A small peptide regulates muscle thermogenesis through interaction with the sarcoplasmic reticulum calcium ATPase.
Abstract
The sarcoplasmic reticulum (SR) Ca2+-ATPase (SERCA) plays a central role in muscle contractility and nonshivering thermogenesis. SERCA is regulated by sarcolipin (SLN), a single-pass membrane protein that uncouples Ca2+ transport from ATP hydrolysis, promoting futile enzymatic cycles and heat generation. The molecular determinants for regulating heat release by the SERCA/SLN complex are unclear. Using thermocalorimetry, chemical cross-linking, and solid-state NMR spectroscopy in oriented phospholipid bicelles, we show that SERCA’s functional uncoupling and heat release rate are dictated by specific SERCA/SLN intramembrane interactions, with the carboxyl-terminal residues anchoring SLN to the SR membrane in an inhibitory topology. Systematic deletion of the carboxyl terminus does not prevent the SERCA/SLN complex formation but reduces uncoupling in a graded manner. These studies emphasize the critical role of lipids in defining the active topology of SLN and modulating the heat release rate by the SERCA/SLN complex, with implications in fat metabolism and basal metabolic rate.
INTRODUCTION
In mammals, skeletal muscle constitutes approximately 40% of the body mass, playing an important role in metabolism and shivering and nonshivering thermogenesis (1). Recent studies have shown that sarcolipin (SLN), a 31–amino acid single-pass membrane protein, is a critical regulator of nonshivering thermogenesis and is involved in fat metabolism and basal metabolic rate (2–7). SLN belongs to a class of single-pass membrane proteins called regulins (8) that modulate the activity of the sarco(endo)plasmic reticulum Ca2+-ATPase (adenosine triphosphatase) (SERCA). SLN (9) and phospholamban (PLN) (10) were the first regulins to be identified. While PLN expression is mostly localized to the heart, SLN is primarily expressed in skeletal muscle (11, 12).
SERCA is a 110-kDa P-type ATPase that uses energy from adenosine triphosphate (ATP) hydrolysis for transporting nearly 70% of Ca2+ ions from the cytosol into the lumen of the sarcoplasmic reticulum (SR) during muscle relaxation, safeguarding Ca2+ homeostatic balance in both skeletal and cardiac muscle (13). Under physiological conditions, regulins keep SERCA’s apparent Ca2+ affinity within a defined physiological window, and regulation outside these limits leads to muscle disease (14). Structurally, SERCA is a mushroom-shaped enzyme with a sizeable cytoplasmic domain composed of an actuator domain (A-domain), nucleotide-binding domain (N-domain), and autophosphorylation domain (P-domain) (15). The membrane-embedded portion contains 10 transmembrane (TM) domains with TM4, TM5, TM6, and TM8, forming a channel for two Ca2+ binding sites (Fig. 1A) (16, 17). SERCA’s enzymatic mechanism involves at least six different states, interconverting between two major conformations: E1-SERCA (Ca2+-bound) and E2-SERCA (Ca2+-free) (18, 19), which differ in the arrangement of the cytoplasmic domains and topology (i.e., tilt and rotation angles relative to the membrane normal) of the TM helices. Mechanistic studies revealed that for each ATP molecule hydrolyzed, SERCA transports two Ca2+ ions in exchange for two or three H+ (20). SLN’s ability to uncouple SERCA’s ATP hydrolysis and Ca2+ transport has been linked to thermogenesis (7). Upon binding SERCA, SLN creates futile enzymatic cycles producing heat and contributing to the muscle’s overall energy balance (21). In lipid membranes, SLN’s TM region (domains Ib and II; Fig. 1B) adopts a helical conformation, with short N and C termini protruding into the cytoplasm and lumen, respectively (22–24). Similar to PLN, SLN is thought to bind the E2-SERCA conformation preferentially and inhibit the ATPase via an unknown allosteric mechanism (25). However, the interactions between E2-SERCA and SLN are still uncharacterized because of the lack of structural studies.
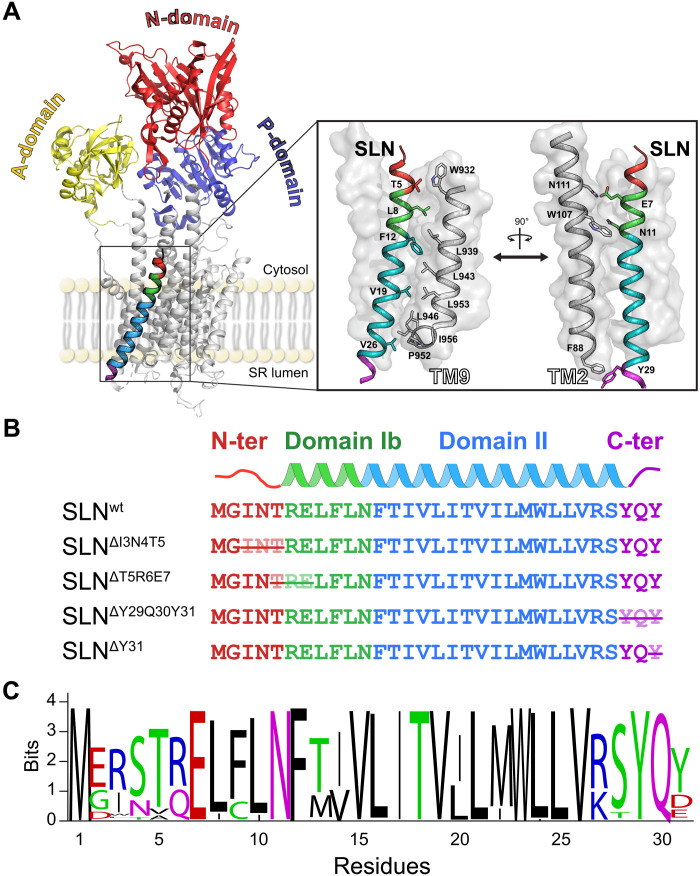
Fig. 1. SERCA/SLN complex and SLN primary structure.
(A) X-ray structure of E1-SERCA in complex with SLNwt (Protein Data Bank 4H1W). SLN’s binding groove in the TM domain of SERCA is highlighted in the inset. (B) Design of SLN truncations. The crossed residues were deleted to test the role of the N terminus, C terminus, and domain Ib on SERCA inhibition. (C) Sequence conservation of SLN.
Two identical crystal structures of the E1-SERCA/SLN complex obtained from lipid preparations revealed SLN’s binding groove and identified the critical residues responsible for intramembrane interactions and inhibition (16, 17). The hydrophobic residues of SLN’s conserved TM domain, which are highly homologous to those of PLN, interact with TM2, TM6, and TM9 of SERCA (Fig. 1A). In the x-ray structures, the variable N terminus and the conserved C terminus of SLN (Fig. 1C) do not diffract well, suggesting the presence of motion/disorder (26). Nonetheless, spectroscopic (27) and truncation/mutation (3, 28) studies in conjunction with functional assays indicated that both termini play a critical role in uncoupling ATP hydrolysis from Ca2+ transport via direct physical interactions with SERCA. In addition, theoretical and experimental studies have proposed that lipids may influence SLN’s inhibitory potency (29, 30). Furthermore, the E1-SERCA/SLN complexes were crystallized with an inhibitory concentration of Mg2+ ions, which traps SERCA in an E1-Mg2+ state rather than the physiologically relevant E1-Ca2+ state. These incongruent data leave several critical questions about the uncoupling mechanism of ATP hydrolysis/Ca2+ transport and the role of SLN’s termini in modulating SERCA function unanswered.
Using a combination of calorimetric measurements, functional assays, chemical cross-linking, and oriented sample solid-state nuclear magnetic resonance (OS-ssNMR) spectroscopy, we determined the factors that regulate the thermogenesis by the SERCA/SLN complex. First, we confirmed that SLN remains bound to SERCA’s major conformational states (i.e., E1 and E2), modulating Ca2+ transport throughout the enzymatic cycle. Second, we found that the juxtamembrane domain Ib plays a critical role in the ATPase inhibition, and removing its conserved residues abrogates SLN function. Last, we found that the N and C termini of SLN are not directly involved in the ATPase’s physical interactions. While the variable N terminus does not affect SLN’s function, the conserved juxtamembrane or C-terminal residues are responsible for anchoring SLN to the SR membrane, stabilizing SLN’s inhibitory orientation, thereby regulating thermogenesis. The elucidation of the factors determining heat release by the SERCA/SLN complex offers new insights into strategies for enhancing fat metabolism and basal metabolic rate to counteract the ongoing obesity crisis (31).
RESULTS
SLN termini affect SERCA’s inhibition and uncoupling of ATP hydrolysis
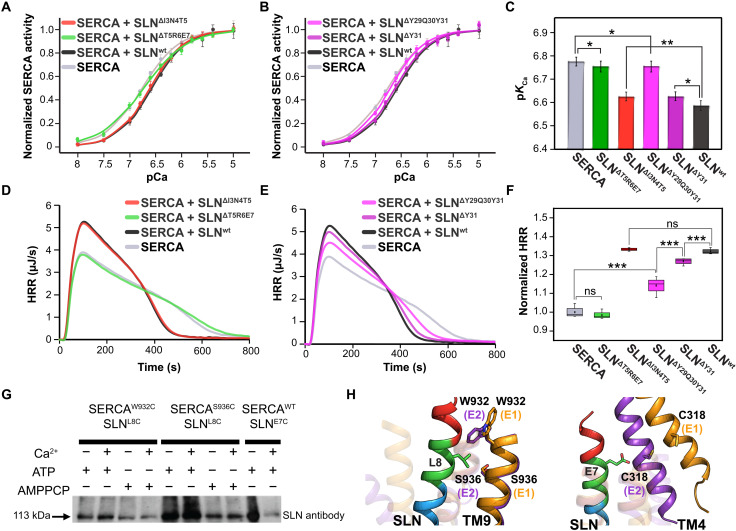
Initial mutagenesis studies have helped to identify the residues of SLN responsible for interactions with SERCA (26). In addition, truncations or mutations have led to the hypothesis that the N and C termini of SLN may play distinct roles in modulating Ca2+ affinity and heat generation (28, 32). However, the recent crystal structures of E1-SERCA/SLN show that both termini do not physically interact with the ATPase (16, 17). To assess the functional relevance of the N- and C-terminal residues and domain Ib, we generated a series of SLN deletion mutations and reconstituted each variant with SERCA to test the inhibitory potency (Fig. 1B). In parallel, we performed calorimetric measurements to determine the heat release rates (HRRs) upon addition of ATP to the SERCA/SLN complex (21, 33). In agreement with previous measurements, we found that wild-type SLN (SLNwt) reduces SERCA’s apparent Ca2+ affinity (pKCa) by approximately 0.2 U (Fig. 2, A to C). In addition, the increase in HRR measured translates into an increase in ATP hydrolysis rate (Vmax) by 30% (Fig. 2, D to F). Note that the integration of the calorimetric curves versus the moles of ATP injected gives rise to ~8.6 kcal/mol, a value close to the theoretical heat release of ATP hydrolysis. This value is constant for all our titrations and is invariant upon co-reconstitution of SERCA with SLN. Therefore, both the pKCa decrease and HRR increase confirm that SLN augments the enzyme’s Vmax, uncoupling ATP hydrolysis and Ca2+ transport (3, 21). Upon deletion of I3, N4, and T5 at the N terminus (SLNΔI3N4T5), we observed only a slight increase in pKCa, while the HRR remained essentially the same. In contrast, deletion of T5, R6, and E7 (SLNΔT5R6E7) in domain Ib abolishes the functional uncoupling, supporting the critical role of the highly conserved R6 and E7 in SERCA’s regulation (34). Removing the C-terminal residues affects SLN’s function in a graded manner (Fig. 2, B and E). Specifically, deletion of Y31 (SLNΔY31) causes a slight increase in pKCa with a concomitant reduction in HRR compared with SLNwt. These effects become more pronounced when Y29 and Q30 are also deleted (SLNΔY29Q30Y31). These data confirm that the two functions of SERCA (ATP hydrolysis and Ca2+ transport) are interdependent (coupled) and that the generation of heat is concomitant to a reduction in Ca2+ transport. Overall, our data suggest that the N terminus of SLN is only marginally involved in SERCA regulation, whereas domain Ib and the C-terminal residues play the most critical role.
Fig. 2. Regulation of SERCA by SLN: Effects on ATP hydrolysis and Ca2+ transport.
(A to C). ATPase-coupled enzymatic assays. Normalized SERCA activity versus pCa2+ for SERCA alone (gray), SERCA/SLNΔT5R6E7 (green), SERCA/SLNΔI3N4T5 (red), SERCA/SLNΔY29Q30Y31 (pink), SERCA/SLNΔY31 (purple), and SERCA/SLNwt (black, control). Four data points of velocity of ATP hydrolysis were acquired for each Ca2+ concentration, and the average values were used to fit the Hill equation. The apparent Ca2+ affinity (pKCa) extracted from fits with errors is summarized in bar graph for all constructs. (D to F) HRR curves versus time for ATP hydrolysis measured for SERCA alone (gray), SERCA/SLNΔT5R6E7 (green), SERCA/SLNΔI3N4T5 (red), SERCA/SLNΔY29Q30Y31 (pink), SERCA/SLNΔY31 (purple), and SERCA/SLNwt (black). Four independent measurements were performed for each condition, and the normalized maximum HRR extracted from the curves is summarized in the box plot. Unpaired Student’s t test was used, and two-tailed P values were calculated to compare the means of two groups. *P < 0.05, **P < 0.01, and ***P < 0.001; ns indicates that the P value is greater than 0.05. The values of pKCa and HRR with errors are summarized in table S1. (G and H) Chemical cross-linking carried out with BMH to detect the formation of the SERCA/SLN complex under different experimental conditions. The gel features the cross-linking of SLNL8C with SERCAW932C and SERCAS936C in the presence or absence of Ca2+, AMPPCP (β,γ-Methyleneadenosine 5′-triphosphate), and ATP. The cross-linking of SLNE7C to SERCAwt is included as a reference. The full gel image is shown in fig. S1.
SLN binds to both E1 and E2 conformations of SERCA
MacLennan and Kranias (35) put forth a dissociative model for SERCA regulation, hypothesizing a complete dissociation of SLN upon increasing Ca2+ concentration. Initial crystallographic studies have shown that SERCA’s binding groove (TM2, TM6, and TM9) is mostly occluded in E1-SERCA but open in E2-SERCA. The latter has led to the hypothesis that SLN preferentially binds E2-SERCA. However, the two recent crystal structures revealed that SLN binds to an E1-Mg2+ state, challenging the dissociative model (16, 17). In addition, spectroscopic data in vitro and in vivo suggest that SLN remains bound to SERCA irrespective of the Ca2+ concentration. These data helped formulate a nondissociative or subunit model of SERCA regulation by SLN (34). We reasoned that SLN may interact with both E1-SERCA and E2-SERCA and that the conformational dynamics of SERCA may modulate the extent of interactions. Therefore, we expressed SLNE7C and cross-linked with SERCAwt using bismaleimidohexane (BMH) in microsomal preparations. In agreement with previous studies (36), we found that SLNE7C cross-links with C318 in TM4 of SERCA, supporting the orientation of SLN relative to SERCA found in the x-ray structures (Fig. 2G and fig. S1). We then repeated the cross-linking reaction of the SLNL8C mutant with two cysteine mutants of the ATPase located in TM9 (SERCAW932C and SERCAS936C). In this case, we observed strong bands in the Western blots for both E1-SERCA/SLN and E2-SERCA/SLN complexes, corroborating that SLN binds SERCA under both low and high Ca2+ concentration.
Structural topology of SLN free and bound to E1- and E2-SERCA
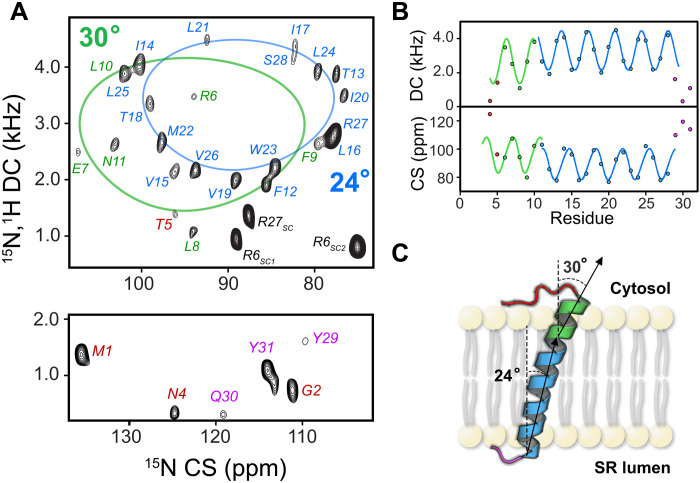
Because SLN binds both E1 and E2 states of SERCA, we surmised that the dynamic transition of SERCA causes structural and dynamic rearrangements of SLN, weakening the intramembrane SERCA/SLN interactions and facilitating Ca2+ transport. To test this hypothesis, we analyzed the structural and topological changes of SLN free and in the presence of both E1- and E2-SERCA. Previous structural studies revealed the predominant helical structure and orientation of SLN in lipid membranes (37, 38). However, the membrane mimetic systems used in these studies prevented the characterization of the large SERCA/SLN membrane protein complex. Therefore, we reconstituted SLN or the SERCA/SLN complex in magnetically aligned lipid bicelles (39). Our lipid composition enables the uniform alignment of the bicelles at a lower temperature (25° to 30°C) than conventional bicelles, as assessed by 31P NMR experiments (fig. S2). The high hydration levels and the lower temperature of alignment maintain SERCA’s stability during data acquisition. We then performed separated local field experiments to image SLN structural topology in membranes. To reduce sample heating, we used a version of the two-dimensional (2D) SE-SAMPI4 (sensitivity enhancement) pulse sequence (40, 41) with heat compensation (hcSE-SAMPI4) (42). When a helical membrane protein is aligned in bicelles, the 1H-15N dipolar couplings (DCs) and 15N chemical shift (CS) of the amide groups give rise to regular patterns of resonances, termed PISA wheels (43). The analysis of these patterns gives an approximate topology (overall tilt and rotation angles) of helical domains relative to the lipid membrane (44, 45). We first analyzed the structural topology of SLN free in lipid bicelles. Figure 3A shows the fully assigned 2D hcSE-SAMPI4 spectrum of SLNwt. Resonance-specific assignment was carried out using both a 3D SE-SAMPI4-(PDSD) (proton driven spin diffusion) experiment and selectively labeled samples as reported previously (46). Using the PISA-SPARKY module (47), we fit the resonances to an ideal helical model. The best fit identified two helical domains with slightly different tilt angles relative to the bicelle normal (Fig. 3A): domains Ib and II, spanning from R6 to N11 and F12 to S28, respectively. These calculations resulted in tilt angles for domains Ib and II of 30° and 24°, respectively (Fig. 3B) (43). The resonances for residues at the N (M1 to T5) and C (Y29 to Y31) termini and the arginine guanidinium resonances are well resolved. Because cross-polarization (CP)–based experiments, e.g., hcSE-SAMPI4, detect dynamically restricted protein moieties, the resolved residues of the two termini are not entirely flexible and interact with the lipid headgroups (Fig. 3C) (48). Peaks assigned to both termini resonate near the isotropic value of the 15N CS [~120 parts per million (ppm)], indicating the presence of disorder. Overall, our NMR data reveal that the TM helix of SLN comprises two helical domains with different tilt angles, not identified previously, with two motionally restricted termini.
Fig. 3. Topology of SLNwt in magnetically aligned bicelles.
(A). The hcSE-SAMPI4 spectrum of SLNwt with residue-specific assignment of the amide resonances. The spectral regions with low (top) and high (bottom) values of the CS are shown separately. The PISA wheel fitting of the resonances for domain II and domain Ib was carried out separately (blue and green PISA wheels). The labels for the resonances of the N terminus, domain Ib, domain II, and the C terminus are indicated in red, green, blue, and pink, respectively. (B) DC and CS wave plots indicating the oscillatory nature associated with the helical conformation for domain Ib and domain II. (C) Cartoon representation of the topology of SLNwt in lipid bilayers.
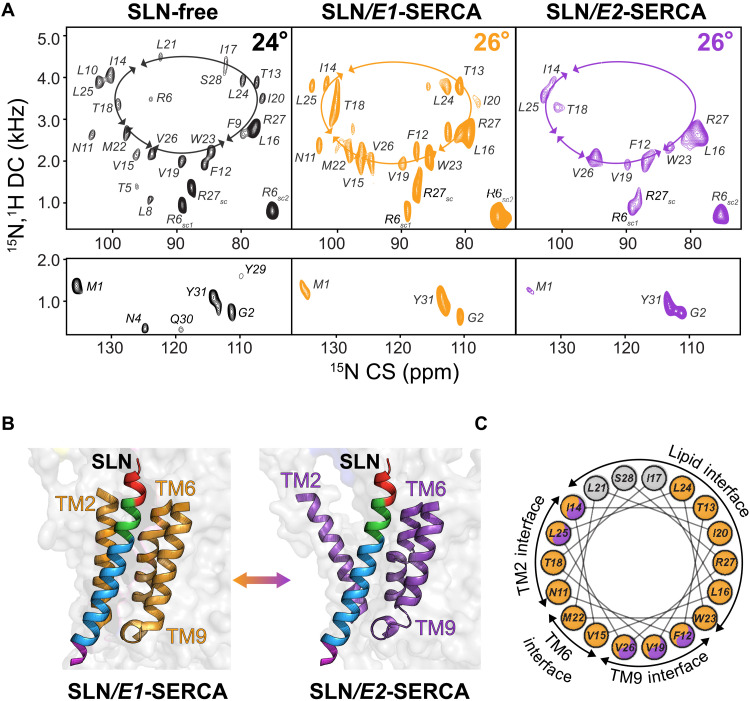
Upon binding the E1-SERCA, the overall helical tilt of domain II of SLNwt transitions to ~26° with no apparent change in the azimuthal (rotational) angle (Fig. 4A). Relatively broader resonance peaks are observed compared to the unbound SLNwt spectrum, indicating a slower rotational diffusion of the SERCA/SLN complex in the lipid bicelles. The resonances corresponding to the N and C termini remain virtually unperturbed, with marginal CS perturbations for well-resolved M1 and G2 resonances, the R6 and R27 side-chain peaks, and the amide resonance of Y31 (Fig. 4A, inset). In contrast, all amide resonances belonging to domains Ib and II are either substantially shifted or broadened, suggesting that these residues are the most affected upon binding E1-SERCA. Similarly, the overall topology of SLN bound to E2-SERCA remains invariant relative to the E1-bound form (Fig. 4, A and B, and fig. S3). Likewise, the amide resonances of domain II are severely broadened. Specifically, the resonances at the interfaces of SLN and SERCA’s TM2 and TM6, as well as the SLN/lipid interface, are broadened beyond detection, suggesting a notable structural and dynamic heterogeneity (Fig. 4C and fig. S4). Unlike the E1-SERCA/SLN complex, the resonances of the residues involved in the SLN/TM9 interface are sharper and more intense than the remainder. A plausible explanation is that SLN may interact more strongly with TM9 of SERCA at low Ca2+ concentrations, with the residues at the protein-protein interface becoming rigid. This interpretation agrees with the chemical cross-linking results (Fig. 2G) and is also supported by the positioning of SLN within the SERCA/SLN complex with domain II closer to TM9 than TM2 and TM6. The hydrophobic interaction between SLN and TM9 of SERCA is one of the most prominent features observed in x-ray structures (17). Similar to the E1-SERCA/SLN complex, the resonances corresponding to residues at the N and C termini of SLN bound to E2-SERCA do not change CS or DCs, and their relative intensities remain essentially the same, indicating that they are not involved in the protein-protein interface. The CS changes and resonance broadening effects observed for SLN in complex with SERCA irrespective of Ca2+ concentration support the subunit model in which SLN remains bound to the ATPase throughout the enzymatic cycle.
Fig. 4. OS-ssNMR spectra of E1-SERCA/SLN and E2-SERCA/SLN complexes.
(A) hcSE-SAMPI4 spectra of free SLNwt (black) and SLNwt bound to E1-SERCA (orange) or E2-SERCA (purple). The calculated tilt angles of domain II are 24° or 26° for free SLN or SLN bound to SERCA, respectively. The overlay of the three spectra is shown in fig. S3 for comparison. (B) Cartoon representation of SLN in the binding groove of SERCA in the E1 (4H1W) and E2 (1IWO) states formed by TM2, TM6, and TM9. (C) Helical wheel representation showing the orientation of SLN with respect to SERCA and lipid bilayers. The colors indicate the relative intensity for domain II resonances of SLN. Orange color indicates SLN resonances with high intensities when bound to E1-SERCA. Orange/purple color indicates resonances with high intensities when bound E1-SERCA or E2-SERCA. Gray color indicates resonances broadened beyond detection in both complexes. A plot with the relative peak intensities is reported in fig. S4.
Deletions alter SLN’s structural topology and dynamics, abrogating functional uncoupling
To determine the structural role of the two termini and domain Ib of SLN in ATPase uncoupling, we first analyzed the topologies of the truncated SLN variants in their free states in oriented bicelles. For all truncations analyzed, the helical conformation of SLN is preserved as demonstrated by the PISA wheel patterns in the hcSE-SAMPI4 spectra (fig. S5). For the truncated SLNΔI3N4T5 variant, we observed a shift toward greater DC and smaller CS values, corresponding to an overall decrease in the tilt angle of ~3° (total tilt angle of ~21° relative to the bilayer) and an unchanged azimuthal angle (fig. S5A). In contrast, deletion of residues in domain Ib (SLNΔT5R6E7) does not alter the tilt angle but causes a clockwise rotation of the TM domain of ~10° (fig. S5B). In this case, the intensities of domain II resonances remain unperturbed, while the N-terminal resonances are broadened out. Therefore, removing R6 at the lipid/water interface affects SLN’s conformational dynamics (48).
For SLNΔY31, we observed a noticeable counterclockwise rotation of ~6°, with line broadening and minor populations for selected peaks (fig. S5C). For the SLNΔY29Q30Y31 truncation, we detected more accentuated line broadening and the presence of a second population of peaks for most of the resolved amide sites (fig. S5D). Numerical simulations of the hcSE-SAMPI4 spectra (49) suggest that the C-terminal truncation causes SLN to interconvert between multiple topologies with tilt angles spanning ±10° (fig. S6). In addition, this truncation causes a rotation of the TM helix of ~14°. We conclude that the C terminus plays a role in maintaining SLN’s topology and provides topological stability through interactions with the lipid headgroups.
To determine the effects of the truncations on the formation of the complexes, we reconstituted the variants of SLN with SERCA in both the E1 and E2 states in oriented lipid bicelles. When reconstituted with SERCA, all SLN variants displayed a pronounced resonance broadening, confirming their physical interactions with the ATPase (27). The SLNΔI3N4T5 truncation shows a substantial change in orientation upon binding either E1- or E2-SERCA conformations, with a ~5° increase in tilt angle (fig. S7A). Upon binding to E2-SERCA, we observed an apparent broadening of the amide resonances similar to the SERCA/SLNwt complex. Because there is only a minor change in pKCa and no change in Vmax or HRR, these observations suggest that removing the N-terminal residues I3-T5 does not affect the topology of bound SLN or its ability to uncouple the ATPase’s functions.
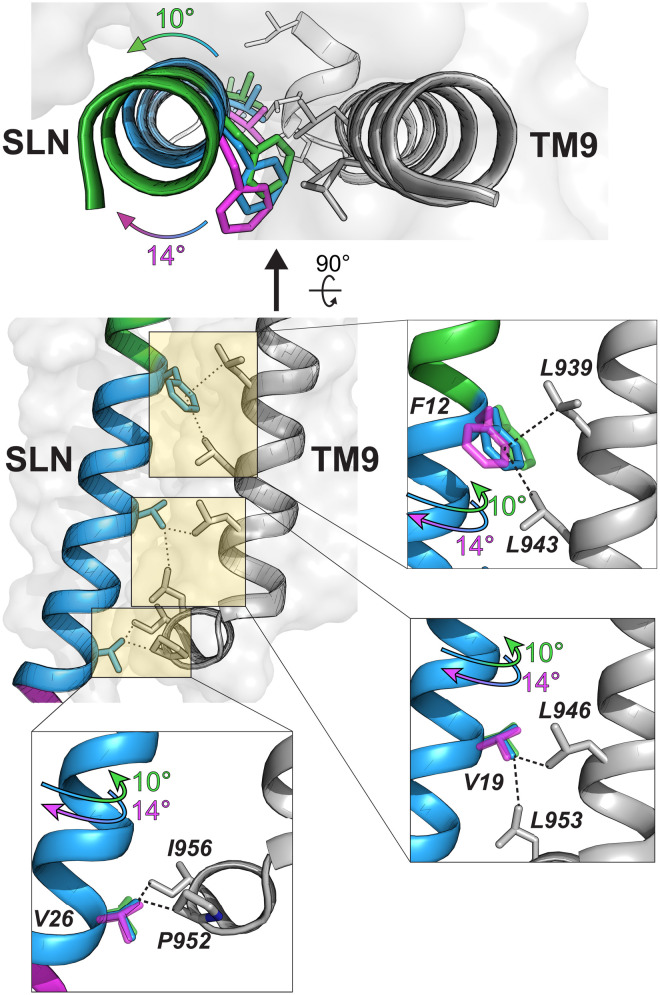
Upon binding to either E1- or E2-SERCA, the orientation of SLNΔT5R6E7 remains unchanged relative to its free form, and its azimuthal angle differs by ~10° relative to SLNwt bound (fig. S7B). However, in the E1-bound state, all amide resonances of SLNΔT5R6E7 become uniformly broad, indicating that all residues are equally affected by the ATPase binding. In contrast, the spectrum of SLNΔT5R6E7 bound to E2-SERCA shows a broadening of the resonances for residues located at the TM9 helix interface (Fig. 5) (16, 17). Overall, the deletion of T5, R6, and E7 in domain Ib causes a ~10o rotation of SLN, hindering the formation of inhibitory interactions with SERCA. This rotation of SLN away from the binding groove and the concomitant disruption of its interactions with TM4 explain the complete loss-of-function character of SLNΔT5R6E7. These data agree with molecular dynamics calculations showing the importance of the functional interactions of E7 with R324 and K328 of SERCA’s TM4 (34).
Fig. 5. The rotation of SLN in the binding groove disrupts the interactions of SLN with TM9 of SERCA.
Hydrophobic interactions between SLN and TM9 of SERCA. Deletion of domain Ib (green) or C terminus (pink) causes a rotation of SLN’s TM helix (blue), disrupting the binding interface.
Among the C-terminal truncations, SLNΔY29Q30Y31 shows the most pronounced effects on structural dynamics (fig. S5C); therefore, we chose this construct to investigate the effects on the SERCA-bound topology. Upon binding E1-SERCA, the resonances of SLNΔY29Q30Y31 become sharper, and the multiple populations of the free form disappear (fig. S7C). In contrast, upon binding E2-SERCA, the majority of the resonances of SLNΔY29Q30Y31 become severely broadened, and several amide resonances display multiple peaks. The latter suggests that SLNΔY29Q30Y31 interactions with E2-SERCA are weaker, likely due to rotating and wobbling motions of SLNΔY29Q30Y31 within the binding groove (Fig. 5). In summary, these data confirm the role of the C terminus in maintaining the inhibitory orientation of the SLN within the binding groove.
DISCUSSION
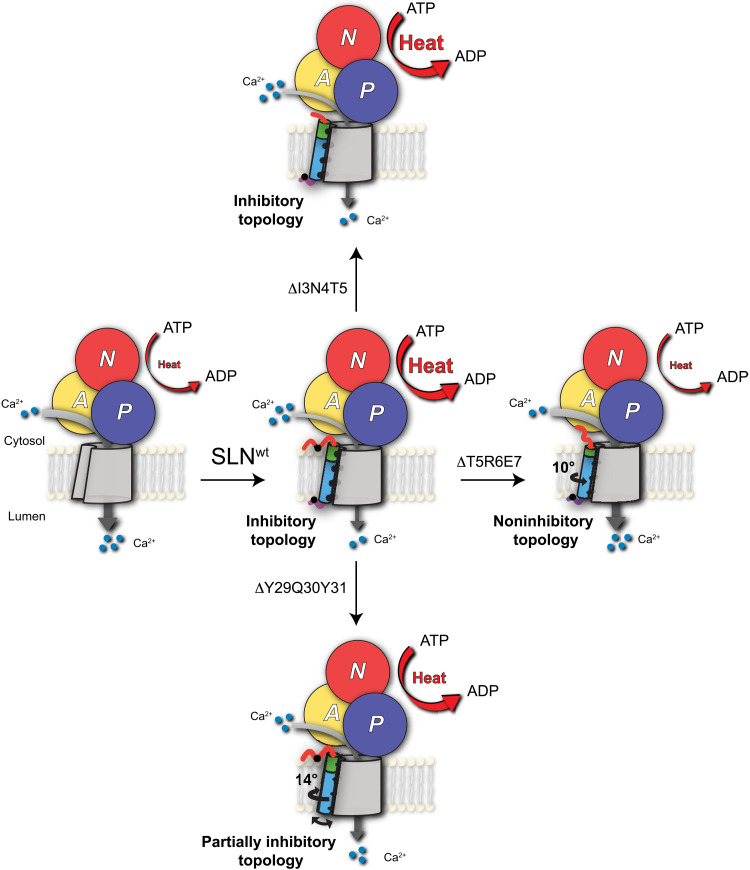
The combination of our structural and functional studies supports the subunit model of SERCA regulation, with SLN remaining bound to both E1 and E2 conformations of the enzyme during the enzymatic cycle. Upon binding to E1-Ca2+ and E2 states of SERCA, the helical secondary structure of SLN remains essentially unchanged. While SLN’s topology changes only slightly upon binding, its conformational dynamics are affected by the different conformational states of the ATPase. These motions manifest as an extensive broadening of the amide resonances in the E2-SERCA/SLN complex. However, the residues at SLN/TM9 of SERCA interface display relatively sharper resonance, highlighting their critical role in stabilizing the E2-SERCA/SLN complex. In the absence of endogenous or exogenous inhibitors, E2-SERCA is quite dynamic (50, 51), and these enzyme’s motions are likely to influence the conformational dynamics of SLN within the complexes. Nonetheless, the two termini invisible in crystal structures, which were deemed to directly interact with SERCA and affect its function, are instead anchored to the lipid membranes in both E1 and E2 conformations, keeping SLN’s TM domain in an inhibitory topology to ensure the most effective interactions with SERCA (Fig. 6). Deletions of either termini or removal of three residues of domain Ib do not prevent the formation of the SERCA/SLN complex; however, the spectroscopic signatures of truncated SLN within these complexes are somewhat different. Unexpectedly, the deletion at the N terminus does not cause rotation of TM domain of SLN in the binding groove, playing only a marginal role in complex formation. However, the N terminus comprises a putative phosphorylation site (T5), which, upon phosphorylation, reverses SERCA inhibition and uncoupling of ATP hydrolysis (11). The x-ray structures suggest that the T5 residue is spatially close to W932 in SERCA and could act as a potential interaction site upon phosphorylation (16). Mutagenesis studies in vivo support the functional relevance of T5, showing that this residue affects cardiac contractility, and its mutation results in cardiac remodeling and diastolic dysfunction (52). However, our study indicates that the T5 residue of SLN is likely to modulate SERCA’s function by affecting the inhibitory topology of SLN instead of interacting directly with SERCA. Despite the C terminus not directly interacting with SERCA, its deletion causes extensive topological heterogeneity and a mismatch of the electrostatic and hydrophobic interactions that hold the SERCA/SLN complex together at the SLN/TM9 interface (partially inhibitory topology). Therefore, on the basis of the ssNMR spectra, we conclude that the C-terminal residues anchor SLN to the lipid membrane in the inhibitory topology and, at the same time, act as membrane sensors for its localization in the SR membrane (53). Removing T5, R6, and E7 in domain Ib, on the other hand, generates a complete loss-of-function mutant of SLN, supporting the critical role of this domain as for PLN (Fig. 6).
Fig. 6. Role of structural topology of SLN in SERCA regulation.
Binding of SLNwt to SERCA increases heat release, featuring an inhibitory topology. Removal of the N-terminal residues does not affect the binding or the uncoupling function of SLN. Removal of the conserved C-terminal residues of SLN causes a rotation of the TM domain and notable conformational/dynamic disorder and generating a partially inhibitory SLN. Deletion of T5, R6, and E7 of domain Ib induces a noninhibitory topology, with a counterclockwise rotation of the TM domain and a complete loss-of-function character.
There is compelling evidence that SERCA’s function is directly affected by SR lipid composition (54). Both in vitro and in vivo studies have shown that changing either lipid headgroups or saturation can increase or decrease SERCA’s function and muscle efficiency (31). SERCA’s thermodynamic efficiency has been estimated to be well below 50% (55). Therefore, Ca2+ leaks and uncoupling pathways are responsible for altering the ATPase’s efficiency. Because skeletal muscle constitutes ~40% of the overall mammalian body mass and SERCA contributes approximately 30% to the isometric contraction, small changes in its enzymatic activity may have repercussions on general metabolism and body temperature (56). Although this could be viewed to have a negative impact on the fitness of the organism, recent evidence suggests that muscle inefficiency may protect mammals against obesity, making the SERCA/SLN complex a possible drug target to tackle this serious health burden (56).
Mini-membrane proteins such as SLN are emerging as widespread regulators of ATPases and some cell surface receptors (8, 57). On the basis of their homology, one would expect similar regulatory mechanisms, i.e., binding in the intramembrane groove and acting as physiological breaks to SERCA’s function. SLN and its homolog PLN share most of the TM domain residues with other regulins, although their inhibitory effects are quite distinct (8, 58, 59). The apparent simplicity of these mini-proteins is deceiving, as they encode specific functions in each residue (60). Therefore, we anticipate that the length and nature of the cytoplasmic and luminal domains of these proteins and their putative posttranslational modifications may hide different regulatory mechanisms (60). From our structural analysis, it emerges that the regulatory mechanism of SERCA not only is a prerogative of SERCA-regulin intermolecular interactions but also involves protein-lipid interactions that play a critical role in modulating the inhibitory function (29, 31, 61).
MATERIALS AND METHODS
SERCA purification
SERCA1a was purified from crude SR extracted from rabbit skeletal muscle using a Reactive Red 120 affinity column (Sigma-Aldrich) (62). Briefly, the muscles were blended with SR preparation buffer solution [20 mM Mops and 100 mM KCl (pH 7.0)] for 1 min and centrifuged at 4200g for 20 min. The supernatant was filtered using cheesecloth and further centrifuged at 12,000g for 20 min to remove mitochondria. The supernatant was filtered using cheesecloth into a beaker, and KCl was added to make a final concentration of 0.6 M KCl. The solution was centrifuged at 22,000g for 1 hour, and the supernatant was removed. The pellet was resuspended in 100 ml of sucrose buffer [20 mM Mops and 300 mM sucrose (pH 7.0)]. The homogenate was centrifuged at 110,000g for 45 min, and the supernatant was removed. The pellets, which mainly contained crude SR, were resuspended with 20 ml of sucrose buffer, homogenized with a Potter-Elvehjem homogenizer, and stored at −80°C in cryovials. To obtain SERCA1a, 10 ml of crude SR solution was thawed and added into 40 ml of extraction buffer [50 mM Mops, 8 mM CaCl2, 1 mM MgCl2, 20% glycerol, and 1% C12E8 (pH 7.0)] with vigorous stirring. The detergents in the extraction buffer extracted SERCA1a from the crude SR into the supernatant. After stirring for 30 min, the solution was centrifuged at 92,000g for 45 min, and the supernatant was loaded on the column with 6 ml of Reactive Red resin. The column was then washed with 50 ml of wash buffer [20 mM Mops, 1 mM CaCl2, 1 mM MgCl2, 20% glycerol, and 0.1% C12E8 (pH 7.0)], and purified SERCA1a was eluted with 50 ml of elution buffer [20 mM Mops, 1 mM CaCl2, 1 mM MgCl2, 8 mM adenosine diphosphate, 20% glycerol, and 0.1% C12E8 (pH 7.0)]. The yield of SERCA was evaluated by a bicinchoninic acid protein assay (63).
Purification of SLN wild type and truncations
Uniformly 15N-labeled SLNwt was expressed as a fusion protein with maltose-binding protein (MBP) in Escherichia coli. Specifically, NEBExpress cells were transformed with SLNwt fusion construct. A 1-liter starter culture containing LB growth medium supplemented with ampicillin (100 μg/ml) was inoculated with a single SLNwt colony and agitated for ~16 hours at 225 rpm at 30°C to OD600 (optical density at 600 nm) of ~2.0. The bacteria were centrifuged and then transferred into 2 liters of standard M9 minimal medium containing 15N-labeled NH4Cl and grown at 30°C until OD600 of ~0.8. Protein expression was induced with isopropyl-β-d-thiogalactopyranoside (1 mM), and cells were harvested after 16 hours of induction by centrifugation at 6370g for 20 min at 4°C. Pellets were flash-frozen and stored at −20°C. For purification, the pellets were resuspended on ice in 200 ml of lysis buffer [20 mM phosphate (pH 7.3), 120 mM NaCl, 8 mM EDTA, 0.1 mM dithiothreitol (DTT), 0.5% glycerol, and 0.5% Triton X-100]. The suspension was homogenized with a cell grinder and sonicated using Branson Sonifier 450 (output of 4; duty cycle, 40%) for 10 min. Cell debris was pelleted by centrifugation at 45,700g for 20 min at 4°C, and the pooled supernatant containing MBP fusion protein was applied to an amylose resin affinity chromatography column at 4°C. The column was washed with 400 ml of buffer [20 mM phosphate and 120 mM NaCl (pH 7.3)]. The fusion protein was eluted with ~100 ml of maltose buffer [46 mM maltose in 20 mM phosphate and 120 mM NaCl (pH 7.3)]. The cleavage was performed using tobacco etch virus protease at 34°C for 6 hours. The cleaved proteins were dialyzed against aqueous buffer until the formation of a precipitate. The precipitate was collected by centrifuge at 18,000g and further purified using reversed-phase high-performance liquid chromatography with a C4 column. The purified proteins were lyophilized overnight and stored at −20°C. All SLN variants were generated using the QuikChange Lightning Mutagenesis Kit (Agilent Genomics) using SLNwt as the template. The expression and purification of all SLN variants followed the same protocol of SLNwt as described above.
SERCA activity measurement
Coupled enzyme assays were used to evaluate the SERCA activity with different regulators according to a well-established protocol (64). Briefly, DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) and DOPC (1,2-Dioleoyl-sn-glycero-3-phosphocholine) [4:1 (mol/mol)] solubilized in chloroform were dried under N2 gas and high vacuum for at least 2 hours to remove the residual organic solvents. The dried lipids were then redissolved into C12E8 solution at 3:1 (w/w) C12E8:lipid ratio. The solutions with SERCA and its regulators were co-solubilized into the mixed micelle solution to a final lipid:SERCA:SLN ratio of 900:1:2 (mol/mol/mol). The samples were incubated with Bio-Beads SM-2 [20:1 (w/w) to C12E8] for 3 hours at 4°C to remove the detergents. A coupled enzyme assay was used to monitor SERCA activity at 25°C as a function of calcium concentration. The enzyme activity rate was measured as a linear decrease of NADH (reduced form of nicotinamide adenine dinucleotide) absorption at 340 nm using a SpectraMax Plus 384 plate reader using 96-well plates. Each well contained 200 μl of buffer solution with 0.8 μg of SERCA and the corresponding amount of the regulators based on the ratio mentioned above. The buffer solution contained 50 mM Hepes, 100 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.2 mM NADH, 0.5 mM phosphoenolpyruvate, 5 mM ATP, 10 IU of pyruvate kinase, 10 IU of lactate dehydrogenase, and 6 μg of calcium ionophore (A23187). The concentration of Ca2+ added for each cell was calculated using MaxChelator (65). To determine the maximum activity (Vmax), Hill coefficient (n), and calcium concentration needed to achieve half-maximal activity (pKCa), data were fit using the Hill equation below
HRR measurements
The HRR measurements were performed with a low-volume Nano ITC (TA Instruments). SERCA and SLN variants were reconstituted into vesicles similarly to coupled enzyme assays. DOPC and DOPE [4:1 (mol/mol)] solubilized in chloroform were dried under N2 gas and high vacuum for at least 2 hours to remove the residual organic solvents. The dried lipids were then redissolved in C12E8 solution at 3:1 (w/w) C12E8:lipid molar ratio. SERCA was dialyzed overnight into a sample buffer consisting of 20 mM Hepes, 100 mM KCl, 5 mM MgCl2, 1 mM EGTA, 1.156 mM CaCl2, 0.02% NaN3, and 2.5% glycerol at pH 7.0. The dialyzed SERCA and SLN variants were added into the mixed micelle solution to a final lipid:SERCA:SLN ratio of 900:1:8 (mol/mol/mol). The mixed micelle solution was incubated with Bio-Beads SM-2 [20:1 (w/w) to C12E8] for 3 hours at 4°C to remove the detergents. For the calorimetry experiments, 100 μl of the above solution containing 18 μg of SERCA was diluted into 250 μl of sample buffer. Calcium ionophore (A23187) was added at this stage to a final concentration of 50 μM. The final volume of the sample solution was ~350 μl, and the free Ca2+ concentration was 79.4 μM, which corresponds to a saturating pCa value of 4.1. Sample solution (300 μl) was loaded into the cell. An injection buffer was prepared by dissolving ATP into the identical sample buffer and transferred to the syringe. The concentration of ATP in the injection buffer was 2.5 mM. The assembled cell and syringe were incubated together at 25°C until a constant baseline was seen. ATP solution (16 μl) was then injected into the cell, and the change in baseline was monitored. Enough time was given to allow the return of the baseline to the original value, which indicated complete hydrolysis of injected ATP.
Mutagenesis and expression of SLN and SERCA in human embryonic kidney 293 cells
The SERCA1a complementary DNA (cDNA) sequence was cloned into the pcDNA3.1 (+) vector. SLN cDNA was polymerase chain reaction–amplified and cloned into pcDNA3.1 (+) vector. Desired mutagenesis of SERCA1a and SLN was done using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). All cDNA clones and mutated constructs were confirmed by direct sequencing. Human embryonic kidney (HEK) 293 cells were cotransfected with SERCA and SLN construct cDNA using Lipofectamine 2000 (Invitrogen). Coexpression was done at a 1:2 ratio of SERCA:SLN. Forty-eight hours after transfection, cells were harvested in phosphate-buffered saline, and the pellet was stored at −80°C after flash freezing in liquid nitrogen. Microsomes from transfected cells were prepared as described previously (36). Briefly, cells were resuspended in a hypotonic solution containing 10 mM tris and 0.5 mM MgCl2 (pH 7.5) for 20 min. Protease inhibitor was added, and cells were homogenized by 30 strokes in a Dounce homogenizer on ice. Homogenates were diluted by an equal volume of solution containing 10 mM tris, 0.5 M sucrose, and 300 mM KCl (pH 7.5). The cell extracts were centrifuged at 10,000g to pellet cell debris. The supernatants were diluted with KCl to a final concentration of 0.6 M and centrifuged at 100,000g for 1 hour at 4°C. The pellet was resuspended in storage buffer containing 10 mM Mops and 10% sucrose (pH 7.0) and stored at −80°C in small aliquots.
Chemical cross-linking assays
The cross-linking reactions were carried out by incubating microsomes under different buffering conditions to mimic the different conformational states of SERCA as reported previously (3, 36). Chemical cross-linking was accomplished by adding 0.1 mM BMH at 25°C for 1 hour and analyzed using SDS–polyacrylamide gel electrophoresis and immunoblotted using anti-SLN antibodies.
Preparation of bicelles for OS-ssNMR experiments
The complex of SERCA and SLNwt or its variants was reconstituted into magnetically aligned anisotropic bicelles formed by long- and short-chain lipids. The long-chain lipids contained 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] ammonium salt (DMPE-PEG2000), and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid copper salt (DMPE-DTPA-Cu2+). The molar ratio of DMPC:POPC:DMPE-PEG2000:DMPE-DTPA-Cu2+ is 4:1:0.125:0.25. The short-chain lipids were 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC). The q ratio (molar ratio of total long-chain lipids versus short-chain lipids) is ~3.8. The DMPE-DTPA-Cu2+ lipid was used to speed up the acquisition of the OS-ssNMR experiments by shortening the 1H T1 relaxation as described previously (46). The lipids were prepared using two different preparations. For the first sample, chloroform was used to solubilize 22.0 mg of DMPC and 6.2 mg of POPC, while for the second sample, chloroform was used to solubilize 5.3 mg of DHPC, 1.0 mg of DMPE-PEG2000, and 2.4 mg of DMPE-DTPA-Cu2+. To eliminate any trace of organic solvents, all lipids were dried under a flux of N2 gas and high vacuum for at least 2 hours. DMPC and POPC were dissolved into C12E8 [detergent/lipid ratio of 1:1 (w/w)] to form a clear suspension of mixed micelles. SLN (0.15 mg) was then solubilized separately with 1% C12E8 solution and added to the mixed micelle/lipid preparation. Purified SERCA (5 mg) was dialyzed overnight into the NMR buffer consisting of 20 mM Hepes, 50 mM KCl, 5.0 mM MgCl2, 0.25 mM DTT, 0.02% NaN3, and 2.5% glycerol. The dialyzed SERCA solution was then added to the mixed micelle solutions. The mixture containing SERCA, SLN, long-chain lipids, and C12E8 was incubated with Bio-Beads SM-2 [20:1 (w/w) ratio to C12E8] for 3 hours to completely remove the detergent. The cloudiness of the suspension confirmed the formation of the DMPC/POPC vesicles containing the SERCA/SLN complex. After Bio-Beads SM-2 removal, we adjusted the pH to 7.0 and centrifuged DMPC/POPC vesicles containing the SERCA/SLN complex at a speed of 50,000g for 50 min. The DHPC/DMPE-PEG2000/DMPE-DTPA-Cu2+ lipids were dissolved into 100 μl of NMR buffer, and the pH was readjusted slightly back to 7.0 and added to the vesicles. Note that 10 mM Ca2+ or 5 mM EGTA was added into the NMR buffer to keep SERCA in the E1 or E2 state, respectively. The protein/lipid mixture was vortexed for a few minutes at 4°C to ensure that the suspension was homogeneous. The bicelles were formed by carrying out three to five cold/warm cycles from 0° to 30°C. Last, the sample was concentrated to a volume of 200 μl before loading into a 5-mm flat-bottom glass tube (New Era) for spectroscopy. For the samples containing only SLN or its variants, the preparation was the same as the SERCA/SLN complex, but 1.5 mg of SLN or its variants was used instead.
OS-ssNMR measurements
All NMR experiments were performed on a Varian VNMRS spectrometer operating at a 1H Larmor frequency of 600 MHz equipped with a Black Fox low-E static bicelle probe built by the RF program at the National High Magnetic Field Laboratory in Florida. For the hcSE-SAMPI4 experiment (40, 42), 15N spin polarization was prepared with adiabatic CP using a linear ramped pulse at the 15N channel and a rectangular pulse for the 1H channel. The 15N RF (radio frequency) field strength was swept from 15 to 25 kHz with the average RF field at 20 kHz, while the 1H RF field amplitude was kept constant at 20 kHz. The CP contact time was 500 μs. During the SAMPI4 t1 evolution (40), the homonuclear 1H decoupling was achieved by applying 31.25-kHz magic sandwich pulses on 1H, and simultaneous phase switched spin-lock pulses on 15N with RF amplitude set to 31.25 kHz, matching the Hartmann-Hahn condition. The dwell time of the t1 acquisition was 96 μs. After the t1 acquisition, two 90° pulses were applied on the 1H and 15N channels simultaneously, followed by a 35.3° pulse on the 1H channel only. For the sensitivity enhancement (SE) element (41), a 100-μs delay of the SE scheme was used, corresponding to four cycles of phase-modulated Lee-Goldberg with an effective RF amplitude of 80 kHz on the 1H channel. After the delay, a 180° inversion pulse was used on the 15N channel followed by another delay period with a 50-kHz SPINAL decoupling on the 1H channel. A final 90° pulse was applied on the 15N channel to prepare the 15N polarization for t2 detection. The t2 acquisition was set to 10 ms for all the NMR experiments. An extra heat compensation block was added after the t2 acquisition, which mirrors the homonuclear decoupling applied during the t1 acquisition period. Note that the length of the heat compensation scheme is complementary with the length of the t1 acquisition to ensure that their sum gives a constant time, which equals to the maximum t1 acquisition time (42). The recycle delay was set to 1 s for all the NMR measurements in this study because of the fast 1H T1 relaxation recovery introduced by Cu2+-chelated lipids. For all the hcSE-SAMPI4 spectra acquired in this study, the temperature of VT (variable temperature) unit was set to 20°C. The actual sample temperature reached equilibrium at 26°C after applying 256 dummy scans of the heat compensation pulse, which was calibrated using the 1H CS from H2O as described previously (42). For hcSE-SAMPI4 experiments on free SLN and its variants, 36 increments were used in the indirect dimension with 256 scans. The total experimental time was ~20 hours. For hcSE-SAMPI4 experiments on SERCA/SLN complexes, 20 increments were used in the indirect dimension with 2048 scans. The total experimental time was ~68 hours. All spectra were processed using NMRPipe. A 40-Hz exponential window function was applied, and 8192 zero filling points were used before the Fourier transformation on the direct dimension. A total of 2048 zero filling points were used without applying a window function before the Fourier transformation on the indirect dimension.
Study approval
The crude SR was prepared from skeletal muscle harvested from New Zealand white rabbits immediately following euthanasia. The protocol is approved by Institutional Animal Care and Use Committee (IACUC Protocol 2104-38975A).
Statistical analysis
For coupled enzymatic assays, four data points of velocity of ATP hydrolysis were acquired at each Ca2+ concentration, and the average values were used to fit the Hill equation using Origin 8.1 software. Numerical values and corresponding errors of pKCa were determined from fitting. For calorimetry measurements, the maximum HRR values are averages of four independent titrations with errors calculated as the SD between these four measurements. Student’s unpaired t test was used to compare the means of two groups. All statistical analyses were performed using Origin 8.1 software. P values <0.05 were considered to be statistically significant.
Acknowledgments
Funding: This work was supported by the NIH (GM 64742 to G.V.) and the American Heart Association (19POST34420009 to D.K.W. and 834110 to S.W.).
Author contributions: G.V. and M.P. conceived and directed all the research. S.W. collected and analyzed NMR, calorimetry, and activity assay data. T.G. assisted NMR data acquisition. S.W., E.K.L., D.K.W., and V.R.U. expressed and purified proteins. C.W. designed the primers of mutations. K.R.M. performed the preliminary calorimetry measurement. S.K.S. executed the chemical cross-linking experiment. G.V. and S.W. prepared the final draft of the manuscript, with input and feedback from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
REFERENCES AND NOTES
- 1.Frontera W. R., Ochala J., Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 96, 183–195 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Maurya S. K., Periasamy M., Sarcolipin is a novel regulator of muscle metabolism and obesity. Pharmacol. Res. 102, 270–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahoo S. K., Shaikh S. A., Sopariwala D. H., Bal N. C., Bruhn D. S., Kopec W., Khandelia H., Periasamy M., The N terminus of sarcolipin plays an important role in uncoupling sarco-endoplasmic reticulum Ca2+-ATPase (SERCA) ATP hydrolysis from Ca2+ transport. J. Biol. Chem. 290, 14057–14067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowland L. A., Bal N. C., Kozak L. P., Periasamy M., Uncoupling protein 1 and sarcolipin are required to maintain optimal thermogenesis, and loss of both systems compromises survival of mice under cold stress. J. Biol. Chem. 290, 12282–12289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurya S. K., Bal N. C., Sopariwala D. H., Pant M., Rowland L. A., Shaikh S. A., Periasamy M., Sarcolipin is a key determinant of the basal metabolic rate, and its overexpression enhances energy expenditure and resistance against diet-induced obesity. J. Biol. Chem. 290, 10840–10849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bombardier E., Smith I. C., Gamu D., Fajardo V. A., Vigna C., Sayer R. A., Gupta S. C., Bal N. C., Periasamy M., Tupling A. R., Sarcolipin trumps β-adrenergic receptor signaling as the favored mechanism for muscle-based diet-induced thermogenesis. FASEB J. 27, 3871–3878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bal N. C., Maurya S. K., Sopariwala D. H., Sahoo S. K., Gupta S. C., Shaikh S. A., Pant M., Rowland L. A., Bombardier E., Goonasekera S. A., Tupling A. R., Molkentin J. D., Periasamy M., Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat. Med. 18, 1575–1579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson D. M., Makarewich C. A., Anderson K. M., Shelton J. M., Bezprozvannaya S., Bassel-Duby R., Olson E. N., Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 9, ra119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odermatt A., Taschner P. E., Scherer S. W., Beatty B., Khanna V. K., Cornblath D. R., Chaudhry V., Yee W. C., Schrank B., Karpati G., Breuning M. H., Knoers N., MacLennan D. H., Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: Absence of structural mutations in five patients with Brody disease. Genomics 45, 541–553 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Tada M., Kirchberger M. A., Katz A. M., Phosphorylation of a 22,000-dalton component of the cardiac sarcoplasmic reticulum by adenosine 3′:5′-monophosphate-dependent protein kinase. J. Biol. Chem. 250, 2640–2647 (1975). [PubMed] [Google Scholar]
- 11.Gramolini A. O., Trivieri M. G., Oudit G. Y., Kislinger T., Li W. P., Patel M. M., Emili A., Kranias E. G., Backx P. H., MacLennan D. H., Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 103, 2446–2451 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhupathy P., Babu G. J., Periasamy M., Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca2+ ATPase. J. Mol. Cell. Cardiol. 42, 903–911 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bers D. M., Cardiac excitation-contraction coupling. Nature 415, 198–205 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Vangheluwe P., Raeymaekers L., Dode L., Wuytack F., Modulating sarco(endo)plasmic reticulum Ca2+ ATPase 2 (SERCA2) activity: Cell biological implications. Cell Calcium 38, 291–302 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Toyoshima C., Nakasako M., Nomura H., Ogawa H., Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature 405, 647–655 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C., Iwasawa S., Ogawa H., Hirata A., Tsueda J., Inesi G., Crystal structures of the calcium pump and sarcolipin in the Mg2+−bound E1 state. Nature 495, 260–264 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Winther A.-M. L., Bublitz M., Karlsen J. L., Møller J. V., Hansen J. B., Nissen P., Buch-Pedersen M. J., The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature 495, 265–269 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Olesen C., Picard M., Winther A. M. L., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P., The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Dyla M., Kjærgaard M., Poulsen H., Nissen P., Structure and mechanism of P-type ATPase ion pumps. Annu. Rev. Biochem. 89, 583–603 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Hao L., Rigaud J. L., Inesi G., Ca2+/H+ countertransport and electrogenicity in proteoliposomes containing erythrocyte plasma membrane Ca-ATPase and exogenous lipids. J. Biol. Chem. 269, 14268–14275 (1994). [PubMed] [Google Scholar]
- 21.Mall S., Broadbridge R., Harrison S. L., Gore M. G., Lee A. G., East J. M., The presence of sarcolipin results in increased heat production by Ca2+-ATPase. J. Biol. Chem. 281, 36597–36602 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Mascioni A., Karim C., Barany G., Thomas D. D., Veglia G., Structure and orientation of sarcolipin in lipid environments. Biochemistry 41, 475–482 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Buffy J. J., Buck-Koehntop B. A., Porcelli F., Traaseth N. J., Thomas D. D., Veglia G., Defining the intramembrane binding mechanism of sarcolipin to calcium ATPase using solution NMR spectroscopy. J. Mol. Biol. 358, 420–429 (2006). [DOI] [PubMed] [Google Scholar]
- 24.De Simone A., Mote K. R., Veglia G., Structural dynamics and conformational equilibria of SERCA regulatory proteins in membranes by solid-state NMR restrained simulations. Biophys. J. 106, 2566–2576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waggoner J. R., Huffman J., Froehlich J. P., Mahaney J. E., Phospholamban inhibits Ca-ATPase conformational changes involving the E2 intermediate. Biochemistry 46, 1999–2009 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Morita T., Hussain D., Asahi M., Tsuda T., Kurzydlowski K., Toyoshima C., Maclennan D. H., Interaction sites among phospholamban, sarcolipin, and the sarco(endo)plasmic reticulum Ca(2+)-ATPase. Biochem. Biophys. Res. Commun. 369, 188–194 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Hughes E., Clayton J. C., Kitmitto A., Esmann M., Middleton D. A., Solid-state NMR and functional measurements indicate that the conserved tyrosine residues of sarcolipin are involved directly in the inhibition of SERCA1. J. Biol. Chem. 282, 26603–26613 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Gorski P. A., Glaves J. P., Vangheluwe P., Young H. S., Sarco(endo)plasmic reticulum calcium ATPase (SERCA) inhibition by sarcolipin is encoded in its luminal tail. J. Biol. Chem. 288, 8456–8467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drachmann N. D., Olesen C., Moller J. V., Guo Z., Nissen P., Bublitz M., Comparing crystal structures of Ca2+-ATPase in the presence of different lipids. FEBS J. 281, 4249–4262 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Hossain K. R., Clarke R. J., General and specific interactions of the phospholipid bilayer with P-type ATPases. Biophys. Rev. 11, 353–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verkerke A. R. P., Ferrara P. J., Lin C. T., Johnson J. M., Ryan T. E., Maschek J. A., Eshima H., Paran C. W., Laing B. T., Siripoksup P., Tippetts T. S., Wentzler E. J., Huang H., Spangenburg E. E., Brault J. J., Villanueva C. J., Summers S. A., Holland W. L., Cox J. E., Vance D. E., Neufer P. D., Funai K., Phospholipid methylation regulates muscle metabolic rate through Ca2+ transport efficiency. Nat. Metab. 1, 876–885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhupathy P., Babu G. J., Ito M., Periasamy M., Threonine-5 at the N-terminus can modulate sarcolipin function in cardiac myocytes. J. Mol. Cell. Cardiol. 47, 723–729 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Meis L., Uncoupled ATPase activity and heat production by the sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 276, 25078–25087 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Autry J. M., Thomas D. D., Espinoza-Fonseca L. M., Sarcolipin promotes uncoupling of the SERCA Ca2+ pump by inducing a structural rearrangement in the energy-transduction domain. Biochemistry 55, 6083–6086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLennan D. H., Kranias E. G., Phospholamban: A crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 4, 566–577 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Sahoo S. K., Shaikh S. A., Sopariwala D. H., Bal N. C., Periasamy M., Sarcolipin protein interaction with sarco(endo)plasmic reticulum Ca2+ATPase (SERCA) is distinct from phospholamban protein, and only sarcolipin can promote uncoupling of the SERCA pump. J. Biol. Chem. 288, 6881–6889 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buffy J. J., Traaseth N. J., Mascioni A., Gor’kov P. L., Chekmenev E. Y., Brey W. W., Veglia G., Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry 45, 10939–10946 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Mote K. R., Gopinath T., Veglia G., Determination of structural topology of a membrane protein in lipid bilayers using polarization optimized experiments (POE) for static and MAS solid state NMR spectroscopy. J. Biomol. NMR 57, 91–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durr U. H. N., Gildenberg M., Ramamoorthy A., The magic of bicelles lights up membrane protein structure. Chem. Rev. 112, 6054–6074 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevzorov A. A., Opella S. J., Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. J. Magn. Reson. 185, 59–70 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Gopinath T., Verardi R., Traaseth N. J., Veglia G., Sensitivity enhancement of separated local field experiments: Application to membrane proteins. J. Phys. Chem. B 114, 5089–5095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., Gopinath T., Veglia G., Improving the quality of oriented membrane protein spectra using heat-compensated separated local field experiments. J. Biomol. NMR 73, 617–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesleh M. F., Veglia G., DeSilva T. M., Marassi F. M., Opella S. J., Dipolar waves as NMR maps of protein structure. J. Am. Chem. Soc. 124, 4206–4207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denny J. K., Wang J., Cross T. A., Quine J. R., PISEMA powder patterns and PISA wheels. J. Magn. Reson. 152, 217–226 (2001). [DOI] [PubMed] [Google Scholar]
- 45.D. K. Weber, E. K. Larsen, T. Gopinath, G. Veglia, in Solid-State NMR (IOP Publishing, 2020), pp. 12-11–12-21. [Google Scholar]
- 46.Wang S., Gopinath T., Veglia G., Application of paramagnetic relaxation enhancements to accelerate the acquisition of 2D and 3D solid-state NMR spectra of oriented membrane proteins. Methods 138-139, 54–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber D. K., Wang S., Markley J. L., Veglia G., Lee W., PISA-SPARKY: An interactive SPARKY plugin to analyze oriented solid-state NMR spectra of helical membrane proteins. Bioinformatics 36, 2915–2916 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L., Cembran A., Gao J., Veglia G., Tilt and azimuthal angles of a transmembrane peptide: A comparison between molecular dynamics calculations and solid-state NMR data of sarcolipin in lipid membranes. Biophys. J. 96, 3648–3662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page R. C., Kim S., Cross T. A., Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure 16, 787–797 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyla M., Terry D. S., Kjaergaard M., Sorensen T. L., Lauwring Andersen J., Andersen J. P., Rohde Knudsen C., Altman R. B., Nissen P., Blanchard S. C., Dynamics of P-type ATPase transport revealed by single-molecule FRET. Nature 551, 346–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabashima Y., Ogawa H., Nakajima R., Toyoshima C., What ATP binding does to the Ca2+ pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 117, 18448–18458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shanmugam M., Li D., Gao S. M., Fefelova N., Shah V., Voit A., Pachon R., Yehia G., Xie L. H., Babu G. J., Cardiac specific expression of threonine 5 to alanine mutant sarcolipin results in structural remodeling and diastolic dysfunction. PLOS ONE 10, e0115822 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gramolini A. O., Kislinger T., Asahi M., Li W., Emili A., MacLennan D. H., Sarcolipin retention in the endoplasmic reticulum depends on its C-terminal RSYQY sequence and its interaction with sarco(endo)plasmic Ca(2+)-ATPases. Proc. Natl. Acad. Sci. U.S.A. 101, 16807–16812 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gustavsson M., Traaseth N. J., Veglia G., Activating and deactivating roles of lipid bilayers on the Ca2+-ATPase/phospholamban complex. Biochemistry 50, 10367–10374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lervik A., Bresme F., Kjelstrup S., Rubi J. M., On the thermodynamic efficiency of Ca2+-ATPase molecular machines. Biophys. J. 103, 1218–1226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bal N. C., Periasamy M., Uncoupling of sarcoendoplasmic reticulum calcium ATPase pump activity by sarcolipin as the basis for muscle non-shivering thermogenesis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 375, 20190135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makarewich C. A., The hidden world of membrane microproteins. Exp. Cell Res. 388, 111853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson D. M., Anderson K. M., Chang C. L., Makarewich C. A., Nelson B. R., McAnally J. R., Kasaragod P., Shelton J. M., Liou J., Bassel-Duby R., Olson E. N., A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson B. R., Makarewich C. A., Anderson D. M., Winders B. R., Troupes C. D., Wu F. F., Reese A. L., McAnally J. R., Chen X. W., Kavalali E. T., Cannon S. C., Houser S. R., Bassel-Duby R., Olson E. N., A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young H. S., Ceholski D. K., Trieber C. A., Deception in simplicity: Hereditary phospholamban mutations in dilated cardiomyopathy. Biochem. Cell Biol. 93, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S. M., Ivanov A. R., Hotamisligil G. S., Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stokes D. L., Green N. M., Three-dimensional crystals of CaATPase from sarcoplasmic reticulum. Symmetry and molecular packing. Biophys. J. 57, 1–14 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C., Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 (1985). [DOI] [PubMed] [Google Scholar]
- 64.Reddy L. G., Cornea R. L., Winters D. L., McKenna E., Thomas D. D., Defining the molecular components of calcium transport regulation in a reconstituted membrane system. Biochemistry 42, 4585–4592 (2003). [DOI] [PubMed] [Google Scholar]
- 65.D. M. Bers, C. W. Patton, R. Nuccitelli, Chapter 1—A practical guide to the preparation of Ca2+ buffers, in Methods in Cell Biology, M. Whitaker, Ed. (Academic Press, 2010), vol. 99, pp. 1–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1