Abstract
The outbreak of the COVID-19 pandemic, caused by Severe Acute Respiratory Syndrome of Coronavirus 2 (SARS-CoV-2), has fueled the search for diagnostic tests aiming at the control and reduction of the viral transmission. The main technique used for diagnosing the Coronavirus disease (COVID-19) is the reverse transcription-polymerase chain reaction (RT-PCR) technique. However, considering the high number of cases and the underlying limitations of the RT-PCR technique, especially with regard to accessibility and cost of the test, one does not need to overemphasize the need to develop new and less expensive testing techniques that can aid the early diagnosis of the disease. With that in mind, we developed an ultrasensitive magneto-assay using magnetic beads and gold nanoparticles conjugated to human angiotensin-converting enzyme 2 (ACE2) peptide (Gln24-Gln42) for the capturing and detection of SARS-CoV-2 Spike protein in human saliva. The technique applied involved the use of a disposable electrochemical device containing eight screen-printed carbon electrodes which allow the simultaneous analysis of eight samples. The magneto-assay exhibited an ultralow limit of detection of 0.35 ag mL−1 for the detection of SARS-CoV-2 Spike protein in saliva. The magneto-assay was tested in saliva samples from healthy and SARS-CoV-2-infected individuals. In terms of efficiency, the proposed technique – which presented a sensitivity of 100.0% and specificity of 93.7% for SARS-CoV-2 Spike protein-exhibited great similarity with the RT-PCR technique. The results obtained point to the application potential of this simple, low-cost magneto-assay for saliva-based point-of-care COVID-19 diagnosis.
Keywords: Electrochemical Device, SARS-CoV-2, COVID-19 Diagnosis, Spike Protein, Saliva
Graphical Abstract

1. Introduction
It has been almost two years now since SARS-CoV-2 was first described, yet reports in the literature show that there has been little improvement in the control of the SARS-CoV-2 viral shedding [1]. With more than 230 million cumulative number of reported cases, the Coronavirus 2019 disease (COVID-19) has evolved into a pandemic health crisis since March 2020, posing a considerable burden on public life and healthcare worldwide [2]. In this context, the development and application of rapid and easily accessible diagnostic tests have proven to be a critical component of the holistic approach toward the prevention and control of COVID-19.
According to the World Health Organization (WHO), nucleic acid amplification tests (NAAT) are the most sensitive and specific testing technique for bacterial or viral detection, and are therefore recommended as the reference standard test for the diagnosis of acute SARS-CoV-2 infection [2]. In spite of its proven efficacy, the NAAT testing method has some underlying non-negligible limitations. Among the major limitations of the NAAT testing technique include the following: i) the high costs of the test impede its wide availability, especially in low and middle-income countries; ii) the test is relatively time-consuming [3] and the analysis requires the use of highly qualified personnel; and iii) the test is of limited use when it comes to qualitative analyses in most cases [4]. In light of these limitations, as an alternative to the NAAT tests, WHO recommends the use of viral antigen tests for the detection of SARS-CoV-2 infection because these tests are relatively cheaper and simpler to operate, in addition to providing quicker results; this allows policymakers and healthcare providers to ensure significantly higher test coverage of the population in a relatively short period of time [5].
The detection of viral gene or antigen in swab or saliva samples has been used for diagnosis of COVID-19 in the early stage and human antibodies have been used for screening of previous exposure to the virus, or collection of epidemiological data, determination of the immune status of asymptomatic individuals [6], [7]. The detection of viral RNA by reverse transcription polymerase chain reaction (RT-PCR) is considered the gold standard technique for COVID-19 diagnosis [6]. However, RT-PCR requires expensive equipment and trained technicians at certified laboratories and long times to generate results for the diagnosis. Therefore, several studies published in the literature have investigated the use of SARS-CoV-2 Spike protein as one of the suitable tools for the development of new alternative tests for the detection of the viral antigens [8], [9] The Spike protein of SARS-CoV-2 is constituted by (i) a receptor-binding domain (RBD), which binds to human angiotensin-converting enzyme 2 (ACE2) receptor and mediates the entry of Coronavirus into host cells [10], and (ii) a fusion domain, which fuses host cell and viral membranes through the C-terminal S2 subunit [11]. Considering that it plays a fundamental role in the infection and dissemination of SARS-CoV-2, the Spike protein of SARS-CoV-2 has become the main tool employed in the development of new tests aimed at the detection of SARS-CoV-2 antigens [12]. Recently, several studies reported in the literature have shown that Spike detection-based diagnostic tests exhibit high sensitivity and specificity when applied for the diagnosis of SARS-CoV-2 infection [13], [14]. However, most of the Spike detection-based diagnostic tests reported in the literature are highly expensive and complex to perform, apart from their being incapable of providing quick results.
Over the past few years, there has been a gradual rise in the development of simple, portable, and low-cost devices for protein immunodetection using screen-printed carbon electrodes (SPCE) [15], [16]. These devices can provide valuable information on the incidence, prevalence, severity, and transmissibility of diseases that are primarily caused by viruses and bacteria [17], [18]. The incorporation of magnetic beads in these detection devices can amplify the electrochemical signal for biomolecules, such as proteins and peptides [19], [20], facilitating the separation and washing procedures and enabling the pre-concentration of the analyte, in addition to increasing the selectivity and sensitivity of the method [21], [22]. These improvements in the functional properties of the detection devices allow for substantial gains in the diagnosis of bacterial or viral infections. In particular, nanomaterials, such as gold nanoparticles, can be applied as electrochemical markers with a view toward improving the sensitivity of the analytical method and for the indirect detection of various biomolecules [23], including DNA and RNA [24], [25], antibodies [26], [27], and proteins [28], [29].
The present work reports the development of a magneto-assay based on a mimotope of ACE2 for the detection of Spike protein in saliva samples for the diagnosis of COVID-19. The detection device was constructed using magnetic beads (MBs) and gold nanoparticles (AuNPs) modified with ACE2 peptide; this device was used for capturing and separating the Spike protein in saliva samples and for the conduct of electrochemical detection analysis. The Spike protein bioconjugate was incorporated into an unmodified SPCE, and the redox properties of the gold in AuNPs enabled the detection of the Spike protein. The application of the magneto-assay helped differentiate the saliva samples of healthy subjects from those of the subjects infected by SARS-CoV-2 with high sensitivity and specificity.
2. Experimental
The Supplementary Material (SM) describes the reagents, materials, and equipment used in the study.
2.1. Synthesis of the ACE2 peptide and the peptide immobilization using MBs and AuNPs
The ACE2 peptide (24QAKTFLDKFNHEAEDLFYQ42C), corresponding to the region of interaction between the human ACE2 and the Spike protein [30], was synthesized using a cysteine residue at the C-terminal (see details in Fig. S1 of the SM), using standard solid-phase synthesis strategy and Fluorenylmethyloxycarbonyl (Fmoc) chemistry. The ACE2 peptide was immobilized on MBs and AuNPs, and this led to the formation of the conjugated materials named MBs-ACE2 (Fig. S2) and ACE2-AuNPs (Fig. S3), respectively. The trimeric Spike protein of SARS-CoV-2 in the prefusion conformation, which was produced in HEK293 cells [31], was kindly provided by the Cell Culture Engineering Lab of COPPE/UFRJ (Federal University of Rio de Janeiro). The Spike proteins were cross-linked to form poly-Spike proteins, as described in the SM.
2.2. Sample Collection
The saliva samples were collected from the UFSCar University Hospital; all the ethical procedures involving the participation of the research subjects were approved by the UFSCar Research Ethics Committee (Number: 66076017.3.0000.5504). All the participants who took part in the research provided their written informed consent prior to their inclusion in the study. The RT-PCR tests conducted confirmed that the subjects were infected by SARS-CoV-2. Prior to performing the tests, the saliva samples of the subjects were heated at 80 °C for 15 min in order to inactivate the virus [32], [33], [34].
2.3. Steps involved in the capture and separation of the Spike protein in saliva samples under the magneto-assay
The detection of the SARS-CoV-2 Spike protein in the saliva samples employed pointed to the presence of SARS-CoV-2. Fig. 1A presents the steps involving the magnetic capture of the SARS-CoV-2 Spike protein in saliva samples using the magneto-assay proposed in this study. Initially, the saliva samples were diluted 1000 times in phosphate buffer saline solution (PBS) with pH 7.4; the PBS solution was composed of the following: 10 mmol L—1 phosphate, 2.7 mmol L—1 KCl, and 137 mmol L—1 NaCl; 1.0 mmol L—1 CaCl2 and 0.5 mmol L—1 MgCl2 (PBS-Ca-Mg).
Fig. 1.
Steps involved in the magneto-assay for the detection of SARS-CoV-2 Spike protein in saliva sample. A) SARS-CoV-2 Spike protein capture and formation of MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate. B) Placing the magnet under the working electrode. C) Electrochemical detection of the SARS-CoV-2 Spike protein using unmodified SPCE. D) Representation of the chronoamperometric oxidation of AuNPs following Au (III) reduction by DPV.
After that, an amount of 450 µL of the diluted sample was transferred to a microtube containing 20 µL of MBs-ACE2 and 30 µL of ACE2-AuNPs, as shown in Fig. 1A (Step 1). The mixture containing the saliva sample and the bioconjugates were incubated under slow shaking for 1 h at 24ºC; this led to the formation of the MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate (Fig. 1A - Step 2). After the incubation time, the MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate was separated from the sample solution, keeping the mixture in a magnetic rack for 2 min and discarding the supernatant (Fig. 1A, Step 3). The MBs-ACE2/ SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate was then washed with PBS containing 0.05% of Tween 20 (PBS-TW); the washing procedure was repeated three times. Finally, the bioconjugate was redispersed in 400 µL of PBS pH 7.4 and applied on the SPCE, which was subsequently used for the electrochemical detection of the SARS-CoV-2 Spike protein.
2.4. Electrochemical Detection of SARS-CoV-2 Spike Protein in Saliva Samples
The electrochemical detection of the SARS-CoV-2 Spike protein in the saliva samples was performed using a SPCE array with 8 independent cells. The detection was carried out without the modification of the working electrodes (WEs). A 3D printed support was constructed containing a keyboard which was used to position the magnets exactly on each working electrode; this was done for the magnetic particles of the bioconjugate to be retained on the electrode surface (see Fig. 1B and S4). The SPCE array was connected to a portable potentiostat, and the electrochemical detection of the Spike protein was conducted by adding an aliquot of 25 µL of MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs dispersion in each of the 8-WEs of the independent electrochemical cells (Fig. 1C). Subsequently, a volume of 25 µL of 0.2 mol L—1 HCl was added to each cell as a supporting electrolyte for the detection analysis. The electrochemical detection of the Spike protein was performed by differential pulse voltammetry (DPV) (Fig. 1D), based on the parameters evaluated previously by Afonso et al. (2013) [35], using the following conditions: deposition potential (E dep) = + 1.25 V, deposition time (t dep) = 120 s, scan rate (ν) = 34 mV s−1, step potential (ΔE s) = 10 mV, and pulse amplitude (a) = 50 mV.
3. Results and discussion
3.1. Characterization of the nanomaterials and bioconjugates
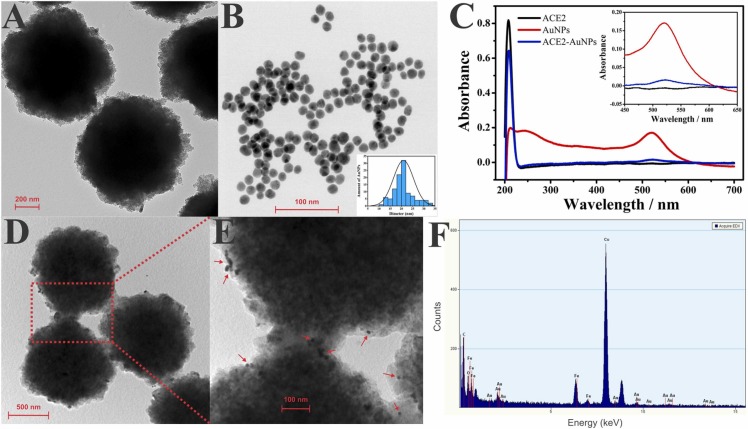
The nanoparticles and bioconjugates obtained in this study were characterized using transmission electron microscopic (TEM), ultraviolet-visible spectrophotometry (UV-Vis), and energy-dispersive X-ray (EDX) analysis. As can be observed in the TEM image in Fig. 2A, the commercial MBs deliver uniform and highly defined spherical beads. The AuNPs (Fig. 2B) also have an aspheric shape with an average diameter of 21 nm, as described in the literature [36].
Fig. 2.
A) TEM image of the MBs and B) AuNPs. C) UV–vis spectra recorded for ACE2 solution, AuNPs dispersion, and ACE2-AuNPs bioconjugate. D) and E) MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate. F) EDX analysis of MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate.
UV-Vis was applied to characterize the ACE2 solution and the AuNPs and ACE2-AuNPs bioconjugate dispersions (Fig. 2C). The absorption band of the ACE2 peptide (black line) was found in the UV region at 209 nm; this is typically characteristic of the amino acids, including histidine and cysteine, present in the peptide structure [37]. About the AuNPs dispersion, the absorption spectra were found in the region that corresponded to the surface plasmon resonance absorbance (ASPR) at 520 nm - see Fig. 2B (red line); this region indicated stability and was characterized by a spherical shape, as previously reported in the literature through TEM analysis [38]. By contrast, the ACE2-AuNPs exhibited a decrease in ASPR, see Fig. 2B (blue line). The thiol group present in the ACE2 cysteine reacted with the AuNPs, presumably through a covalent bond [39], [40], and this led to a significant decrease in ASPR through the formation of the bioconjugate [41]. In addition, at 209 nm, the band that corresponded to the peptide remained present though at a lower intensity, indicating its presence after binding with AuNPs [42], [43]. These results point to the formation of the AuNPs-ACE2 bioconjugate.
Figs. 2D and 2E show the TEM images of MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate formed following the procedure reported. As can be seen, AuNPs are presented on the surface of the MBs demonstrating that the SARS-CoV-2 Spike had been successfully captured by the ACE2 conjugated with AuNP. This result was also confirmed by EDX analysis, as can be seen in Fig. 2F. Fe and Au signals confirm their presence in the bioconjugate. These findings agreed with studies reported in the literature using similar strategies [44], [45]. In addition, the intense Cu peak was related to the grid used in the TEM measurements [44], [46]. These results demonstrate that the magneto-assay proposed was successfully applied, indicating that AuNPs can be effectively used as a redox probe for electrochemical detection of Spike protein.
3.2. Analysis of the interaction between ACE2 peptide and SARS-CoV-2 Spike protein by quartz crystal microbalance (QCM)
Previous studies published in the literature reported the binding of SARS-CoV-2 Spike protein to human ACE2 with high affinity [47], [48]; in other words, the pathway by which SARS-CoV-2 enters the cell [49], [50]. To show that the ACE2 peptide synthesized in this study is able to bind to the SARS-CoV-2 Spike protein, the ACE2 peptide was covalently immobilized on the gold surface of quartz crystal via thiol-gold interaction [51]; saliva samples from healthy individuals which were fortified with SARS-CoV-2 Spike protein were used for the analysis of the interaction by quartz crystal microbalance (QCM) (Fig. S7A and B). Although a similar pattern of behavior was observed for the curves in the presence and absence of the SARS-CoV-2 Spike protein, there was a negative variation in the resonance frequency when the SARS-CoV-2 Spike protein-containing saliva was analyzed; this result points to an increase in mass of the quartz crystal (Fig. S7A and B). Saliva is a complex matrix with high content of proteins including mucins; mucins are heavily glycosylated high molecular weight glycoproteins which are responsible for the viscoelastic properties of saliva [52], [53]. The increase in mass of the quartz crystal observed when the non-fortified saliva was injected can be linked to the unspecific absorption of the proteins in saliva on the crystal surface which were not been completely removed after the washing procedure. Interestingly, in the presence of the SARS-CoV-2 Spike protein, the variation observed in the resonance frequency was significantly higher than that observed when the non-fortified saliva sample was applied (Fig. S7B). This outcome points to the successful binding of the SARS-CoV-2 Spike protein to the ACE2 peptide immobilized on the crystal surface [54], [55]. In essence, the results obtained here clearly show that the ACE2 peptide interacts effectively with the SARS-CoV-2 Spike protein in the saliva samples.
3.3. Electrochemical detection of SARS-CoV-2 Spike protein
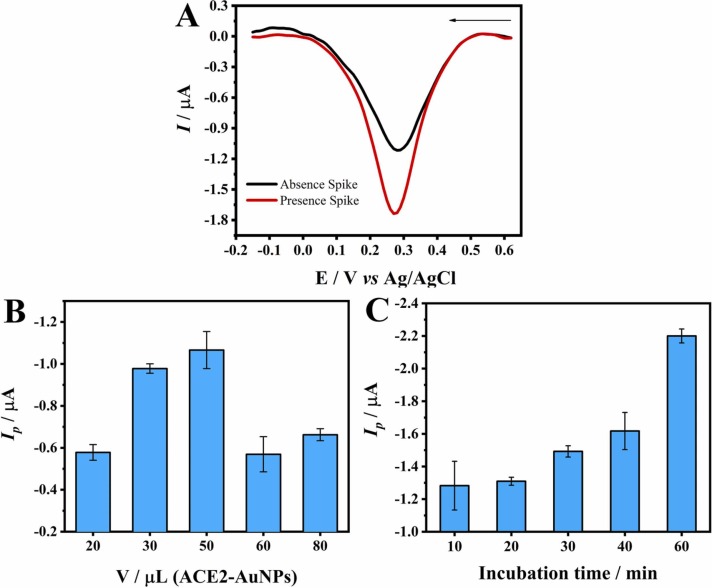
The electrochemical detection of the Spike protein was performed in standard PBS-Ca-Mg solutions with or without 360 fg mL−1 of SARS-CoV-2 Spike protein to mimic the viral structure with many Spike proteins on the surface. The MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate obtained was placed on the surface of an unmodified working electrode with a magnet positioned externally under the electrode. After that, a diluted HCl solution was added to the bioconjugate mixture, and the potential of + 1.25 V was applied for 120 s in order to oxidize the Au present in the AuNPs, leading to the formation of the complex [AuCl4]-. Subsequently, the application of DPV led to the generation of a cathodic peak at approximately 0.3 V ( Fig. 3A) due to the reduction of [AuCl4]- to Au [56]. The peak current observed for the sample without the SARS-CoV-2 Spike protein can be linked to the non-specific binding of the ACE2-AuNPs and the adsorption of the saliva proteins on the surface of the MBs during incubation, which influenced the analytical response of the control material [57], [58]. By contrast, a more intense peak current was observed in the presence of the SARS-CoV-2 Spike protein; this clearly points to the interaction between the SARS-CoV-2 Spike protein and the ACE2 peptide [55], [59].
Fig. 3.
A) DPV curves obtained from the application of the MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate, generated in the presence and absence of 360 fg mL−1 SARS-CoV-2 Spike protein, along with standard PBS-Ca-Mg solutions; incubation time: 30 min Edep = + 1.25 V, tdep = 120 s, ν = 34 mV s−1, ΔEs = 10 mV, and a = 50 mV. B) Peak current for different volumes of ACE2-AuNPs in the presence of 78 fg mL−1 SARS-CoV-2 Spike protein in the MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate; incubation time: 30 min C) Peak current for different incubation times based on the application of 78 fg mL−1 SARS-CoV-2 Spike protein in the MBs-ACE2/SARS-CoV-2 Spike protein/ACE2-AuNPs bioconjugate dispersion containing 20 µL MBs-ACE2 and 30 µL of ACE2-AuNPs.
A thorough analysis was also performed in order to evaluate the influence of incubation time on the capture of the SARS-CoV-2 Spike protein and the quantity of ACE2-AuNPs employed in the magneto-assay (Figs. 3B and 3C). The quantity of ACE2-AuNPs was controlled by adding different volumes of a standard dispersion of the conjugate prepared as described in the SM. The volume of 50 µL ACE2-AuNPs presented the highest peak current in terms of the detection of the SARS-CoV-2 Spike protein; however, the volume of 30 µL of ACE2-AuNPs was selected for conduct of analysis because it exhibited slightly lower values with better standard deviation (Fig. 3B). With regard to incubation, the application of 60 min incubation time yielded the highest peak response (Fig. 3C).
3.4. Analytical performance of the magneto-assay
After selecting the optimal experimental parameters, the magneto-assay was applied using standard solutions containing SARS-CoV-2 Spike protein at concentrations ranging from 0.0009 to 360.00 fg mL—1, as shown in Fig. 4A. Peak current values were obtained as a function of the SARS-CoV-2 Spike protein concentrations and were used to construct the calibration curve (Fig. 4B). The calculated linear regression equation obtained was as follows: −I p (μA) = 0.0887 + 0.0918 log [SARS-CoV-2 Spike protein] (fg mL—1), with a linear correlation coefficient of 0.998; this value indicates an excellent linear correlation between the current response and the SARS-CoV-2 Spike protein concentration. The limit of detection (LOD) obtained was 0.35 ag mL−1; this value was calculated based on the following equation: 3.3 ×SD/θ, where SD is the standard deviation of the intercept of the analytical curve, and θ is the slope of the analytical curve [60].
Fig. 4.
A) DPV curves obtained from the application of the magneto-assay using standard solutions containing SARS-CoV-2 Spike protein in the concentration range of 0.0009–360.00 fg mL—1. DPV parameters applied: Edep= + 1.25 V, tdep= 120 s, ν = 34 mV s−1, ΔEs = 10 mV, and a = 50 mV; B) Cathodic peak current values obtained as a function of the SARS-CoV-2 Spike protein concentration; Repeatability study using the magneto-assay for C) intraday analysis and D) interday analysis using a standard solution of SARS-CoV-2 Spike protein at 78 fg mL−1.
The repeatability of the magneto-assay was analyzed using SARS-CoV-2 Spike protein in PBS standard solution at a concentration of 78 fg mL−1 and using different arrays. The relative standard deviation (RSD) obtained for the intra-day (n = 4) (Fig. 4C) and inter-day (n = 5) (Fig. 4D) analyses were 2.62% and 3.81%, respectively; these results point to the precision of the analytical method proposed in this study.
3.5. Analysis of saliva samples from the cohorts of SARS-CoV-2-free healthy and SARS-CoV-2-infected subjects using the magneto-assay
Sixteen saliva samples were collected from SARS-CoV-2–free healthy subjects (negative group) and sixteen saliva samples were collected from SARS-CoV-2 infected subjects (positive group). The saliva samples were diluted 1000 times in PBS-Ca-Mg aqueous solution before evaluating the analytical performance of the magneto-assay.
A comparative analysis was performed using the results obtained from the analysis of the saliva samples of SARS-CoV-2–free healthy subjects and SARS-CoV-2 infected subjects under the magneto-assay technique and the results obtained from the application of the RT-PCR technique using nasopharyngeal and oropharyngeal swab samples collected from the same subjects. The cycle threshold (Ct) values obtained from the RT-PCR test for the N1 and N2 genes were correlated with the subjects’ viral load. Overall, the subjects with higher viral loads (consequently, lower Ct values) in the nasopharyngeal and oropharyngeal swab samples also presented higher SARS-CoV-2 Spike protein levels in the magneto-assay ( Fig. 5A). These results clearly point to the satisfactory selectivity and specificity of the magneto-assay as an efficient analytical method for the detection of SARS-CoV-2 Spike protein in human saliva samples.
Fig. 5.
A) Peak current values obtained from the analysis of saliva samples from SARS-CoV-2-free healthy (negative) and SARS-CoV-2-infected (positive) subjects using the magneto-assay and RT-PCR Ct values for N1 and N2 genes of swab samples obtained from the same subjects. B) ROC curve for SARS-CoV-2 Spike protein detection using the magneto-assay (healthy vs. infected subjects). C) Interactive point diagram of peak current responses obtained for SARS-CoV-2 Spike protein in saliva samples for SARS-CoV-2-free healthy and SARS-CoV-2-infected subjects; * t-test, p < 0.0001.
The performance of the magneto-assay was evaluated in terms of discriminating healthy saliva samples from SARS-CoV-2-infected saliva samples. The analysis of the area under the receiver operating characteristic (ROC) curve demonstrated that the application of the magneto-assay technique yielded an area under the curve (AUC) of 0.996 (p < 0.001) (Fig. 5B) - which is clearly an excellent value [61]. It is worth noting that there are noticeable trade-offs between sensitivity and specificity; in other words, as sensitivity increases, specificity decreases and vice versa. A high sensitivity rules out the presence of the disease if a person's test is negative. Meanwhile, a test with high specificity confirms the presence of the disease if a person's test is positive [62]. The interactive dot diagram between the negative and positive subjects showed an ideal cutoff point of > 81 fg mL−1, based on the Youden index. This cut-off point differentiated saliva samples of SARS-CoV-2-free healthy subjects from those of SARS-CoV-2-infected subjects with sensitivity of 100.0% and specificity of 93.7% (Fig. 5C). These findings show that the magneto-assay proposed in this study has a good degree of accuracy comparable to that of the RT-PCR technique [63], [64], [65], [66].
One needs to point out that the use of saliva samples without thermal treatment may improve the efficiency of the magneto-assay through the application of a fully automated system, such as labs-on-chips or microfluidic devices [67], [68]. Heating saliva to high temperatures can denature the SARS-CoV-2 Spike protein and prevent its receptor-binding domain from interacting with the ACE2 peptide. Thus, the use of thermal treatment can be a limiting factor when analyzing human saliva samples under the method proposed in this study [69].
4. Conclusions
This work demonstrated that the magneto-assay proposed in the study is able to discriminate SARS-CoV-2-free healthy subjects from SARS-CoV-2-infected subjects; the results obtained showed the application potential of the proposed technique in terms of the successful diagnosis of SARS-CoV-2 infection. Compared to the RT-PCR detection technique, the magneto-assay was found to present outstanding advantages, which include the following: short analysis time (approximately 60 min); simple sample preparation without requiring long, tedious steps - such as RNA extractions, reserve transcription, and thermal cycling; and low-cost equipment. The outstanding advantages of the magneto-assay technique for the detection of SARS-CoV-2 make the detection method relevant for public health policy in the sense that the implementation of the technique can help expand the capacity to detect SARS-CoV-2 infection and be a useful aid in the fight against the spread of the COVID-19 disease.
CRediT authorship contribution statement
Ronaldo C. Faria, Vitor M. Faça, Henrique Pott-Junior, Felipe R. Teixeira; Conceptualization, Ronaldo C. Faria; Project administration, Ronaldo C. Faria.; Funding acquisition, Ronaldo C. Faria, Evair D. Nascimento, Wilson T. Fonseca and Tássia R. de Oliveira,; Method, Ronaldo C. Faria, Vitor M. Faça, Camila R. S. T. B. de Correia, Beatriz P. de Morais, Virginia C. Silvestrini, Henrique Pott-Junior and Felipe R. Teixeira; Resources, Evair D. Nascimento, Wilson T. Fonseca, Tássia R. de Oliveira, and Ronaldo C. Faria.; Investigation, Evair D. Nascimento, Wilson T. Fonseca, Tássia R. de Oliveira and Ronaldo C. Faria.; Writing, Ronaldo C. Faria.; Supervision.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Ronaldo Censi Faria reports was provided by State of Sao Paulo Research Foundation. Ronaldo Censi Faria reports a relationship with State of Sao Paulo Research Foundation that includes:. Ronaldo Censi Faria has patent #BR 10 2021 005125 6 issued to Assignee.
Acknowledgments
The authors thank Prof. Leda Castilho from COPPE/UFRJ (Universidade Federal do Rio do Janeiro) for the trimeric Spike protein of SARS-CoV-2 produced in HEK293 cells. This work was financially supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/Brazil ENDEMIAS Proc No 88881.504385/2020–01, CAPES Finance Code 001), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP/Brazil Proc. No. 2020/07539–6, 2020/04635–4); C.R.S.T.B.C (FAPESP: 2019/03943–0); F.R.T. (FAPESP: 2016/25798–3), V.M.F. (FAPESP: 2020/12920–0).
Biographies
Evair D. Nascimento received his graduation and M.Sc. from Federal University of sul e sudeste do Pará and currently, he is doctoral researcher in the Chemistry Department at Federal University of São Carlos, São Carlos, Brazil. Her works with the development of disposable electrochemical devices and analytical methods for the detection of biomarkers for disease diagnosis.
Wilson T. Fonseca is Postdoctoral Researcher at the Chemistry Department of the Federal University of São Carlos. In 2014 obtained B.S. Degree in Chemistry from the Federal University of Uberlândia (Ituiutaba, Brazil). In 2016 he received his M.Sc in Chemistry from the Federal University of Uberlândia (Uberlândia, Brazil). He received his PhD degree in Analytical Chemistry at the Federal University of São Carlos (São Carlos, Brazil). He works with the development of disposable electrochemical devices and analytical methods for the diagnosis of cancer and paper-based sensors with application in clinical and pharmaceutical areas.
Tássia R. de Oliveira received her M.Sc. from Federal University of Mato Grosso and PhD in Analytical Chemistry from Federal University of São Carlos. Currently, she is postdoctoral researcher fellow in the Chemistry Department at Federal University of São Carlos, São Carlos, Brazil. Her research focuses on the development of new electrochemical/electrochemiluminescence biosensors for disease diagnosis.
Camila R. S. T. B. de Correia is undergraduate student in Biological Science at Federal University of São Carlos. Has experience in the field of Biochemistry, with emphasis in Molecular Biology. Member of the Laboratory of Cellular Biochemistry (UFScar), as a Scientific initiation student with a FAPESP Scholarship, and previous CNPq scholarship in 2018–2019.
Vitor M. Faça Bachelor in Chemistry and Industrial Chemistry from the University of São Paulo (1995), Master (1998) and Doctorate (2002) in Biological Sciences (Molecular Biology) from the Federal University of São Paulo. Post-doctorate at the University of São Paulo (2003–2004) and worked as an associate researcher at the Fred Hutchinson Cancer Research Center (2004–2009) focused on developing strategies and applications of clinical proteomics in various types of cancer. He also served as a visiting researcher at Cornell University (2019) in the area of phosphoproteomics. It has extensive experience in protein chemistry and proteomic applications in biological problems.
Beatriz P. de Morais undergraduate student in biomedical sciences since 2018 at the University of São Paulo, participating in research with viral proteomics of SARS-CoV-2 in the laboratory of cancer proteomics at the Ribeirão Preto medical school.
Virginia C. Silvestrini received a degree in Biochemistry from the Federal University of São João Del Rei, Centro-Oeste Dona Lindu Campus and a M.Sc in Biochemistry from the University of São Paulo, Faculty of Medicine of Ribeirão Preto. Doctoral student in Biochemistry at the University of São Paulo, Faculty of Medicine of Ribeirão Preto. She is currently developing research in analisis proteomics and phosphoproteomics applied to investigation of cell survival and resistance mechanisms in Chronic Myeloid Leukemia.
Henrique Pott-Junior received his Doctorate in Infectious Diseases from the Federal University of São Paulo (UNIFESP), Brazil. Currently, he is a professor of the Department of Medicine at the Federal University de São Carlos (UFSCar), Brazil. His research projects are focused primarily on applying clinical epidemiological methods to study the relationship between infectious diseases and frailty among vulnerable populations. Moreover, he works on developing ultrasensitive immunosensors for the early detection of specific biomarkers in infectious diseases.
Felipe R. Teixeira received Bachelor in Biochemistry from the Federal University of Viçosa (2004), M.Sc. in Biochemistry from the Faculty of Medicine of Ribeirão Preto, University of São Paulo (FMRP/USP) (2007), Doctor in Biochemistry from FMRP/USP (2011) and Postdoctoral BEPE-FAPESP at the University of Cambridge under the supervision of Phd Heike Laman (2012–2013). He is currently Adjunct Professor I at the Department of Genetics and Evolution at UFSCar. He is currently working on the functional characterization of E3 ubiquitin ligases of the SCF type (SKP, Cullin, F-box) or CRLs (Cullin RING Ligases).
Ronaldo C. Faria received a Master degree in Chemistry and PhD in Analytical Chemistry from Federal University of São Carlos. Currently, he is an associate professor of Analytical Chemistry in the Department of Chemistry at Federal University of São Carlos, São Carlos, Brazil. His broad spectrum of research interests includes development of novel analytical instrumentation, electrochemical sensors and immunosensors and their application in determination of analytes in the fields of clinical, environmental, pharmacological, and food science. Currently, he is working on the development of ultrasensitive immunosensors for the detection of biomarkers in early diagnostics.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2021.131128.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO, COVID-19 Weekly Epidemiological Update 35, World Heal. Organ., 2021, pp. 1–3. https://www.who.int/docs/default-source/coronaviruse/situation-reports/weekly_epidemiological_update_22.pdf.
- 3.Axell-House D.B., Lavingia R., Rafferty M., Clark E., Amirian E.S., Chiao E.Y. The estimation of diagnostic accuracy of tests for COVID-19: a scoping review. J. Infect. 2020;81:681–697. doi: 10.1016/j.jinf.2020.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krupp K., Madhivanan P., Perez-Velez C.M. Should qualitative RT-PCR be used to determine release from isolation of COVID-19 patients? J. Infect. 2020 doi: 10.1016/j.jinf.2020.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., Tambe M., Mina M.J., Parker R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021;7:1–11. doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis — a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Q., Mu J., Lei Y., Ge J., Aryee A.A., Zhang X., Li Z. Simultaneous detection of the spike and nucleocapsid proteins from SARS-CoV-2 based on ultrasensitive single molecule assays. Anal. Bioanal. Chem. 2021:1–10. doi: 10.1007/s00216-021-03435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized tio2 nanotube-based electrochemical biosensor for rapid detection of sars-cov-2. Sens. (Switz. ) 2020;20:1–10. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16:1–19. doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idili A., Parolo C., Alvarez-Diduk R., Merkoçi A. Rapid and efficient detection of the SARS-CoV-2 Spike protein using an electrochemical aptamer-based sensor. ACS Sens. 2021;6:3093–3101. doi: 10.1021/acssensors.1c01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavrikou S., Tsekouras V., Hatziagapiou K., Paradeisi F., Bakakos P., Michos A., Koutsoukou A., Konstantellou E., Lambrou G.I., Koniari E., Tatsi E., Papaparaskevas J., Iliopoulos D., Chrousos G.P., Kintzios S. Clinical application of the novel cell-based biosensor for the ultra-rapid detection of the SARS-CoV-2 S1 Spike protein antigen. A Pract. Approach. 2021;2:1–14. doi: 10.3390/bios11070224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J.H., Choi M., Jung Y., Lee S.K., Lee C.S., Kim J., Kim J., Kim N.H., Kim B.T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-COV-2 S1 spike protein antigen. Sens. (Switz. ) 2020;20 doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Miranda Ferrari A., Rowley-Neale S.J., Banks C.E. Screen-printed electrodes: transitioning the laboratory in-to-the field. Talanta Open. 2021;3 doi: 10.1016/j.talo.2021.100032. [DOI] [Google Scholar]
- 17.Uliana C.V., de Oliveira T.R., Cominetti M.R., Faria R.C. Label-free evaluation of small-molecule–protein interaction using magnetic capture and electrochemical detection. Anal. Bioanal. Chem. 2019;411:2111–2119. doi: 10.1007/s00216-019-01636-1. [DOI] [PubMed] [Google Scholar]
- 18.Cortina M.E., Melli L.J., Roberti M., Mass M., Longinotti G., Tropea S., Lloret P., Serantes D.A.R., Salomón F., Lloret M., Caillava A.J., Restuccia S., Altcheh J., Buscaglia C.A., Malatto L., Ugalde J.E., Fraigi L., Moina C., Ybarra G., Ciocchini A.E., Comerci D.J. Electrochemical magnetic microbeads-based biosensor for point-of-care serodiagnosis of infectious diseases. Biosens. Bioelectron. 2016;80:24–33. doi: 10.1016/j.bios.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Kabe Y., Sakamoto S., Hatakeyama M., Yamaguchi Y., Suematsu M., Itonaga M., Handa H. Application of high-performance magnetic nanobeads to biological sensing devices. Anal. Bioanal. Chem. 2019;411:1825–1837. doi: 10.1007/s00216-018-1548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews W.T., Skube S.B., Hummon A.B. Magnetic bead-based peptide extraction methodology for tissue imaging. Analyst. 2018;143:133–140. doi: 10.1039/c7an00757d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S.E., Van Tieu M., Hwang S.Y., Lee M.H. Magnetic particles: Their applications from sample preparations to biosensing platforms. Micromachines. 2020;11:1–20. doi: 10.3390/mi11030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandhu A., Handa H., Abe M. Synthesis and applications of magnetic nanoparticles for biorecognition and point of care medical diagnostics. Nanotechnology. 2010;21 doi: 10.1088/0957-4484/21/44/442001. [DOI] [PubMed] [Google Scholar]
- 23.Quesada-González D., Merkoçi A. Nanomaterial-based devices for point-of-care diagnostic applications. Chem. Soc. Rev. 2018;47:4697–4709. doi: 10.1039/c7cs00837f. [DOI] [PubMed] [Google Scholar]
- 24.Draz M.S., Shafiee H. Applications of gold nanoparticles in virus detection. Theranostics. 2018;8:1985–2017. doi: 10.7150/thno.23856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing X., Cao X., Wang L., Lan T., Li Y., Xie G. DNA-AuNPs based signal amplification for highly sensitive detection of DNA methylation, methyltransferase activity and inhibitor screening. Biosens. Bioelectron. 2014;58:40–47. doi: 10.1016/j.bios.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 26.Liu P., Li C., Zhang R., Tang Q., Wei J., Lu Y., Shen P. An ultrasensitive electrochemical immunosensor for procalcitonin detection based on the gold nanoparticles-enhanced tyramide signal amplification strategy. Biosens. Bioelectron. 2019;126:543–550. doi: 10.1016/j.bios.2018.10.048. [DOI] [PubMed] [Google Scholar]
- 27.Liu G., Liu J., Davis T.P., Gooding J.J. Electrochemical impedance immunosensor based on gold nanoparticles and aryl diazonium salt functionalized gold electrodes for the detection of antibody. Biosens. Bioelectron. 2011;26:3660–3665. doi: 10.1016/j.bios.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Quinchia J., Echeverri D., Cruz-Pacheco A.F., Maldonado M.E., Orozco J.A. Electrochemical biosensors for determination of colorectal tumor biomarkers. Micromachines. 2020;11:1–46. doi: 10.3390/MI11040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordeiro M., Carlos F.F., Pedrosa P., Lopez A., Baptista P.V. Gold nanoparticles for diagnostics: Advances towards points of care. Diagnostics. 2016;6 doi: 10.3390/diagnostics6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvim R.G.F., Lima T.M., Rodrigues D.A.S., Marsili F.F., Bozza V.B.T., Higa L.M., Monteiro F.L., Abreu D.P.B., Leitão I.C., Carvalho R.S., Galliez R.M., Castineiras T.M.P.P., Nobrega A., Travassos L.H., Tanuri A., Ferreira O.C., Vale A.M., Castilho L.R. An affordable anti-SARS-COV-2 spike protein ELISA test for early detection of IgG seroconversion suited for large-scale surveillance studies in low-income countries. MedRxiv. 2020:1–19. doi: 10.1101/2020.07.13.20152884. [DOI] [Google Scholar]
- 32.Il Kim Y., Casel M.A.B., Kim S.M., Kim S.G., Park S.J., Kim E.H., Jeong H.W., Poo H., Choi Y.K. Development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) thermal inactivation method with preservation of diagnostic sensitivity. J. Microbiol. 2020;58:886–891. doi: 10.1007/s12275-020-0335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kampf G., Voss A., Scheithauer S. Inactivation of coronaviruses by heat. J. Hosp. Infect. 2020;105:348–349. doi: 10.1016/j.jhin.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonso A.S., Pérez-López B., Faria R.C., Mattoso L.H.C., Hernández-Herrero M., Roig-Sagués A.X., Maltez-da Costa M., Merkoçi A. Electrochemical detection of Salmonella using gold nanoparticles. Biosens. Bioelectron. 2013;40:121–126. doi: 10.1016/j.bios.2012.06.054. [DOI] [PubMed] [Google Scholar]
- 36.Ibrahim H.M., Reda M.M., Klingner A. Preparation and characterization of green carboxymethylchitosan (CMCS) – Polyvinyl alcohol (PVA) electrospun nanofibers containing gold nanoparticles (AuNPs) and its potential use as biomaterials. Int. J. Biol. Macromol. 2020;151:821–829. doi: 10.1016/j.ijbiomac.2020.02.174. [DOI] [PubMed] [Google Scholar]
- 37.Prasad S., Mandal I., Singh S., Paul A., Mandal B., Venkatramani R., Swaminathan R. Near UV-Visible electronic absorption originating from charged amino acids in a monomeric protein. Chem. Sci. 2017;8:5416–5433. doi: 10.1039/c7sc00880e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moris S., Silva N., Saitz C., Jara P., Chornik B. Nanodecoration of single crystals of 5,11,17,23-tetra-tert-butyl-25,27- bis(cyanomethoxy)-26,28-dihydroxycalix[4]arene. J. Chil. Chem. Soc. 2017;62:3772–3778. doi: 10.4067/s0717-97072017000403772. [DOI] [Google Scholar]
- 39.Zhang L., Xu C., Song G., Li B. Self-assembly of l-cysteine-gold nanoparticles as chiral probes for visual recognition of 3,4-dihydroxyphenylalanine enantiomers. RSC Adv. 2015;5:27003–27008. doi: 10.1039/c5ra01271f. [DOI] [Google Scholar]
- 40.Acres R.G., Feyer V., Tsud N., Carlino E., Prince K.C. Mechanisms of aggregation of cysteine functionalized gold nanoparticles. J. Phys. Chem. C. 2014;118:10481–10487. doi: 10.1021/jp502401w. [DOI] [Google Scholar]
- 41.Hormozi-Nezhad M.R., Seyedhosseini E., Robatjazi H. Spectrophotometric determination of glutathione and cysteine based on aggregation of colloidal gold nanoparticles. Sci. Iran. 2012;19:958–963. doi: 10.1016/j.scient.2012.04.018. [DOI] [Google Scholar]
- 42.Boussoufi F., Gallón S.M.N., Chang R., Webster T.J. Synthesis and study of cell-penetrating peptide-modified gold nanoparticles. Int. J. Nanomed. 2018;13:6199–6205. doi: 10.2147/IJN.S168720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majzik A., Fülöp L., Csapó E., Bogár F., Martinek T., Penke B., Bíró G., Dékány I. Functionalization of gold nanoparticles with amino acid, β-amyloid peptides and fragment. Colloids Surf. B Biointerfaces. 2010;81:235–241. doi: 10.1016/j.colsurfb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Fonseca W.T., Cincotto F.H., Lourencao B.C., de Almeida S.V., Moraes F.C., Fatibello-Filho O., de Carvalho A.C., Carvalho A.L., Melendez M.E., Faria R.C. Ultrasensitive Magnetogenoassay for Detection of microRNA for Diagnosis of Metastatic Lymph Nodes in Head and Neck Cancer Using Disposable Electrodes. Sens. Actuators B Chem. 2021;352 doi: 10.1016/j.snb.2021.131040. [DOI] [Google Scholar]
- 45.de la Escosura-Muñiz A., Maltez-da Costa M., Sánchez-Espinel C., Díaz-Freitas B., Fernández-Suarez J., González-Fernández Á., Merkoçi A. Gold nanoparticle-based electrochemical magnetoimmunosensor for rapid detection of anti-hepatitis B virus antibodies in human serum. Biosens. Bioelectron. 2010;26:1710–1714. doi: 10.1016/j.bios.2010.07.069. [DOI] [PubMed] [Google Scholar]
- 46.Shittu K.O., Bankole M.T., Abdulkareem A.S., Abubakre O.K., Ubaka A.U. Application of gold nanoparticles for improved drug efficiency. Adv. Nat. Sci. Nanosci. Nanotechnol. 2017;8 doi: 10.1088/2043-6254/aa7716. [DOI] [Google Scholar]
- 47.Lu J., Sun P.D. High affinity binding of SARS-cov-2 spike protein enhances ACE2 carboxypeptidase activity. J. Biol. Chem. 2020;295:18579–18588. doi: 10.1074/jbc.RA120.015303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D.M.E., van der Voort P.H.J., Mulder D.J., van Goor H. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonso A.S., Zanetti B.F., Santiago A.C., Henrique-Silva F., Mattoso L.H.C., Faria R.C. QCM immunoassay for recombinant cysteine peptidase: A potential protein biomarker for diagnosis of citrus canker. Talanta. 2013;104:193–197. doi: 10.1016/j.talanta.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Kämäräinen S., Mäki M., Tolonen T., Palleschi G., Virtanen V., Micheli L., Sesay A.M. Disposable electrochemical immunosensor for cortisol determination in human saliva. Talanta. 2018;188:50–57. doi: 10.1016/j.talanta.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 53.Frenkel E.S., Ribbeck K. Salivary mucins in host defense and disease prevention. J. Oral. Microbiol. 2015;7:29759. doi: 10.3402/jom.v7.29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.John T., Abel B., Martin L.L. The quartz crystal microbalance with dissipation monitoring (QCM-D) technique applied to the study of membrane-active peptides. Aust. J. Chem. 2018;71:543–546. doi: 10.1071/CH18129. [DOI] [Google Scholar]
- 55.Mercurio I., Tragni V., Busto F., De Grassi A., Pierri C.L. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. Cell. Mol. Life Sci. 2021;78:1501–1522. doi: 10.1007/s00018-020-03580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pumera M., Aldavert M., Mills C., Merkoçi A., Alegret S. Direct voltammetric determination of gold nanoparticles using graphite-epoxy composite electrode. Electrochim. Acta. 2005;50:3702–3707. doi: 10.1016/j.electacta.2005.01.035. [DOI] [Google Scholar]
- 57.Zhu M., Tang Y., Wen Q., Li J., Yang P. Dynamic evaluation of cell-secreted interferon gamma in response to drug stimulation via a sensitive electro-chemiluminescence immunosensor based on a glassy carbon electrode modified with graphene oxide, polyaniline nanofibers, magnetic beads, and gold n. Microchim. Acta. 2016;183:1739–1748. doi: 10.1007/s00604-016-1804-9. [DOI] [Google Scholar]
- 58.AFONSO A.S., ULIANA C.V., MARTUCCI D.H., FARIA R.C. Simple and rapid fabrication of disposable carbon-based electrochemical cells using an electronic craft cutter for sensor and biosensor applications. Talanta. 2016;146:381–387. doi: 10.1016/j.talanta.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hussein H.A., El Nashar R.M., El-Sherbiny I.M., Hassan R.Y.A. High selectivity detection of FMDV- SAT-2 using a newly-developed electrochemical nanosensors. Biosens. Bioelectron. 2021;191 doi: 10.1016/J.BIOS.2021.113435. [DOI] [PubMed] [Google Scholar]
- 61.Mandrekar J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010;5:1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 62.Zweig M.H., Campbell G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993;39:561–577. doi: 10.1093/clinchem/39.4.561. [DOI] [PubMed] [Google Scholar]
- 63.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin. Microbiol. Infect. 2020;2019 doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Padhye N.S. Reconstructed diagnostic sensitivity and specificity of the RT-PCR test for COVID-19. MedRxiv. 2020 doi: 10.1101/2020.04.24.20078949. [DOI] [Google Scholar]
- 65.Böger B., Fachi M.M., Vilhena R.O., Cobre A.F., Tonin F.S., Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control. 2021;49:21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babady N.E., McMillen T., Jani K., Viale A., Robilotti E.V., Aslam A., Diver M., Sokoli D., Mason G., Shah M.K., Korenstein D., Kamboj M. Performance of Severe Acute Respiratory Syndrome Coronavirus 2 Real-Time RT-PCR Tests on Oral Rinses and Saliva Samples. J. Mol. Diagn. 2021;23:3–9. doi: 10.1016/j.jmoldx.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian F., Liu C., Deng J., Han Z., Zhang L., Chen Q., Sun J. A fully automated centrifugal microfluidic system for sample-to-answer viral nucleic acid testing. Sci. China Chem. 2020;63:1498–1506. doi: 10.1007/s11426-020-9800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arshavsky-Graham S., Segal E. Lab-on-a-Chip Devices for Point-of-Care Medical. Diagnostics. 2020 doi: 10.1007/10_2020_127. [DOI] [PubMed] [Google Scholar]
- 69.Rath S.L., Kumar K. Investigation of the effect of temperature on the structure of SARS-Cov-2 Spike Protein by Molecular Dynamics Simulations. BioRxiv. 2020 doi: 10.1101/2020.06.10.145086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material