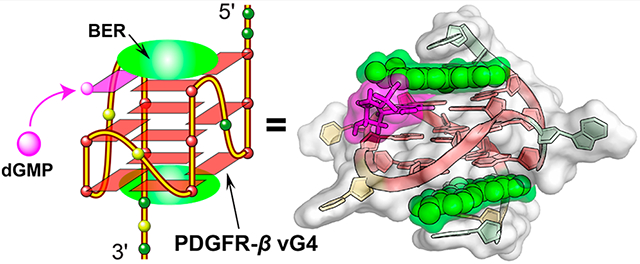
Abstract
The G-quadruplexes (G4s) formed in the PDGFR-β gene promoter are transcriptional modulators and amenable to small-molecule targeting. Berberine (BER), a clinically important natural isoquinoline alkaloid, has gained increasing attention due to its potential as anticancer drug. We previously showed that the PDGFR-β gene promoter forms a unique vacancy G4 (vG4) that can be filled in and stabilized by guanine metabolites, such as dGMP. Herein, we report the high-resolution NMR structure of a ternary complex of berberine bound to the dGMP-fill-in PDGFR-β vG4 in potassium solution. This is the first small-molecule complex structure of a fill-in vG4. This ternary complex has a 2:1:1 binding stoichiometry with a berberine molecule bound at each the 5′-and 3′-end of the 5′-dGMP-fill-in PDGFR-β vG4. Each berberine recruits the adjacent adenine residue from the 5′- or 3′-flanking sequence to form a “quasi-triad plane” that covers the external G-tetrad of the fill-in vG4, respectively. Significantly, berberine covers and stabilizes the fill-in dGMP. The binding of berberine involves both π-stacking and electrostatic interactions, and the fill-in dGMP is covered and well-protected by berberine. The NMR structure can guide rational design of berberine analogues that target the PDGFR-β vG4 or dGMP-fill-in vG4. Moreover, our structure provides a molecular basis for designing small-molecule guanine conjugates to target vG4s.
Graphical Abstract
INTRODUCTION
Guanine-rich nucleic acid sequences are able to form G-quadruplexes (G4s), which are new types of DNA or RNA secondary structures characterized by Hoogsteen hydrogen-bonded G-tetrads and stabilized by K+ or Na+.1,2 G4-Forming sequences are most abundant in genome key functional sites, such as oncogene promoters, 5′- and 3′-untranslated regions of mRNAs, and telomeres.2–8 The biological function of G4s are implicated in gene expression, genomic instability, epigenetics, and telomere maintenance.2,3,9–14 Targeting DNA G4 structures in the promoter regions of MYC, K-Ras, Bcl-2, and PDGFR-β oncogenes with G4-interactive small molecules causes downregulation of gene transcription, suggesting that G4s are promising DNA targets for therapeutics.14–22
The platelet-derived growth factor receptor beta (PDGFR-β) is a cell-surface-receptor tyrosine kinase and is overexpressed in many types of human diseases, including cancers, atherosclerosis, and fibrotic disorders.23–25 Inhibition of the PDGFR-β signaling pathway by G4-interactive small molecules has been well-documented as an attractive strategy for therapy of these diseases;21,25,26 however, no selective PDGFR-β G4-interactive compound has been entered in clinical trials. Recently, we found that the PDGFR-β gene promoter forms a distinct vacancy G4 (vG4), which lacks a guanine in one tetrad.27–31 We determined the NMR solution structure of a novel type of dGMP-fill-in PDGFR-β vG4 using a 19-mer PDGFR-β promoter DNA sequence (Figure 1a), which provides insights into the possibility that cellular guanine derivatives can take part in the G4 formation and gene regulation.31 This unique fill-in vG4 raises new opportunities for selective targeting of PDGFR-β gene, i.e., by small molecules that specifically bind to this distinct type of fill-in vG4.
Figure 1.
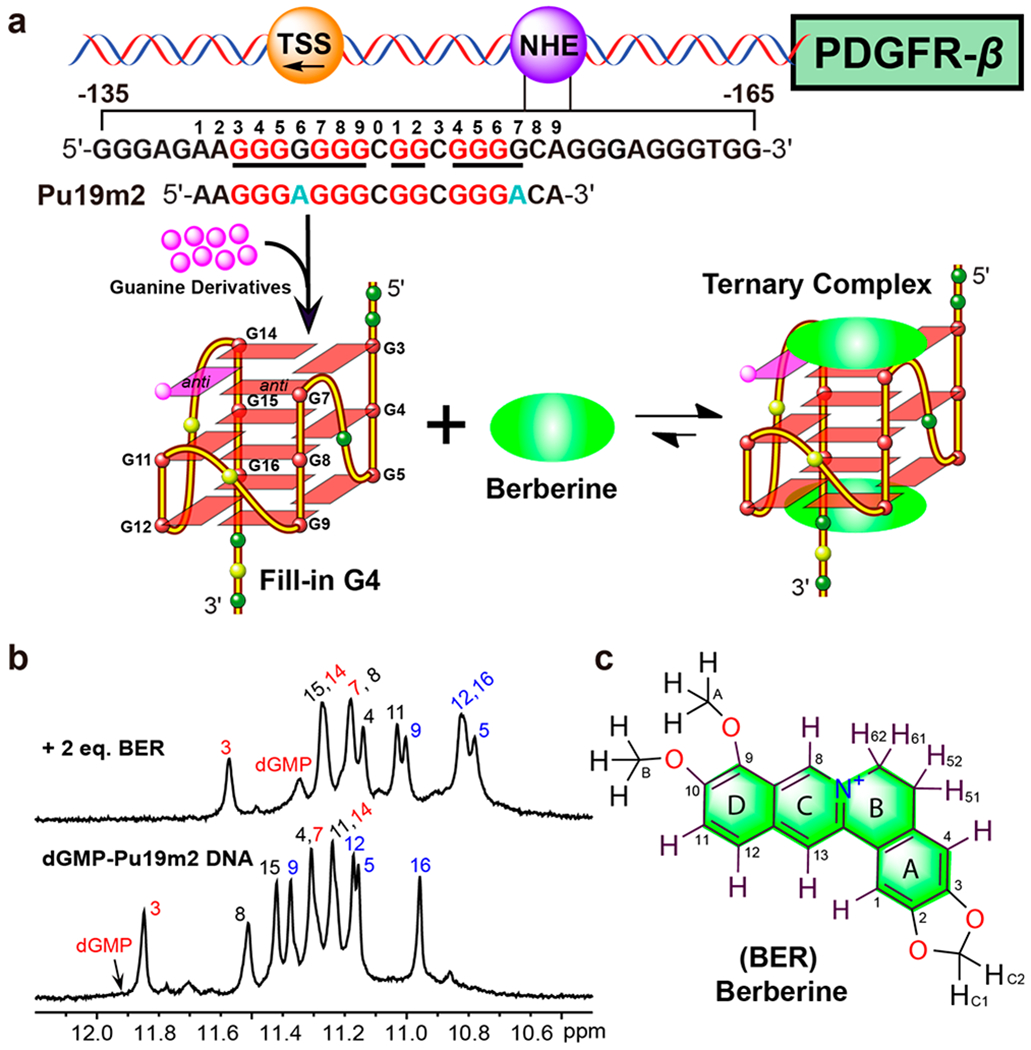
(a) Schematic of the human PDGFR-β gene promoter and the formation of a dGMP-fill-in vG4 as well as the berberine–dGMP–vG4 ternary complex. The vG4-forming region of the NHE sequence and its modifications are shown. The guanine residues involved in the formation of the fill-in vG4 are colored in red and mutations in cyan, respectively. (b) 1D 1H NMR spectra of dGMP–Pu19m2 DNA in the absence and presence of berberine with complete imino proton assignment. Conditions: 1.5 mM Pu19m2 DNA, 3:1 dGMP/DNA, 2:1 drug/DNA, pH 7, 50 mM K+ solution, 25 °C. (c) Chemical structure of berberine with numbering.
Berberine (BER), a natural occurring isoquinoline alkaloid, is characterized by a variety of pharmacological effects related to its nucleic acid and protein interactions, including anticancer, antimicrobial, anti-inflammatory, and antidiabetic activities.32–34 Many species of medicinal plants are rich in berberine, which has been used as folk medicine for hundreds of years in China.32,35 Berberine and its derivatives have been shown to stabilize G4s and represent an attractive G4-interactive natural scaffold for chemical modifications.36–40 Structural studies on berberine-G4 complexes have been reported, using NMR spectroscopy, X-ray crystallography, and computational simulations.38,40–42 Specifically, a previous crystal structure study reported that berberine binds as dimer to a parallel human telomeric G4 in a 6:2 molar ratio.38
In this study, we show that berberine stabilizes the dGMP-fill-in PDGFR-β gene promoter vG4 and determine the first high-resolution NMR solution structure of this unique ternary complex. Using CD spectroscopy and a fluorescence-based binding assay, the binding activity of berberine to the dGMP-fill-in vG4 is characterized. On the basis of the obtained complex structure, rational improvement on affinity and/or selectivity of berberine analogues for a specific vG4 are proposed.
RESULTS AND DISCUSSION
Berberine Binds and Stabilizes dGMP Fill-In vG4 Formed in the PDGFR-β Gene Promoter.
The PDGFR-β gene promoter Pu19m2 DNA forms a dGMP-fill-in vG4 (dGMP–vG4) in K+-containing solution as indicated by 11 well-resolved imino proton peaks at 25 °C, from 11 tetradguanines of the fill-in vG4 (Figures 1b and 2a).31 Noteworthy, the dGMP-fill-in vG4 is also observed for extended PDGFR-β promoter sequences.31 Both 5′- and 3′-end extended sequences can form vG4s that bind dGMP, as indicated by the emergence of a new set of imino protons in the NMR titration data (Figure S1). While the imino proton of the filled-in dGMP is detectable at 5 °C, it is broadened beyond detection due to its exchange between free- and bound-state at 25 °C.31 Upon addition of berberine to this fill-in vG4, signals in the 1H NMR spectrum first became broader and then a new set of distinct signals appeared at 2.0 mol equiv of berberine (Figures 1b and 2a), indicating a medium-to-fast exchange rate of berberine binding to the dGMP–vG4 binary complex on the NMR time-scale. The imino proton of dGMP in the ternary complex was detectable at 25 °C, unambiguously assigned by 15N-edited experiments using 13C,15N-labeled dGMP and indicative of the involvement of dGMP in G-tetrad formation (Figure S2). Observation of the imino proton of the dGMP at 25 °C (Figure 1b) indicates its stabilization by berberine binding. As only one set of 12 imino protons was observed, we concluded that only one dominant conformation of the berberine–dGMP–vG4 ternary complex was present.
Figure 2.
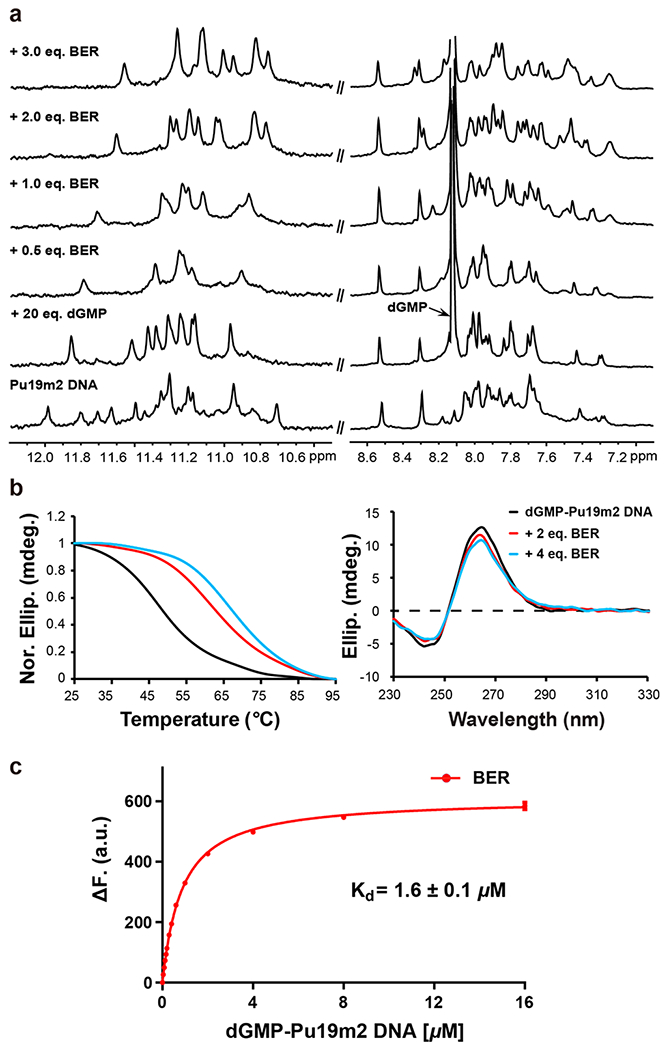
(a) 1D 1H NMR titration of Pu19m2 DNA with dGMP and berberine. Conditions: 0.15 mM Pu19m2 DNA, pH 7, 50 mM K+ solution, 25 °C. (b) CD thermal melting curves (left) and CD spectra (right) of dGMP–Pu19m2 DNA (15 μM) in the absence and presence of 2 and 4 equiv of berberine. Conditions: pH 7, 50 mM K+. (c) Fluorescence intensity change of berberine (0.2 μM) upon titration with dGMP–Pu19m2 DNA. The dissociation constant (Kd) for a 2:1 binding stoichiometry is determined. Conditions: 25 °C, pH 7, 50 mM K+. The experiment was conducted in duplicate.
Biophysical Characterization of Berberine Binding to the dGMP–vG4.
The CD spectra of both dGMP–vG4 and its berberine complex showed the characteristic bands for a parallel G4 (Figure 2b).43 Berberine increased the melting temperature of dGMP–vG4 by 14 °C at 2 equiv and 18 °C at 4 equiv, indicating two dominant binding sites of berberine to dGMP–vG4 as much smaller Tm change was observed between 2 and 4 equiv of drug (Figure 2b).
A fluorescence-based binding assay was used to quantify the binding affinity of berberine to the dGMP–vG4. A preformed dGMP–vG4 binary complex was gradually titrated to a berberine solution and the induced fluorescence signal was recorded to calculate the dissociation constant (Kd) (Figure 2c). A Kd value of 1.6 μM was obtained by data fitting following a 2:1 binding stoichiometry, which is comparable with the reported binding affinity of berberine to a human telomeric G4.44,45
NMR Structure Determination of the Berberine–dGMP–Pu19m2 Ternary Complex.
The high-quality of the 1D NMR data suggested a well-defined berberine–dGMP–vG4 ternary complex that is suitable for structure determination (Figures 1b and 2a). A set of 2D NMR spectra was recorded for this ternary complex including NOESY, TOCSY, and 1H–13C HSQC experiments at different mixing times and temperatures in 90% H2O/10% D2O (Figures 3, 4, S3, and S4). Comparison of cross-peak fingerprints between NOESY spectra of berberine–dGMP–vG4 ternary complex (Figures 3 and S4) and dGMP–vG4 DNA31 (Figure S5) revealed a great degree of similarity. All imino, aromatic, and sugar resonances of the berberine–dGMP–vG4 complex were assigned with the aid of the previous dGMP–vG4 spectral assignment.31 Protons of the free and bound berberine (Table S3) were assigned using 1D 1H NMR (Figure S6) and 2D NOESY spectra, consistent with the previous data.46 Both NMR and CD spectra showed that the berberine–dGMP–vG4 complex maintained the parallel topology (Figures 1a, 2a,b, and 3). The guanine H1–H8 and H1–H1 NOE cross-peaks determined the arrangement of three G-tetrad planes: G7–dGMP–G14–G3, G15–G4–G8–G11, and G16–G5–G9–G12, which are the same as in the dGMP–vG4 binary complex (Figures 1a, 3, S4, and S5). All the tetrad, loop, and flanking residues adopt anti-glycosidic torsion angles except for the 5′-terminal A1 which adopts a syn conformation, as shown by a strong intraresidue H1′–H8 NOE cross-peak and the corresponding downfield C8 chemical shift (Figures 3 and S3).31,47,48 The largest Δδ values were observed for the H1 protons of guanines at the 5′-end G-tetrad (including dGMP) and 3′-end tetrad (Figure S7). Additionally, the H8/H6 protons of flanking A1, A2, A17, and C18 showed much larger Δδ values than the protons of A6, C10, and C13 in three loops, indicating berberine stacks the two external G-tetrads (Figure S8). Moreover, the H1 and H8 protons of dGMP had large Δδ values, suggesting the dGMP is involved in the ternary complex interactions (Figures S7 and S8).
Figure 3.
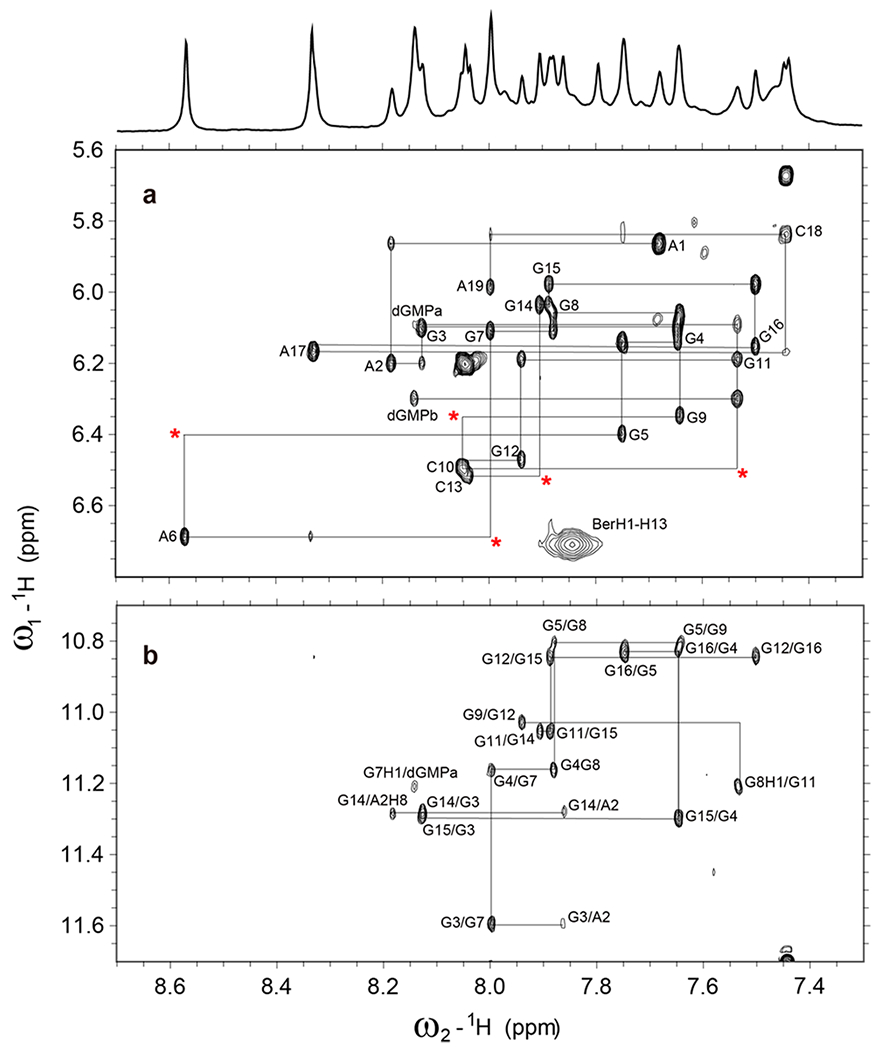
(a) The H1′-H6/H8 region and (b) H1–H8 region from the 2D-NOESY spectrum of 2:1:1 berberine–dGMP–vG4 ternary complex in H2O with sequential assignment pathway. Missing connectivities are labeled with asterisks. Condition: 1.5 mM Pu19m2 DNA, 3:1 dGMP/DNA, 2:1 drug/DNA, pH 7, 50 mM K+, 25 °C, mixing time of 300 ms.
Figure 4.
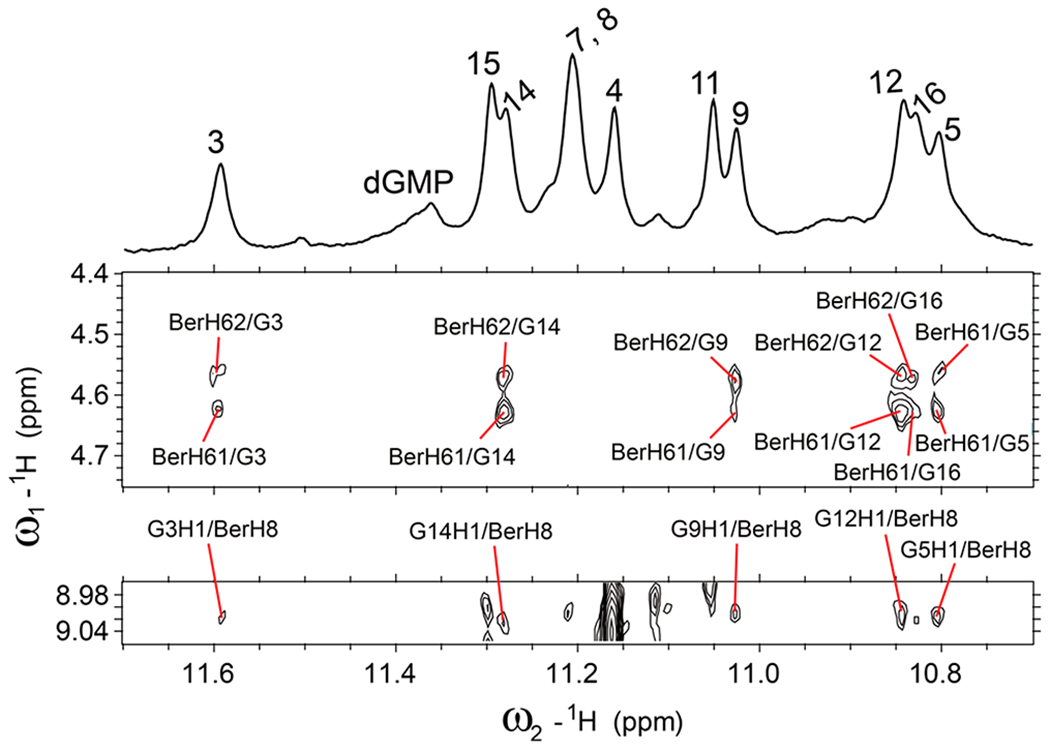
Select regions of the 2D-NOESY spectrum of 2:1:1 berberine–dGMP–vG4 ternary complex in H2O showing intermolecular cross peaks between berberine and DNA imino protons. Condition: 1.5 mM Pu19m2 DNA, 3:1 dGMP/DNA, 2:1 drug/DNA, pH 7, 50 mM K+, 25 °C, mixing time of 300 ms.
Numerous intra- and intermolecular NOEs were observed (Figures 3, 4, and S4, Tables S4–S7) that clearly define the berberine binding sites and the overall complex structures. The high-resolution structure of the 2:1:1 berberine–dGMP–vG4 ternary complex in K+-containing solution was thus calculated using molecular dynamics (MD) simulations based on NOE-derived distances, H-bond, and torsion-angle restraints (Table 1, Figures 5 and 6). A total of 456 NOE distance restraints including 43 berberine–DNA and 8 dGMP–vG4 intermolecular restraints for the berberine–dGMP–vG4 ternary complex were used for the simulations (Tables 1, S4, and S5, and Figures 3, 4, and S4). The position of the berberine at both external G-tetrads is well-defined and supported by 43 intermolecular NOE restraints (Figures 5 and 6, Tables 1 and S5). The final 10 lowest energy NMR structures are well-converged and exhibit an overall root-mean-square-deviation (RMSD) of 0.98 ± 0.28 Å for all atoms and 0.71 ± 0.20 Å for the G-tetrad core (Figure 5 and Table 1).
Table 1.
NMR Restraints and Structural Statistics for the 2:1:1 Berberine–dGMP–Pu19m2 Vacancy G-Quadruplex
| NOE-Based Distance Restraints | |
|---|---|
| total | 456 |
| intraresidue | 254 |
| inter-residue | 151 |
| sequential | 113 |
| long range | 38 |
| dGMP–Pu19m2 | 8 |
| berberine–Pu19m2 | 43 |
| Other Restraints | |
| hydrogen bond restraints | 48 |
| torsion angle restraints | 20 |
| G-tetrad planarity restraints | 48 |
| Structural Statistics | |
| pairwise heavy atom RMSD (Å) | |
| overall | 0.98 ± 0.28 |
| G4 without berberine | 1.01 ± 0.30 |
| G-tetrad core | 0.71 ± 0.20 |
| Violations | |
| max. NOE restraint violation (Å) | 0.14 |
| mean NOE restraint violation (Å) | 0.003 ± 0.012 |
|
| |
Figure 5.
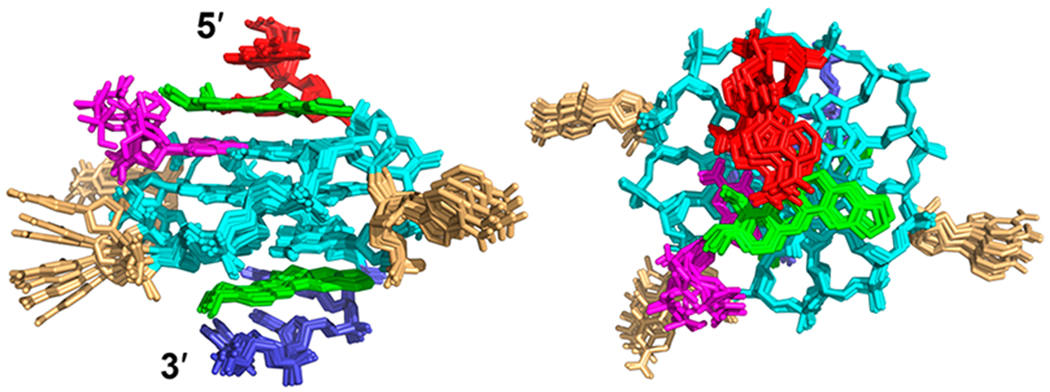
Superposition of the 10 lowest energy NMR structures of the berberine–dGMP–vG4 ternary complex by NOE-restrained structure calculation: side view (left) and top view (right). Green, berberine; magenta, dGMP; cyan, guanine; red, 5′-end flanking; blue, 3′-end flanking; and yellow, loop.
Figure 6.
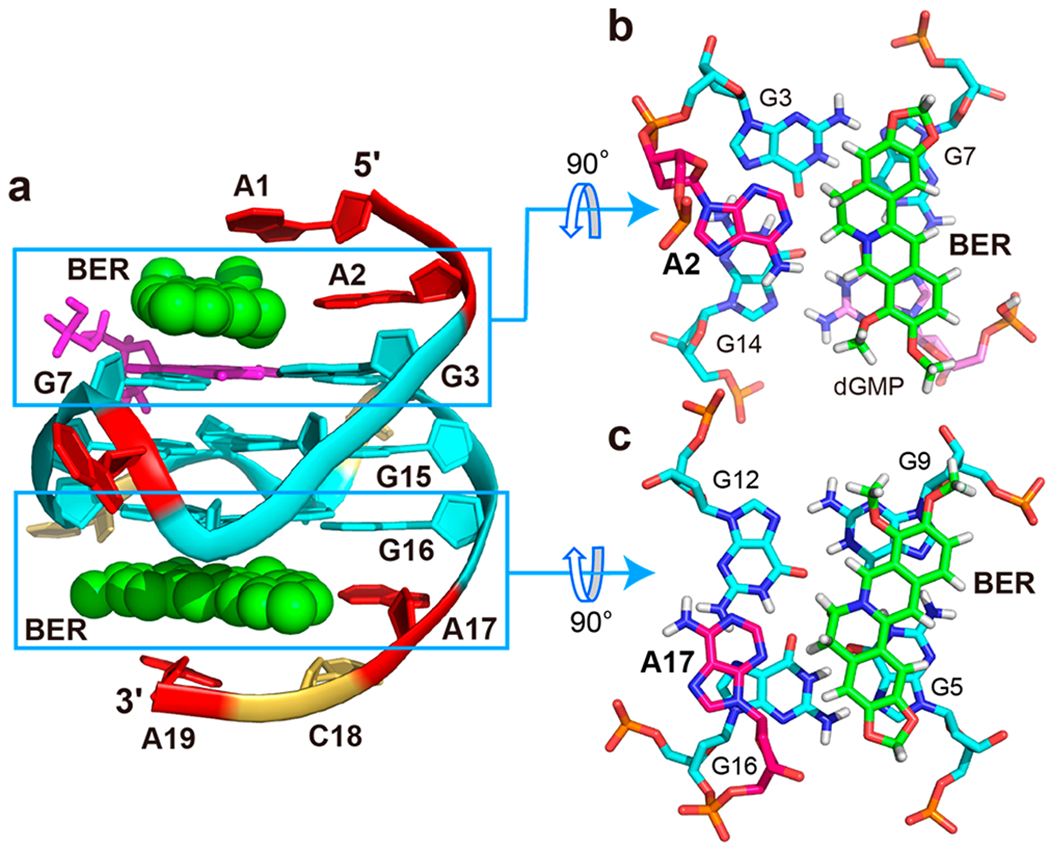
(a) Cartoon representation of the 2:1:1 berberine–dGMP–vG4 ternary complex (Protein Data Bank ID: 7MSV). (b) 5′-End and (c) 3′-end top views of the berberine/A2 and berberine/A17 “quasi-triad” planes. Cyan, guanine; magenta, dGMP; red, adenine; yellow, cytosine; and green, berberine.
The NMR complex structure in K+ solution showed that the berberine binding induced significant rearrangement for the flanking segments at both ends (Figure S9 and S10). At the 5′-end, berberine recruits A2, the flanking (−1) adenine, to form a “quasi-triad plane” stacking over the 5′-tetrad including the fill-in dGMP, which is covered by berberine, as supported by numerous NOE cross-peaks, such as A2H8/G14H1, A2H2/G14H1, A2H2/G3H1, BerH8/G3H1, BerH8/G14H1, BerH5/G14H1, BerH6/G14H1, and BerH6/G3H1 (Figures 4, 6b, and S9, Tables S5 and S6). The “quasi-triad plane” is further covered by the A1 residue that adopts a syn-conformation as observed in the dGMP–Pu19m2 fill-in-vG4 complex (Figures 6a and S9). Compared to the dGMP–vG4 complex, the flanking residue A2 is flipped and follows the right-handed backbone of the G-tetrad core upon binding to the berberine (Figure S11). As a result, the Watson–Crick edge of A2 points toward the tetrad center and a A2-berberine plane is formed (Figure S11). Similar to the 5′-end conformation, berberine recruits the flanking A17 to cover the 3′-tetrad, as supported by numerous NOE contacts, including BerH8/G5H1, BerH8/G9H1, BerH8/G12H1, BerH6/G5H1, BerH6/G9H1, BerH6/G12H1, BerH6/G16H1, A17H2/G12H1, A17H2/G16H1, and A17H8/A16H8 (Figures 4, 6c, and S10, Tables S5 and S7). The 3′-end-capping structure was ended by the covering of C18 and A19 residues, as shown by numerous NOE cross-peaks (Figures 6c and S10, Table S7). In contrast to the 5′-end site, the 3′-binding of berberine shows less change of the recruited A17. Upon berberine binding, the A17 base rotates ca. 10° and moves toward the G12/G16 edge, so berberine can replace A19 as binding partner (Figure S11). Again, berberine recruits A17 to form a base–ligand plane to maximize the stacking over the 3′-tetrad. This base-recruiting mechanism appears to be important for specific ligand recognition of G4s, such as that formed in the MYC promoter and telomere.22,39,49–51
In addition to the stacking interactions between the polyaromatic core of berberine and guanine bases of the external G-tetrads, potential electrostatic interactions can occur between the positively charged quaternary nitrogen of the berberine and the negatively polarized tetrad-guanine carbonyl groups, analogous to a K+ cation. In both the 5′- and 3′-end complexes, the positively charged BerN7 (Figure 1c) is positioned over the central channel above the external tetrad (Figure 6b,c). Notably, BerHB and BerHC show several extra NOE cross-peaks to aromatic protons of both the 3′- and 5′-tetrad guanines at high threshold levels, indicating additional minor binding modes of berberine. However, insufficient intermolecule NOE contacts were observed to define the structures of any minor species.
Designing New Berberine Analogues with Enhanced Affinity and/or Selectivity for Fill-in G4s.
The resolved structure herein suggests potential positions on the berberine scaffold that could permit substituents to enable additional interaction with the dGMP-fill-in vG4. First, the H1, H12, and H13 of the berberine have no direct interaction with dGMP vG4, while they are pointed toward grooves at both 3′- and 5′-sites, and can be used to attach a functional group for groove interactions (Figure 7). In contrast, one can design a guanine and G4–ligand conjugate to selectively target the vG4 by using the G-vacancy site as an anchor point based on the obtained structural information. The H11 and 10-OMe group of the berberine are pointed to the sugar–phosphate moiety of the dGMP (Figure 7, left) and define the most likely positions to make conjugates of berberine analogues and guanine derivatives. By this strategy, the guanine moiety can facilitate the selectivity toward a vG4 and the berberine analogue can enhance the binding affinity by stacking to the external tetrad. A similar strategy was proposed for conjugating a G4-binding peptide and a guanine moiety, which lead to enhanced affinity and selectivity for a vG4.52
Figure 7.
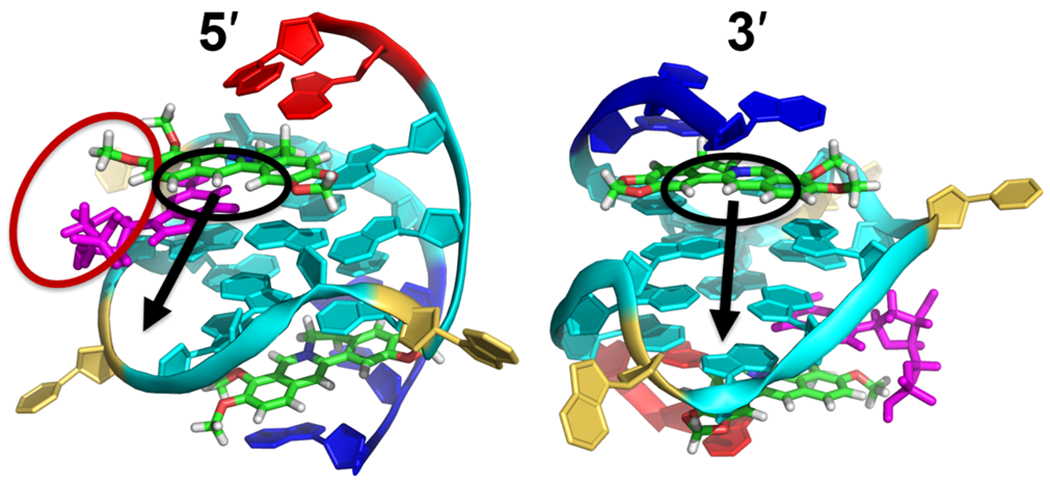
Suggested modifications of berberine to enable additional interaction with the dGMP–Pu19m2 vG4. The black circles suggest the positions (H1, H12, and H13) to be modified by introducing side chains. The black arrows indicate the grooves at 5′- and 3′-sites that the attached side chain will locate in. The red circle suggests the positions (H11 and 10-OMe) to make conjugate of berberine and guanine moiety. The Pu19m2 vG4 is shown in cartoon representation. Cyan, guanine; magenta, dGMP; red, 5′-end flanking; blue, 3′-end flanking; yellow, loop; and green, berberine.
CONCLUSION
In conclusion, the study herein presents the first high-resolution NMR solution structure of a ternary complex of a small molecule and a dGMP-fill-in vG4, i.e., berberine bound to a dGMP-fill-in vG4 from the PDGFR-β gene promoter. This ternary complex has a 2:1:1 binding stoichiometry, which differs from a previous study that reported a 6:2 molar ratio of berberine to a parallel human telomeric G4 in a crystal structure and a 1:1 berberine–RET-G4 solution structure.38,41 Whereas these studies showed a dimeric binding mode of berberine or preferred 3′-end binding both without ligand-flanking interactions, our results show a monomeric binding mode with ligand induced binding pockets involving both 5′-and 3′-end flanking residues. In our ternary complex structure, each berberine recruits the adjacent flanking adenine residue to form a “quasi-triad plane” that stacks over the external G-tetrad at each end and the binding involves both π-stacking and electrostatic interactions. On the basis of the determined structure, enhancing affinity and/or selectivity of berberine analogues for the PDGFR-β vG4 is proposed by introducing side chains and/or guanine conjugate. Our study thus provides a new platform to specifically target vG4s by designing new G4-interactive small molecules.
MATERIAL AND METHODS
Sample Preparation.
DNA oligonucleotides were synthesized and purified as described previously.20 The 15N–13C-labeled dGMP (>98%) was obtained from Cambridge Isotope Laboratories, Inc. The DNA was dissolved in potassium phosphate buffer (37.5 mM potassium chloride, 12.5 mM potassium phosphate, pH 7, 10/90% D2O/H2O). Final concentrations of the NMR samples between 0.015–1.5 mM were determined based on the UV absorption at 260 nm. NMR was used to monitor the binary and ternary complex formation. For 1D NMR study, we used low-concentration DNA samples (0.15 mM) and 15–20 fold dGMP for binary complex preparation. For 2D NMR study, we used higher concentration DNA samples (1.5 mM) and ~3 fold dGMP for binary complex preparation. It is noted that at a higher sample concentration a lower ratio of dGMP is needed to form the dGMP-fill-in–vG4 binary complex. Unlabeled dGMP (Sigma-Aldrich, ≥98.5%) and berberine (Shanghai Standard Technology Co., Ltd., >98%) were dissolved in 50 mM K+-containing buffer or DMSO-d6 to 40 or 75 mM.
Nuclear Magnetic Resonance (NMR) Experiments.
NMR spectra were obtained with a Bruker AV-800 spectrometer (with QCI cryoprobe) or a Bruker AV-500 spectrometer (with Prodigy cryoprobe). The w5 water suppression was used for 1D and 2D NOESY experiments. The NMR spectra were processed and analyzed using Topspin 3.5 (Bruker) and Sparky (UCSF) software. The 15N-edited 1D GE-JRSE HMQC experiment were carried out to identify the dGMP H1 proton.31 NOESY spectra were collected in 90% H2O/10% D2O with mixing times of 80, 150, 300, and 400 ms at temperatures of 15, 25, and 35 °C. TOCSY experiment was collected with a 80 ms mixing time at 25 °C. HSQC experiments were collected using a 3–9–19 water suppression with a 1J(C,H) of 180 Hz.
NOE-Based Structure Calculation.
The structure determination was done as described previously for the dGMP system with a few adjustments.31 Three repulsive restrains of 7.5 ± 1.5 Å were applied to prevent the loop residues 6 and 10 from a position in the grooves not in line with experimental data. Berberine–DNA intermolecular cross-peaks were defined as very weak (6.0 ± 1.5 Å), weak (5.0 ± 1.5 Å), and medium (4.0 ± 1.5 Å). Berberine parameter files were obtained from ChemDraw 18.2 and further optimized and calculated with the Gaussian09 program.53 PyMOL and the VMD software were used for analyzing and visualization.54,55
Circular Dichroism (CD) Experiments.
CD experiments were conducted using a Jasco-1100 spectropolarimeter (Jasco Inc.). DNA samples were dissolved in potassium phosphate buffer (37.5 mM potassium chloride, 12.5 mM potassium phosphate, pH 7) with a concentration of 15 μM in the presence and absence of the dGMP. CD spectra were collected with a 1 mm path length quartz cuvette, 1 s response time, and 1 nm bandwidth at 25 °C, and subsequently blank corrected with a buffer spectrum.
Fluorescence Measurements.
Fluorescence experiments were collected with a Jasco-FP8300 spectrofluorometer (Jasco Inc., Easton, MD). Emission spectra were measured from 520 to 600 using a 1 cm path length quartz cell. The selected excitation wavelength was 377 nm. dGMP–Pu19m2 binary complex (20:1) was titrated to a 0.2 μM berberine solution in potassium phosphate buffer (50 mM K+) and incubated for 2 min before each measurement. The Kd value was calculated by fitting the data to an equation: F = Fmin + (Fmax – Fmin) [(PT + CT + Kd) – [((DT + CT + Kd)2 – (4DTCT))1/2]/(2CT), where F represents the ligand-induced fluorescence intensity, CT is the ligand concentration, and DT is the ternary complex concentration.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health R01CA177585 (D.Y.), P30CA023168 (Purdue Center for Cancer Research), and the Deutsche Forschungsgemeinschaft (German Research Foundation) - Projektnummer 427347592 (J.D.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c06200.
Tables of proton chemical shifts and NOEs, 15N-edited NMR spectra, 1H–13C-HSQC, 2D-NOESY, proton chemical shift, 5′- and 3′-end-capping structures (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.1c06200
The authors declare no competing financial interest.
Contributor Information
Kai-Bo Wang, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University, West Lafayette, Indiana 47907, United States.
Jonathan Dickerhoff, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University, West Lafayette, Indiana 47907, United States.
Danzhou Yang, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue Center for Cancer Research, Department of Chemistry, and Purdue Institute for Drug Discovery, Purdue University, West Lafayette, Indiana 47907, United States.
REFERENCES
- (1).Yang D; Okamoto K Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem 2010, 2 (4), 619–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Balasubramanian S; Hurley LH; Neidle S Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discovery 2011, 10 (4), 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rhodes D; Lipps HJ G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43 (18), 8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Raguseo F; Chowdhury S; Minard A; Di Antonio M Chemical-biology approaches to probe DNA and RNA G-quadruplex structures in the genome. Chem. Commun 2020, 56 (9), 1317–1324. [DOI] [PubMed] [Google Scholar]
- (5).Chen MC; Tippana R; Demeshkina NA; Murat P; Balasubramanian S; Myong S; Ferré-D’Amaré AR Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558 (7710), 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Parkinson GN; Lee MP; Neidle S Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417 (6891), 876–880. [DOI] [PubMed] [Google Scholar]
- (7).Biffi G; Tannahill D; McCafferty J; Balasubramanian S Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem 2013, 5 (3), 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Biffi G; Di Antonio M; Tannahill D; Balasubramanian S Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem 2014, 6 (1), 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Brooks TA; Hurley LH Targeting MYC expression through G-quadruplexes. Genes Cancer 2010, 1 (6), 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hänsel-Hertsch R; Di Antonio M; Balasubramanian S DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol 2017, 18 (5), 279–284. [DOI] [PubMed] [Google Scholar]
- (11).Hänsel-Hertsch R; Beraldi D; Lensing SV; Marsico G; Zyner K; Parry A; Di Antonio M; Pike J; Kimura H; Narita M; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet 2016, 48 (10), 1267–1272. [DOI] [PubMed] [Google Scholar]
- (12).Wu G; Xing Z; Tran EJ; Yang D DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (41), 20453–20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Guilbaud G; Murat P; Recolin B; Campbell BC; Maiter A; Sale JE; Balasubramanian S Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem 2017, 9 (11), 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Asamitsu S; Bando T; Sugiyama H Ligand design to acquire specificity to intended G-quadruplex structures. Chem. - Eur. J 2019, 25 (2), 417–430. [DOI] [PubMed] [Google Scholar]
- (15).Neidle S Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem 2016, 59 (13), 5987–6011. [DOI] [PubMed] [Google Scholar]
- (16).Siddiqui-Jain A; Grand CL; Bearss DJ; Hurley LH Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A 2002, 99 (18), 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cogoi S; Xodo LE G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34 (9), 2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Marquevielle J; Robert C; Lagrabette O; Wahid M; Bourdoncle A; Xodo LE; Mergny JL; Salgado GF Structure of two G-quadruplexes in equilibrium in the KRAS promoter. Nucleic Acids Res. 2020, 48 (16), 9336–9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang KB; Elsayed MS; Wu G; Deng N; Cushman M; Yang D Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J. Am. Chem. Soc 2019, 141 (28), 11059–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Onel B; Carver M; Wu G; Timonina D; Kalarn S; Larriva M; Yang D A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc 2016, 138 (8), 2563–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Brown RV; Wang T; Chappeta VR; Wu G; Onel B; Chawla R; Quijada H; Camp SM; Chiang ET; Lassiter QR; et al. The consequences of overlapping G-quadruplexes and i-motifs in the platelet-derived growth factor receptor β core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J. Am. Chem. Soc 2017, 139 (22), 7456–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Liu W; Lin C; Wu G; Dai J; Chang TC; Yang D Structures of 1:1 and 2:1 complexes of BMVC and MYC promoter G-quadruplex reveal a mechanism of ligand conformation adjustment for G4-recognition. Nucleic Acids Res. 2019, 47 (22), 11931–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Andrae J; Gallini R; Betsholtz C Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22 (10), 1276–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chen P-H; Chen X; He X Platelet-derived growth factors and their receptors: structural and functional perspectives. Biochim. Biophys. Acta, Proteins Proteomics 2013, 1834 (10), 2176–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Qin Y; Fortin JS; Tye D; Gleason-Guzman M; Brooks TA; Hurley LH Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-beta expression. Biochemistry 2010, 49 (19), 4208–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chen Y; Agrawal P; Brown RV; Hatzakis E; Hurley L; Yang D The major G-quadruplex formed in the human platelet-derived growth factor receptor beta promoter adopts a novel broken-strand structure in K+ solution. J. Am. Chem. Soc 2012, 134 (32), 13220–13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Li XM; Zheng KW; Zhang JY; Liu HH; He YD; Yuan BF; Hao YH; Tan Z Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (47), 14581–14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Heddi B; Martín-Pintado N; Serimbetov Z; Kari TMA; Phan AT G-quadruplexes with (4n-1) guanines in the G-tetrad core: formation of a G-triad·water complex and implication for small-molecule binding. Nucleic Acids Res. 2016, 44 (2), 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Winnerdy FR; Das P; Heddi B; Phan AT Solution structures of a G-quadruplex bound to linear- and cyclicdinucleotides. J. Am. Chem. Soc 2019, 141 (45), 18038–18047. [DOI] [PubMed] [Google Scholar]
- (30).Li XM; Zheng KW; Hao YH; Tan Z Exceptionally selective and tunable sensing of guanine derivatives and analogues by structural complementation in a G-quadruplex. Angew. Chem. Int. Ed 2016, 55 (44), 13759–13764. [DOI] [PubMed] [Google Scholar]
- (31).Wang KB; Dickerhoff J; Wu G; Yang D PDGFR-β Promoter Forms a Vacancy G-Quadruplex that Can be Filled-in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs. J. Am. Chem. Soc 2020, 142 (11), 5204–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhang C; Zhao L; Yang W; Yao X; Sun L; Cui R Effects of berberine and its derivatives on cancer: A systems pharmacology review. Front. Pharmacol 2019, 10, 1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tillhon M; Guamán Ortiz LM; Lombardi P; Scovassi AI Berberine: new perspectives for old remedies. Biochem. Pharmacol 2012, 84 (10), 1260–1267. [DOI] [PubMed] [Google Scholar]
- (34).Kuo CL; Chi CW; Liu TY The anti-inflammatory potential of berberine in vitro and in vivo. Cancer Lett. 2004, 203 (2), 127–137. [DOI] [PubMed] [Google Scholar]
- (35).Sun Y; Xun K; Wang Y; Chen X A systematic review of the anticancer properties of berberine, a natural product from Chinese herbs. Anti-Cancer Drugs 2009, 20 (9), 757–769. [DOI] [PubMed] [Google Scholar]
- (36).Ma Y; Ou TM; Hou JQ; Lu YJ; Tan JH; Gu LQ; Huang ZS 9-N-Substituted berberine derivatives: stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc. Bioorg. Med. Chem 2008, 16 (16), 7582–7591. [DOI] [PubMed] [Google Scholar]
- (37).Zhang L; Liu H; Shao Y; Lin C; Jia H; Chen G; Yang D; Wang Y Selective lighting up of epiberberine alkaloid fluorescence by fluorophore-switching aptamer and stoichiometric targeting of human telomeric DNA G-quadruplex multimer. Anal. Chem 2015, 87 (1), 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bazzicalupi C; Ferraroni M; Bilia AR; Scheggi F; Gratteri P The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013, 41 (1), 632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Lin C; Wu G; Wang K; Onel B; Sakai S; Shao Y; Yang D Molecular recognition of the hybrid-2 human telomeric G-quadruplex by epiberberine: insights into conversion of telomeric G-quadruplex structures. Angew. Chem. Int. Ed 2018, 57 (34), 10888–10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bessi I; Bazzicalupi C; Richter C; Jonker HR; Saxena K; Sissi C; Chioccioli M; Bianco S; Bilia AR; Schwalbe H; Gratteri P Spectroscopic, molecular modeling, and NMR-spectroscopic investigation of the binding mode of the natural alkaloids berberine and sanguinarine to human telomeric G-quadruplex DNA. ACS Chem. Biol 2012, 7 (6), 1109–19. [DOI] [PubMed] [Google Scholar]
- (41).Wang F; Wang C; Liu Y; Lan W; Wang R; Huang S; Cao C NMR studies on the interaction between oncogene RET G-quadruplex and berberine. Chin. J. Chem 2020, 38 (12), 1656–1662. [Google Scholar]
- (42).Moraca F; Amato J; Ortuso F; Artese A; Pagano B; Novellino E; Alcaro S; Parrinello M; Limongelli V Ligand binding to telomeric G-quadruplex DNA investigated by funnel-metadynamics simulations. Proc. Natl. Acad. Sci U. S. A 2017, 114 (11), e2136–e2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).del Villar-Guerra R; Trent JO; Chaires JB G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem. Int. Ed 2018, 57 (24), 7171–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Arora A; Balasubramanian C; Kumar N; Agrawal S; Ojha RP; Maiti S Binding of berberine to human telomeric quadruplex-spectroscopic, calorimetric and molecular modeling studies. FEBS J. 2008, 275 (15), 3971–83. [DOI] [PubMed] [Google Scholar]
- (45).Bhadra K; Kumar GS Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: A comparative spectroscopic and calorimetric study. Biochim. Biophys. Acta, Gen. Subj 2011, 1810 (4), 485–496. [DOI] [PubMed] [Google Scholar]
- (46).Tripathi AN; Chauhan L; Thankachan PP; Barthwal R Quantum chemical and nuclear magnetic resonance spectral studies on molecular properties and electronic structure of berberine and berberrubine. Magn. Reson. Chem 2007, 45 (8), 647–55. [DOI] [PubMed] [Google Scholar]
- (47).Greene KL; Wang Y; Live D Influence of the glycosidic torsion angle on 13C and 15N shifts in guanosine nucleotides: investigations of G-tetrad models with alternating syn and anti bases. J. Biomol. NMR 1995, 5 (4), 333–338. [DOI] [PubMed] [Google Scholar]
- (48).Dickerhoff J; Weisz K Flipping a G-tetrad in a unimolecular quadruplex without affecting its global fold. Angew. Chem. Int. Ed 2015, 54 (19), 5588–5591. [DOI] [PubMed] [Google Scholar]
- (49).Dai J; Carver M; Hurley LH; Yang D Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc 2011, 133 (44), 17673–17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Calabrese DR; Chen X; Leon EC; Gaikwad SM; Phyo Z; Hewitt WM; Alden S; Hilimire TA; He F; Michalowski AM; et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun 2018, 9 (1), 4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Dickerhoff J; Dai J; Yang D Structural recognition of the MYC promoter G-quadruplex by a quinoline derivative: insights into molecular targeting of parallel G-quadruplexes. Nucleic Acids Res. 2021, 49, 5905–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).He Y; Zheng K; Wen C; Li X; Gong J; Hao Y; Zhao Y; Tan Z Selective Targeting of Guanine-vacancy-bearing G-quadruplexes by G-quartet complementation and stabilization with a guanine-peptide conjugate. J. Am. Chem. Soc 2020, 142 (26), 11394–11403. [DOI] [PubMed] [Google Scholar]
- (53).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Mennucci B; Petersson GA; Nakatsuji H; Caricato M; Li X; Hratchian HP; Izmaylov AF; Bloino J; Zheng G; Sonnenberg JL; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark M; Heyd JJ; Brothers E; Kudin KN; Staroverov VN; Kobayashi R; Normand J; Raghavachari K; Rendell A; Burant JC; Iyengar SS; Tomasi J; Cossi M; Rega N; Millam JM; Klene M; Knox JE; Cross JB; Bakken V; Adamo C; Jaramillo J; Gomperts R; Stratmann RE; Yazyev O; Austin AJ; Cammi R; Pomelli C; Ochterski JW; Martin RL; Morokuma K; Zakrzewski VG; Voth GA; Salvador P; Dannenberg JJ; Dapprich S; Daniels AD; Farkas O; Foresman JB; Ortiz JV; Cioslowski J; Fox DJ Gaussian 09, revision D.01; Gaussian, Inc.: Wallingford, CT, 2009. [Google Scholar]
- (54).Schrödinger L PyMOL Molecular Graphics System, version 2.1; Schrödinger, 2018. [Google Scholar]
- (55).Humphrey W; Dalke A; Schulten K VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14 (1), 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


