qRT-PCR. The x-axis represents the different tissue samples. The values are expressed as mean ± standard deviation of three replicates.
3. Discussion
J. mandshurica is a well-known and valuable tree species for its elite hardwood, edible seed kernel and extensive medicinal value of walnut exocarp [46,47]. However, only a few studies have been conducted on its growth, development, and stress response at molecular level due to the lack of genome sequence and RNA-seq data. Previous studies have found that TFs are powerful tools for regulating plant development and enhancing plant tolerance to environmental stresses such as drought, low temperature, high salinity, and high temperature. NAC TFs are one of the largest plant-specific TF families in plants and play a vital role in regulating plant senescence, metabolite synthesis, fruit development and signal transduction. Genome-wide identification and analysis are indispensable methods for investigating specific functions of the NAC gene family. To date, some NAC family members have been identified and characterized in plant species such as A. thaliana [21,48], Pyrus bretschneideri [49], Jatropha curcas [50], and Sorghum bicolor [51]. However, genome-wide analysis of the gene family of J. mandshurica has not been performed, and the regulatory function of JmNAC genes remains unclear. In the present study, the identification and expression analysis of the NAC family at the genome level provided genetic information and a better understanding of the development of walnut exocarp and embryo.
The number and structural features of TFs in a gene family are related not only to the genome size of the species but also to the influence of long-term evolution of plants. In this study, we identified a total of 114 genes encoding NAC protein in the J. mandshurica genome (Table S1). The number of NAC genes was significantly higher in J. mandshurica than that in Panax ginseng (89 PgNACs genes) [43], Phyllostachys heterocycla (94 PeNACs genes) [52], Xanthoceras sorbifolia (103 XsNACs genes) [26] and J. regia (102 JrNACs genes) [53] but was the same as in Betula pendula (114 BpNACs genes) [54] and Dimocarpus longan (114 DlNACs genes) [44], indicating high conservation of NAC gene family in J. mandshurica, which may be related to gene duplication during species formation and evolution. With regard to subcellular localization, all 114 JmNACs were predicted and located in the nucleus, which may be closely related to the expression regulation of target genes. Furthermore, the exons and introns of identified JmNAC genes varied from 3–13 and 1–2, respectively, wherein genes on the same branch displayed similar organization of exons and introns (Figure 3). Most JmNAC genes consist of three exons, similar to J. regia [53], Cucumis sativus [55] and Chenopodium quinoa [56], which may be related to the functional differentiation and structural diversity of the JmNAC gene family. Although there were some differences in the arrangement of conserved motifs, numerous JmNAC genes shared similar motif composition, indicating similar structure and biological function. These results confirm the characteristics of the JmNAC gene family and facilitate further study on the function of JmNAC genes.
Investigating the origin and differentiation of gene families is a prime research focus of evolutionary studies. Identifying a gene family based on the whole genome data is of great significance for understanding gene evolution history, gene function and species differentiation [57,58]. According to the phylogenetic tree analysis of NAC genes in J. mandshurica and A. thaliana, 114 JmNACs were significantly clustered into eight subgroups (Figure 2), wherein Group I contained the largest number of JmNAC and AtNAC genes, and the genes from Group I mainly involved in DNA-binding TF activity, embryo development, leaf senescence, signal transduction, oxidative stress response, and xylem formation, indicating various functions of JmNAC genes in plant growth and development as well as stress response. Also, the findings showed that the NAC members from the same subfamily share a similar functional characteristic, however, further functional feature need to be confirmed. Differences in the NAC family members between J. mandshurica and A. thaliana in different subgroups may be owing to gene differentiation caused by the continuously changing environment during evolution.
Previous studies have confirmed that gene duplication plays a crucial role in adaptive evolution of plants. Many plants have experienced evolutionary events such as large fragments and tandem and whole genome duplication, which contribute to the formation and rapid expansion of gene families. Furthermore, to adapt to the drastically changing external environment and avoid destructive extinction, high-level gene duplication events may occur in many gene families such as WRKY, MYB, and MADS-box TFs. In the present study, four tandemly duplicated gene pairs were identified, suggesting that the expansion of the NAC gene family might have been originated from genome polyploidy events that contributed to diversify gene function. In addition, among the identified 114 JmNACs, we obtained 50 orthologous gene pairs using synteny analysis (Figure 5), indicating that J. mandshurica may undergo multiple selecting evolutionary directions. JmNAC93, JmNAC106 and JmNAC107 had more than one orthologous gene pair, which further supports the fact that they play a crucial role in the adaptation of plants to the changing terrestrial environment.
Gene family members have formatted protein complexes and diverse PPIs during long evolutionary processes. Analysis of the PPT network indicated that JmNACs were widely involved in tissue formation (Figure 4). A total of five key hub genes including JmNAC055, JmNAC085, JmNAC097, JmNAC011, and JmNAC102 were found in the interaction network, and they could be used as candidate gene for studying tissue development and formation of J. mandshurica. JmNAC085 and JmNAC097 were significantly upregulated in the S4 stage, indicating that they may be positive regulators and exhibited tissue-specific function for regulating the development and maturation of walnut exocarp and embryo and could be used as candidate genes in future study on fruit development of J. mandshurica. These results further enriched the molecular mechanism of NAC transcription factor for regulating J. mandshurica fruit development.
Fruit development is a complex biological process that involves nutrient formation and secondary product metabolism, directly affecting fruit quality formation and commodity value [59]. Previous studies have found that the development, maturation, senescence and quality formation of fruits are specifically regulated by many environmental and regulatory factors, especially NAC TFs [38]. A lot of attention has been paid to the role of the NAC gene family members during fruit development and ripening. A recent study found that PpNAC1 regulated the expression of ripening-related PpAAT1 and contributed to volatile ester formation in Prunus persica [60], indicating a key role during fruit ripening. In papaya (Carica papaya), CpNAC1 was reported to regulate carotenoid accumulation during fruit ripening [61]. Similar results were reported in studies on Actinidia eriantha [45], Pyrus pyrifolia [62], Malus domestica [63], Eriobotrya japonica [64], and other fruit tree species. In addition, NAC TFs also regulate fruit development and ripening through multiple hormone such as ethylene and abscisic acid biosynthesis and signal transduction pathways [65]. For instances, in tomato, SNAC4 and SNAC9 could positively regulate the fruit ripening process by interacting with ethylene synthesis genes [66]. Also, the expression patterns of the SNAC genes were affected by abscisic acid in tomato [67]. However, the regulatory function of NAC TFs during fruit development in J. mandshurica remains unknown. The expression profile analysis of NAC gene in different tissues and developmental stages of J. mandshurica provides new data for understanding the potential biological functions of NAC gene in J. mandshurica. In the present study, the process of J. mandshurica fruit ripening is accompanied by the gene expression and TFs regulation. Expression analyses of all identified 114 JmNACs during different developmental stages in J. mandshurica revealed that most genes may have possible functions during fruit development and ripening. Particularly, according to the expression analysis, we found that most of the same subfamily genes displayed the similar expression patterns in two tissues, but small amounts of genes from the same subfamilies also showed differential expression level, which may be caused by the functional differentiation of JmNACs. A total of 72 (63.16%) and 75 (65.79%) JmNAC genes were specifically expressed during fruit development in walnut exocarp and embryos (Figure 6). Furthermore, 39 and 25 key JmNAC genes were significantly upregulated in the S4 stage in walnut exocarp and embryos, respectively, indicating that they specifically regulated the development of walnut exocarp and embryos. We also identified 12 key hub JmNAC genes that exhibited constitutive high expression levels during ripening stages in walnut exocarp and embryos, and RT-qPCR analysis also confirmed the results (Figure 7), inferring a strict regulation of fruit development. Therefore, we predict that these key JmNAC genes above will offer new avenues for exploring the role of NAC TFs in fruit growth and development, and their genetic functions require further characterization. Further validation of their functions will greatly advance our understanding of fruit development in J. mandshurica.
4. Materials and Methods
4.1. Plant Materials
The fruit of J. mandshurica used in this study was collected from the Northeast Forestry University (126°37′57.28″ E, 45°43′6.53″ N), Harbin, Heilongjiang province, China. From June to September, the fruits with strong growth and no pests or diseases were sampled and used for RNA extraction. Walnut exocarp and embryos were collected from four stages, at 30 days (S1 stage), 50 days (S2 stage), 70 days (S3 stage), and 90 days (S4 stage) after natural pollination, and were used for real-time reverse transcription polymerase chain reaction (RT-qPCR). In which, S1, S2, and S3 indicate the different developmental stages, while the S4 represents the ripening stages of Walnut fruits. For each stage, the sampling time (2:00 p.m.) of collected fruits was consistent. All sampled fruits were rapidly frozen in liquid nitrogen and subsequently stored at −80 °C for RNA extraction.
4.2. Identification of JmNAC Genes in J. mandshurica Genome
To perform genome-wide identification of the JmNAC gene family, the whole genome sequence and annotation data were accessed from our previous genomic study, and the accession number of the Genome Warehouse in National Genomics Data Center (NGDC) (https://ngdc.cncb.ac.cn/) (accessed on 6 October 2021) was PRJCA006358. We assembled a high-quality chromosome-scale reference genome assembly and annotation for J. mandshurica (n = 16) with a contig N50 of 21 Mb and BUSCO complete gene percentage of 98.3% by combining PacBio high-fidelity (HiFi) reads with high-throughput chromosome conformation capture (Hi-C) data. The genome obtained is of sufficient quality for genome-wide studies and expression analysis of NAC gene families and gene function in J. mandshurica. The A. thaliana NAC full-length protein sequences were obtained from The Arabidopsis Information Resource database (TAIR; https://www.arabidopsis.org/) (accessed on 4 August 2021). First, JmNAC genes in J. mandshurica were identified using two BLAST methods. A total of 96 AtNAC genes were used as reference sequences to search the possible JmNAC genes using BLAST (e-value, 1 × e−5) in the TBtools software (version 1.087) [68]. Subsequently, all genes retrieved using BLAST were further identified using Batch NCBI CD-Search Tools (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (accessed on 4 August 2021) to obtain the conserved NAC domains. Lastly, the Pfam (http://pfam.xfam.org/) (accessed on 6 August 2021) and SMART (http://smart.embl-heidelberg.de/) (accessed on 6 August 2021) databases were used to verify the final candidate JmNAC genes.
4.3. Sequence Analysis and Gene Structural Characterisation
The number of amino acids and length of coding sequences (CDS) were calculated using the TBtools software [68]. Molecular weight (MW) and isoelectric point (pI) of each NAC protein were analysed using the online ExPASy program (https://web.expasy.org/protparam/) (accessed on 8 August 2021). Subcellular localisation of all JmNAC proteins was performed using the Cell-PLoc 2.0 web tool (http://www.friendbio.com/meansMore/id/52) (accessed on 8 August 2021). To investigate the gene motifs of JmNAC proteins, MEME (http://meme-suite.org/tools/meme) (accessed on 10 August 2021) was used, with the parameters set at maximum eight motifs. To further visualise the motif composition and gene structure, integration analysis was conducted using TBtools [68].
4.4. Chromosomal Location and Evolutionary Analysis of JmNAC Genes
For identifying chromosomal locations, the TBtools software was used to locate and visually map JmNAC genes on 16 chromosomes in J. mandshurica based on the General Feature Format (GFF) information. Furthermore, tandemly duplicated gene pairs were marked with a red line. The names of candidate JmNAC genes were encoded according to the physical location information of chromosomes. To further analyse gene replication, duplicated and orthologous pairs of JmNAC genes were obtained by constructing a dual synteny plot in MCscanX, and the Advanced Circos software (https://github.com/CJ-Chen/TBtools) (accessed on 15 August 2021) was used to display the collinearity circle plot. Homology among NAC genes in J. mandshurica and five representative plant species including J. regia, J. sigillata, P. trichocarpa, A. thaliana and V. vinifera was inferred using the multiple collinearity scan toolkit (MCScanX, http://chibba.pgml.uga.edu/mcscan2/) (accessed on 15 August 2021) with default parameters of the TBtools software.
4.5. Chromosomal Location and Evolutionary Analysis of JmNAC Genes
To further demonstrate the evolutionary relationship between NAC proteins of J. mandshurica and A. thaliana, multiple sequence alignment was performed using the ClustalW algorithm in MEGA (version 7.0) software [69] with the following parameters: pairwise deletion and 1000 replicates for bootstrap analysis. Subsequently, the alignment results were used as input files to construct a phylogenetic tree using neighbour-joining methods. For graphic display, the final phylogenetic tree of the JmNAC gene family was further processed using the Interactive Tree Of Life (iTOL) (https://itol.embl.de/) (accessed on 20 August 2021) web tools. NAC proteins from J. mandshurica and A. thaliana were eventually divided into different subgroups.
4.6. Interaction Network Construction of JmNAC Genes
To predict the function of JmNAC proteins, protein–protein interaction (PPI) networks were constructed on the STRING website (https://www.string-db.org/) (accessed on 20 August 2021). Briefly, the amino acid sequences of JmNAC proteins were used to scan the representative AtNAC proteins using BLASTP in TBtools. Subsequently, the candidate sequences were uploaded on the STRING website to construct an interaction network of JmNAC genes with default parameters.
4.7. Expression Analysis of JmNAC Genes during the Development and Ripening of Fruits
To analyze the expression patterns of JmNAC genes in different developmental stages of fruits of J. mandshurica, the raw RNA-seq reads of two tissues including walnut exocarp and embryos were obtained from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) (accessed on 9 October 2021) sequence read archive (SRA) database (PRJNA733587). After quality control, alignment and quantitative analysis, the count matrix was obtained and used to calculate the expression of JmNAC genes in reads per kilobase per million (RPKM). Subsequently, the normalized data were further used to obtain the expression heatmap of all JmNAC genes involved in the development and ripening of fruits in J. mandshurica using the Heatmap program in TBtools. Furthermore, the common JmNAC genes in walnut exocarp and embryos were selected and annotated to the Gene Ontology (GO) database using the eggnog database (http://eggnog-mapper.embl.de/) (accessed on 5 October 2021).
4.8. RNA Extraction and RT-qPCR
The total RNA in four developmental stages of walnut exocarp and embryos was extracted using the plant total RNA extraction kit (Takara, Beijing, China) according to the manufacturer’s instructions. Subsequently, the integrity and quality of total RNA of all tissue samples were assessed using agarose gel electrophoresis and the K5500 plus microspectrophotometer (KAIAO Technology Development Co., Ltd., Beijing, China), respectively. Approximately 1 μg of total RNA was used for synthesising cDNA using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Kyoto, Japan), and the amplification products were used for qRT-PCR analysis. The qRT-PCR primers were designed using Primer3web tools (version 4.1.0; https://primer3.ut.ee/) (accessed on 5 September 2021). Furthermore, 18s RNA was used as an internal reference gene. A total of 12 key DEGs were selected and used for RT-qPCR analysis with specific primers (Table S2). Real-time PCR was performed in triplicates on the ABI 7500 Fast Real-Time Detection System using the TaKaRa SYBR Green Mix kit (TaKaRa, Beijing, China). The PCR reaction was performed in a solution of 20 µL, which contained 10 µL of 2× SYBR Premix Ex Taq, 6 µL of double-distilled water (ddH2O), 2 µL of cDNA template, 0.8 µL of upstream and downstream primers (10 μmol/L) and 0.4 µL of ROX reference dye. The PCR reaction conditions were set as follows: 95 °C for 30 s, 40 cycles at 95 °C for 5 s, 60 °C for 35 s and 95 °C for 15 s, 60 °C for 1 min, followed by 95 °C for 15 s. The relative expression level was assessed using the 2−∆∆CT method. A histogram was created based on the expression data of qRT-PCR.
Acknowledgments
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Abbreviations
| NAC | NAM, ATAF and CUC |
| TFs | transcription factors |
| CDS | coding sequence |
| M W | molecular weight |
| pI | isoelectric point |
| GO | Gene Ontology |
| RPKM | reads per kilobase per million |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222212414/s1.
Author Contributions
X.Z. and C.D. conceived and designed the research; X.L., K.C. and Y.L. performed the experiments; X.P., Y.H., F.M. and X.S. analyzed the date; X.L. wrote the manuscript and M.T. revised the manuscript; X.Z. and C.D. provided the funding. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Fundamental Research Funds for the Central Universities (2572020DR01 and 2572020AW07) and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The transcriptome raw datasets (two tissues for four stages with triplicates) were deposited in the National Center for Biotechnology Information (NCBI) Sequences Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna733587) (accessed on 29 May 2021) under the accession number PRJNA733587 (including embryos and exocarp).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang Y.J., Zhang Y., Fan C.J., Wei Y.C., Meng J.C., Li Z., Zhong C.L. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021;21:328. doi: 10.1186/s12870-021-03083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai X.L., VegaLéon R., Hugouvieux V., BlancMathieu R., Wal F.v.d., Lucas J., Silva C.S., Jourdain A., Muino J.M., Nanao M.H., et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021;12:4760. doi: 10.1038/s41467-021-24978-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D., Li Y.Y., Zhou Z.C., Xiang X.H., Liu X., Wang J., Hu Z.R., Xiang S.P., Li W., Xiao Q.Z., et al. Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidase NtRbohE expression. Plant Physiol. 2021;186:1706–1720. doi: 10.1093/plphys/kiab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z.M., Wong D.C.J., Wang Y., Xu G.Z., Ren C., Liu Y.F., Kuang Y.F., Fan P.G., Li S.H., Xin H.P., et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021;186:1660–1678. doi: 10.1093/plphys/kiab142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L.M., Leng C.Y., Luo H., Wu X.Y., Liu Z.Q., Zhang Y.M., Zhang H., Xia Y., Shang L., Liu C.M., et al. Sweet Sorghum Originated through Selection of Dry, a Plant-Specific NAC Transcription Factor Gene. Plant Cell. 2018;30:2286–2307. doi: 10.1105/tpc.18.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souer E., Van H.A., Kloos D., Mol J., Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 7.Aida M., Ishida T., Fukaki H., Tasaka F.M. Genes Involved in Organ Separation in Arabidopsis: An Analysis of the cup-shaped cotyledon Mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ooka H., Satoh K., Doi K., Nagata T., Kikuchi S. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2004;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 9.Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Ernst H.A., Olsen A.N., Larsen S., Lo L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen M.K., Rung J.H., Gregersen P.L., Gjetting T., Fuglsang A.T., Hansen M., Joehnk N., Lyngkjaer M.F., Collinge D.B. The Hv NAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007;65:137–150. doi: 10.1007/s11103-007-9204-5. [DOI] [PubMed] [Google Scholar]
- 12.Trupkin S.A., Astigueta F.H., Baigorria A.H., García M.N., Delfosse V.C., González S.A., Torre M.C.P.d.l., Moschen S., Lía V.V., Fernández P., et al. Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrida. Plant Sci. 2019;287:110195. doi: 10.1016/j.plantsci.2019.110195. [DOI] [PubMed] [Google Scholar]
- 13.Sun Q.W., Huang J.F., Guo Y.F., Yang M.M., Guo Y.J., Li J., Zhang J., Xu W.L. A cotton NAC domain transcription factor, GhFSN5, negatively regulates secondary cell wall biosynthesis and anther development in transgenic Arabidopsis. Plant Physiol. Biochem. 2020;146:303–314. doi: 10.1016/j.plaphy.2019.11.030. [DOI] [PubMed] [Google Scholar]
- 14.Ha K.M., Anh T.T.N., Seong C.J., Jun P.E., Hyoshin L., Gwan K.D., Seongbin H., Heung K.J. Wood transcriptome analysis of Pinus densiflora identifies genes critical for secondary cell wall formation and NAC transcription factors involved in tracheid formation. Tree Physiol. 2021;41:1289–1305. doi: 10.1093/treephys/tpab001. [DOI] [PubMed] [Google Scholar]
- 15.Wang H.L., Zhang Y., Wang T., Yang Q., Yang Y.L., Li Z., Li B.S., Wen X., Li W.Y., Lun Y.W., et al. An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell. 2021;33:1594–1614. doi: 10.1093/plcell/koab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmen M.P., José G.V., Sonia O., Victoriano M., María U., Jeremy P., Ana C., Catharina M., Iraida A., Lothar W., et al. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell. 2021;33:1574–1593. doi: 10.1093/plcell/koab070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zoë M., Trevor H.Y., Sophie W., Jun S., Charles F.F., Karen B.M., Somers D.J., Gong Y., Zhang Z., Julia V., et al. Apple Ripening Is Controlled by a NAC Transcription Factor. Front. Genet. 2021;12:671300. doi: 10.3389/fgene.2021.671300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anat H., Ran S., Hanita Z., Tzahi A. The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. J. Exp. Bot. 2013;64:5497–5507. doi: 10.1093/jxb/ert324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong X.F., Yan Q., Wu F., Ma Q., Zhang J.Y. Genome-Wide Analysis of the Role of NAC Family in Flower Development and Abiotic Stress Responses in Cleistogenes songorica. Genes. 2020;11:927. doi: 10.3390/genes11080927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S.S., Wu J.D., Zhang Y.F., Zhao Y., Jie X.W., Li Y., Xie J.B. Genome-Wide Analysis of Coding and Non-coding RNA Reveals a Conserved miR164–NAC–mRNA Regulatory Pathway for Disease Defense in Populus. Front. Genet. 2021;12:668940. doi: 10.3389/fgene.2021.668940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W.P., Zhao B.G., Chao Q., Wang B.C., Zhang Q., Zhang C.X., Li S.F., Jin F.X., Yang D.G., Li X.H. Function analysis of ZmNAC33, a positive regulator in drought stress response in Arabidopsis. Plant Physiol. Biochem. 2019;145:174–183. doi: 10.1016/j.plaphy.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 22.Liu X.H., Lyu Y.S., Yang W.P., Yang Z.T., Lu S.J., Liu J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020;18:1317–1329. doi: 10.1111/pbi.13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q., Yan S.J., Huang W.J., Yang J.Y., Dong J.F., Zhang S.H., Zhao J.L., Yang T.F., Mao X.X., Zhu X.Y., et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018;98:289–302. doi: 10.1007/s11103-018-0768-z. [DOI] [PubMed] [Google Scholar]
- 24.Li M., Chen R., Jiang Q.Y., Sun X.J., Zhang H., Hu Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2020;105:333–345. doi: 10.1007/s11103-020-01091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang L.P., Peng Z.Y., Xu P.L., Tang G.Y., Ma C.L., Zhu J.Q., Shan L., Wan S.B. Genome-Wide Identification of NAC Transcription Factors and Their Functional Prediction of Abiotic Stress Response in Peanut. Front. Genet. 2021;12:630292. doi: 10.3389/fgene.2021.630292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang Y.H. Genome-Wide Identification and Characterization of Yellow Horn (Xanthoceras sorbifolia Bunge) NAC Transcription Factor Gene Family against Diverse Abiotic Stresses. Russ. J. Genet. 2021;57:423–437. doi: 10.1134/S1022795421040062. [DOI] [Google Scholar]
- 27.Han D.G., Du M., Zhou Z.Y., Wang S., Li T.M., Han J.X., Xu T.L., Yang G.H. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020;21:1198. doi: 10.3390/ijms21041198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., Wang J.W., Sun L.J., Song X.S. The molecular cloning and functional characterization of ChNAC1, a NAC transcription factor in Cerasus humilis. Plant Growth Regul. 2019;89:331–343. doi: 10.1007/s10725-019-00536-9. [DOI] [Google Scholar]
- 29.Wang Y., Cao S.J., Guan C.J., Kong X., Wang Y.P., Cui Y., Liu B., Zhou Y.W., Zhang Y.N. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020;149:96–110. doi: 10.1016/j.plaphy.2020.01.036. [DOI] [PubMed] [Google Scholar]
- 30.Ping H., Min Z.K., Ping Y.C. BpNAC012 Positively Regulates Abiotic Stress Responses and Secondary Wall Biosynthesis. Plant Physiol. 2019;179:700–717. doi: 10.1104/pp.18.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Y.F., Wu K., Chen J.F., Liu Q., Wu Y.J., Liu B.M., Fu X.D. OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice. 2018;11:1–14. doi: 10.1186/s12284-018-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan W., Chen J.Y., Kuang J.F., Lu W.J. Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against C olletotrichum musae. Mol. Plant Pathol. 2016;17:330–338. doi: 10.1111/mpp.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F., Chen H.W., Li Q.T., Wei W., Li W., Zhang W.K., Ma B., Bi Y.D., Lai Y.C., Liu X.L., et al. Gm WRKY 27 interacts with Gm MYB 174 to reduce expression of Gm NAC 29 for stress tolerance in soybean plants. Plant J. 2015;83:224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- 34.Koltunow A.M., Brennan P. Paternal transmission of a seed size reduction gene varies with age of a primary transformant and seed set is also influenced by gene expression in maternal tissues. Mol. Breed. 1998;4:253–261. doi: 10.1023/A:1009615103408. [DOI] [Google Scholar]
- 35.Giovannoni J. Molecular Biology Of Fruit Maturation And Ripening. Annu. Rev. Plant Biol. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- 36.Yamasaki K., Kigawa T., Inoue M., Watanabe S., Tateno M., Seki M., Shinozaki K., Yokoyama S. Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol. Biochem. 2008;46:394–401. doi: 10.1016/j.plaphy.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 37.Wang J.F., Wang Y.P., Zhang J., Ren Y., Li M.Y., Tian S.W., Yu Y.T., Zuo Y., Gong G., Ying Z.H., et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 2021;8:214. doi: 10.1038/s41438-021-00649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sara F., Chiara M., Simona M. The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 2021;21:238. doi: 10.1186/s12870-021-03029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerstin D., Johanna W.J., Miguel N.G., Almuth H., Karl L., Ines E., Malin E. Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 2017;17:6. doi: 10.1186/s12870-016-0952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S.L., Dong R.Z., Wang Y.W., Li X.M., Ji M.M., Wang X.P. NAC domain gene VvNAC26 interacts with VvMADS9 and influences seed and fruit development. Plant Physiol. Biochem. 2021;164:63–72. doi: 10.1016/j.plaphy.2021.04.031. [DOI] [PubMed] [Google Scholar]
- 41.Li B., Fan R.Y., Yang Q.S., Hu C.H., Sheng O., Deng G.M., Dong T., Li C.Y., Peng X.X., Bi F.C., et al. Genome-Wide Identification and Characterization of the NAC Transcription Factor Family in Musa Acuminata and Expression Analysis during Fruit Ripening. Int. J. Mol. Sci. 2020;21:634. doi: 10.3390/ijms21020634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min X.Y., Jin X.Y., Zhang Z.S., Wei X.Y., Ndayambaza B., Wang Y.R., Liu W.X. Genome-Wide Identification of NAC Transcription Factor Family and Functional Analysis of the Abiotic Stress-Responsive Genes in Medicago sativa L. J. Plant Growth Regul. 2020;39:324–337. doi: 10.1007/s00344-019-09984-z. [DOI] [Google Scholar]
- 43.Liu Q., Sun C.Y., Han J.Z., Li L., Wang K.Y., Wang Y.F., Chen J., Zhao M.Z., Wang Y., Zhang M.P. Identification, characterization and functional differentiation of the NAC gene family and its roles in response to cold stress in ginseng, Panax ginseng C.A. Meyer. PLoS ONE. 2020;15:e0234423. doi: 10.1371/journal.pone.0234423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nigarish M., Chen Y.K., Chen X.H., Azher N.M., Junaid I., Muhammad R.H., Shen X., Lin Y.L., Xu X.H., Lai Z.X. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol. Biochem. 2020;157:169–184. doi: 10.1016/j.plaphy.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Feng J.D., Qiang J.Z., Hui F.H., Chen L., Liao G.L., He Y.Q., Huang C.H., Xu X.B. Genome-wide identification and comprehensive analysis of NAC family genes involved in fruit development in kiwifruit (Actinidia) BMC Plant Biol. 2021;21:44. doi: 10.1186/s12870-020-02798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luan F., Wang Z.Y., Yang Y., Ji Y.F., Lv H.Z., Han K., Heng L.D., Shang X.F., He X.R., Zeng N. Juglans mandshurica Maxim.: A Review of Its Traditional Usages, Phytochemical Constituents, and Pharmacological Properties. Front. Pharmacol. 2021;11:569800. doi: 10.3389/fphar.2020.569800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umarov A.U., Burnasheva S.N., Makhmudova K.S. Oil from the seeds of Juglans mandshurica and Anabasis Aphylla. Chem. Nat. Compd. 1970;6:258–259. doi: 10.1007/BF00941697. [DOI] [Google Scholar]
- 48.Lee M.H., Jeon H.S., Kim H.G., Park O.K. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 2017;214:343–360. doi: 10.1111/nph.14371. [DOI] [PubMed] [Google Scholar]
- 49.Gong X., Zhao L.Y., Song X.F., Lin Z.K., Gu B.J., Yan J.X., Zhang S.L., Tao S.T., Huang X.S. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri) BMC Plant Biol. 2019;19:161. doi: 10.1186/s12870-019-1760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z.Y., Xu X.Q., Xiong W.D., Wu P.Z., Chen Y.P., Li M.R., Wu G.J., Jiang H.W. Genome-Wide Analysis of the NAC Gene Family in Physic Nut (Jatropha curcas L.) PLoS ONE. 2017;10:e0131890. doi: 10.1371/journal.pone.0131890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadier Y., Zu Y.Y., Dai Q.M., Song G., Lin S.W., Sun Q.P., Pan J.B., Lu M. Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench] Plant Growth Regul. 2017;83:301–312. doi: 10.1007/s10725-017-0295-y. [DOI] [Google Scholar]
- 52.Shan X.M., Yang K.B., Xu X.R., Zhu C.L., Gao Z.M. Genome-Wide Investigation of the NAC Gene Family and Its Potential Association with the Secondary Cell Wall in Moso Bamboo. Biomolecules. 2019;9:609. doi: 10.3390/biom9100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan H., Yan F., Yan Y.J., Chen P., Xi R.M., Ullah I., Peng X.B., Luo X., Yue M., Zhao P. Genome-wide analysis of evolution and expression profiles of NAC transcription factor gene family in Juglans regia L. J. Fr. Natl. Inst. Agric. Food Environ. (INRAE) 2020;77:381–390. doi: 10.1007/s13595-020-00983-9. [DOI] [Google Scholar]
- 54.Chen S., Lin X., Zhang D.W., Li Q., Zhao X.Y., Chen S. Genome-Wide Analysis of NAC Gene Family in Betula pendula. Forests. 2019;10:741. doi: 10.3390/f10090741. [DOI] [Google Scholar]
- 55.Liu X.W., Wang T., Bartholomew E., Black K., Dong M.M., Zhang Y.Q., Yang S., Cai Y.L., Xue S.D., Weng Y.Q., et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.) Hortic. Res. 2018;5:31. doi: 10.1038/s41438-018-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li F., Guo X., Liu J., Zhou F., Wen R. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Transcription Factor in Chenopodium quinoa. Genes. 2019;10:500. doi: 10.3390/genes10070500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan K., Mao Z.J., Ye F.T., Pan X.F., Li Z.W., Lin W.W., Zhang Y.Q., Huang J.W., Lin W.X. Genome-wide identification and molecular evolution analysis of the heat shock transcription factor (HSF) gene family in four diploid and two allopolyploid Gossypium species. Genomics. 2021;113:3112–3127. doi: 10.1016/j.ygeno.2021.07.008. [DOI] [PubMed] [Google Scholar]
- 58.Wu W.H., Zhu S., Zhu L.M., Wang D.D., Liu Y., Liu S.Q., Zhang J.J., Hao Z.D., Lu Y., Cheng T.L., et al. Characterization of the Liriodendron Chinense MYB Gene Family and Its Role in Abiotic Stress Response. Front. Plant Sci. 2021;12:641280. doi: 10.3389/fpls.2021.641280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Wu X.F., Tang Y., Li J.G., Zhao M.L. RNA-Seq Provides New Insights into the Molecular Events Involved in “Ball-Skin versus Bladder Effect” on Fruit Cracking in Litchi. Int. J. Mol. Sci. 2021;22:454. doi: 10.3390/ijms22010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao X.M., Wei C.Y., Duan W.Y., Gao Y., Kuang J.F., Chun L.M., Chen K.S., Klee H., Zhang B. Transcriptional and epigenetic analysis reveals that NAC transcription factors regulate fruit flavor ester biosynthesis. Plant J. 2021;106:785–800. doi: 10.1111/tpj.15200. [DOI] [PubMed] [Google Scholar]
- 61.Fu C.C., Han Y.C., Fan Z.Q., Chen J.Y., Chen W.X., Lu W.J., Kuang J.F. The Papaya Transcription Factor CpNAC1 Modulates Carotenoid Biosynthesis through Activating Phytoene Desaturase Genes CpPDS2/4 during Fruit Ripening. J. Agric. Food Chem. 2016;64:5454–5463. doi: 10.1021/acs.jafc.6b01020. [DOI] [PubMed] [Google Scholar]
- 62.Li M.T., Cheng C.X., Zhang X.F., Zhou S.P., Wang C.H., Ma C.H., Yang S.L., Crestini C. PpNAC187 Enhances Lignin Synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘Hard-End’ Fruit. Molecules. 2019;24:4338. doi: 10.3390/molecules24234338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q.J., Li T., Zhang L.J., Dong W.X., Wang A.D. Expression analysis of NAC genes during the growth and ripening of apples. Hortic. Sci. 2018;45:1–10. doi: 10.17221/153/2016-HORTSCI. [DOI] [Google Scholar]
- 64.Ge H., Zhang J., Zhang Y.J., Li X., Yin X.R., Grierson D., Chen K.S. EjNAC3 transcriptionally regulates chilling-induced lignification of loquat fruit via physical interaction with an atypical CAD-like gene. J. Exp. Bot. 2017;68:5129–5136. doi: 10.1093/jxb/erx330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kou X.H., Zhou J.Q., Wu C.E., Yang S., Liu Y.F., Chai L.P., Xue Z.H. The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: A review. Plant Mol. Biol. 2021;106:223–238. doi: 10.1007/s11103-021-01128-w. [DOI] [PubMed] [Google Scholar]
- 66.Kou X.H., Liu C., Han L.H., Wang S., Xue Z.H. NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genom. 2016;291:1205–1217. doi: 10.1007/s00438-016-1177-0. [DOI] [PubMed] [Google Scholar]
- 67.Kou X.H., Wang S., Wu M.S., Guo R.Z., Xue Z.H., Meng N., Tao X.M., Chen M.M., Zhang Y.F. Molecular Characterization and Expression Analysis of NAC Family Transcription Factors in Tomato. Plant Mol. Biol. Report. 2014;32:501–516. doi: 10.1007/s11105-013-0655-3. [DOI] [Google Scholar]
- 68.Chen C.J., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y.H., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 69.Sudhir K., Glen S., Koichiro T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:187–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome raw datasets (two tissues for four stages with triplicates) were deposited in the National Center for Biotechnology Information (NCBI) Sequences Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna733587) (accessed on 29 May 2021) under the accession number PRJNA733587 (including embryos and exocarp).