Whole-organism perturbations reveal single-cell responses across the Clytia medusa.
Abstract
We present an organism-wide, transcriptomic cell atlas of the hydrozoan medusa Clytia hemisphaerica and describe how its component cell types respond to perturbation. Using multiplexed single-cell RNA sequencing, in which individual animals were indexed and pooled from control and perturbation conditions into a single sequencing run, we avoid artifacts from batch effects and are able to discern shifts in cell state in response to organismal perturbations. This work serves as a foundation for future studies of development, function, and regeneration in a genetically tractable jellyfish species. Moreover, we introduce a powerful workflow for high-resolution, whole-animal, multiplexed single-cell genomics that is readily adaptable to other traditional or nontraditional model organisms.
INTRODUCTION
Single-cell RNA sequencing (scRNA-seq) is enabling the survey of complete transcriptomes of thousands to millions of cells (1), resulting in the establishment of cell atlases across whole organisms (2–6), exploration of the diversity of cell types throughout the animal kingdom (3, 7–9), and investigation of transcriptomic changes under perturbation (10, 11). However, scRNA-seq studies involving multiple samples can be costly and may be confounded by batch effects resulting from multiple distinct library preparations (12, 13). Recent developments in scRNA-seq multiplexing technology expand the number of samples, individuals, or perturbations that can be incorporated within runs, facilitating well-controlled scRNA-seq experiments (11, 14–18). These advances have created an opportunity to explore systems biology of whole organisms at single-cell resolution, merging the concepts of cell atlas surveys with multiplexed single-cell experimentation.
Here, we apply this powerful experimental paradigm to a planktonic model organism. We examine the medusa (free-swimming jellyfish) stage of the hydrozoan Clytia hemisphaerica, with dual motivations. First, Clytia is a powerful, emerging model system spanning multiple fields, from evolutionary and developmental biology to regeneration and neuroscience (19–24). While previous work has characterized a number of cell types in the Clytia medusa (21), a whole-organism atlas of transcriptomic cell types has been lacking. Such an atlas is a critical resource for the Clytia community and an important addition to the study of cell types across animal phylogeny.
Second, emerging multiplexing techniques present new opportunities for system-level studies of cell types and their changing states at unprecedented resolution in whole organisms. The Clytia medusa offers an appealing platform for pioneering these studies. It is small, transparent, and has simple tissues and organs, stem cell populations actively replenishing many cell types in mature animals, and remarkable regenerative capacity (19, 22, 24–27). Furthermore, the 1-cm-diameter adult medusae used in this study contain on the order of 105 cells, making it possible to sample cells comprehensively across a whole animal in a cost-effective manner using current scRNA-seq technology (fig. S1 and tables S1 and S2). In this study, we generate a cell atlas for the Clytia medusa while simultaneously performing a whole-organism perturbation study, providing the first medusa single-cell dataset and an examination of changing cell states across the organism. Our work also provides a proof-of-principle for perturbation studies in nontraditional model organisms, using multiplexing technology and a reproducible workflow with lessened reliance on functional annotation, from the experimental implementation to the data processing and analysis.
RESULTS
We compared control versus starved animals, as this strong, naturalistic stimulus was likely to cause notable, interpretable changes in transcription across multiple cell types. Laboratory-raised, young adult, female medusae were split into two groups of five animals, one deprived of food for 4 days, and the second fed daily (see Materials and Methods). We observed numerous phenotypic changes in starved animals, including a marked size reduction reflecting two- to threefold fewer cells (Fig. 1, fig. S2, and see Materials and Methods) (28), and a notable reduction in gonad size. Correspondingly, the number of eggs released per day decreased (fig. S3) (29).
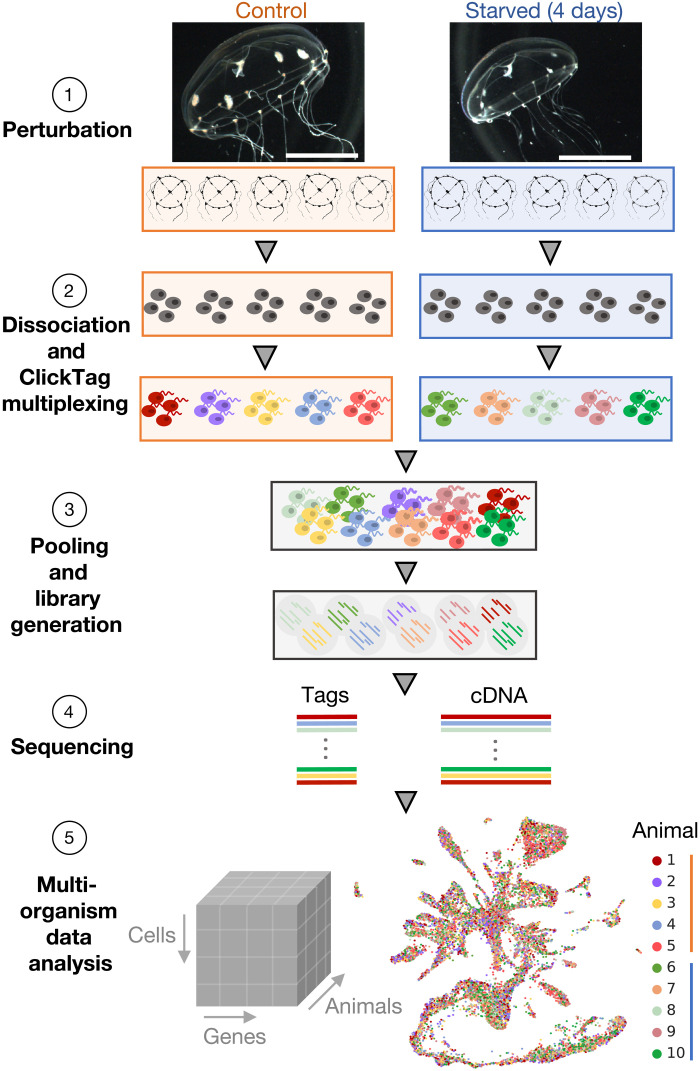
Fig. 1. Overview of whole-organism multiplexed experimentation.
Experimental design of the starvation experiment showing (1) images of control versus 4-day starved animals (scale bars, 0.5 cm), (2) dissociation of individual medusa and chemical tagging of cells with ClickTags to enable multiplexed scRNA-seq, (3) pooling of cells and library generation from lysed cells to generate (4) sequencing libraries for the multiplexed cDNA and ClickTag data and create (5) single-cell resolved gene expression count matrices from all animals (see Materials and Methods).
For scRNA-seq, single-cell suspensions were prepared from each whole medusa and individually labeled with unique ClickTag barcodes (14) using a seawater (SW) compatible workflow (see Materials and Methods, tables S2 and S3, Supplementary Methods, and fig. S4). All labeled suspensions were pooled and processed with the 10X Genomics V2.0 workflow and Illumina sequencing, allowing construction of a combined dataset across organisms and treatments, without requiring batch correction (Fig. 1, fig. S4, A to D, and table S1). A total of 13,673 single-cell profiles derived from 10 individuals (5 control and 5 starved) passed quality control, with high concordance in cell type abundance and gene expression among animals in the same treatment condition (see Materials and Methods and fig. S5). From this gene expression matrix, we (i) derived a Clytia medusa cell atlas and (ii) generated a high-resolution resource of the transcriptional impact of starvation across all observed cell types.
To validate the cell atlas and assess technical variability across and within multiplexed experiments, we performed an additional, independent round of sequencing from 12 individuals. We found that cell types were highly concordant between experiments and confirmed a reduction of batch effect–induced variability within multiplexed experiments (see below). During this second sequencing run, we took advantage of our multiplexing approach to perform an experiment designed both to search for transcripts with “immediate early gene (IEG)”–like behavior in Clytia and to test its sensitivity for detecting more rapid or subtle gene changes than those of the extreme starvation perturbation. For this, we exposed Clytia medusae to multiple transient, ionic stimuli and dissociated ~1 hour later. This paradigm allowed us to identify candidate genes with IEG-like properties across many cell types, including neurons (figs. S6 to S8 and table S4) (30). IEGs are valuable tools in neuroscience, to identify neurons that are active following a specific stimulus or behavior (30). This methodology is thus able to detect transcriptional responses across diverse stimulus-response paradigms (table S5).
A Clytia cell atlas
To generate the cell atlas, we clustered the cells using the gene expression matrix, extracting 36 cell types and their corresponding marker genes (see Materials and Methods; Fig. 2, A and B; figs. S9 to S11; and table S5). Each of the cell types was present in each of the individual animals sequenced (fig. S12). We then generated a low-dimensional representation (31, 32) of these cell types (Fig. 2A). We could group the cell types into seven broad classes (Fig. 2A) that correspond to the outer epidermis, the inner gastrodermis, and to likely derivatives of the multipotent interstitial stem cell population (i-cells). I-cells are a specific feature of hydrozoans, and are particularly well characterized in Hydra, where they generate neural cells, gland cells, and stinging cells (nematocytes), as well as germ cells (8, 20, 33). Our dataset was derived from female medusae so it lacks male germ cells, and late stage oocytes are expected to be too large for capture by the dissociation procedure.
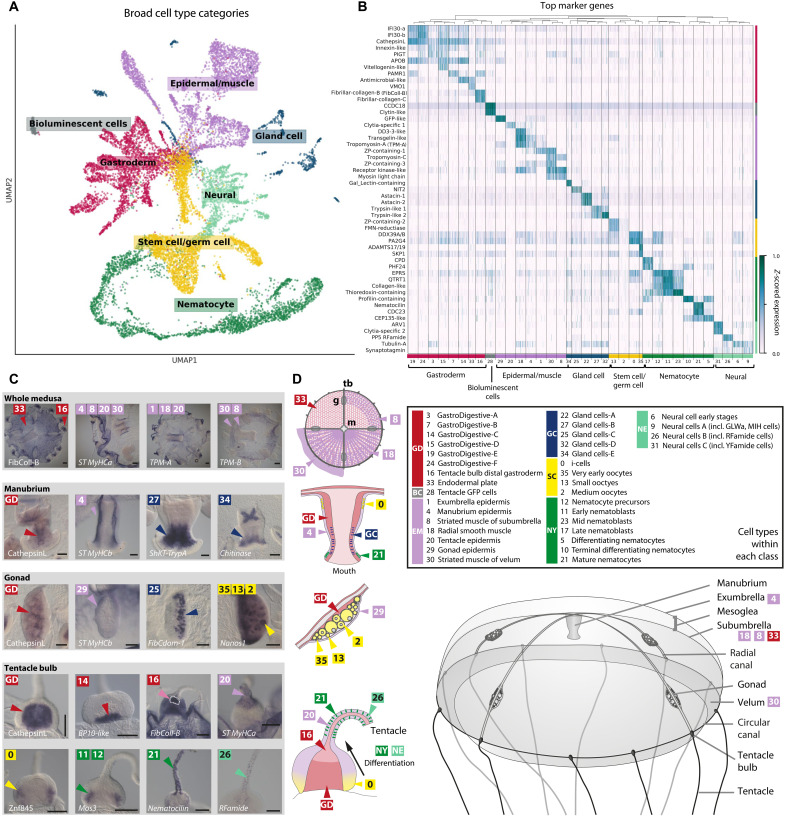
Fig. 2. The Clytia Medusa Cell Atlas.
(A) Two-dimensional UMAP embedding of cells labeled by seven cell type classes. Class colors are retained in (B) to (D). (B) Heatmap of top marker genes from the sequencing data with 36 Louvain clusters comprising the seven cell type classes. (C) In situ hybridization patterns for a selection of cluster marker genes providing spatial location on the animal (comprehensive set in fig. S14). The label GD denotes general markers for GastroDigestive cell types. Scale bars, 100 μm. (D) Schematics of Clytia medusa, manubrium, gonad, and tentacle bulb showing the main cell types. Abbreviations of cell class names: GD, GastroDigestive; BC, bioluminescent cells; EM, epidermal/muscle; GC, gland cells; SC, stem cell/germ cell; NY, nematocytes; NE, neural cells.
The 36 cell types (see Materials and Methods and Fig. 2, B to D) were concordant between the two separate multiplexed experiments (see “Starvation” and “Stimulation” sections in Materials and Methods) and robust to different transcriptome annotations (figs. S6 and S13). For some of them, cell type identity could be assigned on the basis of published information on gene expression in Clytia and/or of homologous genes in other animals, while for the others we performed in situ hybridization for selected marker genes (Fig. 2C, figs. S11 and S14, and table S3). Previously known cell types apparent in our data included i-cells (34) and nematocytes at successive stages of differentiation (35–37), as well as oocytes (38), gonad epidermis, manubrium epidermis, and bioluminescent cells in the tentacles that each express specific endogenous green fluorescent proteins (GFPs) (39).
In situ hybridization for a selection of diagnostic muscle cell type genes allowed us to describe cell types making up the smooth and striated muscles, for instance, distinguishing the striated muscle cells lining the bell (subumbrella) and velum (Fig. 2, C and D, and fig. S14) (23, 27). Within known cell types, clustering revealed an unappreciated degree of cell heterogeneity, yielding novel subtypes. For example, eight cell types could be distinguished within the gastrodermis, six of which were designated gastro-digestive (GD A-F) on the basis of a largely shared set of marker genes (Fig. 2B), including enzymes associated with intracellular digestion, such as CathepsinL (40). Unlike most other clusters, the GD clusters differ primarily in their relative levels of gene expression, rather than by unique marker genes. They thus likely represent variations on a similar digestiveabsorptive epithelial cell type with different functional specializations, distributed across the main digestive compartments of the gastrodermis—the manubrium, gonad, and tentacle bulb—and the gastrovascular canals that link them (figs. S11 and S14). Comparison of gene modules discriminating these gastrodermal clusters (fig. S15) indicates that GD-B may have a particular role in transforming growth factor–β signaling, likely involving the ligands BMP2/4 and BMP5/8. GD-D is enriched for a module associated with cell-cell junctions, while GD-F shows relative depletion, suggesting poorer integration into the gastrodermal epithelium (fig. S15) and possible involvement in GD cell mobilization during starvation and regeneration (19). GD-C cells, localized closest to the endodermal plate, are enriched for transcripts associated with extracellular matrix and mesoglea (jelly) production (Fig. 2, C and D, and figs. S11 and S14). Expression of these and other genes implicated in mesoglea production, such as fibrillar collagens, is also a characteristic of endodermal plate cells (cluster 33) and proximal tentacle-bulb endoderm cells (cluster 16).
Digestive gland cells fell into five types expressing different mixtures of enzymes for extracellular digestion. These showed overlapping distributions in the mouth and stomach regions of the manubrium. Two subtypes of gland cells (type C and E) were also present within the gonad gastroderm. Four broad clusters corresponding to neural cells each appeared to represent mixed populations and could be subdivided by further analyses to define 14 likely subpopulations of neurons (see below). Seven major clusters could be assigned identities as nematocytes at different developmental stages, comprising two groups with highly distinct transcriptional signatures. Four of these we designate “nematoblasts” on the basis of high levels of transcripts related to formation of the nematocyst (stinging capsule) (35, 36, 41). The other three, designated as differentiating and mature “nematocytes,” show no enrichment of these nematocyst transcripts but strongly express highly conserved proteins of the actin-rich “stereovilli” of vertebrate hair cells, including Whirlin, Harmonin, and Sans/USH-IG. This is consistent with observations of similar actin-based protrusions surrounding a central cilium in many of the mechanosensory cell types described in other cnidarian species (42). Related but more elaborate actin structures are associated with the cnidocil of mature nematocytes, but it had not previously been known to share functional hair-cell components (42, 43). Nematocilin, a hydrozoan-specific component of the nematocil (ciliary trigger for nematocyte discharge) (44), is also expressed in these clusters (fig. S14, table S5, and see below). In situ hybridizations revealed marker expression in morphologically distinguishable nematocytes, notably including two lines along the oral face of each tentacle (Fig. 3E and fig. S14), a notable arrangement overlooked in previous studies.
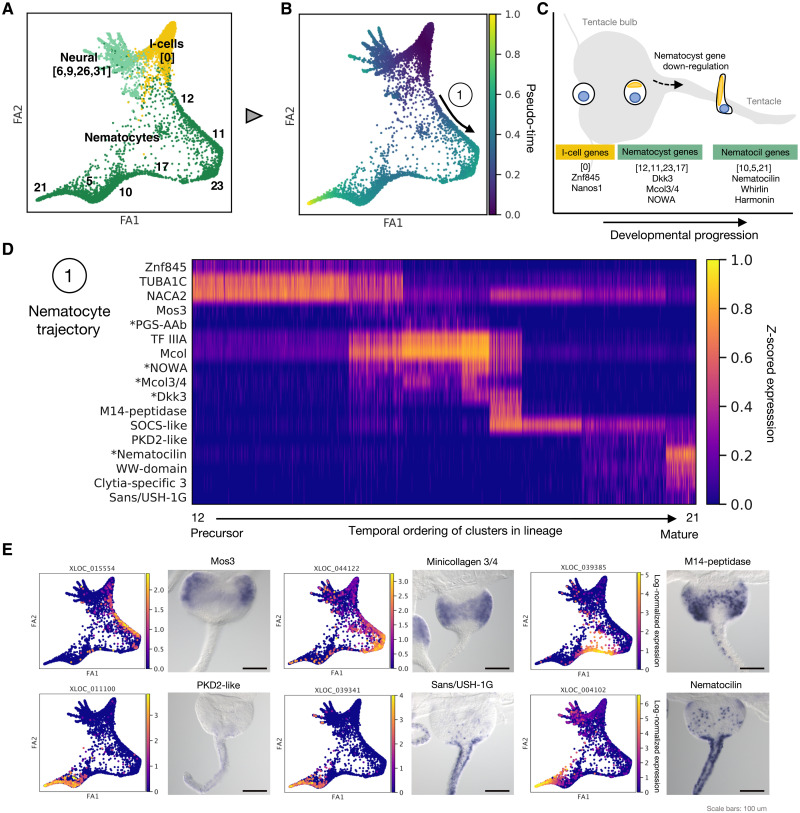
Fig. 3. Developmental and hierarchical analysis of nematocytes.
(A) Subset of i-cell, neural, and nematocyte populations for trajectory analysis with respective clusters denoted. (B) Pseudo-time trajectories for differentiating nematocytes. (C) Diagram indicating the two main phases of nematogenesis with the corresponding “nematocyst” and “nematocil” stages and representative genes. (D) Heatmap of expression of dynamic genes in nematocyte development across the maturing populations. * denotes published nematogenesis genes. (E) Trajectory embeddings for selected nematogenesis markers and corresponding in situs. Scale bars, 100 μm.
A remarkable feature of the Clytia medusa is that it constantly generates many cell types, notably neural cells and nematocytes from prominent i-cell pools in the tentacle bulb epidermis (36) and at other sites (34). Within our dataset, we thus expected to be able to capture dynamic information relating to the development of i-cell–derived cell types, similar to that extracted from Hydra polyp single-cell transcriptome data (8). As in Hydra, our cell atlas revealed clear connections between the neuronal and nematocyte populations and the i-cell population (Fig. 2A and figs. S11 and S14) (3, 8), likely corresponding to differentiation trajectories (35, 36). In contrast, we found no clear developmental connection between i-cells and gland cells and little to no expression of markers of the common neuronal-gland cell precursors identified in Hydra (8) (fig. S16). In Hydra, gland cells are generated not only from the i-cell lineage but also by processes of self-renewal and position-dependent transdifferentiation (8). In the Clytia medusa, digestive gland cells show widespread distribution across distinct regions of the manubrium and gonad compartments of the gastrovascular system, spatially separated from i-cell populations positioned proximally in both these organs (fig. S14). It is possible that these alternative pathways may dominate over direct differentiation from the i-cell lineage in this system.
To address the developmental relationships between the different neural and nematocyte clusters and identify developmental markers, we assigned pseudo-time values to the cells and ranked genes in each trajectory (see Materials and Methods and Fig. 3A). This revealed trajectories consistent with these cell types both deriving from i-cells (Fig. 3, A and B) (8). Examination of the nematocyte trajectory revealed the early expression of genes previously not associated with this process, including Znf845 and Mos3 (Fig. 3, C and D) (45). Nematocyst-related genes—such as minicollagens, polyglutamate synthases, Dkk3, and NOWA—were then expressed during a first major phase of nematogenesis, consistent with previous reports (Fig. 3, C and D; corresponding expression domains in Fig. 3E and fig. S14) (35–37, 41). The trajectory analysis confirmed continuity between the “nematoblast” clusters and the distinct and underappreciated nematocyte differentiation phase, characterized by expression of putative nematocil structural proteins and nematocilin expression at the end of the trajectory (Fig. 3, C and D, and figs. S11 and S14) (see above). The two phases of nematogenesis were linked by the expression of rare specific marker genes for cluster 17 (e.g., M14 peptidase in Fig. 3E and fig. S14). Consistent with this linking of the nematoblast and differentiation phases revealed in trajectory analysis, in situ markers showed distinct expression territories in the tentacle bulb and tentacle, respectively (Fig. 3E). Furthermore, we found that markers of both phases and their respective orthologs, including the “hair cell” gene set, were appropriately distributed among transcriptomes derived from dissected Clytia bulb and tentacle regions (35) and between developing and mature nematocyte scRNA-seq clusters in Hydra (8).
Cnidarian nervous systems represent both valuable points of phylogenetic comparison and tractable platforms for systems neuroscience (3, 8, 20). However, the molecular heterogeneity of neural cell types and their developmental progression remains largely unexplored, particularly in the more complex medusa forms. We therefore extracted genes expressed during neural development that included those encoding bHLH, Sox, and other transcription factors with potential roles in neurogenesis or fate specification and numerous other genes of interest in neuronal development, such as cell adhesion molecules (Fig. 4, A to C, and table S5) (36, 46–48). In mature neurons, neuropeptides are thought to be the dominant neurotransmitters in cnidarians (49, 50) but are challenging to identify because of rapid sequence evolution (51, 52). In conjunction with sequence-based analysis, we were able to identify 10 new likely neuropeptides on the basis of their inclusion as marker genes for the four basic neural clusters (6, 9, 26, and 31 in Fig. 2, B and D), increasing the number of predicted Clytia neuropeptides to 21 (table S3). Our pseudo-time ranking revealed that many of these predicted neuropeptides mark the later stages of neural cluster trajectories, likely defining distinct, mature neural subpopulations (Fig. 4D).
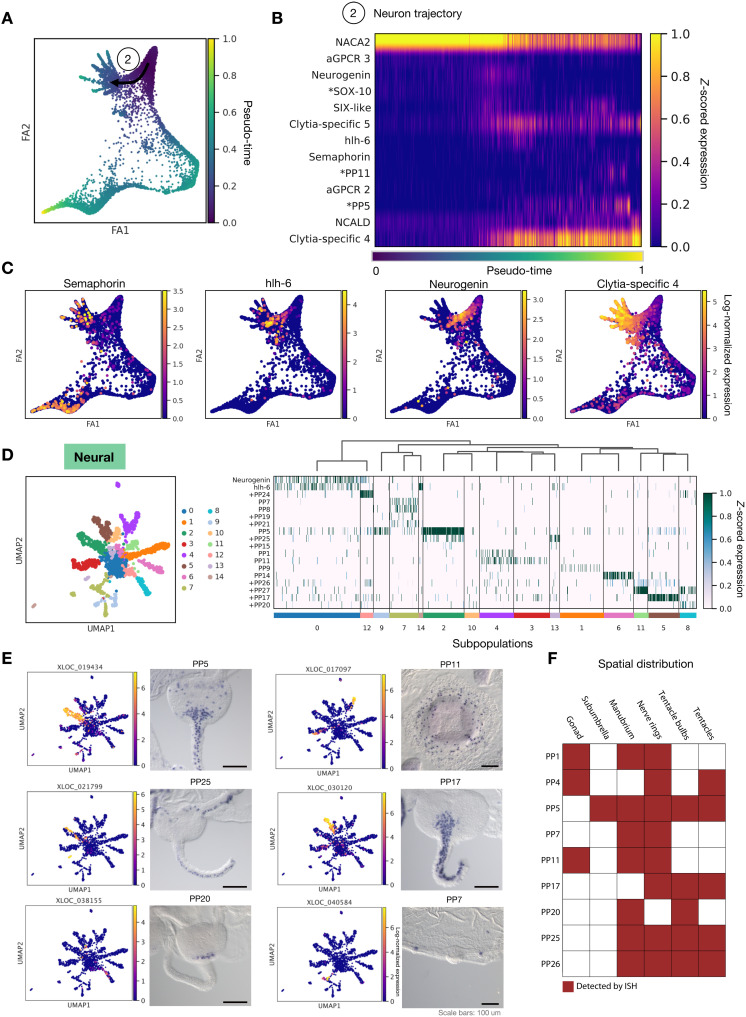
Fig. 4. Developmental and hierarchical analysis of neural cell types.
(A) Pseudo-time trajectories for differentiating neurons. (B) Heatmap of expression of dynamic genes in neural development over pseudo-time. * denotes previously reported genes. (C) Embeddings show gene expression of markers for neural development across the neurons. (D) Diagram of the 14 neural subpopulations and progenitor cells along with a heatmap of neuropeptide markers and the bHLH transcription factors Neurogenin and Hlh6 across these subpopulations. Hlh6, a member of the Atonal superfamily, corresponds to Nematostella Ath-like (82). + denotes previously unpublished neuropeptide candidates. XLOC identifiers and accession numbers in table S3. (E) Embedding for neural subpopulation neuropeptide marker expression and corresponding in situs. Scale bars, 100 μm. (F) Summary of subpopulation marker expression between medusa territories.
We extracted and reclustered the neural supergroup (“Neural;” Fig. 2, A and B) to characterize neural subtypes. This distinguished 14 subpopulations of neurons and a progenitor population, expressing cell cycle and conserved neurodevelopmental genes including the bHLH transcription factor Neurogenin (subcluster 0; Fig. 4D). Notably, the neuronal subpopulations show combinatorial neuropeptide precursor expression, often with a distinct and identifying neuropeptide (Fig. 4D). Expression of putative neuropeptide processing enzymes was detected across the subpopulations and in the nematocytes and gland cells (fig. S17). The UMAP (uniform manifold approximation and projection) expression embedding for neuropeptide precursors suggest that further complexity remains to be discovered within these subpopulations; for instance, subpopulation 4 includes cells either expressing pp1 or pp11. In situ hybridization for a set of neuropeptide precursors indicated that some were broadly distributed across the animal, while others had highly specific spatial locations, suggestive of distinct functions in regulating physiology and behaviors relating to swimming, feeding, and orientation (Fig. 4E and fig. S18). For example, pp5+ (GRFamide precursor) cells were widely detected across the tentacles, bulbs, nerve rings, subumbrella, and mouth, while pp11+ (GLWamide precursor) cells were detected predominantly in the manubrium and nerve ring. Pp7+ cells were located both around the rim of the mouth and in patches of the nerve ring, close to the statocysts (vestibular organs). Subpopulations of neurons also occupied different expression domains within the tentacles: pp25, which generates distinct RFamide family neuropeptides (fig. S18 and table S3), distinguishes a subpopulation of the pp5+ neurons positioned on the aboral side of the tentacle, pp17 labels distinct cells in the same region, and pp20 labels a small group of cells at the base of each tentacle (Fig. 4, E and F, and fig. S18).
Unlike cells using neuropeptides, the transcripts for which are directly assayable in scRNA-seq data, cells using classical chemical neurotransmitters are identified by enzymatic or transporter proxies. Cells using glutamate as a neurotransmitter are usually inferred via the presence of vesicular glutamate transporter markers (vGluts; in human SLC17A6/7/8) (53). Of the closest Clytia homologs of human vGluts, one was expressed in neurons and non-neural cell types, and several were marker genes for nematocytes. However, in common with nearly all cnidarian genes annotated in silico as vesicular glutamate transporters, Clytia sequences lack an arginine residue, conserved in all bilaterian vGluts, and recently shown to be required for vGlut function (54). Furthermore, we detected no neuronal expression of the closest homologs of glutamate decarboxylase (GAD), a marker for gamma-aminobutyric acid-ergic (GABAergic) neurons in Bilateria. GAD homologs were instead detected in some gastrodermal subtypes. There were a number of other interesting neuron subtype-specific genes potentially involved in “chemical” neurotransmission, for example, a possible nitric oxide synthase, a choline (SLC5A7-like) transporter, a gene encoding a taurine dioxygenase–like domain, and a member of the Slc6 family of transporters (table S5).
Cell state shifts in response to starvation across the cell atlas
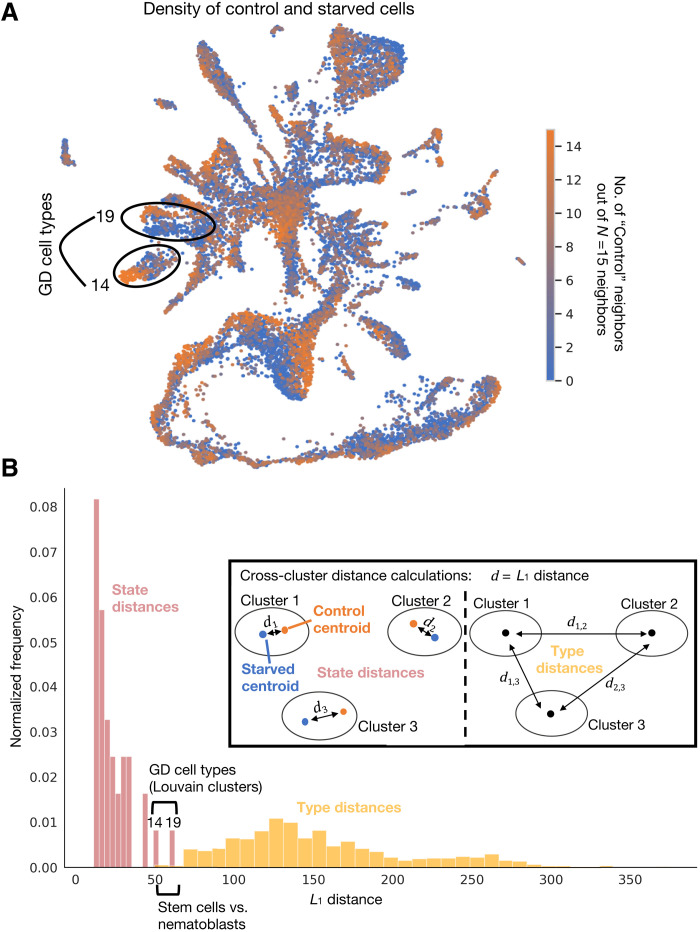
To assess the transcriptional impact of starvation, we mapped individual cells to their corresponding control or starved labels. As there are around 60% fewer cells in a starved animal (fig. S2), we first asked whether there were significantly different numbers of cells per cluster between control and starved conditions. We found that only one cluster had a significant difference (cluster 11, early nematoblasts in fig. S5B), suggesting a nearly uniform reduction across cell types in the starved condition. In contrast, the distribution of cells from control versus starved animals across the atlas embedding showed marked shifts in the local density of cells from the two conditions within most clusters (Fig. 5A).
Fig. 5. Starvation-induced shifts in cell types.
(A) Density map of control and starved cells across cell types (see cell types across conditions in fig. S5B), with each cell colored by the number of its 15 nearest neighbors that are control cells to highlight the clusters most affected by starvation, i.e., most clearly separated from their control neighbors. Clusters 14 and 19, with particularly dense and separated control/starved regions are circled. (B) Histogram of L1 distances between centroids of control and starved cells within cell types versus pairwise L1 distances between centroids of the 36 cell types. Clusters 14 and 19, with largest internal distances, are highlighted, and clusters with small pairwise distances are also noted for comparison. Inset illustrates inter- and intra-L1 distance calculations.
Given the cell type resolution of the atlas, as determined operationally by clustering, we then asked how marked the transcriptional changes incurred by perturbation were in comparison to the transcriptional differences defining the cell types, i.e., are the perturbation-induced changes encompassed within these cell type designations or are they larger in magnitude. We thus compared distances between control and starved cells within clusters to the distances between clusters. As a metric, we used the L1 distance (see Materials and Methods), the sum of the absolute differences between centroid coordinates in principal components analysis (PCA)–reduced space. We found that the L1 distances between control and starved cells within a cell type, versus between cell types (regardless of condition), formed nearly nonoverlapping distributions (see Materials and Methods and Fig. 5B). This suggests that, overall, in Clytia, the transcriptional responses to starvation are defined by cell state shifts, and their cell type repertoire is well represented by the original clusters. However, the impact of starvation was variable across cell types, as reflected by the range of internal (state) distances (Fig. 5B). Starvation produced the largest perturbations in cells of the gastrovascular system, causing control-versus-starved distances large enough to overlap with the smallest inter-type distance, i.e., that between the stem cells and nematocyte precursors (Fig. 5B). This distinction between state shifts and type was also clearly visible in the lack of overlap between the distributions of inter- and intracluster distances within the second multiplexed experiment (fig. S6E). Although classification and distinction of cell state and type is a complex task (55), this analysis, based on relative distance in transcriptional space, provides a quantitative basis for delineation of type/state effects that may be useful in other contexts. We additionally validated the ability of this method to recapitulate the magnitude of state shifts in response to graded stimuli and state-versus-type distinctions, on two other published, multiperturbation datasets (see Materials and Methods; fig. S19).
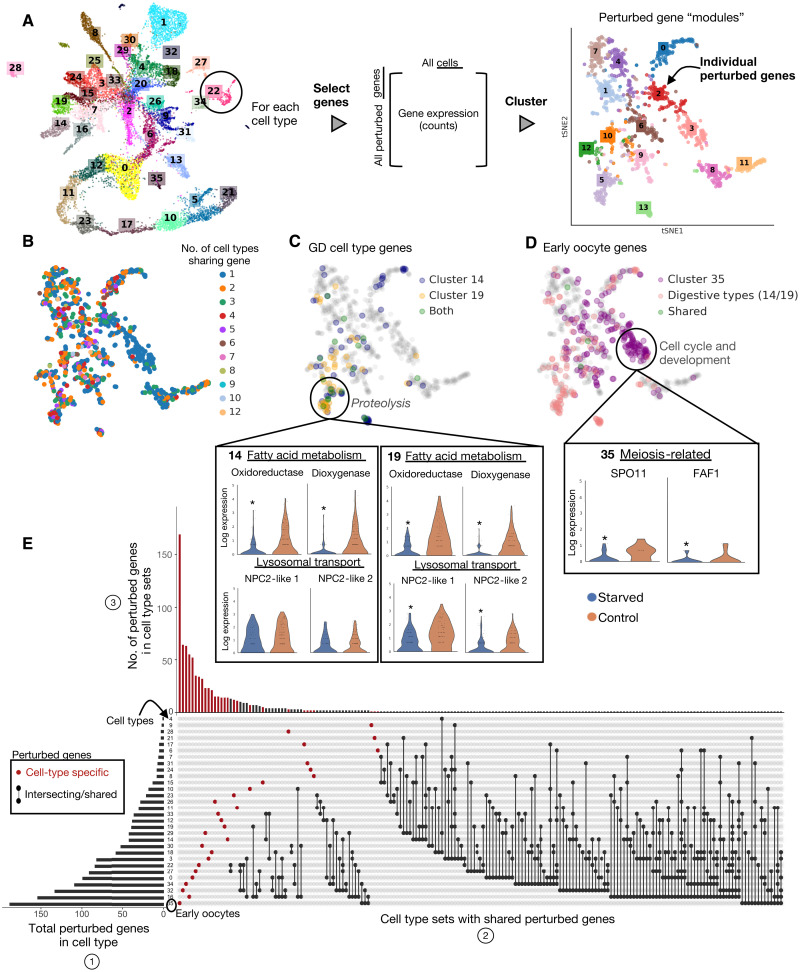
To characterize gene-level responses underlying these starvation-induced shifts, we then asked whether responses are shared or unique across the cell types and compared the extent of the responses, in terms of gene quantity and expression level, across the atlas. For each cell type, we collected genes that were differentially expressed under starvation (“perturbed genes”; Fig. 6A) (see Materials and Methods). For a high-level view of the general functions and processes affected by starvation and their cell type specificity, we clustered perturbed genes into apparent “gene modules” (56) by their patterns of coexpression across cells (see Materials and Methods and Fig. 6A). We assigned putative functions to these gene modules through Gene Ontology (GO) term enrichment, giving a global view of affected processes (fig. S20), and examined the distribution of cell types across modules by asking in how many cell types is a given gene a perturbed gene (Fig. 6B). We found that certain gene modules were broadly shared across cell types, while others were almost entirely cell type–specific (fig. S20). Notable examples include gene module 5, which is enriched in proteolytic genes (Fig. 6C and fig. S20) and has shared expression across multiple GD cell types (Fig. 6C). Notably, there is also divergent gene expression between GD types (Fig. 6C). In comparison, gene module 3 is largely composed of early oocyte gene expression (~70%) and is enriched in cell cycle and developmental genes, which are commonly enriched in growing oocytes (Fig. 6D). Changes in expression of these genes likely reflect the processes of oocyte phagocytosis activated in the gonads of starving animals (see below). Overall, these modules thus provide an overview of which processes affected by starvation are shared across cell types and reveal divergent expression potentially reflecting different motility (19), locations, or as-yet-undescribed functional differences between the cell types (fig. S21).
Fig. 6. Perturbation responses in the oocyte and digestive cell types.
(A) Workflow for extracting perturbed genes per cell type and clustering on genes to extract coexpressed groups or modules of genes (fig. S20). (B) Each gene in module embedding colored by number of cell types in which the gene was differentially expressed under starvation. (C) Visualization of locations of perturbed genes (among the embedded modules) of the two highly perturbed GD cell types (clusters 14 and 19) and (D) early oocytes (cluster 35). Violin plots showing expression profiles for several perturbed (differentially expressed) genes in “functional” categories of interest. *P < 0.05 from nonparametric Wilcoxon test. Horizontal lines show quartiles and width of violins denote density of points in that region. (E) UpSet plot visualization for intersecting sets of perturbed genes. The barplot in (1) shows the number of genes that are differentially expressed under starvation (perturbed genes) for each cell type. This is the cardinality or size of each cell type’s set of perturbed genes. (2) The connected dots represent intersections (overlaps) between the sets of each cell type’s perturbed genes. Genes with starved differential expression in only one cell type are denoted by the red, unconnected dots. (3) The top bar plot is the number of perturbed genes within these sets of shared perturbed genes, i.e., the cardinality of the intersections in (2).
To examine how individual perturbed genes are distributed across cell types, we visualized, for each cell type, how many perturbed genes it had, and how many of these genes are unique versus shared with other cell types (Fig. 6E). We found a large number of perturbed genes (~72%) were cell type specific (Fig. 6E). For the most perturbed cell types, we examined whether the state shifts that we had observed were due to changes in a large or small number of genes, and how highly these genes were expressed. Consistent with the marked shrinkage of the gonads during starvation treatment (Fig. 1), early oocytes contained the highest number of perturbed genes, which were spread across many gene modules (Fig. 6D). In contrast, the GD cell types had fewer perturbed genes that were expressed at higher levels and localized to more specific gene modules (Fig. 6C), highlighting the diverse logic used by cell types under starvation (fig. S22).
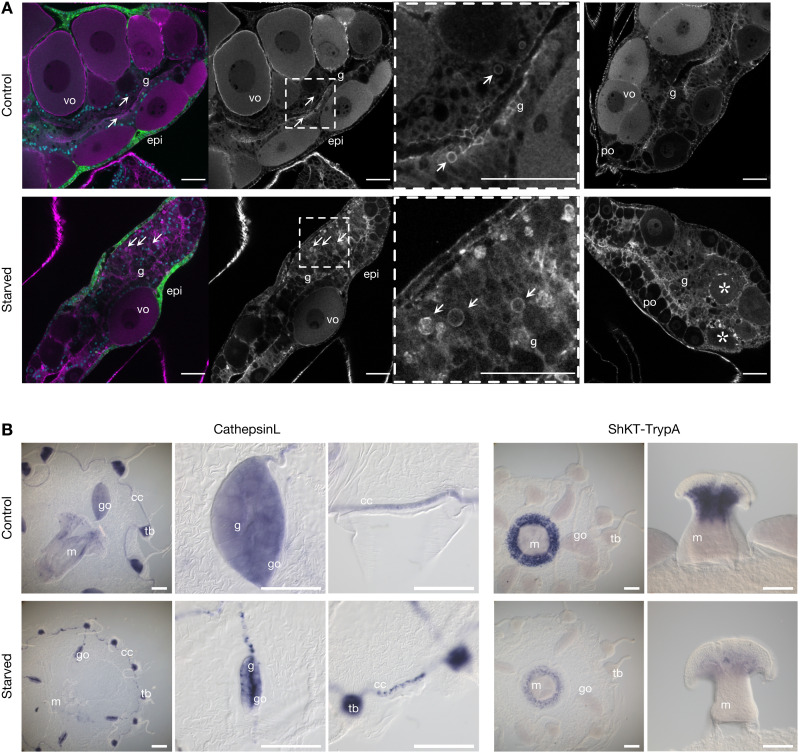
In accordance with these distinct responses in GD cells and oocytes, comparison of the cellular organization of gonads from control and starved medusae revealed major reorganization of both the gastrodermis and the oocyte populations (Fig. 7). Most notably, the population of midsized, growing oocytes, which progress daily through vitellogenesis under conditions of normal feeding (29), was largely depleted following starvation, leaving a majority of previtellogenic oocytes (Fig. 7A). A sparse population of large oocytes in starved gonads likely results from growth of a minor subpopulation of oocytes fueled by recycling of somatic tissue and oocytes (disintegration and phagocytosis of smaller oocytes visible in Fig. 7A, asterisks). Consistently, GD cells in many parts of the gonad lost their regular epithelial organization and, despite the absence of any external food supply, showed evidence of active phagocytosis involving variably sized vesicles (arrows in Fig. 7A). Changes in organization and activity of the gonad gastrodermis were also evident from in situ hybridization images for the GD cell marker CathepsinL, while reduced expression was confirmed for a protease (ShKT-TrypA) expressed in gland cell types A and B positioned within the manubrium gastroderm, which is down-regulated during the starvation treatment (Fig. 7B). Shifts between gonad gastrodermis organization and transcriptional profiles induced by starvation thus accompany activation of tissue autodigestion programs and likely the mobilization of GD cells [termed MGD for mobilizing gastro digestive cells (19)] from the gonad through the gastrovascular canal system, which has been observed both under conditions of starvation and during regeneration of the feeding organ (19).
Fig. 7. Perturbations to gastroderm cell types in response to starvation.
(A) Confocal sections through gonads from control and starved medusae with cell morphology revealed by phalloidin staining of cell boundaries (magenta/gray). The first panel of each row shows costaining of nuclei with Hoechst (blue) and endogenous GFP4 (green) in the outer epidermis (epi). Vitellogenic oocytes (vo) are largely absent during starvation, leaving mostly previtellogenic oocytes (po). The gastroderm (g) is heavily reorganized, with evidence of active phagocytosis (vesicles arrowed) and disintegrating oocytes (asterisks). The third panel in each row is a higher magnification of the boxed area in the second panel, and the fourth panel shows a second example gonad for each condition. Scale bars, 50 μm. (B) In situ hybridization visualization of example cell-types (left) and genes (right) affected by starvation. The general GD cell marker CathepsinL confirms extensive reorganization of the digestive gastroderm in starved animals, especially its collapse into central regions of the strongly reduced, oocyte-depleted, gonad, and fragmentation in the circular canal; expression of the gland cell A/B marker “ShKT and trypsin domain protein A” (ShKT-TrypA) in the manubrium consistent with its strong down-regulated following starvation. Gonads, go; manubrium, m; tentacle bulbs, tb; gastroderm, g; circular canal, cc. Scale bars, 200 μm.
DISCUSSION
The Clytia medusa single-cell atlas presented here is an important addition to the growing number of single-cell atlases across the animal tree of life. It is available for easy interrogation by the community through the UCSC Genome Browser (see Data and materials availability). This provides the first cell-level transcriptomic characterization of a pelagic medusa stage, the most complex of the life cycle forms within the large and diverse phylum Cnidaria. Reflecting this complexity, we found greater cell type diversity in the Clytia medusa than in its polyp-only hydrozoan cousin Hydra (8). The outer, epidermal body layer could be subdivided into seven clusters encompassing all of the described Clytia muscle types, including two types of fast-contracting striated swimming muscle (23, 27). Rich diversity was also uncovered in the inner gastroderm layer, which is elaborated in the medusa into distinct digestive compartments (mouth, stomach, gonad, and tentacle bulb) and generates the thick mesoglea (jelly) characteristic of the medusa form. Of the eight gastroderm cell clusters, four could be mapped to distinct sites by marker gene in situ hybridization including three likely involved in mesoglea/extracellular matrix production and thus in modulating the medusa structure. Only two of the four clusters belong to the “GastroDigestive” cluster set, characterized by a largely shared transcriptomic profile (Fig. 2B and table S5), with the other GD clusters distributed more uniformly across the digestive compartments (fig. S14), suggesting a dominant role in intracellular digestion but with functional specializations that remain to be fully understood. Our starvation experiment analyses revealed that these clusters were maintained operationally as distinct “cell types” rather than “cell states” between the two extreme conditions tested, but we cannot rule out that responses to other environmental or physiological perturbations may reveal plasticity between these clusters; for instance, transdifferentiation between muscle and nerve cell types is well documented in hydrozoan medusae [overview in (27)].
In addition to the epithelial cell types of the epidermis and gastroderm, our single-cell atlas confirms the presence of an i-cell population in Clytia providing a similar set of somatic cell types to that described in Hydra, as well as the germ cells (8, 34). In these medusa data, we do not find strong evidence for direct progression from i-cells to gland cells or for the shared neural-gland cell progenitors described in (8). In contrast, our pseudo-time analyses provide transcriptional signatures of the progressive stages of nematogenesis and neurogenesis from i-cells that will guide future studies of their developmental regulation. The large representation of nematogenic stages in this Clytia medusa scRNA dataset allowed us to link two distinct phases of nematocyte formation with extremely different transcriptional profiles. The initial phase covering nematocyst formation has been the focus of many studies (35–37, 41), but the terminal phase has been largely overlooked in previous transcriptomics studies, likely due to the relatively low mRNA content (8, 35) and the extremely abrupt degradation of nematocyst-related mRNAs before the terminal phase (37). We uncovered 14 mature neuronal subtypes in Clytia, which is similar to the number reported in Hydra and Nematostella (3, 8). It is likely that further heterogeneity exists within these 14 subpopulations. Spatial expression analysis of neuropeptides that contributed to the signatures of one or more subpopulations revealed a wide variety of neuronal populations either associated with specific anatomical structures—such as the tentacles, nerve rings, and manubrium—or distributed across the medusa (Fig. 4, D and E, and fig. S18). How molecular cell type maps to function both within and across body parts, the roles of these peptides as primary transmitters and/or neuromodulators, and the uses, if any, of classical, small-molecule neurotransmission remain unknown. Moving forward, with this cell atlas as the foundation, the ability to perform whole-organism, multiplexed scRNA-seq, in combination with emerging genetic tools and advantageous life history traits, makes Clytia a powerful, tractable platform for high-resolution systems biology.
This work further serves as a case study in using multiplexed single-cell transcriptomics to assess cellular responses to whole-organism perturbations and provides a guide for deployment in other organisms. We anticipate that whole animal multiplexed scRNA-seq (WHAM-seq) will benefit researchers studying various biologies, from developing embryos to organoids to nonmodel organisms. The techniques for multiplexed experimentation that underlie this study are also well suited to large-scale perturbation studies (such as temperature, pH, or other environmental disturbances) in other marine organisms given the SW compatible workflow. Although the inclusion of multiple animals and conditions may currently limit the detection of very rare cell populations (fig. S1), as sequencing costs drop and cell throughput in scRNA-seq grows, WHAM-seq should become tractable for larger, more complex systems. The lack of library-induced batch effects demonstrates how large-scale experiments can be conducted without introduction (or minimizing introduction) of confounding factors from multiple experiments, which can be highly nonlinear and difficult to account for (13). The second perturbation dataset also demonstrates both how batch effect variability is reduced within multiplexed experiments (see Materials and Methods; fig. S11) and the utility of this multiplexed approach in discerning cell type–specific activity markers (see Materials and Methods; fig. S11).
The fully reproducible framework we have presented, which includes code that can be run on a laptop or for free in the cloud (see “Code availability” in Acknowledgements), will further assist in extending this expression-based analysis to other organisms. By relying on expression, our strategy reduces the reliance on prior gene functional annotation using specificity of expression to identify genes of interest, allowing for targeted annotation. This includes determination of strong diagnostic markers for cell type definition, cell type–specific and shared transcriptional responses to starvation, and “modules” of coexpressed genes underlying these responses. The extent of these expression-based changes additionally highlights areas of the organism’s biology that are strongly or uniquely affected by a perturbation. By applying simple and interpretable quantitative analyses to the various cell type–specific perturbation responses, we revealed the large-scale down-regulation of gene expression in two GastroDigestive cell types and severe disruption of oocyte development under starvation. Together, this approach markedly lowers the barriers for working with nontraditional models and affords opportunities to match uniquely suited organisms to specific questions. Moving forward, the combination of scRNA-seq and other sequencing-based genomics techniques with multiplexing and annotation-agnostic analyses could foster comprehensive high-resolution molecular studies of diverse organisms and their responses to numerous environmental perturbations.
MATERIALS AND METHODS
All animal experimentation performed did not require Institutional Animal Care and Use Committee approval.
Animal culture and experimental setup
Starvation
Culture of the Clytia life cycle was carried out as previously described (57), with some modifications to the tank design. The system used in this study to culture polyps uses zebrafish tanks (Pentair), with polyp slides held in glass slide racks (Fisher, catalog no. 02-912-615). Medusae used in this experiment were raised in 4-liter beakers with a circular current generated by stirring with a constant speed of 5 rpm DC motor (Uxcell), attached to the lid of a multiwell tissue culture plate. Artificial SW for culture and experiments was made using Red Sea Salts (Bulk Reef Supply, catalog no. 207077) diluted into building deionized (DI) water to 36 parts per thousand. Experiments used ~1-cm female medusa of the Z4B strain.
For the experiment, baby medusae were collected overnight and then cultured together until they reached ~1 cm (about 2 weeks). Animals were fed once per day using 2- to 4-day old brine shrimp. Before the experiment, animals were split into two beakers. Feeding continued as before for one beaker, while the other was starved for 4 days. The “control” group was not fed on the day of the experiment. The 4-day time point was chosen as animals show strong phenotypic changes (Fig. 1), but it is far from their survival limit following starvation, as Clytia medusae can survive for more than 3 weeks with no food.
Stimulation
Rearing of the medusae before the experiment was performed as described above. On the day before the experiment, each Clytia medusa (3 to 5 weeks old) was placed in a separate container (~150 ml of SW), which was covered with foil and moved to the experimentation area to acclimate overnight in the dark. The morning following overnight acclimation, lights were turned on for ~2.5 hours to allow for spawning to complete (58). Each animal was then given repeated bouts of stimulation over a period of 30 min, with each stimulus administered every 2 min. One hundred microliters of each stimulant [150 mM KCl, DI water, or SW as a control] was gently added just below (or just above for KCl) each medusa by pipette (fig. S7A). Stimuli were chosen on the basis of their ability to reliably induce crumpling behavior, a protective response in which the bell is drawn in toward the mouth using the radial muscle (59). Animals were dissociated 30 min following the last stimulation.
Single-cell suspension and multiplexing
For the starvation experiment, animals were washed with hypertonic phosphate-buffered saline [PBS; 500 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4 (pH 7.4)] by serial transfer from SW through three successive containers each with 150 ml of hypertonic PBS to prepare cells for fixation and to avoid precipitation of SW salts in methanol. The animals were then thoroughly homogenized with a dounce. After homogenization, cells were collected by centrifugation at 500g for 5 min and resuspended in 100 μl of hypertonic PBS. Cells were then fixed by addition of 400 μl of ice-cold methanol and stored at −80°C until sample indexing and library preparation. Each sample was labeled according to the ClickTag labeling procedure described previously (14). ClickTags used for each animal are outlined in table S2. Each sample was labeled with two distinct and unique ClickTags, and samples were then pooled after addition of the “blocking” oligo. Control and starved samples were counted on a Countess, both to estimate cell numbers per animal and to determine concentrations for 10× loadings. A total of 200,000 cells/ml were counted for the starved sample and 1,000,000 cells/ml for the control. Around 120 to 150,000 cells of the starved and 200,000 cells of the control were then pooled. A loading of 20,000 pooled cells was then used as input into two lanes of the 10X Chromium Controller with v2 chemistry. Sample tag libraries were separated and processed after an SPRI size-selection step as previously described (14). cDNA samples were run on two lanes of HiSeq 4000 (two HiSeq 3000/4000 SBS 300 cycle kits), and tag libraries were run on two lanes of MiSeq (using MiSeq v3 150 cycle kits).
For cell counting (fig. S2), the starvation experiment was repeated with four animals per condition with animals dissociated and resuspended in 500 μl of SW, and then cells were counted on a hemocytometer (InCyto DHC-B02) with a 10× objective. Two 16-square grids were counted per sample.
The same protocol was followed for fixing and labeling cells from the stimulation experiment. Each animal in this case was assigned one unique ClickTag and one ClickTag per condition (table S2). After the separation of the cDNA and ClickTag samples, ClickTags were added at a 3% final concentration to the cDNA samples sequenced on the HiSeq, in addition to separate ClickTag sequencing on the MiSeq. The full protocol is described in Supplementary Methods.
In situ hybridization
Colorimetric in situ hybridization (Figs. 2C, 3E, and 4E) was performed as previously (60) with minor modifications. Briefly, 2-week-old medusae (Z4B strain) were relaxed in 0.4 mM menthol in SW, and tentacles were trimmed before fixation in a prechilled solution of 3.7% formaldehyde and 0.2% glutaraldehyde in PBS on ice for 40 min. Specimens were then washed thoroughly with PBST (PBS + 0.1% Tween 20), dehydrated in methanol stepwise, and stored in 100% methanol at −20°C. Hybridization (at 62°C for 72 hours) and washing steps were performed with a robot (Intavis AG, Bioanalytical Instruments) using 20× saline-sodium citrate pH adjusted to 4.7 throughout. Acetylation steps using 0.1 M triethanolamine in PBST (2× 5 min) and then 0.25% acetic anhydride in 0.1 M triethanolamine (2× 5 min) followed by PBST washes (3× 10 min) were included before prehybridization to reduce probe nonspecific binding. Incubation with 1:2000 anti-DIG (digoxigenin)AP in 1× blocking solution was performed for 3 hours before washing and the nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl-phosphate (NBT-BCIPP) color reaction at pH 9.5. Following postfixation, washing, and equilibration of samples in 50% glycerol/PBS, images were acquired using a Zeiss Axio Imager A2.
Probes were generated by polymerase chain reaction (PCR) from cDNA clones corresponding to our expressed sequence tag collection (61) or from medusa cDNA; the Elav probe was synthesized as a gBlock by Integrated DNA Technologies (details in table S3). For probes against the Elav, the T3 polymerase recognition site (AATTAACCCTCACTAAAGGG) was added to the 3′-end of the PCR product, or gBlock, respectively. Products were TOPO cloned (Thermo Fisher Scientific, catalog no. K280020) and sequence verified. All probes were labeled with DIG RNA labeling mix (Sigma-Aldrich, 11277073910) and purified with ProbeQuant G-50 Micro Columns (GE Healthcare Life Sciences, catalog no. 28-9034-08).
Confocal microscopy
Visualization of cell morphology within the gonads of control and starved young adult female medusae (Z4B strain) by confocal microscopy was performed as previously (58). Fixation used 4% EM-grade paraformaldehyde in 0.1 M Hepes (pH 6.9)/50 mM EGTA/10 mM MgSO4/80 mM maltose/0.2% Triton X-100 for 2 hours at room temperature. Specimens were washed 3× for 15 min with PBS/0.02% Triton and 3× for 5 min in PBS. Cell boundaries and nuclei staining were performed by overnight incubation in 1:50 rhodamine-phalloidin (1 mg/ml; Molecular Probes) and 1:5000 Hoechst 33258 (1 mg/ml stock; Sigma-Aldrich) in PBS. Samples were washed 3× for 15 min with PBS/0.02% Triton and 3× for 5 min with PBS and equilibrated in 50% PBS/Citifluor (Citifluor AF1) before imaging using a Leica SP5 confocal microscope. Control medusae were fixed 24 hours after the last feeding, while starved ones were fixed 4 days after the last feeding.
Generation of reference transcriptome
Assignments to PANTHER database entries (version 11) were made using the “pantherScore2.0.pl” script available from the database website (62). Human-Clytia orthologs were assigned using the OMA (orthologous matrix) program (63) as described in (21) and taken from the pairwise human/Clytia orthologs output, rather than the orthologous groups.
To ensure highly sensitive transcriptome alignment, a new transcriptome assembly for C. hemisphaerica was generated from bulk RNA-seq data produced from Clytia medusae (organisms at the same life stage as in the single-cell experiments) (www.ncbi.nlm.nih.gov/sra/ERX2868482%5Baccn%5D). We used the Trinity (64) de novo assembler, with default parameters, to generate a transcriptome (http://dx.doi.org/10.22002/D1.1824) and the Cufflinks Cuffcompare utility (55) to merge the Trinity assembled transcripts with any XLOC annotations from the MARIMBA v.1 (created on 30 May 2016; http://dx.doi.org/10.22002/D1.1830) transcriptome assembly (21). Then, with the CD-HIT (clustering method), assembled sequences with at least 95% were clustered and only one representative sequence was kept for each cluster. With the published MARIMBA v.1 genome sequence as a reference (created on 30 May 2016; http://dx.doi.org/10.22002/D1.1828) (21), the GMAP (genomic mapping and alignment program) aligner converted the collapsed Trinity fasta records to gff3 coordinate file (http://dx.doi.org/10.22002/D1.1824). Most differentially expressed genes found in scRNA-seq data from this study were previously identified and annotated (fig. S13). This annotation was used for the preprocessing and quantification of scRNA-seq described below. Protein sequences were then obtained by running TransDecoder (65) with default settings for the Trinity transcriptome (http://dx.doi.org/10.22002/D1.1827).
Preprocessing and clustering of sequencing data
Initial cell ranger demultiplexing for ClickTags
Initial demultiplexing of ClickTag libraries was done using output from the 10X Cell Ranger pipeline using Cell Ranger 3.0 count and aggr functions with the MiSeq ClickTag fastqs as input () and combining the counts from the two lanes using the denoted sample IDs. A ClickTag count matrix (cell-by-ClickTag) was generated by counting ClickTag barcodes that had high sequence similarity to the designed sequences using the Python fuzzywuzzy package to identify targets within Levenshtein distance 1. We additionally quantified gene expression with the kallisto-bustools workflow (66), which reproduced concordant results described below.
Initial cell ranger demultiplexing and clustering for cDNA
Initial processing of starvation cDNA libraries was performed with the 10X Cell Ranger pipeline using Cell Ranger 3.0 count and aggr functions to align and quantify the HiSeq reads and combining the counts from the two lanes using the denoted sample IDs. This was followed by filtering cells for the high-quality cells chosen during ClickTag analysis, in addition to filtering cells by thresholding the rank-unique molecular identifier (UMI) versus cell barcodes plot. Values were log1p-normalized, mean-centered, and scaled for downstream dimensionality reduction and visualization using Scanpy (67).
We then conducted Louvain clustering (68) on the data mapped to a lower dimensional space by applying PCA to the expression data filtered for highly variable genes, initially using Scanpy’s filter_genes_dispersion on only the log-normalized data. This resulted in the identification of 36 clusters (Fig. 2B and figs. S9 to S11), which we also refer to as cell types. The marker genes were selected by analyzing the top 100 markers extracted by Scanpy’s rank_genes_groups using default settings (P values adjusted with the Benjamini-Hochberg method for multiple testing). The clusters were annotated and validated with marker genes previously identified in the literature and manually categorized into the seven classes in Fig. 2A based on the marker gene patterns and their functional annotations.
Kallisto bustools for demultiplexing and clustering: Standardization of workflow
To integrate and update the analysis using a platform with streamlined ClickTag demultiplexing and count matrix generation workflows, we used the kITE demultiplexing protocol, which is based on the kallisto-bustools workflow and is described in the ClickTag demultiplexing protocol (14). Briefly, MiSeq reads are aligned to possible tag sequences (Hamming distance 1 away from designed oligo sequence whitelist) by building a kallisto index and pseudo-aligning reads to this index. Counts for these sequences were then collapsed into counts for their respective ClickTags, creating a cell-by-tag count matrix. We used Louvain clustering of cell barcodes based on the observed ClickTags to filter for clearly delineated cells (clusters strongly marked by the individual’s two corresponding tags) and exclude sample doublets (14). We also followed similar preprocessing to standard cell-by-gene workflows using the inflection point in rank-UMI versus cell barcodes knee plots to filter cell barcodes based on their tag UMI counts. We do find, similar to findings in the original ClickTag multiplexing publication (14), that the number of ClickTags per fed cell is higher than for the starved counterparts, possibly supporting the previous observation of ClickTag number per cell increasing with cell size (fig. S4E, shown for oocytes).
For the stimulation experiment, we concatenated sequencing data from the MiSeq and HiSeq as input to the previously described kallisto-kITE workflow. With the same clustering procedure, we selected cell barcodes in clusters with strong overlapping expression of both individual and condition ClickTags.
To standardize the cDNA analysis workflow, we reprocessed the starvation data using the kallisto-bustools workflow to generate gene count matrices for each lane, which were then concatenated. Cells were also filtered on the basis of the ClickTag analysis. Values were log1p-normalized, mean-centered and scaled, and filtered for highly variable genes using the same procedure described above for downstream dimensionality reduction and visualization (e.g., PCA) using Scanpy. We found that with the kallisto-processed data (with the same Cell Ranger clustering applied to the cells), the top 100 markers for each of the 36 clusters determined with Scanpy’s rank_genes_groups function (table S5) (using the nonparametric Wilcoxon test) overlap with markers in the Cell Ranger expression data, verifying that the cluster labels were concordant (fig. S10). To ensure that we additionally detected low expression marker genes, we extracted low expression markers in table S5, which consist of genes with at least 10 counts over all cells and with 90% of those counts deriving from the same cell type.
We used the PAGA partition-based graph abstraction method (31) to generate an underlying graph representation of the connectivity between cells (determining connectivity by the number of inter-edges between cell groups compared to the number of interedges under random assignment). We then generated a two-dimensional UMAP (32) embedding initialized with the PAGA graph structure for cell atlas visualization (Fig. 2A) using Scanpy. For Fig. 2B, 100 cells were randomly subsampled from each cell type to generate the heatmap.
The stimulation cDNA data were processed with the same kallisto-bustools workflow and commands as the starvation experiment data. We initially used Louvain clustering to also filter low UMI count clusters that were then removed from downstream analysis.
Distance-based comparative analysis of clusters
We first used L1 distances between starved and control cells (within each cell type) to assess how comprehensive our cell type designations were (Fig. 5). Centroids for a given cell type were calculated for starved and control cells separately in PCA-reduced space (60PC coordinates for each cell as opposed to the raw gene expression matrix). The centroid vectors are represented as cs and cc for the starved and control cells, respectively. The L1 distance (d) between them was calculated as the sum of absolute difference between the centroid coordinates
| (1) |
These intracluster distances were then compared to the pairwise L1 distances between the cell types, with centroids calculated for all cells in a given type (c1 and c2 for cell types 1 and 2) in the same manner as described above. The L1 distances were then calculated for all possible pairs of these cell type centroids using (Eq. 1). The distributions of the inter- and intracluster distances are shown in (Fig. 5B). We chose to use the L1 distance metric as it tends to better retain relative distances in high dimensions, particularly in comparison to the commonly used Euclidean distance or other higher L-norms (69, 70).
We validated this method of transcriptional distance measurement on the multiplexed perturbation of mouse NSCs in (14) and the multiple immunomodulatory drug treatments across heterogeneous cell populations in human peripheral blood mononuclear cells (PBMCs) in (71). For (14), we calculated centroid distances between perturbed and control populations, in PCA space (14), as well as pairwise distances between individual perturbed and control cells in (14) to demonstrate the ability of the L1 distance to recapitulate the graded response to a perturbant, with larger distances representing greater perturbant impact (with respect to the control cells) (fig. S19, A and B). We additionally compared all pairwise L1 distances between perturbed and control cells within the monocyte population or “T cell” PBMC populations in (71) using the oNMF space. We then compared these measurements against the magnitude of cell type distances (between control monocyte and T cell populations) to highlight “state”-versus-“type”-level transcriptional differences (fig. S19C). To then extract the most perturbed monocyte populations, we calculated centroid distances between the centroid of each perturbation condition and the CTRL1 (control) condition to rank populations by perturbation “distance” (fig. S19D).
We then used the stimulation dataset to assess the validity of the clusters/cell types generated with the starvation data. We created a joint representation of the two datasets by using a concatenated cell-by-gene matrix including only the genes highly variable in both the control and starved datasets and used 70% of the starvation data to train a k-nearest neighbor (KNN) classifier (using sklearn’s KNNClassifier with k = 15) to assign cluster labels to the remaining starvation cells and to the stimulation dataset. This showed that the stimulation cells’ labels from their neighbors in starvation data were assigned at the same accuracy as the test starvation data, meaning that clusters from the starvation data are applicable to the stimulation dataset and capture the main features of the stimulation dataset to the same extent (fig. S6A).
We also examined batch effects in the stimulation experiment. As with the starvation experiment, the L1 metric was used to visualize the magnitude of batch effect within the multiplexed experiments compared to between experiments (fig. S8). We used the merged representation (used for the KNN assignment) between both experiments to find the average pairwise distances between cell types of control condition individuals within the starvation experiment, within the stimulation experiment, and across both experiments. We found that cell type distances between organisms were reduced within multiplexed experiments compared to distances across experiments. The merged atlas was also used for determination of strong in situ markers, mainly for gastrodermal cell types, since these highly related types share many marker genes.
RNA-seq analysis and clustering with MARIMBA annotation
To make our dataset more easily searchable with the MARIMBA v.1 (created on 05/30/2016) transcriptome annotation (21), we generated single-cell gene count matrices with respect to transcript sequences distributed via the MARIMBA website (http://dx.doi.org/10.22002/D1.1830). The gene count matrices were generated using the same kallisto-bustools workflow. We compared the application of the previous clustering/cell type assignment (including the overlap in differentially expressed genes delineating these clusters) to validate the quantification derived from the Trinity/Cuffcompare assembled transcriptome (fig. S13).
We also produced a notebook for visualization of gene expression in this dataset to facilitate its use in future studies. This establishes a code base for rapidly and transparently processing and comparing single-cell datasets with future transcriptome annotations.
Neural analysis
We clustered all cells within the broad class of Neural with Louvain clustering to obtain distinct subpopulations (labeled in Fig. 2A). Markers were determined with Scanpy’s rank_genes_groups function (using the Wilcoxon test) for each subpopulation (table S5).
Marker genes from neural clusters, with predicted signal peptides (72), were screened for candidate neuropeptide cleavages sites (regular expression G[KR][KRED]). In cases where a sequence had more than one match to this motif, the six residues immediately N-terminal to the motif were inspected for similarity to each other—when similarity was present, the protein was considered a neuropeptide candidate. Predicted sequences can be found in table S3.
Pseudo-time analysis
We selected cells from cell types of interest (i-cells, nematoblasts, nematocytes, and neural cells) and used diffusion maps (73) to create a reduced dimension representation of cells, along with Scanpy’s dpt function that uses geodesic distance along the graph of cells (in the determined “diffusion component” space) to estimate pseudo-time. We then computed a PAGA-based embedding to visualize the cells in the context of the different trajectories with a ForceAtlas2 layout. To determine which genes constituted the important features in a given pseudo-time trajectory, we implemented a method based on the random forest method used in the dynverse R package for extracting “important” genes (74). A random forest regression model implemented with sklearn’s random:forest_regressor was used to identify genes that were good predictors of the generated pseudo-time values (grouped into quantiles). This was run for each of the two inferred trajectories separately (stem cells to nematocytes, and stem cells to neurons). The training set consisted of 80% of the genes’ expression data, within which 80% was used for optimizing the model, and 20% were used to evaluate the mean squared error (MSE). Both trajectory models had an R2(coefficient of determination) of 0.85 or greater. The remaining 20% of data from the full dataset were used to calculate the gene-wise permutation importance scores, providing a ranking of each feature (gene) in terms of its contribution to the model’s predictive capabilities using sklearn’s permutation_importance. In the ranking, positive scores indicate importance, with the importance ijfor gene j being the difference between the original score s (MSE) calculated from the original model, and the average score across K = 5 random permutations of the feature columns in the validation dataset
Genes with nonzero (positive) scores in the permutation test were retained and ranked (table S5).
To assess the relationship of the gland cells to the neural populations, in the investigation of a neural-gland progenitor population, we applied the URD tree construction pipeline (75) to create a pseudo-time–biased dendrogram of the neural and gland cell types beginning at the defined i-cell population. This included the i-cell, gland cell, and neuron clusters. We additionally plotted expression along the generated tree of Myb and Myc3 orthologs corresponding to the neural-gland progenitor markers in (8) and other progenitor/developmental markers from (8) for the gland and neural cell types (fig. S16).
Perturbation response analysis
Extracting DE (perturbed) genes
We used a likelihood ratio test (LRT) with negative binomial–based models of each gene’s expression in DESeq2 (76, 77), with the reduced model not including the condition label (control or starved) and single cells treated as individual replicates. There are other approaches that can be used (78, 79); we found that this method yielded many biologically relevant genes, which we could validate via the in situ experiments. All nonzero-expression genes were used for analysis. From the LRT, we obtained P values (corrected with the Benjamini-Hochberg multiple testing correction method across the number of genes tested) to determine whether the gene’s expression was significantly affected by the condition and thus the perturbation. Genes with α < 0.05 and |log2FC| > 1 were selected as significant. We used the parameters sfType = “poscounts”, minmu = 1 × 106, and minReplicatesForReplace = Inf in the DESeq2 model. Clusters with greater than 100 cells in each condition were subsampled evenly from fed and starved cells to reduce the effect of uneven cluster sizes on differential expression analysis (fig. S5). Clusters with less than 10 cells in any condition were not used for this analysis. To create the UpSet Plot (80) in Fig. 6E, P values from the LRT analysis were corrected for multiple testing (Bonferroni correction with n = number of clusters, since the test is assessing the intersection of genes across all cell types). All genes are included in table S5.
We additionally include low expression DE genes detected by pseudo-bulk analysis in table S5. We applied the same methodology as above, treating each animal as a replicate (five control and five starved replicates) and summing the counts per gene for each organism. For the DESeq2 model, we used the default parameters, as we were simulating bulk results.
Quantitative PCR validation for up-regulated, stimulation-responsive genes
To further validate the gene candidates from the stimulation multiplexed experiment, which displayed immediate early gene-like expression under KCl or DI stimulation, we chose broadly expressed candidates, i.e., DE genes in multiple cell types, for whole-animal, bulk quantitative PCR (qPCR). Here, we looked for up-regulation of these genes as compared to a “housekeeping” collagen marker, XLOC_008048. Two organisms were housed together in 150 ml of SW, with four animals total per each condition (SW, KCl, and DI) treated as described previously in Materials and Methods. The two animals were homogenized together in 1 ml of TRIzol on ice with a syringe. A total of 0.2 ml of chloroform were added per 1 ml of TRIzol and shaken for 15 s. After spinning at 12,000g for 15 min at 4°C, the top phase is removed and, in a new tube, mixed with an equal volume of 60% EtOH. The RNEasy Mini Kit (QIAGEN, catalog no. 74004) was then used for bulk RNA extraction from the sample. Probes for the candidates and housekeeping gene were produced as previously described, and the LightCycler 480 SYBR Green I Master Mix (2X) was used for 20 μl of qPCR reactions with 5 μl of RNA template per reaction, using the standard cycling procedure with a 62°C annealing temperature. For each RNA sample (n = 6), three qPCR replicate reactions were done, resulting in a total of six replicates for each perturbation condition. Fold-change (log2FC) values presented in fig. S7E were calculated by first normalizing Ct values for each replicate to the average housekeeping gene Ct value in the same condition (ΔCtNorm = CtRep − CtHouse_Avg). The log2FC was then calculated as log(2ΔΔCtNorm) where ΔΔCtNorm = ΔCtNorm - avg.(ΔCtNorm:SW), is the difference of each normalized Ct from the average normalized Ct for the SW (control) condition. All primers for qPCR can be found in table S3.
De novo perturbed gene clusters
To cluster genes affected by perturbation (table S5) and to obtain information on their coexpression and possible functional similarity, we transposed the cell-by-gene expression matrix (obtaining a gene-by-cell matrix) for only the aggregated perturbed genes with padj > 0.05 (adjusted across genes and clusters) from the DeSeq2 analysis. We used Louvain clustering on the gene expression matrix, identifying both coexpressed genes and cell type–specific genes, similar to Monocle’s procedure for detecting gene modules (56). We then used the aggregated information from the modules to determine putative functions/response types. The topGO weight algorithm (81) was used to determine GO terms that were significantly enriched in each gene module compared to GO terms in all other groups, with significance threshold α < 0.05. The P values were also adjusted for multiple testing over the number of different gene modules using Bonferroni correction, and only significant GO terms were used to label the response types among the modules (fig. S20 and table S5). This analysis was replicated to determine gene modules distinguishing the GD cell types by clustering groups of coexpressed marker genes across the subtypes.
Acknowledgments
We thank X. Da and X. Wang for technical assistance, T. Momose for assistance with the single-cell experimentation, the Caltech Single-Cell Profiling and Engineering Center for the use of their single-cell and sequencing tools, and the Caltech Bioinformatics Resource Center for transcriptome assembly and annotation analysis. We additionally thank the Caltech Center for Evolutionary Science for the bioinformatics resources to create a local UCSC Genome Browser. We thank A. S. Booeshaghi for help with kallisto, bustools, and the kITE demultiplexing of the ClickTag reads and for rescuing the stimulation experiment sequencing data. We thank J. Malamy for helping to establish Clytia work at Caltech. We thank S. Peron for initial characterization of some of the cell type marker genes, P. Lapébie for identification of novel neuropeptide sequences, M. Jager for valuable advice on the in situ protocol, and J. R. Mateu for providing pp11 probe.
Funding: J.G., M.H., and L.P. were supported in part by a seed grant from the Chen Institute at the California Institute of Technology. T.C., J.G., and L.P. were supported in part by NIH U19MH114830 and NIH RF1AG062324A. We thank the Marine Resources Centre (CRBM and PIV imaging platform) of Institut de la Mer de Villefranche (IMEV), supported by EMBRC-France. The French state funds of EMBRC-France are managed by the ANR within the investments of the Future program. L.L. was supported by the Agence Nationale de la Recherche (ANR-19-CE13-0003). A.F., R.R.C., and E.H. were supported by the H2020/Marie Skłodowska-Curie ITN “EvoCell” Grant agreement no. 766053. B.W. was supported in part by a Howard Hughes Medical Institute Fellowship of the Life Sciences Research Foundation and by NIH K99NS119749. This work was in part supported by the Whitman Center of the Marine Biological Laboratory in Woods Hole, MA and a visiting grant from EMBRC-France. D.J.A. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: Conceived of the experiments: T.C., B.W., J.G., R.R.C., E.H., D.J.A., and L.P. Developed cell dissociation, fixation, and labeling procedures compatible with the 10X Genomics platform: J.G. and M.H. Performed the single-cell experiments: T.C., B.W., and J.G. Performed the in situ hybridization and other microscopy experiments: B.W., A.F., L.L., and S.C. Performed whole-organism qPCR: T.C. Performed bioinformatics analysis including assembly and annotation of the transcriptome: F.G. and R.R.C. Wrote scripts for processing the data and code for the analysis: T.C. and J.G. Developed the Google Colab notebooks: T.C. Analyzed and interpreted the data: T.C., B.W., J.G., A.F., L.L., R.R.C., E.H., D.J.A., and L.P. Writing and editing the manuscript: T.C., B.W., J.G., A.F., L.L., R.R.C., E.H., D.J.A., and L.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All raw sequencing and processed data files used for analysis are available from CaltechData, (https://data.caltech.edu/search?page=1&size=25&ln=en&q=clytia), with links additionally provided via the notebooks in the code repository. The sequencing read alignments are available at http://evolution.caltech.edu/genomebrowser/cgi-bin/hgTracks?db=hub_135_clyHem1&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=scaffold_1%3A1%2D30003&hgsid=3413_LS7OcP5N7VA2rApGOfk8iaX2kVFR, and an interactive browser for gene expression visualization (http://131.215.78.40/cb), is publicly hosted on a UCSC Genome Browser by the Caltech Bioinformatics Resource Center. The softwares used are as follows: Cell Ranger 3.0.1, Trinity-v2.8.4, Cufflinks v2.2.1, kallisto v0.46.2, bustools v0.40.0, anndata 0.7.5, louvain 0.7.0, rpy2 3.4.2, scanpy 1.6.0, biopython 1.78, pysam 0.16.0.1, fuzzywuzzy 0.18.0, numpy 0.19.5, pandas 1.1.5, matplotlib 3.2.2, sklearn 0.0, scipy 1.4.1, seaborn 0.11.1, requests 2.23.0, tqdm 4.41.1, multiprocess 0.70.11.1, DESeq2 1.3.0, topGO 2.42.0, and UpSet 1.4.0. Code availability: All the codes used to perform the analyses and generate the results and figures are available in Google Colab notebooks archived with Zenodo at https://zenodo.org/record/5519756#.YUonytNKgUE and directly available at https://github.com/pachterlab/CWGFLHGCCHAP_2021. The notebooks, which include the complete preprocessing of the raw data and a walkthrough of the code, provide a transparent implementation of the methods and can be run for free in the Google cloud.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
Tables S1 to S5
Supplementary Methods
Legends for tables S3 and S5
Other Supplementary Material for this manuscript includes the following:
Tables S3 and S5
REFERENCES AND NOTES
- 1.Hwang B., Lee J. H., Bang D., Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 50, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vergara H. M., Bertucci P. Y., Hantz P., Tosches M. A., Achim K., Vopalensky P., Arendt D., Whole-organism cellular gene-expression atlas reveals conserved cell types in the ventral nerve cord of Platynereis dumerilii. Proc. Natl. Acad. Sci. U.S.A. 114, 5878–5885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebé-Pedrós A., Saudemont B., Chomsky E., Plessier F., Mailhé M.-P., Renno J., Loe-Mie Y., Lifshitz A., Mukamel Z., Schmutz S., Novault S., Steinmetz P. R. H., Spitz F., Tanay A., Marlow H., Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-seq. Cell 173, 1520–1534.e20 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Cao J., Packer J. S., Ramani V., Cusanovich D. A., Huynh C., Daza R., Qiu X., Lee C., Furlan S. N., Steemers F. J., Adey A., Waterston R. H., Trapnell C., Shendure J., Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., Huang D., Xu Y., Huang W., Jiang M., Jiang X., Mao J., Chen Y., Lu C., Xie J., Fang Q., Wang Y., Yue R., Li T., Huang H., Orkin S. H., Yuan G.-C., Chen M., Guo G., Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Wagner D. E., Weinreb C., Collins Z. M., Briggs J. A., Megason S. G., Klein A. M., Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fincher C. T., Wurtzel O., de Hoog T., Kravarik K. M., Reddien P. W., Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, eaaq1736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siebert S., Farrell J. A., Cazet J. F., Abeykoon Y., Primack A. S., Schnitzler C. E., Juliano C. E., Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365, eaav9314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S., Currie J. D., Camp J. G., Tanaka E. M., Treutlein B., Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixit A., Parnas O., Li B., Chen J., Fulco C. P., Jerby-Arnon L., Marjanovic N. D., Dionne D., Burks T., Raychowdhury R., Adamson B., Norman T. M., Lander E. S., Weissman J. S., Friedman N., Regev A., Perturb-seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFarland J. M., Paolella B. R., Warren A., Geiger-Schuller K., Shibue T., Rothberg M., Kuksenko O., Colgan W. N., Jones A., Chambers E., Dionne D., Bender S., Wolpin B. M., Ghandi M., Tirosh I., Rozenblatt-Rosen O., Roth J. A., Golub T. R., Regev A., Aguirre A. J., Vazquez F., Tsherniak A., Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action. Nat. Commun. 11, 4296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran H. T. N., Ang K. S., Chevrier M., Zhang X., Lee N. Y. S., Goh M., Chen J., A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehring J., Hwee Park J., Chen S., Thomson M., Pachter L., Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat. Biotechnol. 38, 35–38 (2020). [DOI] [PubMed] [Google Scholar]
- 15.McGinnis C. S., Patterson D. M., Winkler J., Conrad D. N., Hein M. Y., Srivastava V., Hu J. L., Murrow L. M., Weissman J. S., Werb Z., Chow E. D., Gartner Z. J., MULTI-seq: Sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619–626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo C., Kong W., Kamimoto K., Rivera-Gonzalez G. C., Yang X., Kirita Y., Morris S. A., CellTag Indexing: Genetic barcode-based sample multiplexing for single-cell genomics. Genome Biol. 20, 90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivatsan S. R., McFaline-Figueroa J. L., Ramani V., Saunders L., Cao J., Packer J., Pliner H. A., Jackson D. L., Daza R. M., Christiansen L., Zhang F., Steemers F., Shendure J., Trapnell C., Massively multiplex chemical transcriptomics at single-cell resolution. Science 367, 45–51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P. K., Swerdlow H., Satija R., Smibert P., Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinigaglia C., Peron S., Eichelbrenner J., Chevalier S., Steger J., Barreau C., Houliston E., Leclère L., Pattern regulation in a regenerating jellyfish. eLife 9, e54868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch T. C. G., Klimovich A., Domazet-Lošo T., Gründer S., Holstein T. W., Jékely G., Miller D. J., Murillo-Rincon A. P., Rentzsch F., Richards G. S., Schröder K., Technau U., Yuste R., Back to the basics: Cnidarians start to fire. Trends Neurosci. 40, 92–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclère L., Horin C., Chevalier S., Lapébie P., Dru P., Peron S., Jager M., Condamine T., Pottin K., Romano S., Steger J., Sinigaglia C., Barreau C., Quiroga Artigas G., Ruggiero A., Fourrage C., Kraus J. E. M., Poulain J., Aury J.-M., Wincker P., Quéinnec E., Technau U., Manuel M., Momose T., Houliston E., Copley R. R., The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle. Nat. Ecol. Evol. 3, 801–810 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Leclère L., Copley R. R., Momose T., Houliston E., Hydrozoan insights in animal development and evolution. Curr. Opin. Genet. Dev. 39, 157–167 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Steinmetz P. R. H., Kraus J. E. M., Larroux C., Hammel J. U., Amon-Hassenzahl A., Houliston E., Wörheide G., Nickel M., Degnan B. M., Technau U., Independent evolution of striated muscles in cnidarians and bilaterians. Nature 487, 231–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamran Z., Zellner K., Kyriazes H., Kraus C. M., Reynier J.-B., Malamy J. E., In vivo imaging of epithelial wound healing in the cnidarian Clytia hemisphaerica demonstrates early evolution of purse string and cell crawling closure mechanisms. BMC Dev. Biol. 17, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galliot B., Schmid V., Cnidarians as a model system for understanding evolution and regeneration. Int. J. Dev. Biol. 46, 39–48 (2002). [PubMed] [Google Scholar]
- 26.A. Amiel, P. Chang, T. Momose, E. Houliston, Clytia hemisphaerica: A cnidarian model for studying oogenesis, in Oogenesis: The Universal Process (John Wiley & Sons, 2010), pp. 81–102. [Google Scholar]
- 27.Leclère L., Röttinger E., Diversity of cnidarian muscles: Function, anatomy, development and regeneration. Front. Cell Dev. Biol. 4, 157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita S., Kuranaga E., Nakajima Y.-I., Cell proliferation controls body size growth, tentacle morphogenesis, and regeneration in hydrozoan jellyfish Cladonema pacificum. PeerJ. 7, e7579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amiel A., Houliston E., Three distinct RNA localization mechanisms contribute to oocyte polarity establishment in the cnidarian Clytia hemisphaerica. Dev. Biol. 327, 191–203 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Sheng M., Greenberg M. E., The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Wolf F. A., Hamey F. K., Plass M., Solana J., Dahlin J. S., Göttgens B., Rajewsky N., Simon L., Theis F. J., PAGA: Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. McInnes, J. Healy, J. Melville, UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv [stat.ML] (2018); http://arxiv.org/abs/1802.03426.
- 33.Hemmrich G., Khalturin K., Boehm A.-M., Puchert M., Anton-Erxleben F., Wittlieb J., Klostermeier U. C., Rosenstiel P., Oberg H.-H., Domazet-Loso T., Sugimoto T., Niwa H., Bosch T. C. G., Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol. Biol. Evol. 29, 3267–3280 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Leclère L., Jager M., Barreau C., Chang P., Le Guyader H., Manuel M., Houliston E., Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev. Biol. 364, 236–248 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Condamine T., Jager M., Leclère L., Blugeon C., Lemoine S., Copley R. R., Manuel M., Molecular characterisation of a cellular conveyor belt in Clytia medusae. Dev. Biol. 456, 212–225 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Denker E., Manuel M., Leclère L., Le Guyader H., Rabet N., Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev. Biol. 315, 99–113 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Sunagar K., Columbus-Shenkar Y. Y., Fridrich A., Gutkovich N., Aharoni R., Moran Y., Cell type-specific expression profiling unravels the development and evolution of stinging cells in sea anemone. BMC Biol. 16, 108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda N., Kon Y., Quiroga Artigas G., Lapébie P., Barreau C., Koizumi O., Kishimoto T., Tachibana K., Houliston E., Deguchi R., Identification of jellyfish neuropeptides that act directly as oocyte maturation-inducing hormones. Development 145, dev156786 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Fourrage C., Swann K., Gonzalez Garcia J. R., Campbell A. K., Houliston E., An endogenous green fluorescent protein–photoprotein pair in Clytia hemisphaerica eggs shows co-targeting to mitochondria and efficient bioluminescence energy transfer. Open Biol. 4, 130206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz P. R. H., A non-bilaterian perspective on the development and evolution of animal digestive systems. Cell Tissue Res. 377, 321–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denker E., Bapteste E., Le Guyader H., Manuel M., Rabet N., Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr. Biol. 18, R858–R859 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Bezares-Calderón L. A., Berger J., Jékely G., Diversity of cilia-based mechanosensory systems and their functions in marine animal behaviour. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 375, 20190376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McPherson D. R., Sensory hair cells: An introduction to structure and physiology. Integr. Comp. Biol. 58, 282–300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang J. S., Takaku Y., Chapman J., Ikeo K., David C. N., Gojobori T., Cilium evolution: Identification of a novel protein, nematocilin, in the mechanosensory cilium of Hydra nematocytes. Mol. Biol. Evol. 25, 2009–2017 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Lapébie P., Ruggiero A., Barreau C., Chevalier S., Chang P., Dru P., Houliston E., Momose T., Differential responses to Wnt and PCP disruption predict expression and developmental function of conserved and novel genes in a cnidarian. PLOS Genet. 10, e1004590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massari M. E., Murre C., Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurki P., Vanderlaan M., Dolbeare F., Gray J., Tan E. M., Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp. Cell Res. 166, 209–219 (1986). [DOI] [PubMed] [Google Scholar]
- 48.Sanes J. R., Zipursky S. L., Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536–556 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Jékely G., The chemical brain hypothesis for the origin of nervous systems. Philos. Trans. R. Soc. B 376, 20190761 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grimmelikhuijzen C. J. P., Hauser F., Mini-review: The evolution of neuropeptide signaling. Regul. Pept. 177 Suppl, S6–S9 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Jékely G., Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 8702–8707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen S. K. D., Koch T. L., Hauser F., Garm A., Grimmelikhuijzen C. J. P., De novo transcriptome assembly of the cubomedusa Tripedalia cystophora, including the analysis of a set of genes involved in peptidergic neurotransmission. BMC Genomics 20, 175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serrano-Saiz E., Poole R. J., Felton T., Zhang F., De La Cruz E. D., Hobert O., Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659–673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F., Eriksen J., Finer-Moore J., Chang R., Nguyen P., Bowen A., Myasnikov A., Yu Z., Bulkley D., Cheng Y., Edwards R. H., Stroud R. M., Ion transport and regulation in a synaptic vesicle glutamate transporter. Science 368, 893–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L., Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lechable M., Jan A., Duchene A., Uveira J., Weissbourd B., Gissat L., Collet S., Gilletta L., Chevalier S., Leclère L., Peron S., Barreau C., Lasbleiz R., Houliston E., Momose T., An improved whole life cycle culture protocol for the hydrozoan genetic model Clytia hemisphaerica. Biol. Open 9, bio051268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quiroga Artigas G., Lapébie P., Leclère L., Takeda N., Deguchi R., Jékely G., Momose T., Houliston E., A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia. eLife 7, e29555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyman L. H., Observations and experiments on the physiology of medusae. Biol. Bull. 79, 282–296 (1940). [Google Scholar]
- 60.Sinigaglia C., Thiel D., Hejnol A., Houliston E., Leclère L., A safer, urea-based in situ hybridization method improves detection of gene expression in diverse animal species. Dev. Biol. 434, 15–23 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Chevalier S., Martin A., Leclère L., Amiel A., Houliston E., Polarised expression of FoxB and FoxQ2 genes during development of the hydrozoan Clytia hemisphaerica. Dev. Genes Evol. 216, 709–720 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., Thomas P. D., Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altenhoff A. M., Levy J., Zarowiecki M., Tomiczek B., Warwick Vesztrocy A., Dalquen D. A., Müller S., Telford M. J., Glover N. M., Dylus D., Dessimoz C., OMA standalone: Orthology inference among public and custom genomes and transcriptomes. Genome Res. 29, 1152–1163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., LeDuc R. D., Friedman N., Regev A., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melsted P., Sina Booeshaghi A., Liu L., Gao F., Lu L., Min K. H., Beltrame E., Hjorleifsson K. E., Gehring J., Pachter L., Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat. Biotechnol. 39, 813–818 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Wolf F. A., Angerer P., Theis F. J., SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.P. De Meo, E. Ferrara, G. Fiumara, A. Provetti, Generalized Louvain method for community detection in large networks, in Proceedings of the 2011 11th International Conference on Intelligent Systems Design and Applications (2011), 88–93. [Google Scholar]
- 69.Ntranos V., Kamath G. M., Zhang J. M., Pachter L., Tse D. N., Fast and accurate single-cell RNA-seq analysis by clustering of transcript-compatibility counts. Genome Biol. 17, 112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.C. C. Aggarwal, A. Hinneburg, D. A. Keim, On the surprising behavior of distance metrics in high dimensional space. Database Theory — ICDT 2001, 420–434 (1973).
- 71.Chen S., Rivaud P., Park J. H., Tsou T., Charles E., Haliburton J. R., Pichiorri F., Thomson M., Dissecting heterogeneous cell populations across drug and disease conditions with PopAlign. Proc. Natl. Acad. Sci. U.S.A. 117, 28784–28794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Armenteros J. J. A., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., von Heijne G., Nielsen H., SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Haghverdi L., Büttner M., Wolf F. A., Buettner F., Theis F. J., Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Saelens W., Cannoodt R., Todorov H., Saeys Y., A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Farrell J. A., Wang Y., Riesenfeld S. J., Shekhar K., Regev A., Schier A. F., Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Love M., Anders S., Huber W., Differential analysis of count data—The DESeq2 package. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang T., Li B., Nelson C. E., Nabavi S., Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics 20, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crowell H. L., Soneson C., Germain P.-L., Calini D., Collin L., Raposo C., Malhotra D., Robinson M. D., muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11, 6077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luecken M. D., Theis F. J., Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 15, e8746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H., UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alexa A., Rahnenführer J., Gene set enrichment analysis with topGO. Bioconduct. Improv. 27, 1–26 (2009). [Google Scholar]
- 82.Richards G. S., Rentzsch F., Regulation of Nematostella neural progenitors by SoxB, Notch and bHLH genes. Development 142, 3332–3342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22
Tables S1 to S5
Supplementary Methods
Legends for tables S3 and S5
Tables S3 and S5