Abstract
The vascular endothelial growth factor (VEGF) family of cytokines plays a key role in vasculogenesis, angiogenesis, and lymphangiogenesis. VEGF-A is the main member of this family, alongside placental growth factor (PlGF), VEGF-B/C/D in mammals, and VEGF-E/F in other organisms. To study the activities of these growth factors under physiological and pathological conditions, resulting in therapeutic applications in cancer and age-related macular degeneration, blocking ligands have been developed. These have mostly been large biomolecules like antibodies. Ligands with high affinities, at least in the nanomolar range, and accurate structural data from X-ray crystallography and NMR spectroscopy have been described. They constitute the main focus of this overview, which evidences similarities and differences in their binding modes. For VEGF-A ligands, and to a limited extent also for PlGF, a transition is now observed towards developing smaller ligands like nanobodies and peptides. These include unnatural amino acids and chemical modifications for designed and improved properties, such as serum stability and greater affinity. However, this review also highlights the scarcity of such small molecular entities and the striking lack of small organic molecule ligands. It also shows the gap between the rather large array of ligands targeting VEGF-A and the general absence of ligands binding other VEGF members, besides some antibodies. Future developments in these directions are expected in the upcoming years, and the study of these growth factors and their promising therapeutic applications will be welcomed.
Keywords: vascular endothelial growth factors, ligands, structures, pharmacological inhibition, macromolecules, peptides
1. Introduction
1.1. Vasculogenesis, Angiogenesis, and Lymphangiogenesis
Vasculogenesis and angiogenesis are two mechanisms of blood vascular networks formation, growth, and remodeling. Vasculogenesis is defined as the process of new blood vessel formation emerging during the embryonic development of the cardiovascular system, which generates the early vascular plexus and subsequently primitive blood vessels [1]. Angiogenesis refers to the process by which new blood vessels take shape from pre-existing ones based on the endothelial cell “sprouting” and intussusceptive microvascular growth [2]. The vascular endothelial growth factor A (VEGF-A) is the main factor responsible for endothelial cell migration, proliferation and tube formation, and other VEGF family members and their receptors play important roles in vasculogenesis and angiogenesis [3,4,5]. Moreover, the VEGF family and their receptors play a role in lymphangiogenesis [6,7,8,9,10]. Blood and lymphatic vessels dysfunction is associated with many pathological conditions such as chronic edema, tumor metastasis, ocular diseases, and impaired immune response [11,12]. Although lymphangiogenesis has been less studied than vasculogenesis and angiogenesis at present, there is no doubt that exploring further the functions and molecular mechanisms of this vascular system must bring some new inspirations for biologists and chemists.
1.2. The VEGF Family of Growth Factors
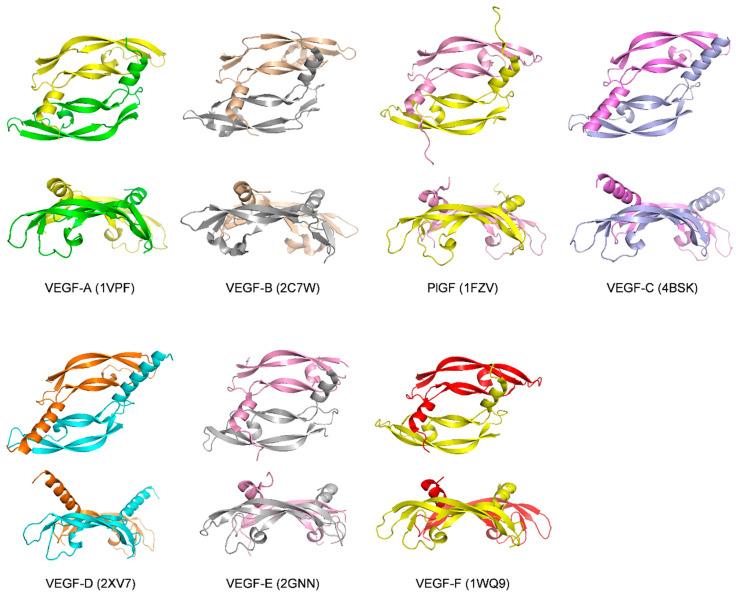
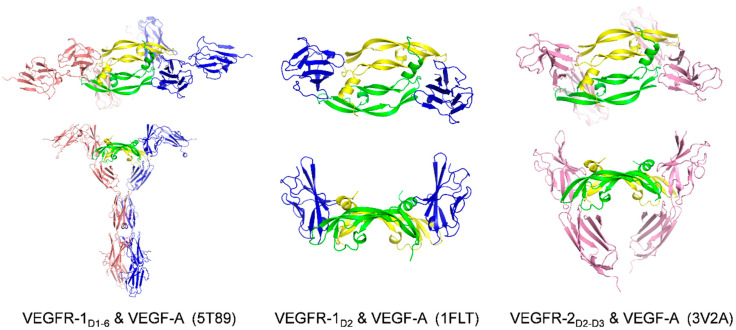
The VEGF family of proteins belongs to the platelet-derived growth factor (PDGF) subgroup of the growth factor cystine knot group [13]. They are characterized by a knotted arrangement of three intramolecular disulfide bridges and the formation of homodimers through two additional disulfide bridges. This VEGF protein family comprises VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placental growth factor (PlGF) in mammals [14]. In addition, VEGF homologs are expressed in Orf viruses, named as VEGF-E, and in snake venom, called VEGF-F [15,16,17,18]. The receptor-binding domains (RBD) of these growth factors, located in the N-terminal part, have been crystallized (Figure 1). Initially designated as “VEGF” in early 1989 by N. Ferrara et al. [19], VEGF-A is the most extensively studied member of the VEGF family. VEGF-A term is used in this review to distinguish it from other family members. Owing to the differences in exon splicing processes (and additional proteolytic processing by plasmin), multiple isoforms of VEGF-A have been identified: VEGF110, VEGF121, VEGF145, VEGF162, VEGF165 (mVEGF164 in mice), VEGF165b, VEGF183, VEGF189, and VEGF206 [20,21,22]. VEGF165 is the most abundant VEGF-A isoform. The most characteristic feature of these isoforms is their different affinities for heparin, resulting from differences in their C-terminal heparin-binding domains (HBD) [22,23]. After the discovery of VEGF-A, four other homologs have been identified successively, and the timeline is PlGF identified in 1991 [24], VEGF-B and VEGF-C identified in 1996 [25,26], and VEGF-D identified in 1998 [27]. All of them have been less explored than VEGF-A. Alternative splicing of mature mRNAs transcribes two isoforms of VEGF-B and four isoforms of PlGF, which consist of VEGF-B167, VEGF-B186 and PlGF-1 (PlGF131), PlGF-2 (PlGF152), PlGF-3 (PlGF203), and PlGF-4 (PlGF224), respectively [28,29,30,31]. The high degree of sequence homology of some VEGFs allows them to bind to the same receptors: VEGF-B, PlGF, and VEGF-A can target VEGFR-1 and the co-receptor neuropilin-1 (NRP-1) [24,32]. VEGF-C and VEGF-D, which are involved in lymphangiogenesis, bind to VEGFR-2, VEGFR-3, and NRP-2. (Figure 2 and Figure 3, and Section 1.3) [7,33]. Unlike other VEGFs, no splice isoforms of VEGF-C and VEGF-D have been reported to date. Their different forms emanate from proteolytic processing [10]. Biological activities of VEGFs in physiological and pathological conditions are still under investigation and have been reviewed [5,13]. Although VEGFs are usually present as homodimers, natural and biologically active heterodimers have been detected, particularly between VEGF-A and PlGF [34,35].
Figure 1.
Structures of the receptor-binding domains of VEGF family members. The top representation shows the view along the two-fold symetry axis of VEGF, while the bottom representation shows a perpendicular view. PDB codes are given in parenthesis. For VEGF-C (4BSK), only the growth factor is shown, while the structure includes a receptor fragment.
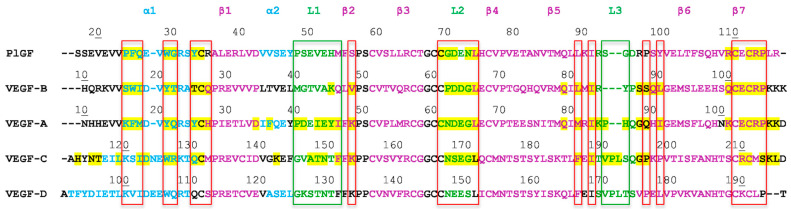
Figure 2.
Sequence alignment of the receptor-binding domains of mammalian VEGF subtypes. The secondary structures are reported as annotated by the PDB: note that the second helix α2 was often a single 3 (10) turn. The main residues implied at the interface with domain 2 and domain 3 of the receptors are in red and green boxes, respectively. Most of them are not conserved through the types. VEGFR-specific binding is associated with sequence features: for example, the presence of aromatic residue in position 26 (PlGF numbering; 17 VEGF numbering) is associated with VEGFR-1 binding. Residues observed at the interfaces with their receptor(s) for each type of VEGF are highlighted in yellow.
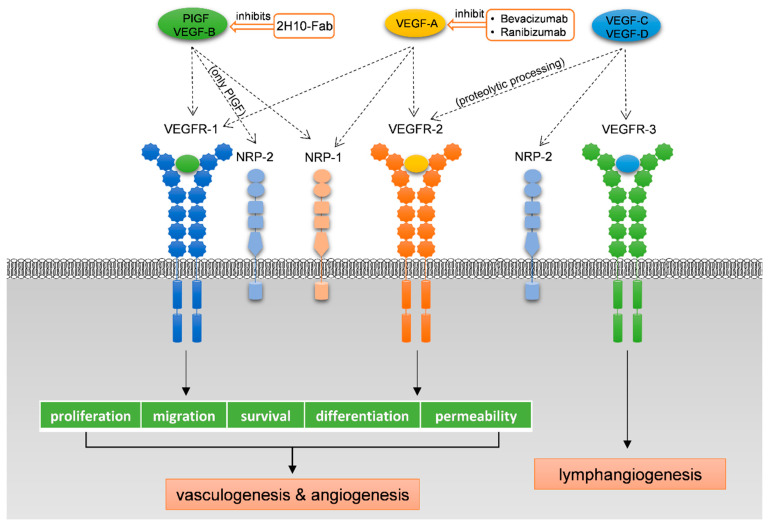
Figure 3.
VEGF signaling pathways and some representative antagonists. VEGF-A binds both VEGFR-1 and VEGFR-2, whereas VEGF-B and PlGF only bind to VEGFR-1. VEGFR-1 modulates the action of VEGFR-2 and acts as a decoy or trap for VEGF-A. These pathways are relevant to vasculogenesis and angiogenesis. On the other hand, VEGF-C and VEGF-D bind to VEGFR-3, thereby regulating lymphangiogenesis and VEGFR-2 after proteolytic processing [36]. VEGF-A and VEGF-B can bind to co-receptor NRP-1, which promotes the activation of VEGFRs but is not essential [37,38]. PlGF isoforms (PlGF-2 and PlGF-4) can bind to both NRP-1 and NRP-2 as they have the insert of the heparin-binding domain [39]. NRP-2 binding of VEGF-C/D could lead to the formation of VEGF-C(D)/VEGFR-3/NRP-2 ternary signaling complexes, subsequently facilitating the physiological or pathological lymphangiogenesis [40].
1.3. The VEGF Receptors and Co-Receptors
Vasculogenesis, angiogenesis, or lymphangiogenesis are all tightly regulated by VEGF receptors (VEGFRs), associated with co-receptors neuropilins [33,41]. VEGFs, VEGFRs, and neuropilins, construct a regular and effective cell signaling network (Figure 3) modulating endothelial cells proliferation, migration, and survival. VEGFRs are homodimeric tyrosine kinase receptors structurally related to the PDGF receptor family. The classic feature of VEGFR is having seven immunoglobulin (Ig)-like domains in the extracellular domain (ECD), a single transmembrane helix, and a tyrosine kinase domain in the intracellular portion [42], which can be activated by transphosphorylation upon binding to VEGF. There are three main types of VEGF receptors related to VEGFs signaling; they are VEGF receptor 1 (VEGFR-1, sometimes also referred to as Flt-1), VEGF receptor 2 (VEGFR-2, sometimes also referred to as Flk-1 in mice and KDR in humans), and VEGF receptor 3 (also referred to as Flt-4, involved in lymphangiogenesis), usually expressed in endothelial cells [33]. VEGF-A can bind to VEGFR-1 and VEGFR-2, VEGF-B and PlGF can bind to VEGFR-1, and VEGF-C and VEGF-D can bind to VEGFR-3 and to VEGFR-2 after proteolytic cleavage [43,44]. NRPs have initially been discovered as independent receptors for class 3 semaphorins, a family of soluble molecules with neuronal guidance functions. Remarkably, they are now identified as co-receptors for the VEGFRs [45] and a wide variety of transmembrane receptors. The extracellular portion of NRPs folds into five domains referred to as a1, a2, b1, b2, and c, followed by a transmembrane helix and a short, approximately 40-residues cytoplasmic tail [46]. VEGF-A and VEGF-B both can bind to co-receptor NRP-1, which can promote the activation of VEGFRs but is not indispensable [37]. PlGF-2 and PlGF-4 can bind to both NRP-1 and NRP-2 as they have the insert of the heparin-binding domain [38,39]. In addition, NRP-2 binding of VEGF-C/D could lead to the formation of VEGF-C(D)/VEGFR-3/NRP-2 ternary signaling complexes, subsequently facilitating the physiological or pathological lymphangiogenesis [40,47]. It is generally considered that VEGFs without a heparin-binding domain are incapable of interacting with NRPs or cannot form ternary complexes even though interactions take place [48,49,50].
1.4. Scope of the Review
As VEGFs are crucial regulators for blood or lymphatic vessel growth and survival, the dysregulation of these cytokines causes some diseases. It has been demonstrated that the VEGFs play an important role in tumor growth and metastasis, age-related macular degeneration (AMD), diabetic and hypertensive retinopathy [51,52,53,54]. Since then, substantial work has been done to produce agents targeting VEGFs (especially VEGF-A), VEGF receptors, or VEGF-regulated pathways. Some agents have already been approved by U.S. FDA and the European Medicines Agency, such as VEGF-A targeting antibodies bevacizumab and ranibizumab, which now are widely used therapeutics in oncology and eye diseases, respectively [44,55,56]. In addition, developing selective and potent ligands targeting VEGFs can give deeper insights into their mechanisms and functions in physiological or pathological conditions, which are not yet completely understood.
This review will provide an exhaustive and up-to-date overview of reported VEGFs ligands, their co-structures, and their binding affinities [13]. A detailed description of the discovery of VEGFs and their biological activities, and of ligands of VEGFRs is outside the scope of this review. The detailed results of clinical trials involving antibodies, proteins, and aptamers ligands, as well as gene therapy of VEGF-A, are also outside the scope of this review. The reader is referred to recent articles on these topics [36,44,57]. The novelty of the present review is the focus on the binding modes and the structures of ligands with high affinity and specificity for VEGFs, for which robust information is available, i.e., X-ray crystallography or NMR data. This gives a unique structural and ligand-based perspective, opposed to the description of biological effects of ligands in other published review articles. The ligands mostly include domains of proteins like receptor fragments, antibodies, and some peptides. They will be classified according to their nature and molecular size because we believe it is important for their pharmacological properties.
In early 1971, the novel concept that anti-angiogenic therapy could be a potential treatment inhibiting tumorigenesis and tumor metastasis had been proposed [58]. Recent results on the biology of the VEGF family indicate that other pathologies can be targeted. The list of known ligands of VEGFs summarized herein should provide chemists, biologists, and pharmacists an up-to-date picture of the current knowledge, and help develop a variety of new molecular structures of reduced sizes that provide novel pharmaceuticals for biological and clinical studies.
2. Ligands of VEGF-A
The structure of the full-length VEGF-A165 has not been determined yet. However, the structures of the two fragments have been successfully solved. The three-dimensional structure of the receptor-binding domain (RBD, residues 1–110, named VEGF110) has been solved by X-ray crystallography (PDB codes: 1VPF and 2VPF) [59,60] and by NMR (1QTY) [61], which proved that VEGF-A was a member of cystine-knot growth factor superfamily. The structure of the heparin-binding domain (HBD, residues 111–165, named VEGF55) has been solved by NMR (PDB codes: 1VGH and 2VGH) [62] and refined further by the same method (PDB code 1KMX) [63].
2.1. Receptors and Receptor Fragments
The receptors binding to VEGFs comprise VEGFR-1, VEGFR-2, VEGFR-3, and co-receptors NRP-1 and NRP-2. As mentioned above, VEGF-A can bind to VEGFR-1 and VEGFR-2 but not VEGFR-3. VEGFR-1 has the highest binding affinity for VEGF-A (Kd value is ≈10–20 pM), while VEGFR-2 has a lower binding affinity (Kd value is ~100–125 pM) [64,65,66]. The extracellular ligand-binding domains of VEGFRs consist of seven Ig-like domains, but the binding site for VEGF-A is mostly located at the second domain of VEGFR-1 (VEGFR-1D2) and the second and third domains of VEGFR-2 (VEGFR-2D2-3) [67,68].
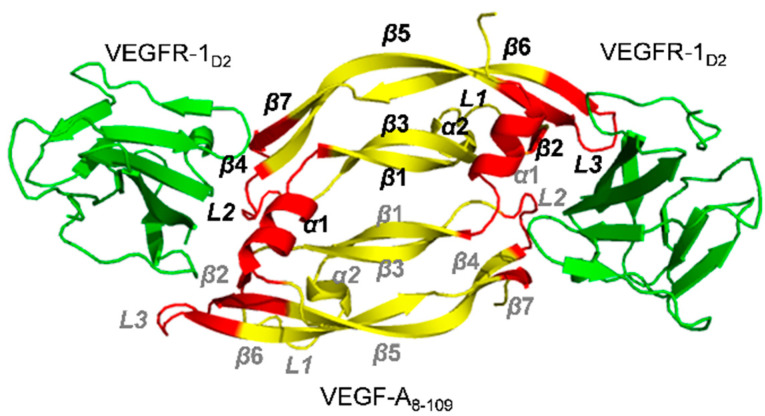
Genentech Inc. disclosed two co-crystal structure of VEGF-A8-109 in complex with VEGFR-1D2 (Figure 4, PDB code: 1FLT and 1QTY) in 1997 and 1999 [61,69]. The epitopes of VEGF-A in contact with the VEGFR-1D2 include residues from one monomer: (1) the N-terminal helix α1 (16–27), (2) the loop 2 connecting β3 to β4 (61–66), (3) the strand β7 (103–106), and residues from the other monomer: (4) the strand β2 (46–48) and (5) strands β5 and β6 together with the connecting loop 3 (79–91). These authors also conducted a domain deletion study of the extracellular domains of VEGFR-1. They showed that the affinity of domain 2 binding to VEGF-A is only 60-fold weaker than that of the full extracellular domain. Moreover, any fragments of VEGFR-1, including domain 2 and domain 3, simultaneously had the same binding affinity as the full ECD [69]. Thus, the second Ig-like domain of VEGFR-1 is necessary and sufficient for high-affinity binding.
Figure 4.
Ribbon representation of the VEGFR-1D2 & VEGF-A complex (PDB 1FLT). VEGF-A is colored in yellow, with the binding epitopes in red, and the VEGFR-1D2 domains are colored in green. Secondary structures are labeled in black for one monomer of VEGF-A and in grey for the other monomer.
Then in 2017, S. Markovic-Mueller et al. reported the X-ray structure of the full-length VEGFR-1 extracellular domain (D1-D6) in complex with VEGF-A (Figure 5 PDB code: 5T89) [70]. They used a combination of X-ray crystallography, single-particle electron microscopy, and molecular modeling for structure determination and validation. The structure revealed some aspects of the mechanism of ligand-induced receptor dimerization and activation through domain interactions. Still, part of the description of molecular contacts was tentative due to the low resolution (d = 4 Å).
Figure 5.
Ribbon representation of the receptor domains/VEGF-A complexes. VEGF-A is colored in green and yellow, and the receptor domains are colored in red and blue. The top representation shows the front view for each complex, while the bottom representation shows the side view. PDB codes are given in parenthesis.
A biochemical assay demonstrated that the binding affinity of VEGFR-2D1-2 to VEGF-A is over 1000-fold weaker than that of VEGFR-2D2-3 to VEGF-A [71]. The high affinity of VEGFR-2D2-3 to VEGF-A was subsequently demonstrated by a thermodynamic and biophysical analysis [72]. Therefore, unlike VEGFR-1, both second and third Ig-like domains of VEGFR-2 are necessary for high-affinity binding. The co-crystal structure of VEGFR-2D2-3 binding to the VEGF-A (PDB code: 3V2A) was solved along with co-structure of VEGF-E and VEGFR-2D2-3 complex (PDB code: 3V6B) in 2012 by M. S. Brozzo et al. Their work contributed to the structural analysis of VEGFR-2 binding to its major ligands VEGF-A, VEGF-C and VEGF-E [72]. Because VEGF-E is not mammalian and not a prevalent member of the VEGF family, we will not discuss it extensively in this review.
Non-natural protein constructs using VEGFR fragments have been produced to bind VEGF-A with high affinity. The first demonstration was achieved with a chimeric protein constructed by joining the ECD of VEGFR-1 with an IgG. Its capability to block VEGF-induced neovascularization in mice eyes was demonstrated [73]. In another study, the VEGFR-1D2 fragment, being sufficient for high-affinity binding, has been fused to the Fc portion of an IgG1 with a glycine linker [74]. Moreover, the oligomerization (dimers and tetramers) of the VEGFR-1D2 with short peptides and PEG linkers improved the binding affinity for VEGF-A by up to 200-fold, mainly by slowing down the dissociation rate [75]. One of such chimeric molecules is aflibercept, which is currently used in the clinic to treat AMD. It is constituted of VEGFR-1D2 and VEGFR-2D3 fused to human Fc region of IgG1 (MW = 96.6 kDa) [76].
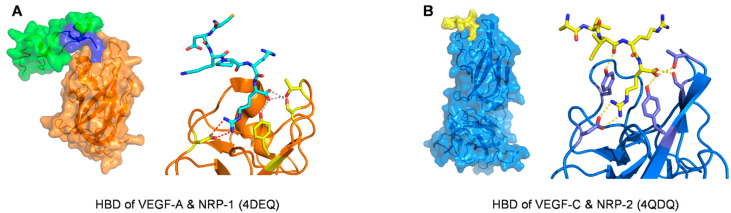
NRP-1 is identified as a co-receptor of VEGF-A, usually collaborating with VEGFRs and facilitating their efficient ligand binding. M. W. Parker et al. reported the first detailed picture of the structural basis for the binding of NRP-1 and VEGF-A. The binding interface involved regions encoded by both exons 7 and 8 of VEGF-A. Exon 7 encoded residues primarily govern selectivity, whereas exon 8 encoded residues mainly mediate high-affinity binding [77]. A fusion of human NRP-1-b1 (residues 274–429) covalently linked to the VEGF-A HBD (115–165) was constructed for crystallization. Human and mouse HBD sequences differ by only a single residue in their N terminus, and mouse residue numbering was used in this structural study (PDB code: 4DEQ, Figure 6).
Figure 6.
(A). Structure of the HBD of VEGF-A (exon 7 in green and exon 8 in blue) in complex with neuropilin-1 (orange) (B). Structure of the C-terminal residues of the HBD of VEGF-C (yellow) in complex with neuropilin-2 (marine). Hydrogen bonds are indicated by dashed lines. PDB codes are given in parenthesis.
2.2. Antibodies and Antibody Fragments
Several antibodies and antibody fragments have been developed to bind VEGF-A. The following section will list these ligands in decreasing size order, from the full antibodies (size ≈ 150–160 kDa) to Fab fragments (Fabs, the antigen-binding fragment, size ≈ 55 kDa), single-chain variable fragments (scFvs, size ≈ 30 kDa), and finally single-domain antibodies (sdAbs, which are also called nanobodies and abbreviated Nbs, size ≈ 12–14 kDa).
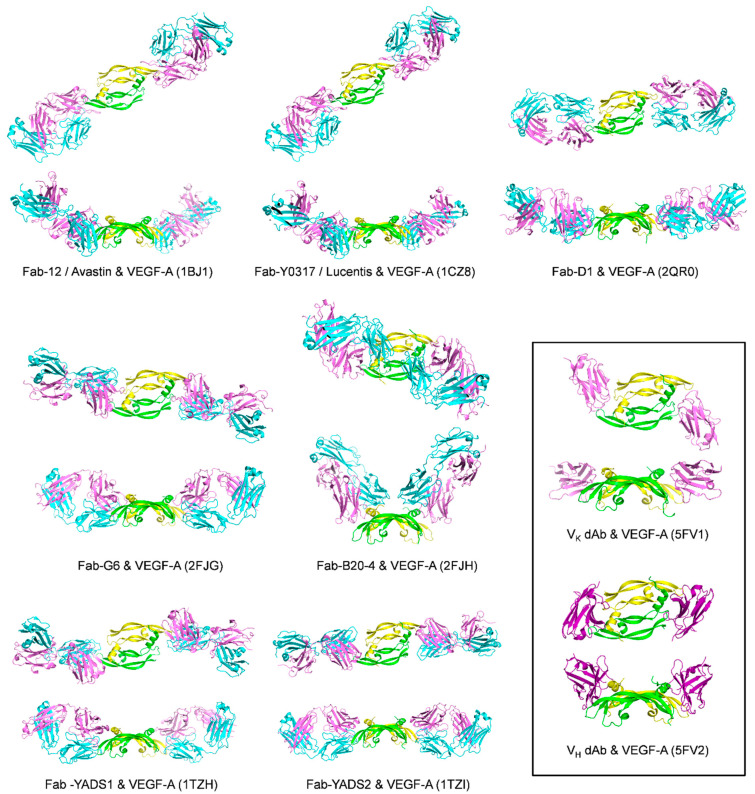
The first demonstration of inhibiting tumor growth by targeting VEGF-A in vivo was achieved with the murine anti-human VEGF-A monoclonal antibody A.4.6.1, in nude mice transplanted with human tumors [78]. This antibody was subsequently humanized and used in the clinic as bevacizumab (Avastin) to treat cancer and later AMD [44,56]. A crystal structure of VEGF-A in complex with the Fab-12 portion of the antibody has been solved (PDB code: 1BJ1) [79]. It binds VEGF-A with high affinity, mainly to the β5-β6 epitope (Figure 7 and Table 1). The binding characteristics and in vitro activities of bevacizumab and other molecules used in the clinic, ranibizumab (see below) and aflibercept, have been extensively studied and compared [80].
Figure 7.
Ribbon representation of antibody/VEGF-A complexes. VEGF-A is colored in green and yellow, and the antibodies are colored in various shades of pink and blue. The top representation shows the front view for each complex, while the bottom representation shows the side view. The insert shows the domain antibody (dAb)/VEGF-A complexes. PDB codes are given in parenthesis.
Table 1.
Structural and affinity data for VEGF ligands for which a co-structure with the growth factor has been published.
| VEGF Member | Ligand | Bound Epitope(s) of VEGF a | Affinity/Method | Reference/PDB Code |
|---|---|---|---|---|
| VEGF-A | VEGFR-1D2 | Monomer one:
|
IC50 = 1.4 nM/ELISA type assay with biotinylated VEGF8-109 IC50 = 3.0 nM (VEGF8-109) SPR competition assay [81] |
1FLT [69] or 1QTY [61] |
| VEGFR-2D2-3 | Idem above (helix α1, loops 2 and 3) + loop 1, which interacts solely with D3 | Kd = 170 nM (VEGF165)/ITC: unfavorable binding enthalpy Kd = 18 ± 5.2 nM (VEGF165)/ITC [82] Kd = 36.7 ± 5.9 nM (VEGF165)/SPR [82] |
3V2A [72] | |
| VEGFR-1D1-6 | Residues interacting with D2 Monomer one:
Monomer one:
|
Kd = 47 nM (VEGFR-1D1-3)/ITC Kd = 1.7 nM (VEGFR-1D1-7)/ITC |
5T89 [70] | |
| Neuropilin-1 |
|
Kd = 3.0 ± 0.2 nM (VEGF164)/ELISA type assay with AP-VEGF164 | 4DEQ [77] | |
| Fab-12 (refer to Y0192 in the article) | Monomer one:
|
IC50 = 4.7 nM/Fab-phage ELISA; Kd = 3.4 ± 0.9 nM (25 °C)/SPR [8] Kd = 13 ± 2.2 nM [37 °C, VEGF109]/BIAcore SPR; Kd = 21 ± 3.8 nM [37 °C, VEGF165]/SPR; IC50 = 9 nM (37 °C)/ELISA assay with Fab12-IgG and Biotin-VEGF109; Kd = 0.433 nM (25 °C)/radiolabeled VEGF binding assay using VEGF competition with [125I] VEGF for binding to Fab [83] |
1BJ1 [79] | |
| Y0317-Fab | Same as Fab-12 (binding site centers on the 80′s loop of VEGF) | Kd ≤ 0.14 nM [25 °C, VEGF109]/SPR; Kd = 0.11 ± 0.02 nM [37 °C, VEGF109]/SPR; Kd = 0.14 ± 0.05 nM [37 °C, VEGF165]/SPR;IC50 = 1 nM/ELISA assay with Biotin-VEGF109 Kd = 0.02 nM (25 °C)/radiolabeled VEGF binding assay using VEGF competition with [125I] VEGF for binding to Fab [9] |
1CZ8 [83] | |
| DutaFab |
|
IC50 = 34 pM/ELISA assay with VEGF165 | 6T9D [84] | |
| Dual dAb | Similar to VEGFR-1D2 | Mammalian cell-derived hVEGF165: Kd = 3.27 pM; E. coli expressed hVEGF165: Kd = 3.14 pM/T200 SPR hVEGF-A165: EC50 = 32 ± 2.7 pM; hVEGF121: EC50 = 127 ± 22.1 pM; mVEGF164: EC50 = 36 ± 4.7 pM/Mesoscale discovery binding assay VEGFR-1 IC50 = 59 ± 11.8 pM;VEGFR-2 IC50 = 22 ± 2.7 pM/receptor binding assay by Mesoscale discovery |
VK·dAb: 5FV1 VH dAb: 5FV2 [85] |
|
| G6-Fab | Monomer one:
|
Anti-mVEGF IC50 = 0.6 nM/Fab-phage ELISA; Kd = 0.91 nM/SPR [86] |
2FJG [87] | |
| B20-4-Fab | Monomer one:
|
Kd = 12 nM | 2FJH [87] | |
| YADS1-Fab | The structural epitopes for binding to YADS1-Fab and YADS2-Fab overlap with each other and also with the structural epitope for binding to VEGFR-1D2 | For hVEGF Kd = 1.8 ± 0.3 nM For mVEGF Kd > 1000 nM/SPR |
1TZH [88] | |
| YADS2-Fab | For hVEGF Kd = 10 ± 2 nM For mVEGF Kd = 5.0 ± 0.8 nM/SPR |
1TZI [88] | ||
| D1-Fab | The structural epitope overlaps with the structural epitope for VEGFR-1D2 | Kd = 7.8 nM/BIAcore SPR | 2QR0 [89] | |
| Peptide v108 | Monomer one:
|
IC50 = 8.2 μM/ELISA biotinylated VEGF8-109 [90] | 1VPP [90] | |
| Peptide v107 | Monomer one:
|
IC50 = 1 μM/ELISA type assay with biotin labeled-v107 | 1KAT [91] | |
| D-RFX001 | Bind to the same region of VEGF-A that interacts with VEGFR-1D2; cover much of the contact surface that VEGF-A uses to interact with VEGFR-1D2; Surface area of the binding interface is 800 Å2 |
Kd = 85 ± 12 nM/SPR, in a ProteOn™ XPR36 Protein Interaction Array System | 4GLN or 4GLS [92] | |
| D-RFX037 | Identical to D-RFX001 Surface area of the binding interface is 1350 Å2 |
Kd = 6.43 ± 0.07 nM/SPR, in a ProteOn XPR36 Protein Interaction Array System | 5HHD or 5HHC [93] | |
| Z-Domain | Overlaps with the VEGFR-1D2 binding interface | IC50 = 343 nM/phage ELISA Kd = 38 nM/Octet binding assay |
3S1K [94] | |
| Mini-Z dimer | Overlaps with the VEGFR-1D2 binding interface | IC50 = 227 nM/phage ELISA Kd = 40 nM/Octet binding assay |
3S1B [94] | |
| Alpha/beta | Overlaps with the VEGFR-1D2 binding interface | Ki = 0.11 μM/FP assay | 4WPB [95] | |
| VEGF-B | VEGFR-1D2 | Monomer one:
|
2XAC [96] | |
| 2H10-Fab | Monomer one:
|
Kd = 113.7 pM (VEGF-B10-108)/SPR IC50 = 3.4 nM (VEGF-B10-108)/cell-based assay |
2VWE [97] | |
| PlGF | VEGFR-1D2 | Monomer one:
|
IC50 = 275 nM (PlGF19-116)/SPR competition assay | 1RV6 [81] |
| VEGF-C | VEGFR-2D2-3 | Monomer one:
|
Kd = 16 ± 6.7 nM/ITC with a VEGF-C mutant C137A Kd = 18.2 ± 5.3 nM/SPR with a VEGF-C mutant C137A |
2X1X or 2X1W [82] |
| VEGFR-3D1-2 | The overall complex architecture is very similar to that of previously reported VEGFR-1 and VEGFR-2 structures VEGF-C binding is limited to D2, with D1 protruding away from VEGF-C |
Kd = 250 nM/ITC with VEGF-C mutant C137A | 4BSK [98] | |
| Neuropilin-2 (with C-terminus of VEGF-C) | R164 and R165 (VEGF165-A numbering) |
ELISA type assay with AP-VEGF-C differential scanning fluorimetry (DSF) thermal shift assay (no data about Kd or IC50 of VEGF-C C-terminus binding to Neuropilin-2) |
4QDQ [99] |
a Numbering corresponding to the sequence alignment in Figure 2.
Further improvements of the Fab 12 portion through complementarity-determining region (CDR) mutations followed by affinity selection using monovalent phage display gave the Fab variant Y0317. This final variant incorporated six mutations and had a more than 100-fold higher affinity for VEGF-A than Fab 12. A crystal structure of VEGF-A and this Y0317 Fab demonstrated that they share similar binding epitopes on VEGF-A and evidenced good correlations between improvements in binding affinity and improved hydrogen bonding and Van der Waals interaction with the growth factor (PDB code: 1CZ8) [83]. The Fab variant Y0317 was called ranibizumab (Lucentis). It is human-specific and is used in the clinic for the treatment of cancer and AMD. A structure of a new bevacizumab Fab mutant in complex with VEGF-A has been deposited in the PDB (accession code 6BFT) but has not been published yet.
Genentech Inc. subsequently developed anti-human VEGF-A Fabs by phage display with restricted amino acid diversity in the CDR region. High-affinity Fab ligands were identified (Kd ≈ 2–10 nM) with only four amino acids possibilities (tyrosine, alanine, aspartate, serine). The residue tyrosine was overwhelmingly present at the binding interface. Crystal structures of two complexes were solved (VEGF-A and antibody YADS1 Fab, PDB code: 1TZH [88] and VEGF-A and antibody YADS2 Fab, PDB code: 1TZI), showing that the binding epitope on VEGF-A was similar to the previously described Fabs (see above and Table 1) [88]. Moreover, in another study with synthetic phage libraries, a large diversity of high-affinity anti-VEGF-A Fabs has been discovered. The process started with Fabs identified with a minimalist binary library (only serine and threonine), further optimized through a gradual increase in amino acid diversity. One of the best ligands, Fab-D1, has been co-crystallized with VEGF-A (PDB code: 2QR0, Figure 7 and Table 1) [89].
Phage-displayed synthetic Fab libraries with a single framework scaffold and variability in the CDR identified new high-affinity antibodies targeting murine VEGF-A [86]. Improvements of this synthetic antibody phage-display library led to discovering two additional antibodies named G6 and B20 (studied both as Fabs or full IgGs) and their variants. In contrast with bevacizumab, these new antibodies can bind both murine and human VEGF-A with high affinities [100]. Their binding epitopes on VEGF-A were studied by alanine-scanning mutagenesis and structural analysis of complexes with VEGF-A (for G6-Fab PDB code: 2FJG, and for B20-4 Fab, PDB code: 2FJH, Figure 7) [87]. Whereas bevacizumab and all Fab variants described above bind to the β5-β6 epitope of VEGF-A, partly overlapping with the receptor binding site, G6 and B20-4 bind in a manner that matches more closely to the receptor-binding epitopes (Figure 7) [87].
Antibody mimetics called Fab-PEG-Fab, constituted of Fab portions of ranibizumab or bevacizumab dimerized with a PEG linker, have been synthesized. SPR and ITC measurements demonstrated similar binding affinities for VEGF as the full antibody bevacizumab (Kd values in the single-digit nanomolar range). However, ranibizumab’s dimerization changed its binding thermodynamic parameters, the dimer having a largely favorable enthalpy and a largely unfavorable entropy [101].
Recently, dual targeting antibody Fab fragments (DutaFabs) have been developed by Roche Inc. Two separate fragments of the CDR of one human Fv region bind specifically to two distinct targets: VEGF-A and PDGF-BB (human PDGF consituted of two B subunits) [84]. Interestingly, the two targets can be bound simultaneously and with high affinity (Table 1). Crystal structures of the complexes have been described (PDB code: 6T9D for the DutaFab-VEGF-A complex).
GlaxoSmithKline pic has developed a new antibody architecture named VEGF dual-domain antibody (dAb). It is composed of two sets of two distinct domain antibodies (named VK and VH) attached to a human IgG1 Fc domain with linkers. The increased binding stoichiometry of four binding sites per dual dAb molecule can explain the observed increase in binding affinity for VEGF-A compared to the monoclonal antibody bevacizumab and the antibody fragment ranibizumab. Each dAb has been co-crystallized with VEGF-A independently: VK·dAb, PDB code: 5FV1 and VH·dAb, PDB code: 5FV2 (Figure 7). Taking these structures into account and using SEC-MALS analysis, a model of VEGF-A binding by a dual dAb was also proposed, in which a single, dual dAb could sequester two VEGF-A homodimers [85]. Studies in rabbits and non-human primates demonstrated the efficacy of a single injection of a formulation of this dual dAb in a model of wet AMD for 6 months [102].
Single chain variable fragments (scFvs) in which only the heavy chain VH was varied and fixed to a light chain VL (kept constant) via a linker were produced by phage-display and selected to bind VEGF-A with affinities of approximately 100 nM [103]. These scFvs were subsequently converted to the larger Fab format described above and constituted starting points to identify smaller entities made of a single domain, the nanobodies (see below). Recently, the first scFv to come to market, brolucizumab (marketed as Beovu), had been approved for the treatment of neovascular AMD in the US in October 2019 and in the EU in February 2020. It is a humanized monoclonal anti-VEGF-A scFv produced in Escherichia coli cells by recombinant DNA technology. Notably, brolucizumab is effective with injections in the eyes every 3 months [104,105,106].
Nanobodies targeting VEGF-A and, more generally, proteins involved in angiogenesis were reviewed in 2017 [107]. Compared to full antibodies, the small size of nanobodies is generally considered an advantage, conferring better properties previously detailed [107]. For example, nanobodies with affinity values for the receptor-binding site of VEGF in the low nanomolar range have been identified, and their high stability has been demonstrated [108]. Oligoclonal nanobodies have also been produced and have shown a synergistic inhibition of VEGF-induced proliferation and tube formation in HUVEC that was more efficient than any individual nanobody [109]. In a recent study, a 25-residues peptide (mimotope) derived from the CDR3 region of a VEGF binding nanobody was designed and was shown to bind VEGF with a similar affinity as the parent nanobody [110]. To our knowledge, only structural data produced by molecular modeling was published regarding the interaction between VEGF-A and nanobodies.
2.3. Aptamers
Many aptamers have been developed to target VEGF-A, and some have been optimized to reach extremely high affinities. Anti-VEGF DNA-based aptamers have been recently extensively reviewed [57]. Therefore, we will only briefly summarize some key aspects regarding affinity and specificity and the scarce structural information. Pegaptanib is a pegylated RNA aptamer able to interact with the heparin-binding domain of VEGF-A with picomolar activity [111]. It was one of the first aptamers targeting VEGF-A to be developed. It has been used in the clinic (as Macugen) to treat AMD before its replacement by the more efficient bevacizumab and aflibercept in 2011 [36]. Most aptamers target the HBD, and few target the RBD. Affinities for VEGF-A in the picomolar range have been often obtained, mostly determined by SPR. In particular, very high affinities were obtained with the use of unnatural DNA bases (with the ExSELEX process) and multimeric constructs (dimers and trimers). Anti-VEGF-A DNA-based aptamers have been used for drug delivery, protein-affinity purification, and for in vitro detection of VEGF-A [57]. A literature search revealed the absence of high-resolution structural data for the complexes formed between aptamers and VEGF-A.
2.4. Small Proteins and Peptides
A few small proteins (of 50–60 amino acids) and peptides (of 15–20 amino acids) have been identified, mostly by phage-display technology. Their structures are not related to receptor or antibody fragments, but their binding sites on VEGF-A overlap with the receptor-binding site. Small proteins and peptides of molecular weights between 500 and 5000 Da occupy a chemical space between small molecules and larger proteins. Peptides are less structured than larger proteins in solution. This leads to a low proportion of the bioactive conformation(s) in the conformational ensemble, and therefore to a generally lower affinity than larger proteins due to an entropic penalty of binding. To avoid this drawback, a cyclization strategy is often chosen to design bioactive peptides by phage-display and chemical synthesis. It results in improved conformational stability and proteolytic resistance [112].
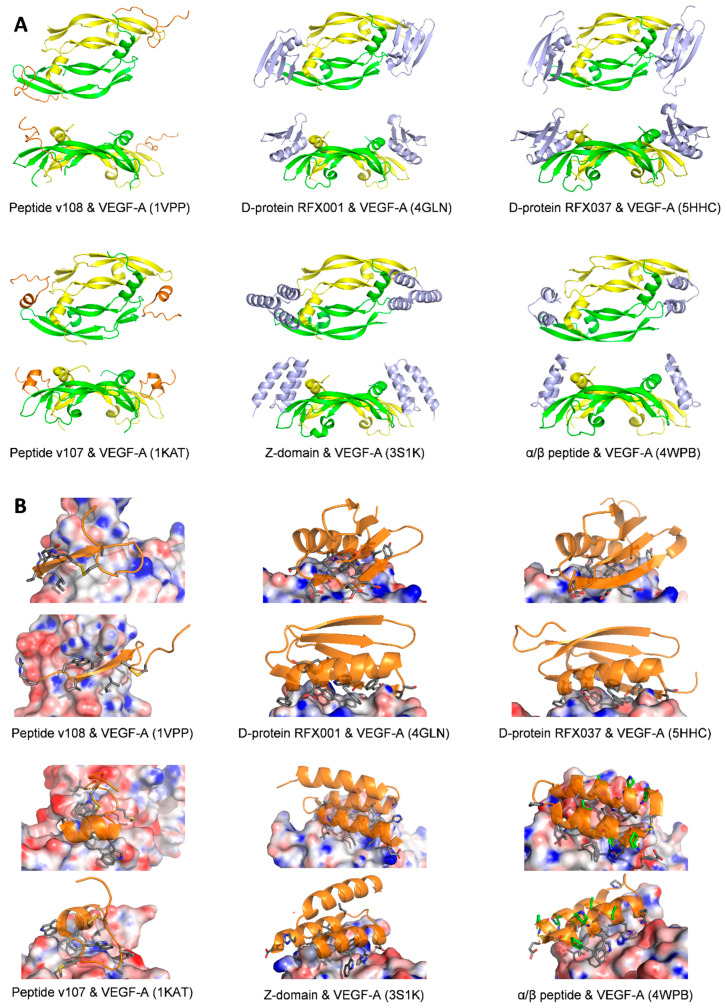
In an example of improved phage display technology, the method of “mirror image phage display” was used, starting from the scaffold of the B1 domain of streptococcal protein G (56 amino acids), against the enantiomer of natural VEGF-A (D-VEGF-A, which was produced by total chemical synthesis with D-amino acids). It allowed the identification of a small protein-ligand of D-VEGF-A with a Kd value of approximately 90 nM. In turn, the total chemical synthesis of this small protein with D amino acids gave ligands of natural (L-) VEGF-A with the same affinity. This D-protein antagonist (D-RFX001) was co-crystallized with VEGF-A by a racemic crystallization process, giving heterochiral complexes solved in two space groups: PDB code 4GLN and 4GLS [92]. The contact surface of D-RFX001 encompasses approximatively 800 Å2 which competes for the binding of VEGF with VEGFR-1D2. It is dominated by a central aromatic cluster surrounded by polar contacts. Further improvement regarding thermal stability, pharmacokinetic properties, and affinity gave a new D ligand RFX037.D (Table 1), which was also co-crystallized with VEGF-A, PDB code: 5HHD and 5HHC. These are also racemic complexes solved in two different space groups (Figure 8A,B). The low immunogenicity of this D-small protein was demonstrated in mice [93].
Figure 8.
(A). Ribbon representation of peptide/VEGF-A complexes. VEGF-A is colored in green and yellow, and the peptides are colored in various shades of orange and blue. The top representation shows the front view for each complex, while the bottom representation shows the side view. PDB codes are given in parenthesis. (B) Details of the interaction of peptides or small protein ligands on the electrostatic surface of VEGF-A, calculated using the Adaptive Poisson-Boltzmann Solver [127]. The side chains of the ligands involved in molecular contacts are in gray sticks and the ribbons in orange. For each complex, two representations show the main interactions. PDB codes are given in parenthesis. For the α/β peptide, the non-natural residues are shown in green.
Ligands of VEGF-A based on the scaffolds of the three-helix 58-residues Z-domain of staphylococcal protein A were identified by phage display by Genentech Inc. Among the Z-domain clones, Z-3B (59 residues) was the best ligand with a Kd value of 55 nM. Another reengineered version named mini-Z, of 34 amino acids structured in two helices stabilized by a disulfide constraint, bound VEGF-A with a Kd of 38 nM. Two crystal structures of complexes of these ligands with VEGF-A were reported (PDB for Z-3B: 3S1K, and mini-Z: 3S1B). Both Z-domain and mini-Z peptides bind to VEGF at the receptor-binding site. The Z-domain buries 744 Å2 of the surface area, mainly composed of van der Waals interactions of aliphatic and aromatic side chains, with five hydrogen bonds and two additional salt-bridges at the site periphery. Imaging in mice xenograft models of colorectal and ovarian cancer with a Z-3B radiolabeled with 18F gave, in 2 h, images of comparable quality to those obtained with 89Zr-radiolabeled B20 antibody 72 h post-injection. This demonstrated the better tumor penetration capacity of the small protein compared to the antibody [94].
Additional studies based on the Z-domain scaffold described above, cyclized by a disulfide bond, were reported. Incorporation of non-natural amino acids to stabilize helices, in particular β-amino acids and 2-aminoisobutyric acid, resulted in ligands with strongly increased stability towards proteases, without significant loss of affinity for VEGF-A. A crystal structure of an α/β peptide in complex with VEGF-A was reported (PDB code: 4WPB, Figure 8A,B) [95,113]. The non-natural amino acids are located at sites that do not contact the VEGF-A surface. Therefore, the interaction surface is very close to that of the Z-domain.
DARPins (designed ankyrin repeat proteins) are genetically engineered antibody mimetics designed to bind protein targets. Abicipar-Pegol, developed by Molecular Partners, Inc., Zurich-Schlieren, Switzerland, is a DARPin constituted of a 14 kDa recombinant protein (4 ankyrin repeats) with a very high affinity for VEGF-A, linked to a 20 kDa PEG tail [114,115]. Three-phase 3 clinical trials have been conducted, but the FDA rejected it in 2020 because of its higher rate of intraocular inflammation than ranibizumab.
Screening of VEGF-A binding peptides by phage-display by Genentech Inc. resulted in identifying three classes of peptide ligands [116]. All of them bound VEGF-A to overlapping epitopes and were able to inhibit receptor binding. These 19 or 20-mer peptides have been extensively studied by NMR and X-ray diffraction [117]. Among them, the peptide v108 was co-crystallized with VEGF-A8-109. (Peptide v108 & VEGF8-109, PDB code: 1VPP, Figure 8A,B) [90]. NMR showed that peptide v108 folds in the presence of VEGF. Its central region of nine amino acids forms a type I β-hairpin loop stabilized by an intramolecular disulfide bridge. The N-terminal half folds in β-strand conformation, which binds to the main chain β6 of VEGF. The C-terminal is flexible and does not interact with VEGF. Overall, it resembles the binding mode of the Fab 12 antibody. However, the extended conformation and the scarcity of specific side-chain interactions led the authors to consider the peptide as not being a promising lead for the development of small molecules.
The structure of peptide v107 in complex with VEGF-A was solved by NMR (PDB code: 1KAT) [91,118]. The peptide folds in the presence of VEGF-A with a C-terminal α-helix and a central β-turn. It wraps around an aromatic core and is stabilized by a disulfide bridge (Figure 8A,B). Although the folding process results in a thermodynamically unfavorable entropy variation [119], the peptide v107 was considered a suitable starting point for further optimization because of its high affinity for VEGF-A, its structuration upon binding, and because it is binding with the growth factor was mainly mediated by specific hydrophobic interactions with its side chains. Additional studies on this v107 peptide include a study in the gas phase of D-amino acids variants [120], the modification with an electrophile chemical function for covalent labeling of VEGF-A [121], the radiolabeling for imaging [122,123], the extension with peptide fragments to improve its affinity as a capture agent [122], the incorporation of non-natural beta-amino acids for increased proteolytic stability (structure of peptide HH4, PDB 6D3O, to be published) [119,124], the shortening of the sequence to 15 amino acids [125], and the stabilization of the α-helix C-terminal end by cyclization for increased affinity (PDB 6Z13, 6ZCD, 6Z3F, 6ZBR, to be published). A bacterial display screening independently identified peptides that were very similar to v107, with comparable affinities for VEGF-A and the same core sequence WE/DWE/D [126].
A cyclic hexapeptide incorporating the Trp/Glu motif found in v107 displayed weak affinity (low millimolar range) for VEGF-A. Still, the demonstration of its specific binding at the receptor-binding site was achieved by NMR spectroscopy [128].
2.5. Small Molecules
To the best of our knowledge, there is no small molecule co-crystallized with VEGF-A. The large hydrophobic and flat interaction surface on receptors constitutes a difficult target for small molecules. Only one study reported attempts to find small molecules as VEGF-A ligands [128]. The authors screened libraries of dipeptides, peptides incorporating unnatural amino acids, and small molecules but did not identify ligands of VEGF-A. Molecular modeling indicated that the most probable binding sites for small molecules on VEGF-A are located outside the receptor-binding interface, in clefts between the two monomers of the growth factor. However, screening by NMR of natural products identified a flavonoid scaffold able to bind VEGF-A at the receptor-binding interface, although with weak affinity (Kd values are in the low millimolar range).
2.6. Analysis of the Binding Modes of VEGF-A Ligands
The VEGFRs domains D2 and D3 bind VEGF-A on several discontinuous epitopes constituting the binding site: the helix α1, the loops L1, L2, and L3, and the strand β7 (Figure 9). All the other known ligands target at least one of these epitopes, thereby inhibiting the growth factor/receptor interaction. The residues of VEGF-A buried at the interface are colored in red in Figure 9. In the literature, we did not find any ligand able to block the interaction by steric hindrance while targeting epitopes outside the binding site.
Figure 9.
Binding sites of artificial ligands on VEGF-A overlap with the binding sites of VEGF-R. Top: Comparison of the VEGF-A residues buried at the interface with VEGFR-1D2 or VEGFR-2D2D3 (yellow boxes) and the residues buried at the interface with the available ligands whose co-structures have been solved (in red letterings). Uppercase letters indicate solvent-accessible residues in the VEGF structure. Bottom: representation of the accessible surface of VEGF-A buried at the ligand interface, as identified with PISA [129]. (a) Red: VEGFRs interface. (b) Blue: ligands interfaces, except v108, v107, and DutaFab. (c) Magenta: DutaFab interface. (d) The residues buried at v108 or v107 interfaces are in yellow and grey, respectively. PDB-ids are indicated in Table 1.
As listed above, the ligands are diverse in sizes and shapes, and their secondary structures are not shared, even at the binding site. However, some specific non-covalent interactions are shared for each epitope across several ligands. For example, ligands targeting the helix α1 make use of π stacking interactions between their aromatic residues and F17, Y21, and Y25 of VEGF-A. Polar interactions are often present with residues of loop L2, hydrogen bonds with N62, and charge/charge with D63.
3. Ligands of PlGF and VEGF-B
Both PlGF and VEGF-B are VEGFR-1 specific ligands and less studied than VEGF-A. Therefore, we will discuss them in the same chapter. The crystal structures of PlGF (PDB code: 1FZV) and VEGF-B (PDB code: 2C7W) were solved by X-ray crystallography in 2001 and 2006, respectively [32,130]. Despite their functional diversities, they showed a highly similar topological structure with VEGF-A (Figure 1) [60].
3.1. Receptors and Receptor Fragments
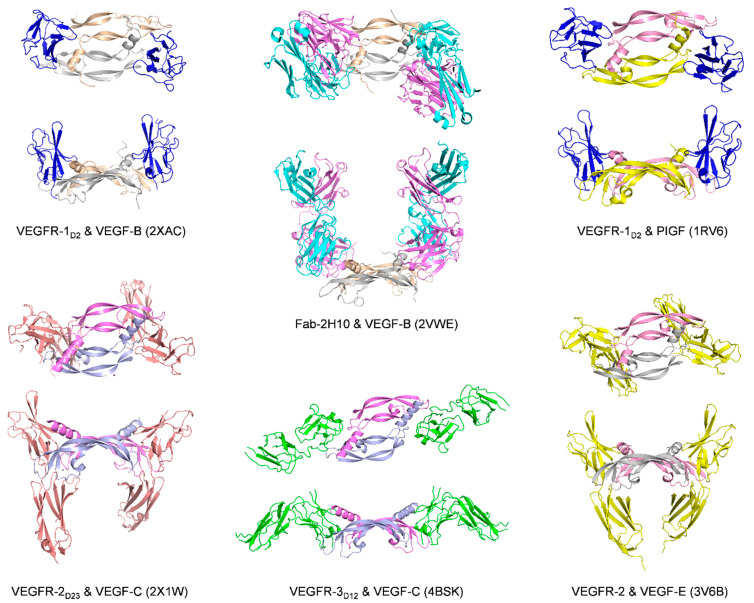
The co-crystal structure of PlGF and VEGFR-1D2 (PDB code: 1RV6, Figure 10) was determined by H.W. Christinger et al. using X-ray crystallography in 2004 [81]. Meanwhile, their research group carried out an SPR competitive binding assay showing that the presence of VEGFR-1D3 has a much greater effect on the binding affinity of PlGF than of VEGF-A. This result strongly indicates that the third Ig-like domain of the VEGFRs plays an important part in the specificity of receptor recognition [81]. Then, in 2010, the crystal structure of VEGF-B in complex with VEGFR-1D2 (PDB code: 2XAC, Figure 10) was disclosed [96], of which the topological structure is essentially similar to that seen in two complexes of this receptor with VEGF-A and PlGF [69,81]. VEGF-B did not require the VEGFR-1D3 for high-affinity binding [131]. Comparing the three co-crystal structures of VEGFR-1D2 with VEGF-A, VEGF-B, and PlGF provides abundant information on how subtle differences influence receptor recognition. Interestingly, the electrostatic surface potentials of the growths factor seem to be complementary to the corresponding surface potential of their cognate receptors [96]. Given that VEGF-A, VEGF-B, and PlGF share the same overall binding mode on VEGFR-1 and some sequence homology, it is intriguing but difficult to explain how receptor recognition and specificity happen and translate into distinct biological activities. In such an attempt, the importance of the L1 loop of the growth factors and its interaction with VEGFR-1D3 has been demonstrated [131].
Figure 10.
Ribbon representation of ligand/VEGF or PlGF complexes. The top representation shows the front view for each complex, while the bottom representation shows the side view. PDB codes are given in parenthesis.
3.2. Antibodies and Antibody Fragments
Inhibition of PlGF activity by antibodies has been the subject of several studies since an initial report demonstrated that αPlGF (clone 5D11D4, a mouse PlGF-2 specific blocking antibody) was able to inhibit the growth of VEGF(R)-inhibitor-resistant tumors in mice [132]. A subsequent study with the same antibody confirmed these results and demonstrated additional beneficial activity by inhibiting angiogenesis in eye diseases [133]. The IC50 value for inhibition of the mPlGF/VEGFR-1 interaction by the full 5D11D4 antibody was 27 pM and in the low nanomolar range with Fab fragments thereof [133]. However, the broad utility of specific PlGF inhibition (or in combination with VEGF-A inhibition) with antibodies in cancer has been challenged, in particular with the observation that only tumors expressing VEGFR-1 are potential targets [134,135]. In addition, the importance of targeting also the PlGF/NPR-1 interaction to inhibit the growth efficiently and spread of brain tumors has been demonstrated. The authors used an antibody blocking only the PlGF/VEGFR-1 interaction (C9.V2) versus antibodies blocking both interactions (TB-403 and 5D11D4) [136]. Therefore, although it is clear that all the antibodies tested bind PlGF with high affinity and selectivity, additional knowledge regarding the precise binding sites and binding modes may help in understanding their different biological activities. To our knowledge, no experimental structural information (X-ray or NMR) regarding the binding modes of anti-PlGF antibodies has been disclosed. The role of PlGF in antitumor immune response and existing anti-PlGF targeting strategies have been recently reviewed [137]. The humanized anti (human) PlGF antibody TB-403 (RO5323441) has been tested in a few clinical trials, initially a phase I clinical trials in patients with advanced solid tumors [138] and more recently in pediatric subjects with relapsed or refractory medulloblastoma, for which the results will be released in 2022 (https://clinicaltrials.gov/ct2/show/NCT02748135, accessed 29 September 2021).
PlGF-specific nanobodies with affinities in the nM range have been identified by phage-display and studied as monomers and dimers [139,140]. Their anti-angiogenic activity was demonstrated with several in vitro angiogenic assays.
Neutralizing monoclonal antibodies targeting VEGF-B specifically (over VEGF-A) and with high affinity and preventing VEGFR-1 binding have been developed [141]. A crystal structure of the complex between VEGF-B and a neutralizing antibody Fab fragment (Fab-2H10) has been described (PDB code: 2VWE) Fab (133). It displays a unique U-shape topology not previously observed in the VEGF family. This antibody binds VEGF-B at its VEGFR-1 binding site, thereby blocking the interaction between the growth factor and the receptor (Figure 10). The 2H10 antibody (by intravitreal injection) and an scFv fragment thereof (by combined topical and sub-conjunctival injection) have been shown to induce regression of pre-existing blood vessels at the back of the eye in mouse and rats models of eye diseases [142,143]. The antibody 2H10 and the scFv have high affinities and selectivities for human, rat, and mouse VEGF-B, which is convenient for preclinical studies [144].
3.3. Small Proteins and Peptides
Using phage display, Pfizer Inc. identified 15-residues peptides binding PlGF-1. Although the affinity of these initial peptides was modest (IC50 ≈ 1 µM), extreme improvement in affinity (103) was demonstrated by dimerization through the covalent binding on a CovX antibody scaffold. The antibody scaffold alone did not bind the growth factor and was only used as a template linker. Therefore, the improvement was explained by possible avidity effects. Additional modifications with the incorporation of non-natural amino acids and optimization of the linker position led to a ligand with a Kd of 0.1 nM for PlGF-1 and increased stability in serum. Moreover, some specificity of the ligands for human PlGF-1 over human PlGF-2, mouse PlGF, and VEGF-A was demonstrated. In the absence of structural information, the authors propose that the specificity may be explained by binding the peptide to an epitope of the RBD accessible only in PlGF-1, which does not possess an HBD and is hindered by the HBD in other growth factors that possess one [145].
4. Ligands of VEGF-C and VEGF-D
The same chapter will discuss VEGF-C and VEGF-D because they share high homology of structure and bind to the same receptors VEGFR-2, VEGFR-3, and NRP-2. They were initially described as having very similar functions. However, recent studies demonstrate that they actually have different binding and regulatory mechanisms in lymphangiogenesis [146]. A crystal structure of VEGF-D alone has been described (PDB code: 2XV7, Figure 1) [147]; however, the crystal structures of VEGF-C are all in complex with fragments of the receptors (see below). A characteristic feature of VEGF-C and VEGF-D compared to other VEGFs is the extended N-terminal α-helix [147]. In VEGF-D (and not VEGF-C), the length of this N-terminal α-helix is critical for VEGFR-2 and VEGFR-3 binding [146].
4.1. Receptors and Receptor Fragments
Crystal structures of VEGF-C with fragments of each of its three receptors have been published. In the complex with VEGFR-2D2-3 (Domains 2 and 3 of VEGFR-2, PDB code: 2 × 1X and 2 × 1W), the binding epitopes of both proteins were identified, as well as critical residues of the receptor explaining its affinity for VEGF-A and -C (Table 1 and Figure 10) [82]. In the complex between VEGF-C and VEGFR-3D1-2 (Domains 1 and 2 of VEGFR-3, PDB code: 4BSK, Figure 10), despite the low resolution and the impossibility to define specific interactions, the binding interface on domain 2 was confirmed, with conserved structural features compared to other members of the VEGF family [98]. Therefore, the presence of domain 3 did not significantly change the dissociation constant of VEGF-C for the receptor fragments: Kd for VEGFR-3D1-2 was 250 nM, and Kd for VEGFR-3D1-3 was 140 nM. However, the Kd for VEGFR-3D1-5 was 3.7 nM because domain 5 dimerized in the presence of VEGF-C, thereby increasing the total affinity. In addition to the RBD, a complex of the HBD of VEGF-C in interaction with NRP-2 has been solved (PDB code: 4QDQ, Figure 6B, from a fusion construct between NRP-2 and the C-terminal amino acids of the HBD of VEGF-C). The structure involves C-terminal residues of the HBD, terminated by two arginine residues in the sequence, with one of them deep into the binding cleft of the b1 subunit. Therefore, the structure demonstrates the importance of the C-terminal proteolytic cleavage of VEGF-C and the shared mode of binding with the VEGF-A/NRP-1 structure [99]. To our knowledge, there are no co-structures of VEGF-D with receptor fragments.
Soluble forms of VEGFR-3 with high affinities for VEGF-C and -D have been used to explore the effect of blocking this pathway on lymphangiogenesis [148]. Recent reports indicate the emerging roles of VEGF-D in several human diseases [148]. A first-in-class drug candidate, the soluble receptor fragment of VEGFR-3 named OPT-302 (VEGFR-3D1-3 fused to the Fc fragment of IgG1), is explored in clinical trials to target VEGF-C and VEGF-D, as monotherapy or combined with VEGF-A blockade, in eye diseases such as neovascular AMD [149].
4.2. Antibodies
Several antibodies binding VEGF-C are commercially available [150]. Some of them can block its activity, like VGX-100, which precipitates all forms of human and mouse VEGF-C. The biological activities of VGX-100 on cancer and ocular diseases have been demonstrated in preclinical models. It has been tested in phase I clinical trial (in combination with bevacizumab) for solid tumors [151,152,153]. However, to our knowledge, there is no specific information regarding the binding epitope or the structures of antibodies in complex with VEGF-C.
In a published study, four antibodies binding VEGF-D have been developed (VD1-4, Kd ≈ 30–61 nM), VD1 being selective for VEGF-D (in particular over VEGF-C) and able to prevent its binding to the VEGFR-2 and -3 [154]. Additional antibodies able to block the interaction of the growth factor with the receptors are now commercially available, like mAB286, and used to study the biological activities of VEGF-D [146]. Although no co-crystal structures have been published, the binding epitope has been accurately identified for VD1 (in the L2 loop), which was explored as a potential therapeutic antibody [155].
5. Ligands of Non-Mammalian VEGFs
Besides the five mammalian VEGFs mentioned above, the VEGF protein family also comprises two non-mammalian VEGFs: VEGF-E and VEGF-F. The research of natural or synthetic ligands is largely unexplored. VEGF-E is an Orf viral homolog of VEGF-A, which contributes to wound re-vascularization and re-epithelialization [156]. Based on sequence similarity technique, the gene encoding a VEGF homolog was discovered in the genome of the Orf virus in 1994 [17]. Orf virus is the type species of the Parapoxvirus genus of the Poxviridae family, which primarily causes acute pustular skin lesions in sheep and goats and can infect humans [157]. The functional activity of VEGF-E was investigated, and it was shown to mediate angiogenesis via signaling through VEGFR-2 (KDR) but not VEGFR-1 [158]. VEGF-E only shows 25–35% amino acid identity to VEGF-A, although it has a comparable affinity for VEGFR-2 [159]. Despite the lack of a heparin-binding domain, some variants of VEGF-E retain the binding to neuropilin-1 [160]. The crystal structure of the Orf virus NZ2 variant of VEGF-E has been solved by X-ray crystallography (PDB code: 2GNN, Figure 1) [16], as well as the co-crystal structure of VEGF-E in complex with VEGFR-2 (PDB code: 3V6B, Figure 10) [72]. These two structures show that the core regions of VEGF-E are very similar to the core regions of VEGF-A.
VEGF-Fs are isolated from snake venoms, for example, from the snake Trimeresurus flavoviridis, and they display a variety of structures and functions among different species [161]. The crystal structures of two VEGF-Fs vammin and VR-1 have been solved (PDB code: 1WQ8 and 1WQ9, Figure 1), and they exhibit significant differences from other known VEGFs, which may explain their high specificity for the VEGFR-2 [15]. A recent study indicated that vammin could induce VEGFR-2/NRP-1 related signaling more efficiently than VEGF-A [162]. VEGF-F has the potential to bind heparin, but it has almost lost the C-terminal heparin-binding domain compared with other heparin-binding VEGFs. Based on this observation, it was found that the C-terminal heparin-binding peptide of VEGF-F is able to inhibit VEGF-A-regulated endothelial cell proliferation [163,164]. As far as we know, no co-crystal structure of a ligand in complex with any VEGF-F has been reported.
6. Conclusions
From this overview of ligands of the VEGF family of growth factors, VEGF-A has been much more studied than the other members have. VEGF-A ligands comprise a variety of receptor fragments, aptamers, antibodies, mini proteins, and peptides. All the available ligand binding sites partially covered the receptor-binding interfaces and consequently acted as competitive inhibitors of VEGFRs. Targeting the receptor-binding epitopes of VEGF-A has been a successful approach and has been achieved through several conserved non-covalent interactions shared across the ligands. The smallest efficient ligands are peptides of 10–15 amino-acid length. Smaller peptides or small molecules may not be able to bind efficiently to the large flat hydrophobic surface of VEGFR binding sites. However, it seems that only a very small number of published studies have been aimed at that purpose. Future studies in this direction may be more successful, as our knowledge of this growth factor improves and new computational and experimental methods are being developed. Therapeutic applications of VEGF-A targeting have been only mediated through macromolecules. Although successful, it has shown some limitations. The use of small molecules or peptides may improve the outcome of this targeting due to their potentially different administration modes and pharmacokinetic properties.
Apart from VEGF-A, the development of ligands of the other members of the VEGF family has been strikingly lacking. Only receptor fragments and antibodies have been described, except a peptide series targeting PlGF. Future development of new ligands with increased potency and specificity and varying pharmacokinetic properties is warranted. It may help decipher further the biological activities of the VEGF family members and reveal or harness potential therapeutic applications.
Author Contributions
Writing—original draft preparation, X.Y. and S.B.; writing—additions and editing, J.-F.G.; J.-F.G., M.V., X.Y. and S.B. reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the CNRS, the INSERM, the University of Paris, and the Chinese Scholarship Council (Ph.D. scholarship to X.Y.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Risau W., Flamme I. Vasculogenesis. Annu. Rev. Cell Dev. Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 2.Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P., Ng Y.S., Nuyens D., Theilmeier G., Brusselmans K., Cornelissen I., Ehler E., Kakkar V.V., Stalmans I., Mattot V., et al. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nat. Med. 1999;5:495–502. doi: 10.1038/8379. [DOI] [PubMed] [Google Scholar]
- 4.Keyt B.A., Nguyen H.V., Berleau L.T., Duarte C.M., Park J., Chen H., Ferrara N. Identification of vascular endothelial growth factor determinants for binding KDR and FLT-1 receptors. Generation of receptor-selective VEGF variants by site-directed mutagenesis. J. Biol. Chem. 1996;271:5638–5646. doi: 10.1074/jbc.271.10.5638. [DOI] [PubMed] [Google Scholar]
- 5.Melincovici C.S., Bosca A.B., Susman S., Marginean M., Mihu C., Istrate M., Moldovan I.M., Roman A.L., Mihu C.M. Vascular endothelial growth factor (VEGF)—Key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. 2018;59:455–467. [PubMed] [Google Scholar]
- 6.Achen M.G., McColl B.K., Stacker S.A. Focus on lymphangiogenesis in tumor metastasis. Cancer Cell. 2005;7:121–127. doi: 10.1016/j.ccr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Alitalo K., Tammela T., Petrova T.V. Lymphangiogenesis in development and human disease. Nature. 2005;438:946–953. doi: 10.1038/nature04480. [DOI] [PubMed] [Google Scholar]
- 8.Tobler N.E., Detmar M. Tumor and lymph node lymphangiogenesis—Impact on cancer metastasis. J. Leukoc. Biol. 2006;80:691–696. doi: 10.1189/jlb.1105653. [DOI] [PubMed] [Google Scholar]
- 9.Warren A.G., Brorson H., Borud L.J., Slavin S.A. Lymphedema: A comprehensive review. Ann. Plast. Surg. 2007;59:464–472. doi: 10.1097/01.sap.0000257149.42922.7e. [DOI] [PubMed] [Google Scholar]
- 10.Tammela T., Alitalo K. Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 2010;140:460–476. doi: 10.1016/j.cell.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 11.Chung A.S., Ferrara N. Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 2011;27:563–584. doi: 10.1146/annurev-cellbio-092910-154002. [DOI] [PubMed] [Google Scholar]
- 12.Maby-El Hajjami H., Petrova T.V. Developmental and pathological lymphangiogenesis: From models to human disease. Histochem. Cell Biol. 2008;130:1063–1078. doi: 10.1007/s00418-008-0525-5. [DOI] [PubMed] [Google Scholar]
- 13.Iyer S., Acharya K.R. Tying the knot: The cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines. FEBS J. 2011;278:4304–4322. doi: 10.1111/j.1742-4658.2011.08350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara N. VEGF and intraocular neovascularization: From discovery to therapy. Transl. Vis. Sci. Technol. 2016;5:10. doi: 10.1167/tvst.5.2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suto K., Yamazaki Y., Morita T., Mizuno H. Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: Insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1. J. Biol. Chem. 2005;280:2126–2131. doi: 10.1074/jbc.M411395200. [DOI] [PubMed] [Google Scholar]
- 16.Pieren M., Prota A.E., Ruch C., Kostrewa D., Wagner A., Biedermann K., Winkler F.K., Ballmer-Hofer K. Crystal structure of the Orf virus NZ2 variant of vascular endothelial growth factor-E. Implications for receptor specificity. J. Biol. Chem. 2006;281:19578–19587. doi: 10.1074/jbc.M601842200. [DOI] [PubMed] [Google Scholar]
- 17.Lyttle D.J., Fraser K.M., Fleming S.B., Mercer A.A., Robinson A.J. Homologs of vascular endothelial growth factor are encoded by the poxvirus orf virus. J. Virol. 1994;68:84–92. doi: 10.1128/jvi.68.1.84-92.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamazaki Y., Morita T. Molecular and functional diversity of vascular endothelial growth factors. Mol. Divers. 2006;10:515–527. doi: 10.1007/s11030-006-9027-3. [DOI] [PubMed] [Google Scholar]
- 19.Ferrara N., Henzel W.J. Pituitary follicular cells secrete a novel heparin-binding growth factor specific for vascular endothelial cells. Biochem. Biophys. Res. Commun. 1989;161:851–858. doi: 10.1016/0006-291X(89)92678-8. [DOI] [PubMed] [Google Scholar]
- 20.Poltorak Z., Cohen T., Sivan R., Kandelis Y., Spira G., Vlodavsky I., Keshet E., Neufeld G. VEGF145, a secreted vascular endothelial growth factor isoform that binds to extracellular matrix. J. Biol. Chem. 1997;272:7151–7158. doi: 10.1074/jbc.272.11.7151. [DOI] [PubMed] [Google Scholar]
- 21.Lei J., Jiang A., Pei D. Identification and characterization of a new splicing variant of vascular endothelial growth factor: VEGF183. Biochim. Biophys. Acta. 1998;1443:400–406. doi: 10.1016/S0167-4781(98)00240-1. [DOI] [PubMed] [Google Scholar]
- 22.Ferrara N. Binding to the extracellular matrix and proteolytic processing: Two key mechanisms regulating vascular endothelial growth factor action. Mol. Biol. Cell. 2010;21:687–690. doi: 10.1091/mbc.e09-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houck K.A., Leung D.W., Rowland A.M., Winer J., Ferrara N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 1992;267:26031–26037. doi: 10.1016/S0021-9258(18)35712-0. [DOI] [PubMed] [Google Scholar]
- 24.Maglione D., Guerriero V., Viglietto G., Delli-Bovi P., Persico M.G. Isolation of a human placenta cDNA coding for a protein related to the vascular permeability factor. Proc. Natl. Acad. Sci. USA. 1991;88:9267–9271. doi: 10.1073/pnas.88.20.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olofsson B., Pajusola K., Kaipainen A., von Euler G., Joukov V., Saksela O., Orpana A., Pettersson R.F., Alitalo K., Eriksson U. Vascular endothelial growth factor B, a novel growth factor for endothelial cells. Proc. Natl. Acad. Sci. USA. 1996;93:2576–2581. doi: 10.1073/pnas.93.6.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joukov V., Pajusola K., Kaipainen A., Chilov D., Lahtinen I., Kukk E., Saksela O., Kalkkinen N., Alitalo K. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J. 1996;15:290–298. doi: 10.1002/j.1460-2075.1996.tb00359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Achen M.G., Jeltsch M., Kukk E., Makinen T., Vitali A., Wilks A.F., Alitalo K., Stacker S.A. Vascular endothelial growth factor D (VEGF-D) is a ligand for the tyrosine kinases VEGF receptor 2 (Flk1) and VEGF receptor 3 (Flt4) Proc. Natl. Acad. Sci. USA. 1998;95:548–553. doi: 10.1073/pnas.95.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauser S., Weich H.A. A heparin-binding form of placenta growth factor (PlGF-2) is expressed in human umbilical vein endothelial cells and in placenta. Growth Factors. 1993;9:259–268. doi: 10.3109/08977199308991586. [DOI] [PubMed] [Google Scholar]
- 29.Olofsson B., Pajusola K., von Euler G., Chilov D., Alitalo K., Eriksson U. Genomic organization of the mouse and human genes for vascular endothelial growth factor B (VEGF-B) and characterization of a second splice isoform. J. Biol. Chem. 1996;271:19310–19317. doi: 10.1074/jbc.271.32.19310. [DOI] [PubMed] [Google Scholar]
- 30.Yang W., Ahn H., Hinrichs M., Torry R.J., Torry D.S. Evidence of a novel isoform of placenta growth factor (PlGF-4) expressed in human trophoblast and endothelial cells. J. Reprod. Immunol. 2003;60:53–60. doi: 10.1016/S0165-0378(03)00082-2. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y., Ji W.R., Qi P., Rosin A., Cao Y. Placenta growth factor: Identification and characterization of a novel isoform generated by RNA alternative splicing. Biochem. Biophys. Res. Commun. 1997;235:493–498. doi: 10.1006/bbrc.1997.6813. [DOI] [PubMed] [Google Scholar]
- 32.Iyer S., Scotney P.D., Nash A.D., Ravi Acharya K. Crystal structure of human vascular endothelial growth factor-B: Identification of amino acids important for receptor binding. J. Mol. Biol. 2006;359:76–85. doi: 10.1016/j.jmb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Lohela M., Bry M., Tammela T., Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr. Opin. Cell. Biol. 2009;21:154–165. doi: 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Cao Y., Chen H., Zhou L., Chiang M.K., Anand-Apte B., Weatherbee J.A., Wang Y., Fang F., Flanagan J.G., Tsang M.L. Heterodimers of placenta growth factor/vascular endothelial growth factor. Endothelial activity, tumor cell expression, and high affinity binding to Flk-1/KDR. J. Biol. Chem. 1996;271:3154–3162. doi: 10.1074/jbc.271.6.3154. [DOI] [PubMed] [Google Scholar]
- 35.Domigan C.K., Ziyad S., Iruela-Arispe M.L. Canonical and noncanonical vascular endothelial growth factor pathways: New developments in biology and signal transduction. Arterioscler. Thromb. Vasc. Biology. 2015;35:30–39. doi: 10.1161/ATVBAHA.114.303215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrara N., Adamis A.P. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discov. 2016;15:385–403. doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 37.Makinen T., Olofsson B., Karpanen T., Hellman U., Soker S., Klagsbrun M., Eriksson U., Alitalo K. Differential binding of vascular endothelial growth factor B splice and proteolytic isoforms to neuropilin-1. J. Biol. Chem. 1999;274:21217–21222. doi: 10.1074/jbc.274.30.21217. [DOI] [PubMed] [Google Scholar]
- 38.Migdal M., Huppertz B., Tessler S., Comforti A., Shibuya M., Reich R., Baumann H., Neufeld G. Neuropilin-1 is a placenta growth factor-2 receptor. J. Biol. Chem. 1998;273:22272–22278. doi: 10.1074/jbc.273.35.22272. [DOI] [PubMed] [Google Scholar]
- 39.Dewerchin M., Carmeliet P. PlGF: A multitasking cytokine with disease-restricted activity. Cold Spring Harb Perspect. Med. 2012;2:a011056. doi: 10.1101/cshperspect.a011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Huang Y., Zhang J., Xing B., Xuan W., Wang H., Huang H., Yang J., Tang J. NRP-2 in tumor lymphangiogenesis and lymphatic metastasis. Cancer Lett. 2018;418:176–184. doi: 10.1016/j.canlet.2018.01.040. [DOI] [PubMed] [Google Scholar]
- 41.Niland S., Eble J.A. Neuropilins in the context of tumor vasculature. Int. J. Mol. Sci. 2019;20:639. doi: 10.3390/ijms20030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park S.A., Jeong M.S., Ha K.T., Jang S.B. Structure and function of vascular endothelial growth factor and its receptor system. BMB Rep. 2018;51:73–78. doi: 10.5483/BMBRep.2018.51.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajusola K., Aprelikova O., Korhonen J., Kaipainen A., Pertovaara L., Alitalo R., Alitalo K. FLT4 receptor tyrosine kinase contains seven immunoglobulin-like loops and is expressed in multiple human tissues and cell lines. Cancer Res. 1992;52:5738–5743. [PubMed] [Google Scholar]
- 44.Apte R.S., Chen D.S., Ferrara N. VEGF in signaling and disease: Beyond discovery and development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachary I. Neuropilins: Role in signalling, angiogenesis and disease. Chem. Immunol. Allergy. 2014;99:37–70. doi: 10.1159/000354169. [DOI] [PubMed] [Google Scholar]
- 46.Pellet-Many C., Frankel P., Jia H., Zachary I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008;411:211–226. doi: 10.1042/BJ20071639. [DOI] [PubMed] [Google Scholar]
- 47.Djordjevic S., Driscoll P.C. Targeting VEGF signalling via the neuropilin co-receptor. Drug Discov. Today. 2013;18:447–455. doi: 10.1016/j.drudis.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Sulpice E., Plouet J., Berge M., Allanic D., Tobelem G., Merkulova-Rainon T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood. 2008;111:2036–2045. doi: 10.1182/blood-2007-04-084269. [DOI] [PubMed] [Google Scholar]
- 49.Pan Q., Chathery Y., Wu Y., Rathore N., Tong R.K., Peale F., Bagri A., Tessier-Lavigne M., Koch A.W., Watts R.J. Neuropilin-1 binds to VEGF121 and regulates endothelial cell migration and sprouting. J. Biol. Chem. 2007;282:24049–24056. doi: 10.1074/jbc.M703554200. [DOI] [PubMed] [Google Scholar]
- 50.Sarabipour S., Mac Gabhann F. VEGF-A121a binding to neuropilins—A concept revisited. Cell Adh. Migr. 2018;12:204–214. doi: 10.1080/19336918.2017.1372878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adamis A.P., Shima D.T. The role of vascular endothelial growth factor in ocular health and disease. Retina. 2005;25:111–118. doi: 10.1097/00006982-200502000-00001. [DOI] [PubMed] [Google Scholar]
- 52.Rapisarda A., Melillo G. Role of the VEGF/VEGFR axis in cancer biology and therapy. Adv. Cancer Res. 2012;114:237–267. doi: 10.1016/B978-0-12-386503-8.00006-5. [DOI] [PubMed] [Google Scholar]
- 53.Barratt S.L., Flower V.A., Pauling J.D., Millar A.B. VEGF (vascular endothelial growth factor) and fibrotic lung disease. Int. J. Mol. Sci. 2018;19:1269. doi: 10.3390/ijms19051269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bry M., Kivela R., Leppanen V.M., Alitalo K. Vascular endothelial growth factor-B in physiology and disease. Physiol. Rev. 2014;94:779–794. doi: 10.1152/physrev.00028.2013. [DOI] [PubMed] [Google Scholar]
- 55.Ferrara N., Hillan K.J., Gerber H.P., Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 56.Presta L.G., Chen H., O’Connor S.J., Chisholm V., Meng Y.G., Krummen L., Winkler M., Ferrara N. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res. 1997;57:4593–4599. [PubMed] [Google Scholar]
- 57.Riccardi C., Napolitano E., Platella C., Musumeci D., Melone M.A.B., Montesarchio D. Anti-VEGF DNA-based aptamers in cancer therapeutics and diagnostics. Med. Res. Rev. 2020;41:464–506. doi: 10.1002/med.21737. [DOI] [PubMed] [Google Scholar]
- 58.Folkman J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 59.Muller Y.A., Li B., Christinger H.W., Wells J.A., Cunningham B.C., de Vos A.M. Vascular endothelial growth factor: Crystal structure and functional mapping of the kinase domain receptor binding site. Proc. Natl. Acad. Sci. USA. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller Y.A., Christinger H.W., Keyt B.A., de Vos A.M. The crystal structure of vascular endothelial growth factor (VEGF) refined to 1.93 A resolution: Multiple copy flexibility and receptor binding. Structure. 1997;5:1325–1338. doi: 10.1016/S0969-2126(97)00284-0. [DOI] [PubMed] [Google Scholar]
- 61.Starovasnik M.A., Christinger H.W., Wiesmann C., Champe M.A., de Vos A.M., Skelton N.J. Solution structure of the VEGF-binding domain of Flt-1: Comparison of its free and bound states. J. Mol. Biol. 1999;293:531–544. doi: 10.1006/jmbi.1999.3134. [DOI] [PubMed] [Google Scholar]
- 62.Fairbrother W.J., Champe M.A., Christinger H.W., Keyt B.A., Starovasnik M.A. Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure. 1998;6:637–648. doi: 10.1016/S0969-2126(98)00065-3. [DOI] [PubMed] [Google Scholar]
- 63.Stauffer M.E., Skelton N.J., Fairbrothe W.J. Refinement of the solution structure of the heparin-binding domain of vascular endothelial growth factor using residual dipolar couplings. J. Biomol. NMR. 2002;23:57–61. doi: 10.1023/A:1015346504499. [DOI] [PubMed] [Google Scholar]
- 64.Millauer B., Wizigmann-Voos S., Schnurch H., Martinez R., Moller N.P., Risau W., Ullrich A. High affinity VEGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis. Cell. 1993;72:835–846. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- 65.Terman B.I., Dougher-Vermazen M., Carrion M.E., Dimitrov D., Armellino D.C., Gospodarowicz D., Bohlen P. Identification of the KDR tyrosine kinase as a receptor for vascular endothelial cell growth factor. Biochem. Biophys. Res. Commun. 1992;187:1579–1586. doi: 10.1016/0006-291X(92)90483-2. [DOI] [PubMed] [Google Scholar]
- 66.de Vries C., Escobedo J.A., Ueno H., Houck K., Ferrara N., Williams L.T. The fms-like tyrosine kinase, a receptor for vascular endothelial growth factor. Science. 1992;255:989–991. doi: 10.1126/science.1312256. [DOI] [PubMed] [Google Scholar]
- 67.Shibuya M. Differential roles of vascular endothelial growth factor receptor-1 and receptor-2 in angiogenesis. J. Biochem. Mol. Biol. 2006;39:469–478. doi: 10.5483/BMBRep.2006.39.5.469. [DOI] [PubMed] [Google Scholar]
- 68.Davis-Smyth T., Chen H., Park J., Presta L.G., Ferrara N. The second immunoglobulin-like domain of the VEGF tyrosine kinase receptor Flt-1 determines ligand binding and may initiate a signal transduction cascade. EMBO J. 1996;15:4919–4927. doi: 10.1002/j.1460-2075.1996.tb00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiesmann C., Fuh G., Christinger H.W., Eigenbrot C., Wells J.A., de Vos A.M. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/S0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 70.Markovic-Mueller S., Stuttfeld E., Asthana M., Weinert T., Bliven S., Goldie K.N., Kisko K., Capitani G., Ballmer-Hofer K. Structure of the full-length VEGFR-1 extracellular domain in complex with VEGF-A. Structure. 2017;25:341–352. doi: 10.1016/j.str.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 71.Fuh G., Li B., Crowley C., Cunningham B., Wells J.A. Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor. J. Biol. Chem. 1998;273:11197–11204. doi: 10.1074/jbc.273.18.11197. [DOI] [PubMed] [Google Scholar]
- 72.Brozzo M.S., Bjelic S., Kisko K., Schleier T., Leppanen V.M., Alitalo K., Winkler F.K., Ballmer-Hofer K. Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization. Blood. 2012;119:1781–1788. doi: 10.1182/blood-2011-11-390922. [DOI] [PubMed] [Google Scholar]
- 73.Aiello L.P., Pierce E.A., Foley E.D., Takagi H., Chen H., Riddle L., Ferrara N., King G.L., Smith L.E. Suppression of retinal neovascularization in vivo by inhibition of vascular endothelial growth factor (VEGF) using soluble VEGF-receptor chimeric proteins. Proc. Natl. Acad. Sci. USA. 1995;92:10457–10461. doi: 10.1073/pnas.92.23.10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pechan P., Rubin H., Lukason M., Ardinger J., DuFresne E., Hauswirth W.W., Wadsworth S.C., Scaria A. Novel anti-VEGF chimeric molecules delivered by AAV vectors for inhibition of retinal neovascularization. Gene Ther. 2009;16:10–16. doi: 10.1038/gt.2008.115. [DOI] [PubMed] [Google Scholar]
- 75.Stefano J.E., Bird J., Kyazike J., Cheng A.W., Boudanova E., Dwyer M., Hou L., Qiu H., Matthews G., O’Callaghan M., et al. High-affinity VEGF antagonists by oligomerization of a minimal sequence VEGF-binding domain. Bioconjug Chem. 2012;23:2354–2364. doi: 10.1021/bc300301m. [DOI] [PubMed] [Google Scholar]
- 76.Holash J., Davis S., Papadopoulos N., Croll S.D., Ho L., Russell M., Boland P., Leidich R., Hylton D., Burova E., et al. VEGF-Trap: A VEGF blocker with potent antitumor effects. Proc. Natl. Acad. Sci. USA. 2002;99:11393–11398. doi: 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parker M.W., Xu P., Li X., Vander Kooi C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012;287:11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim K.J., Li B., Winer J., Armanini M., Gillett N., Phillips H.S., Ferrara N. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 79.Muller Y.A., Chen Y., Christinger H.W., Li B., Cunningham B.C., Lowman H.B., de Vos A.M. VEGF and the Fab fragment of a humanized neutralizing antibody: Crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface. Structure. 1998;6:1153–1167. doi: 10.1016/S0969-2126(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 80.Yang J., Wang X., Fuh G., Yu L., Wakshull E., Khosraviani M., Day E.S., Demeule B., Liu J., Shire S.J., et al. Comparison of binding characteristics and in vitro activities of three inhibitors of vascular endothelial growth factor A. Mol. Pharm. 2014;11:3421–3430. doi: 10.1021/mp500160v. [DOI] [PubMed] [Google Scholar]
- 81.Christinger H.W., Fuh G., de Vos A.M., Wiesmann C. The crystal structure of placental growth factor in complex with domain 2 of vascular endothelial growth factor receptor-1. J. Biol. Chem. 2004;279:10382–10388. doi: 10.1074/jbc.M313237200. [DOI] [PubMed] [Google Scholar]
- 82.Leppanen V.M., Prota A.E., Jeltsch M., Anisimov A., Kalkkinen N., Strandin T., Lankinen H., Goldman A., Ballmer-Hofer K., Alitalo K. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc. Natl. Acad. Sci. USA. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Y., Wiesmann C., Fuh G., Li B., Christinger H.W., McKay P., de Vos A.M., Lowman H.B. Selection and analysis of an optimized anti-VEGF antibody: Crystal structure of an affinity-matured Fab in complex with antigen. J. Mol. Biol. 1999;293:865–881. doi: 10.1006/jmbi.1999.3192. [DOI] [PubMed] [Google Scholar]
- 84.Beckmann R., Jensen K., Fenn S., Speck J., Krause K., Meier A., Roth M., Fauser S., Kimbung R., Logan D.T., et al. DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously. Nat. Commun. 2021;12:708. doi: 10.1038/s41467-021-20949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker A., Chung C.W., Neu M., Burman M., Batuwangala T., Jones G., Tang C.M., Steward M., Mullin M., Tournier N., et al. Novel interaction mechanism of a domain antibody-based inhibitor of human vascular endothelial growth factor with greater potency than ranibizumab and bevacizumab and improved capacity over aflibercept. J. Biol. Chem. 2016;291:5500–5511. doi: 10.1074/jbc.M115.691162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee C.V., Liang W.C., Dennis M.S., Eigenbrot C., Sidhu S.S., Fuh G. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J. Mol. Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 87.Fuh G., Wu P., Liang W.C., Ultsch M., Lee C.V., Moffat B., Wiesmann C. Structure-function studies of two synthetic anti-vascular endothelial growth factor Fabs and comparison with the Avastin Fab. J. Biol. Chem. 2006;281:6625–6631. doi: 10.1074/jbc.M507783200. [DOI] [PubMed] [Google Scholar]
- 88.Fellouse F.A., Wiesmann C., Sidhu S.S. Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc. Natl. Acad. Sci. USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fellouse F.A., Esaki K., Birtalan S., Raptis D., Cancasci V.J., Koide A., Jhurani P., Vasser M., Wiesmann C., Kossiakoff A.A., et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 90.Wiesmann C., Christinger H.W., Cochran A.G., Cunningham B.C., Fairbrother W.J., Keenan C.J., Meng G., de Vos A.M. Crystal structure of the complex between VEGF and a receptor-blocking peptide. Biochemistry. 1998;37:17765–17772. doi: 10.1021/bi9819327. [DOI] [PubMed] [Google Scholar]
- 91.Pan B., Li B., Russell S.J., Tom J.Y., Cochran A.G., Fairbrother W.J. Solution structure of a phage-derived peptide antagonist in complex with vascular endothelial growth factor. J. Mol. Biol. 2002;316:769–787. doi: 10.1006/jmbi.2001.5370. [DOI] [PubMed] [Google Scholar]
- 92.Mandal K., Uppalapati M., Ault-Riche D., Kenney J., Lowitz J., Sidhu S.S., Kent S.B. Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography. Proc. Natl. Acad. Sci. USA. 2012;109:14779–14784. doi: 10.1073/pnas.1210483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uppalapati M., Lee D.J., Mandal K., Li H., Miranda L.P., Lowitz J., Kenney J., Adams J.J., Ault-Riche D., Kent S.B., et al. A potent D-protein antagonist of VEGF-A is nonimmunogenic, metabolically stable, and longer-circulating in vivo. ACS Chem. Biol. 2016;11:1058–1065. doi: 10.1021/acschembio.5b01006. [DOI] [PubMed] [Google Scholar]
- 94.Fedorova A., Zobel K., Gill H.S., Ogasawara A., Flores J.E., Tinianow J.N., Vanderbilt A.N., Wu P., Meng Y.G., Williams S.P., et al. The development of peptide-based tools for the analysis of angiogenesis. Chem. Biol. 2011;18:839–845. doi: 10.1016/j.chembiol.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 95.Checco J.W., Kreitler D.F., Thomas N.C., Belair D.G., Rettko N.J., Murphy W.L., Forest K.T., Gellman S.H. Targeting diverse protein-protein interaction interfaces with alpha/beta-peptides derived from the Z-domain scaffold. Proc. Natl. Acad. Sci. USA. 2015;112:4552–4557. doi: 10.1073/pnas.1420380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iyer S., Darley P.I., Acharya K.R. Structural insights into the binding of vascular endothelial growth factor-B by VEGFR-1(D2): Recognition and specificity. J. Biol. Chem. 2010;285:23779–23789. doi: 10.1074/jbc.M110.130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leonard P., Scotney P.D., Jabeen T., Iyer S., Fabri L.J., Nash A.D., Acharya K.R. Crystal structure of vascular endothelial growth factor-B in complex with a neutralising antibody Fab fragment. J. Mol. Biol. 2008;384:1203–1217. doi: 10.1016/j.jmb.2008.09.076. [DOI] [PubMed] [Google Scholar]
- 98.Leppanen V.M., Tvorogov D., Kisko K., Prota A.E., Jeltsch M., Anisimov A., Markovic-Mueller S., Stuttfeld E., Goldie K.N., Ballmer-Hofer K., et al. Structural and mechanistic insights into VEGF receptor 3 ligand binding and activation. Proc. Natl. Acad. Sci. USA. 2013;110:12960–12965. doi: 10.1073/pnas.1301415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parker M.W., Linkugel A.D., Goel H.L., Wu T., Mercurio A.M., Vander Kooi C.W. Structural basis for VEGF-C binding to neuropilin-2 and sequestration by a soluble splice form. Structure. 2015;23:677–687. doi: 10.1016/j.str.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang W.C., Wu X., Peale F.V., Lee C.V., Meng Y.G., Gutierrez J., Fu L., Malik A.K., Gerber H.P., Ferrara N., et al. Cross-species vascular endothelial growth factor (VEGF)-blocking antibodies completely inhibit the growth of human tumor xenografts and measure the contribution of stromal VEGF. J. Biol. Chem. 2006;281:951–961. doi: 10.1074/jbc.M508199200. [DOI] [PubMed] [Google Scholar]
- 101.Khalili H., Brocchini S., Khaw P.T., Filippov S.K. Comparative thermodynamic analysis in solution of a next generation antibody mimetic to VEGF. RSC Adv. 2018;8:35787–35793. doi: 10.1039/C8RA07059H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Adamson P., Wilde T., Dobrzynski E., Sychterz C., Polsky R., Kurali E., Haworth R., Tang C.M., Korczynska J., Cook F., et al. Single ocular injection of a sustained-release anti-VEGF delivers 6months pharmacokinetics and efficacy in a primate laser CNV model. J. Control. Release Off. J. Control. Release Soc. 2016;244:1–13. doi: 10.1016/j.jconrel.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sidhu S.S., Li B., Chen Y., Fellouse F.A., Eigenbrot C., Fuh G. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J. Mol. Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 104.Dugel P.U., Koh A., Ogura Y., Jaffe G.J., Schmidt-Erfurth U., Brown D.M., Gomes A.V., Warburton J., Weichselberger A., Holz F.G., et al. HAWK and HARRIER: Phase 3, multicenter, randomized, double-masked trials of brolucizumab for neovascular age-related macular degeneration. Ophthalmology. 2020;127:72–84. doi: 10.1016/j.ophtha.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 105.Nguyen Q.D., Das A., Do D.V., Dugel P.U., Gomes A., Holz F.G., Koh A., Pan C.K., Sepah Y.J., Patel N., et al. Brolucizumab: Evolution through preclinical and clinical studies and the implications for the management of neovascular age-related macular degeneration. Ophthalmology. 2020;127:963–976. doi: 10.1016/j.ophtha.2019.12.031. [DOI] [PubMed] [Google Scholar]
- 106.Dugel P.U., Singh R.P., Koh A., Ogura Y., Weissgerber G., Gedif K., Jaffe G.J., Tadayoni R., Schmidt-Erfurth U., Holz F.G. HAWK and HARRIER: Ninety-six-week outcomes from the phase 3 trials of brolucizumab for neovascular age-related macular degeneration. Ophthalmology. 2021;128:89–99. doi: 10.1016/j.ophtha.2020.06.028. [DOI] [PubMed] [Google Scholar]
- 107.Arezumand R., Alibakhshi A., Ranjbari J., Ramazani A., Muyldermans S. Nanobodies As Novel Agents for Targeting Angiogenesis in Solid Cancers. Front. Immunol. 2017;8:1746. doi: 10.3389/fimmu.2017.01746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ebrahimizadeh W., Mousavi Gargari S.L., Javidan Z., Rajabibazl M. Production of novel VHH nanobody inhibiting angiogenesis by targeting binding site of VEGF. Appl. Biochem. Biotechnol. 2015;176:1985–1995. doi: 10.1007/s12010-015-1695-y. [DOI] [PubMed] [Google Scholar]
- 109.Ahadi M., Ghasemian H., Behdani M., Kazemi-Lomedasht F. Oligoclonal selection of nanobodies targeting vascular endothelial growth factor. J. Immunotoxicol. 2019;16:34–42. doi: 10.1080/1547691X.2018.1526234. [DOI] [PubMed] [Google Scholar]
- 110.Karami E., Sabatier J.M., Behdani M., Irani S., Kazemi-Lomedasht F. A nanobody-derived mimotope against VEGF inhibits cancer angiogenesis. J. Enzym. Inhib. Med. Chem. 2020;35:1233–1239. doi: 10.1080/14756366.2020.1758690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjic N. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 112.Empting M. Cyclic Peptides: From Bioorganic Synthesis to Applications. Elsevier; Amsterdam, The Netherlands: 2017. Chapter 1: An introduction to cyclic peptides; pp. 1–14. [Google Scholar]
- 113.Checco J.W., Gellman S.H. Iterative nonproteinogenic residue incorporation yields alpha/beta-peptides with a helix-loop-helix tertiary structure and high affinity for VEGF. Chembiochem A Eur. J. Chem. Biol. 2017;18:291–299. doi: 10.1002/cbic.201600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stahl A., Stumpp M.T., Schlegel A., Ekawardhani S., Lehrling C., Martin G., Gulotti-Georgieva M., Villemagne D., Forrer P., Agostini H.T., et al. Highly potent VEGF-A-antagonistic DARPins as anti-angiogenic agents for topical and intravitreal applications. Angiogenesis. 2013;16:101–111. doi: 10.1007/s10456-012-9302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodrigues G.A., Mason M., Christie L.A., Hansen C., Hernandez L.M., Burke J., Luhrs K.A., Hohman T.C. Functional characterization of Abicipar-Pegol, an anti-VEGF DARPin therapeutic that potently inhibits angiogenesis and vascular permeability. Investig. Ophthalmol. Vis. Sci. 2018;59:5836–5846. doi: 10.1167/iovs.18-25307. [DOI] [PubMed] [Google Scholar]
- 116.Fairbrother W.J., Christinger H.W., Cochran A.G., Fuh G., Keenan C.J., Quan C., Shriver S.K., Tom J.Y., Wells J.A., Cunningham B.C. Novel peptides selected to bind vascular endothelial growth factor target the receptor-binding site. Biochemistry. 1998;37:17754–17764. doi: 10.1021/bi981931e. [DOI] [PubMed] [Google Scholar]
- 117.Pan B., Fairbrother W.J. NMR structural analysis of vascular endothelial growth factor in complex with a phage-derived peptide antagonist. Spectroscopy. 2003;17:169–181. doi: 10.1155/2003/364613. [DOI] [Google Scholar]
- 118.Pan B., Fairbrother W.J. 1H, 13C, and 15N resonance assignment of the vascular endothelial growth factor receptor-binding domain in complex with a receptor-blocking peptide. J. Biomol. NMR. 2002;22:189–190. doi: 10.1023/A:1014297525543. [DOI] [PubMed] [Google Scholar]
- 119.Guryanov I., Korzhikov-Vlakh V., Bhattacharya M., Biondi B., Masiero G., Formaggio F., Tennikova T., Urtti A. Conformationally constrained peptides with high affinity to the vascular endothelial growth factor. J. Med. Chem. 2021;64:10900–10907. doi: 10.1021/acs.jmedchem.1c00219. [DOI] [PubMed] [Google Scholar]
- 120.Dyachenko A., Goldflam M., Vilaseca M., Giralt E. Molecular recognition at protein surface in solution and gas phase: Five VEGF peptidic ligands show inverse affinity when studied by NMR and CID-MS. Biopolymers. 2010;94:689–700. doi: 10.1002/bip.21462. [DOI] [PubMed] [Google Scholar]
- 121.Marquez B.V., Beck H.E., Aweda T.A., Phinney B., Holsclaw C., Jewell W., Tran D., Day J.J., Peiris M.N., Nwosu C., et al. Enhancing peptide ligand binding to vascular endothelial growth factor by covalent bond formation. Bioconjug. Chem. 2012;23:1080–1089. doi: 10.1021/bc300114d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coppock M.B., Warner C.R., Dorsey B., Orlicki J.A., Sarkes D.A., Lai B.T., Pitram S.M., Rohde R.D., Malette J., Wilson J.A., et al. Protein catalyzed capture agents with tailored performance for in vitro and in vivo applications. Biopolymers. 2017;108:e22934. doi: 10.1002/bip.22934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marquez B.V., Ikotun O.F., Parry J.J., Rogers B.E., Meares C.F., Lapi S.E. Development of a radiolabeled irreversible peptide ligand for PET imaging of vascular endothelial growth factor. J. Nucl. Med. 2014;55:1029–1034. doi: 10.2967/jnumed.113.130898. [DOI] [PubMed] [Google Scholar]
- 124.Haase H.S., Peterson-Kaufman K.J., Lan Levengood S.K., Checco J.W., Murphy W.L., Gellman S.H. Extending foldamer design beyond alpha-helix mimicry: Alpha/beta-peptide inhibitors of vascular endothelial growth factor signaling. J. Am. Chem. Soc. 2012;134:7652–7655. doi: 10.1021/ja302469a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reille-Seroussi M., Gaucher J.F., Desole C., Gagey-Eilstein N., Brachet F., Broutin I., Vidal M., Broussy S. Vascular endothelial growth factor peptide ligands explored by competition assay and isothermal titration calorimetry. Biochemistry. 2015;54:5147–5156. doi: 10.1021/acs.biochem.5b00722. [DOI] [PubMed] [Google Scholar]
- 126.Kenrick S.A., Daugherty P.S. Bacterial display enables efficient and quantitative peptide affinity maturation. Protein. Eng. Des. Sel. 2010;23:9–17. doi: 10.1093/protein/gzp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K., et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. Publ. Protein Soc. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bayo-Puxan N., Rodriguez-Mias R., Goldflam M., Kotev M., Ciudad S., Hipolito C.J., Varese M., Suga H., Campos-Olivas R., Barril X., et al. Combined use of oligopeptides, fragment libraries, and natural compounds: A comprehensive approach to sample the druggability of vascular endothelial growth factor. ChemMedChem. 2016;11:928–939. doi: 10.1002/cmdc.201500467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 130.Iyer S., Leonidas D.D., Swaminathan G.J., Maglione D., Battisti M., Tucci M., Persico M.G., Acharya K.R. The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution. J. Biol. Chem. 2001;276:12153–12161. doi: 10.1074/jbc.M008055200. [DOI] [PubMed] [Google Scholar]
- 131.Anisimov A., Leppanen V.M., Tvorogov D., Zarkada G., Jeltsch M., Holopainen T., Kaijalainen S., Alitalo K. The basis for the distinct biological activities of vascular endothelial growth factor receptor-1 ligands. Sci. Signal. 2013;6:ra52. doi: 10.1126/scisignal.2003905. [DOI] [PubMed] [Google Scholar]
- 132.Fischer C., Jonckx B., Mazzone M., Zacchigna S., Loges S., Pattarini L., Chorianopoulos E., Liesenborghs L., Koch M., De Mol M., et al. Anti-PlGF inhibits growth of VEGF(R)-inhibitor-resistant tumors without affecting healthy vessels. Cell. 2007;131:463–475. doi: 10.1016/j.cell.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 133.Van de Veire S., Stalmans I., Heindryckx F., Oura H., Tijeras-Raballand A., Schmidt T., Loges S., Albrecht I., Jonckx B., Vinckier S., et al. Further pharmacological and genetic evidence for the efficacy of PlGF inhibition in cancer and eye disease. Cell. 2010;141:178–190. doi: 10.1016/j.cell.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 134.Bais C., Wu X., Yao J., Yang S., Crawford Y., McCutcheon K., Tan C., Kolumam G., Vernes J.M., Eastham-Anderson J., et al. PlGF blockade does not inhibit angiogenesis during primary tumor growth. Cell. 2010;141:166–177. doi: 10.1016/j.cell.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 135.Yao J., Wu X., Zhuang G., Kasman I.M., Vogt T., Phan V., Shibuya M., Ferrara N., Bais C. Expression of a functional VEGFR-1 in tumor cells is a major determinant of anti-PlGF antibodies efficacy. Proc. Natl. Acad. Sci. USA. 2011;108:11590–11595. doi: 10.1073/pnas.1109029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Snuderl M., Batista A., Kirkpatrick N.D., Ruiz de Almodovar C., Riedemann L., Walsh E.C., Anolik R., Huang Y., Martin J.D., Kamoun W., et al. Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell. 2013;152:1065–1076. doi: 10.1016/j.cell.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Albonici L., Giganti M.G., Modesti A., Manzari V., Bei R. Multifaceted role of the placental growth Factor (PlGF) in the antitumor immune response and cancer progression. Int. J. Mol. Sci. 2019;20:2970. doi: 10.3390/ijms20122970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lassen U., Nielsen D.L., Sorensen M., Winstedt L., Niskanen T., Stenberg Y., Pakola S., Stassen J.M., Glazer S. A phase I, dose-escalation study of TB-403, a monoclonal antibody directed against PlGF, in patients with advanced solid tumours. Br. J. Cancer. 2012;106:678–684. doi: 10.1038/bjc.2011.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arezumand R., Mahdian R., Zeinali S., Hassanzadeh-Ghassabeh G., Mansouri K., Khanahmad H., Namvar-Asl N., Rahimi H., Behdani M., Cohan R.A., et al. Identification and characterization of a novel nanobody against human placental growth factor to modulate angiogenesis. Mol. Immunol. 2016;78:183–192. doi: 10.1016/j.molimm.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 140.Nikooharf A., Arezumand R., Mansouri K., Khoshi A.H., Namdar Ahmadabad H. Development of a recombinant monospecific anti-PLGF bivalent nanobody and evaluation of it in angiogenesis modulation. Mol. Biotechnol. 2020;62:580–588. doi: 10.1007/s12033-020-00275-7. [DOI] [PubMed] [Google Scholar]
- 141.Scotney P.D., MacKenzie A., Maccarone P., Fabri L.J., Scrofani S.D., Gooley P.R., Nash A.D. Human vascular endothelial growth factor B: Characterization of recombinant isoforms and generation of neutralizing monoclonal antibodies. Clin. Exp. Pharmacol. Physiol. 2002;29:1024–1029. doi: 10.1046/j.1440-1681.2002.03769.x. [DOI] [PubMed] [Google Scholar]
- 142.Zhang F., Tang Z., Hou X., Lennartsson J., Li Y., Koch A.W., Scotney P., Lee C., Arjunan P., Dong L., et al. VEGF-B is dispensable for blood vessel growth but critical for their survival, and VEGF-B targeting inhibits pathological angiogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:6152–6157. doi: 10.1073/pnas.0813061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Irani Y.D., Scotney P.D., Klebe S., Mortimer L.A., Nash A.D., Williams K.A. An anti-VEGF-B antibody fragment induces regression of pre-existing blood vessels in the rat cornea. Investig. Ophthalmol. Vis. Sci. 2017;58:3404–3413. doi: 10.1167/iovs.16-21343. [DOI] [PubMed] [Google Scholar]
- 144.Irani Y., Scotney P., Nash A., Williams K.A. Species cross-reactivity of antibodies used to treat ophthalmic conditions. Investig. Ophthalmol. Vis. Sci. 2016;57:586–591. doi: 10.1167/iovs.15-18239. [DOI] [PubMed] [Google Scholar]
- 145.Bower K.E., Lam S.N., Oates B.D., Del Rosario J.R., Corner E., Osothprarop T.F., Kinhikar A.G., Hoye J.A., Preston R.R., Murphy R.E., et al. Evolution of potent and stable placental-growth-factor-1-targeting CovX-bodies from phage display peptide discovery. J. Med. Chem. 2011;54:1256–1265. doi: 10.1021/jm101226k. [DOI] [PubMed] [Google Scholar]
- 146.Davydova N., Harris N.C., Roufail S., Paquet-Fifield S., Ishaq M., Streltsov V.A., Williams S.P., Karnezis T., Stacker S.A., Achen M.G. Differential receptor binding and regulatory mechanisms for the lymphangiogenic growth factors vascular endothelial growth factor (VEGF)-C and -D. J. Biol. Chem. 2016;291:27265–27278. doi: 10.1074/jbc.M116.736801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leppanen V.M., Jeltsch M., Anisimov A., Tvorogov D., Aho K., Kalkkinen N., Toivanen P., Yla-Herttuala S., Ballmer-Hofer K., Alitalo K. Structural determinants of vascular endothelial growth factor-D receptor binding and specificity. Blood. 2011;117:1507–1515. doi: 10.1182/blood-2010-08-301549. [DOI] [PubMed] [Google Scholar]
- 148.Stacker S.A., Achen M.G. Emerging roles for VEGF-D in human disease. Biomolecules. 2018;8:1. doi: 10.3390/biom8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dugel P.U., Boyer D.S., Antoszyk A.N., Steinle N.C., Varenhorst M.P., Pearlman J.A., Gillies M.C., Finger R.P., Baldwin M.E., Leitch I.M. Phase 1 study of OPT-302 inhibition of vascular endothelial growth factors C and D for neovascular age-related macular degeneration. Ophthalmol. Retin. 2020;4:250–263. doi: 10.1016/j.oret.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 150.Timoshenko A.V., Rastogi S., Lala P.K. Migration-promoting role of VEGF-C and VEGF-C binding receptors in human breast cancer cells. Br. J. Cancer. 2007;97:1090–1098. doi: 10.1038/sj.bjc.6603993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kampen K.R., Scherpen F.J.G., Mahmud H., Ter Elst A., Mulder A.B., Guryev V., Verhagen H., De Keersmaecker K., Smit L., Kornblau S.M., et al. VEGFC antibody therapy drives differentiation of AML. Cancer Res. 2018;78:5940–5948. doi: 10.1158/0008-5472.CAN-18-0250. [DOI] [PubMed] [Google Scholar]
- 152.Hajrasouliha A.R., Funaki T., Sadrai Z., Hattori T., Chauhan S.K., Dana R. Vascular endothelial growth factor-C promotes alloimmunity by amplifying antigen-presenting cell maturation and lymphangiogenesis. Investig. Ophthalmol. Vis. Sci. 2012;53:1244–1250. doi: 10.1167/iovs.11-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goyal S., Chauhan S.K., Dana R. Blockade of prolymphangiogenic vascular endothelial growth factor C in dry eye disease. Arch. Ophthalmol. 2012;130:84–89. doi: 10.1001/archophthalmol.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Achen M.G., Roufail S., Domagala T., Catimel B., Nice E.C., Geleick D.M., Murphy R., Scott A.M., Caesar C., Makinen T., et al. Monoclonal antibodies to vascular endothelial growth factor-D block its interactions with both VEGF receptor-2 and VEGF receptor-3. Eur. J. Biochem. 2000;267:2505–2515. doi: 10.1046/j.1432-1327.2000.01257.x. [DOI] [PubMed] [Google Scholar]
- 155.Davydova N., Roufail S., Streltsov V.A., Stacker S.A., Achen M.G. The VD1 neutralizing antibody to vascular endothelial growth factor-D: Binding epitope and relationship to receptor binding. J. Mol. Biol. 2011;407:581–593. doi: 10.1016/j.jmb.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 156.Wise L.M., Inder M.K., Real N.C., Stuart G.S., Fleming S.B., Mercer A.A. The vascular endothelial growth factor (VEGF)-E encoded by orf virus regulates keratinocyte proliferation and migration and promotes epidermal regeneration. Cell. Microbiol. 2012;14:1376–1390. doi: 10.1111/j.1462-5822.2012.01802.x. [DOI] [PubMed] [Google Scholar]
- 157.Fleming S.B., Wise L.M., Mercer A.A. Molecular genetic analysis of orf virus: A poxvirus that has adapted to skin. Viruses. 2015;7:1505–1539. doi: 10.3390/v7031505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Meyer M., Clauss M., Lepple-Wienhues A., Waltenberger J., Augustin H.G., Ziche M., Lanz C., Buttner M., Rziha H.J., Dehio C. A novel vascular endothelial growth factor encoded by Orf virus, VEGF-E, mediates angiogenesis via signalling through VEGFR-2 (KDR) but not VEGFR-1 (Flt-1) receptor tyrosine kinases. EMBO J. 1999;18:363–374. doi: 10.1093/emboj/18.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ogawa S., Oku A., Sawano A., Yamaguchi S., Yazaki Y., Shibuya M. A novel type of vascular endothelial growth factor, VEGF-E (NZ-7 VEGF), preferentially utilizes KDR/Flk-1 receptor and carries a potent mitotic activity without heparin-binding domain. J. Biol. Chem. 1998;273:31273–31282. doi: 10.1074/jbc.273.47.31273. [DOI] [PubMed] [Google Scholar]
- 160.Wise L.M., Veikkola T., Mercer A.A., Savory L.J., Fleming S.B., Caesar C., Vitali A., Makinen T., Alitalo K., Stacker S.A. Vascular endothelial growth factor (VEGF)-like protein from orf virus NZ2 binds to VEGFR2 and neuropilin-1. Proc. Natl. Acad. Sci. USA. 1999;96:3071–3076. doi: 10.1073/pnas.96.6.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yamazaki Y., Matsunaga Y., Tokunaga Y., Obayashi S., Saito M., Morita T. Snake venom Vascular Endothelial Growth Factors (VEGF-Fs) exclusively vary their structures and functions among species. J. Biol. Chem. 2009;284:9885–9891. doi: 10.1074/jbc.M809071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Toivanen P.I., Nieminen T., Laakkonen J.P., Heikura T., Kaikkonen M.U., Yla-Herttuala S. Snake venom VEGF Vammin induces a highly efficient angiogenic response in skeletal muscle via VEGFR-2/NRP specific signaling. Sci. Rep. 2017;7:5525. doi: 10.1038/s41598-017-05876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yamazaki Y., Tokunaga Y., Takani K., Morita T. C-terminal heparin-binding peptide of snake venom VEGF specifically blocks VEGF-stimulated endothelial cell proliferation. Pathophysiol. Haemost. Thromb. 2005;34:197–199. doi: 10.1159/000092423. [DOI] [PubMed] [Google Scholar]
- 164.Yamazaki Y., Tokunaga Y., Takani K., Morita T. Identification of the heparin-binding region of snake venom vascular endothelial growth factor (VEGF-F) and its blocking of VEGF-A165. Biochemistry. 2005;44:8858–8864. doi: 10.1021/bi050197d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.