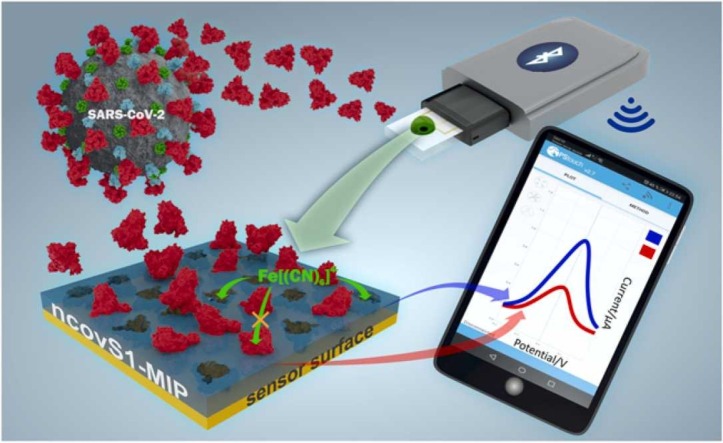
Abstract
The continued spread of the coronavirus disease and prevalence of the global pandemic is exacerbated by the increase in the number of asymptomatic individuals who unknowingly spread the SARS-CoV-2 virus. Although remarkable progress is being achieved at curtailing further rampage of the disease, there is still the demand for simple and rapid diagnostic tools for early detection of the COVID-19 infection and the following isolation. We report the fabrication of an electrochemical sensor based on a molecularly imprinted polymer synthetic receptor for the quantitative detection of SARS-CoV-2 spike protein subunit S1 (ncovS1), by harnessing the covalent interaction between 1,2-diols of the highly glycosylated protein and the boronic acid group of 3-aminophenylboronic acid (APBA). The sensor displays a satisfactory performance with a reaction time of 15 min and is capable of detecting ncovS1 both in phosphate buffered saline and patient’s nasopharyngeal samples with LOD values of 15 fM and 64 fM, respectively. Moreover, the sensor is compatible with portable potentiostats thus allowing on-site measurements thereby holding a great potential as a point-of-care testing platform for rapid and early diagnosis of COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2 spike protein, Molecularly imprinted polymer, Covalent imprinting, Electrochemical sensor, antigen test
Graphical Abstract
1. Introduction
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has strongly encouraged the search for highly sensitive and rapid diagnostic tools. It was found that SARS-CoV-2 is characterized by a high risk for silent spread by asymptomatic persons accounting for a significant percentage of 40–45% of the total infected cases [1]. Therefore, in order to timely detect and isolate the cases and their contacts, the rapid and easy-to-perform diagnostic tools enabling the testing of large numbers of samples in a short period of time are highly demanded.
Currently, molecular assays, i.e. real-time polymerase chain reaction (RT-PCR) has become the diagnostic workhorse in the detection of COVID-19, being able to accurately identify viral RNA. Unfortunately, due to the time-consuming analysis, instrumentation costs, and the need for trained personnel, the use of RT-PCR is limited when the rapid response is needed or for screening purposes. In addition, it was found that due to the high sensitivity of PCR for fragments of viral RNA, the test could be positive for a prolonged period of time in patients previously recovered from COVID-19, but are not associated with effective infectiousness [2], [3], [4]. Therefore, rapid point-of-care diagnostic tests detecting SARS-CoV-2 specific antigen, i.e. a viral protein such as nucleocapsid (N) or spike (S), providing the possibility of early detection of infectious COVID-19 cases, have recently been developed and many of them are now commercially available [5]. Most of these tests are qualitative lateral flow immunochromatographic assays utilizing the antibody-based detection principle and producing results in around 15–30 min. Alternatively, point-of-care diagnostic tools based on electrochemical sensing platforms have been reported [6]. Thus, two papers reported on field-effect transistor (FET)-based biosensors for detecting SARS-CoV-2 S protein with a limit of detection (LOD) of 1 fg/ml in phosphate buffered saline (PBS) [7], [8]. Also, researchers from Denmark developed a graphene-functionalized screen-printed electrode (SPE) immunosensor capable of detecting SARS-CoV-2 S1 subunit at the lowest concentration of 20 µg/ml [9]. Fabiani L. et al. reported on an electrochemical biosensor for SARS-CoV-2 detection in saliva using a magnetic beads-based electrochemical assay combined with carbon black-based SPE sensor [10]. The biosensor was capable of detecting S and N proteins in untreated saliva with LOD of 19 ng/ml and 8 ng/ml, respectively. However, similar to the lateral flow tests, all these electrochemical biosensors utilize a biological receptor i.e. an antibody specific to N or S protein, as a molecular recognition element. This suggests that such sensors would demand a special storage system to preserve their shelf life due to the environmental sensitivity of the biological materials.
Consequently, there is a growing interest in the replacement of labile and expensive biological receptors with a synthetic analogue such as molecularly imprinted polymer (MIP). MIPs are prepared by the molecular imprinting technology that can be defined as the process of template-induced formation of specific molecular recognition sites in a polymer matrix [11]. MIPs represent materials with antibody-like ability to bind and discriminate between molecules. The main benefits of MIPs are related to their synthetic nature, i.e. excellent chemical and thermal stability coupled with their reproducible and cost-effective fabrication. MIPs have been shown to be a promising alternative to biological receptors in biosensors [12], [13], [14], [15]. MIP-based sensors have also been investigated for the detection of viral proteins such as bovine leukemia virus glycoprotein gp51 [16], dengue virus NS1 protein [17], glycoprotein of HIV type 1 [18]. Up to date there are only a few reports on MIP sensors for SARS-CoV-2 viral proteins detection. Cennamo N. et al. reported on a proof of concept for a plasmonic optical fiber sensor coupled with MIP nanolayer and capable of selectively recognizing SARS-CoV-2 S1 subunit [19]. Also, MIP Diagnostics (Colworth Park, Sharnbrook Bedford, UK) has developed a COVID-19 nanoMIP as a synthetic receptor for SARS-CoV-2 S protein that is suitable for application in diagnostics assays.
Very recently, our research group developed a MIP-based electrochemical sensor for detection of SARS-CoV-2 N protein (ncovNP) [20]. Although the sensor demonstrated an outstanding performance, a recent report indicates that the use of N protein as an immunogenic target may lead to false positive results [24], hence the need to develop such a sensor using other protein targets becomes imperative. SARS-CoV-2 S protein is the transmembrane glycosylated protein that forms spikes protruding from the virus envelope. S protein has two domains, named S1 and S2. The S1 subunit contains the receptor binding domain (RBD), which is responsible for host cell receptor binding, while the S2 subunit facilitates membrane fusion between the viral and host cell membranes. In contrast to ncovNP that is highly conserved, S protein, especially its S1 subunit, is more variable sharing around 70% identity with the respective SARS-CoV and could potentially demonstrate less cross-reactivities with other coronaviruses [21], [22], [23]. Therefore, SARS-CoV-2 S protein represents a key target for vaccines, therapeutic antibodies as well as diagnostics. Interestingly, due to the unique feature of molecular imprinting allowing its applicability to a wide variety of target analytes, MIP synthesis strategy can be easily adapted to prepare synthetic receptors against other viral proteins, e.g. SARS-CoV-2 S protein.
In this work we report on the development of an electrochemical sensor for rapid detection of SARS-CoV-2 S protein, where the disposable thin-film metal electrodes (Au-TFME) chip is modified with a MIP film endowed with the selectivity for S protein subunit S1 (ncovS1) and used as a recognition element. The chip is connected to a potentiostat, which measures the ncovS1-specific reduction in the intensity of the charge transfer carried by a redox probe through the MIP film. The performance and selectivity of the sensor was studied in both buffer and in COVID-19 patients' nasopharyngeal swab samples.
2. Experimental
2.1. Material and method
The following reagents were purchased from Sigma-Aldrich including dithiothreitol, DTT (Ottawa, Canada), 4-aminothiophenol, 4-ATP (Hong-Kong, China), sodium fluoride, NaF (Kolkata, India), Immunoglobulin G, IgG and human serum albumin, HSA (St. Louis, MO, USA). 3-aminophenylboronic acid (APBA) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Ethanol (96%) was purchased from Estonian Spirit OÜ (Tallinn, Estonia). 3,3′-dithiobis [sulfosuccinimidyl propionate] (DTSSP) was purchased from Thermo Fisher Scientific Inc. (Rockford, IL, USA) and acetic acid was purchased from Lach-ner (Neratovice, Czechia). Potassium ferricyanide and ferrocyanide were purchased from Riedel-de Haen (Seelze, Germany). MicruX™ gold-based thin-film metal electrodes (Au-TFME) were purchased from Micrux Technologies (Gijón, Spain). SARS-CoV-2 nucleoprotein (ncovNP), spike protein subunit S1 (ncovS1) and its different strains (S1 UK VOC 202012/01, S1 Brazil P1 and S1 South Africa VOC 501.V2) were supplied by Icosagen AS (Tartu, Estonia). Hepatitis C virus (HCV) surface viral antigen (E2) was obtained from the Institute of Macromolecular Compounds of the Russian Academy of Sciences. All chemicals were of analytical grade and were used as received without any further purification. Ultrapure Milli-Q water (resistivity 18.2 MΩ cm at 25 °C, Merck KGaA, Darmstadt, Germany) was used in preparing all aqueous solutions. PBS pH 7.4 (Na2HPO4 10 mM, KH2PO4 1.8 mM, NaCl 137 mM, KCl 2.7 mM) was used to prepare the synthesis and analyte solutions.
2.2. Sensor preparation
The ncovS1 sensor was prepared by synthesizing ncovS1-MIP film directly on Au-TFME adapting the surface imprinting strategy previously developed by our group [25], [26]. Prior to the modification, the Au-TFME was cleaned in ozone for 15 min followed by washing with ethanol, rinsing with MQ and then drying under a nitrogen atmosphere. Modification of the working electrode of Au-TFME was achieved by incubating it for 30 min in 100 mM 4-ATP ethanolic solution and vortexing for 5 min to remove loosely bound ATP molecules. A monolayer of cleavable linker was obtained via the covalent attachment of DTSSP to the ATP modified Au-TFME by drop casting 10 µL (10 mM) DTSSP solution in PBS for 30 min followed by washing with PBS. ncovS1 was immobilized on the DTSSP/ATP-modified Au-TFME by dropping 3 µL of PBS containing 0.33 µM of ncovS1 for 30 min and washed severally with PBS. Poly(3-aminophenylboronic acid), PAPBA was synthesized on the ncovS1-modified Au-TFME in a set-up consisting of an electrochemical cell (ED-AIO-Cell, Micrux Technologies, Spain) connected with the electrochemical workstation (Reference 600TM, Gamry Instruments, USA). PAPBA was electrodeposited from a PBS containing 20 mM APBA and 50 mM NaF by cycling the potential between − 0.2–0.9 V vs Ag/AgCl/KCl. Imprints of ncovS1 were generated in the polymer film by cleaving the S-S bond of DTSSP using 50 mM dithiothreitol (DTT) solution for 30 min, followed by washing for another 30 min in 10% acetic acid to remove the ncovS1. The non-imprinted polymer (NIP) film as reference, was prepared using a similar protocol but without treatment in DTT to preserve the covalently attached ncovS1 in the polymer thereby avoiding the formation of the target imprint within the matrix.
Each step of the sensor preparation was characterized by cyclic voltammetry (CV) in the potential range of −0.2 to 0.2 V at a scan rate of 100 mV/s and square wave voltammetry (SWV) at a potential range of −0.2 to 0.2 V, pulse amplitude of 12.5 mV, frequency of 10 Hz, and a step potential of 5 mV in 1 M KCl solution containing 4 mM redox probe K3[Fe(CN)6]/K4[Fe(CN)6].
2.3. Evaluation of the sensor performance
The rebinding of ncovS1 by the sensor was measured by square wave voltammetry (SWV) in a 1 M KCl solution containing 4 mM redox probe K3[Fe(CN)6]/K4[Fe(CN)6]. The measurements were performed in an electrochemical cell (ED-AIO-Cell, Micrux Technologies, Spain) connected with the electrochemical workstation (Reference 600TM, Gamry Instruments, USA) or a portable potentiostat (EmStat3 Blue, PalmSens BV, The Netherlands). The measurement parameters include a potential range of − 0.2–0.2 V vs open circuit potential (OCP) provided by the pseudo reference Au electrode of the TFME, pulse amplitude of 12.5 mV, frequency of 10 Hz, and a step potential of 5 mV. The sensor’s response to analyte rebinding (I n) was calculated using Eq. 1:
| In = (I0−I)/I0) | (1) |
where I and I 0 are the SWV anodic current peaks measured following sensor incubation in solution containing and without ncovS1, respectively.
The thickness of the sensor was optimized (See section S3) and the optimized thickness was further used in evaluating the sensor performance, both in buffer and in clinical samples. Analytical limits, including the limit of detection (LOD), Eq. 2 and limit of quantification (LOQ), Eq. 3 were determined from the linear regression of the sensor’s response to increasing ncovS1 concentrations in both PBS and in clinical samples of negative COVID-19 patients:
| LOD = 3∙SD/b | (2) |
| LOQ = 10∙SD/b | (3) |
where SD and b represent the standard deviation and the slope of the regression line, respectively.
Sensor selectivity was evaluated by comparing the rebinding of ncovS1 to those of other proteins such as ncovNP (45 kDa, pI 10.07), E2 (47 kDa, pI 8.24), HSA (67 kDa, pI 4.7), and IgG (152 kDa, pI 8.8), at the same concentrations.
The clinical samples used in this work were obtained from SYNLAB Eesti medical laboratory (Estonia). They consist of nasopharyngeal specimens of three negative and five positive COVID-19 patients in sample preservation solution (SPS), (Jiangsu Mole Bioscience Co., Ltd). Their COVID-19 status was previously confirmed with the RT-PCR method. The samples were diluted in PBS (1:99), (see section S5 for details). Calibration plot was made using the sensor’s responses to diluted negative samples spiked with an increasing concentration (0–400 fM) of ncovS1.
3. Results and Discussion
3.1. Strategy of molecular recognition and sensor operating principle
MIP recognition relies on retaining within a polymer the memory of the physicochemical interactions generated between the target molecules and the functional monomer prior to polymerization. Herein, to ensure the high selectivity of MIP synthetic receptor (ncovS1-MIP) towards ncovS1 antigen the covalent molecular imprinting approach was applied. The approach relies on the formation of reversible covalent bonds between 1,2-diols of highly glycosylated ncovS1 and boronic acid groups containing functional monomers such as APBA, (see Fig. S1a). Covalent imprinting, being stoichiometric, ensures that functional monomer residues exist only in the imprinted cavities thereby avoiding any non-specific binding and yielding a homogeneous binding sites distribution. Such interaction has been exploited in fabricating sensors for detecting different analytes including small and large biomolecules as well as whole cells [27], [28], [29], [30].
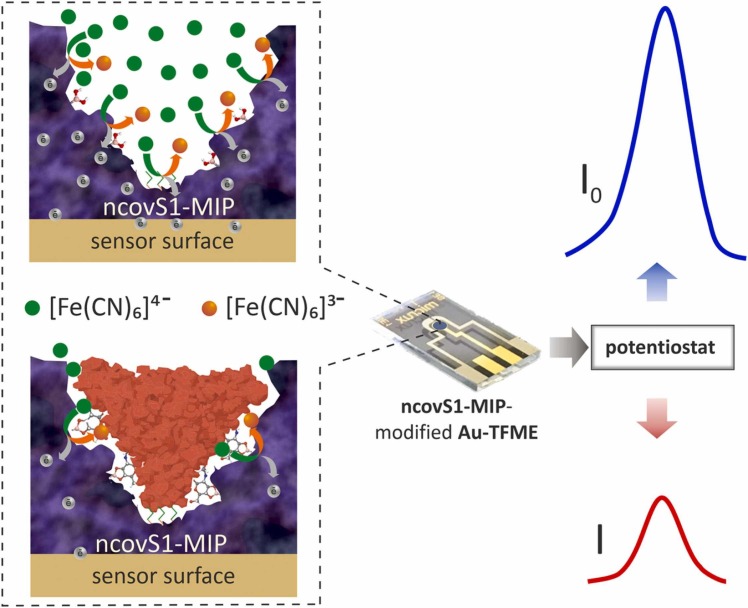
The detection principle of the sensor relies on the measurement of the changes in charge transfer between the Au-TFME and [Fe(CN)6]3-/[Fe(CN)6]4- redox probe, through the imprinted pathways created within ncovS1-MIP film. Upon ncovS1 rebinding following sensor incubation in analyte solution, the charge transfer is greatly obstructed by the non-conductive protein thus, leading to a concentration dependent contraction in the recorded current peak ( Fig. 1).
Fig. 1.
The operating principle of ncovS1 sensor in COVID-19 diagnosis: a) redox probe readily carrying the charge through ncovS1-MIP producing current I0, b) the rebound ncovS1 blocks pathways for redox probe to carry the charge through ncovS1-MIP leading to a concentration dependent contraction in the recorded current I.
3.2. Sensor preparation and performance evaluation
The preparation of ncovS1 sensor follows a surface imprinting strategy described in Section 2.2. The characterization of each preparation step was measured by CV and SWV (Section S2) to confirm successful formation of ncovS1-imprinted film (ncovS1-MIP) on Au-TFME. It is essential to grow the polymer film with the optimal thickness to ensure efficient removal of the protein during the washing step to allow the formation of permeable protein-specific pathways that, upon target analyte rebinding, can selectively be plugged thus restricting charge exchange between the redox probe and Au-TFME (Fig. S1b). With such tailor-made pathways created within the polymer film, it is expected that the sensor will respond selectively after encountering ncovS1. Consequently, ncovS1-MIP thickness was optimized considering both the number of CV scans during electrosynthesis and the corresponding response of the sensor after ncovS1 rebinding. It was found that ncovS1-MIP film electrodeposited from 10 CV cycles endowed the sensor with the best performance and was used for further analysis (See section S3). Furthermore, optimization of the time for rebinding ncovS1 on the sensor indicated that at 15 min a complete rebinding of the protein is achieved as the sensor’s response remains unchanged subsequently (Fig. S5). Hence, 15 min was chosen as the optimal rebinding time. This, when combined with the measurement time, amounts to a total testing duration of about 20 min, that is comparable with other reported electrochemical SARS-CoV-2 antigen tests (Table S3) as well as commercially available lateral flow assays with an average testing time of 15–30 min [5].
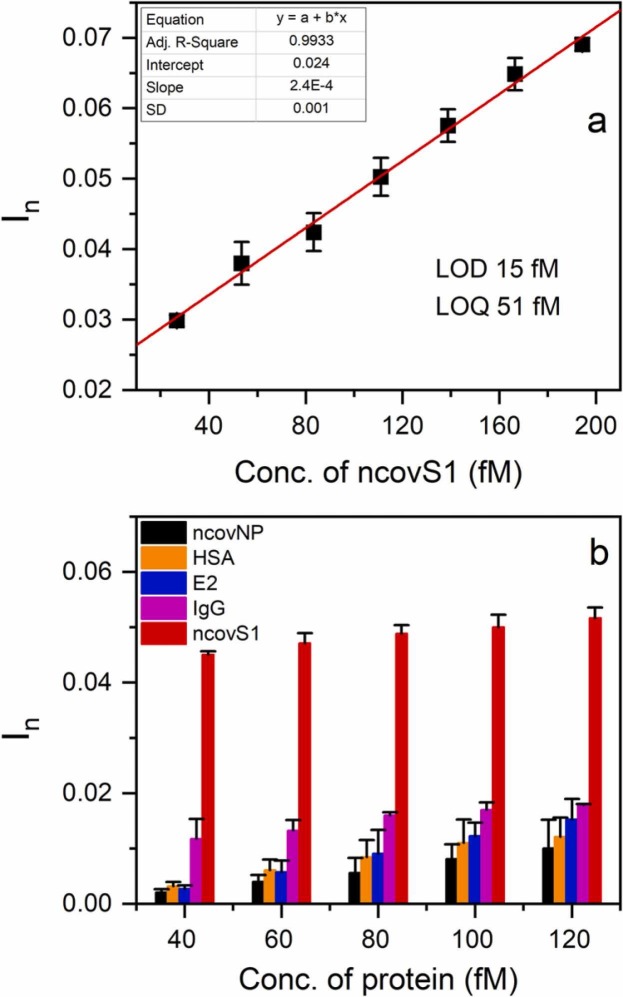
Initially, the sensor was characterized using a series of PBS containing different concentrations of ncovS1. Fig. 2(a) shows that its response linearly rises with ncovS1 concentration up to about 200 fM. LOD and LOQ were obtained as 15 fM (1.12 pg/ml) and 51 fM (3.82 pg/ml) respectively. These detection limits are remarkable since ncovS1 concentration in the nasopharyngeal samples of COVID-19 patients, estimated from the reported amount of RNAs observed (106-109 RNAs/swab) following diagnosis [31], ranges from 0.02 to 18.7 ng/ml. When compared to other electrochemical sensor platforms for SARS-CoV-2 detection reported in the literature (Table S3), ncovS1 sensor shows an interesting performance.
Fig. 2.
(a) Calibration plot of ncovS1 sensor at the low concentration range (26.7–194 fM) of ncovS1 in PBS (b) Selectivity of ncovS1 sensor against increasing concentrations (40, 60, 80, 100, and 120 fM) of different proteins (ncovNP, HSA, E2, IgG and S1) in PBS.
To further characterize the performance of the sensor in PBS, we examined its capability to recognize ncovS1 as compared to other proteins. Thus, four proteins including ncovNP, E2, IgG and HSA (see Fig. S6) were selected. While ncovNP belong to SARS-CoV-2 virus, with an almost certainty of being found together with ncovS1 in the patient sample, E2 was chosen to demonstrate the sensor discrimination against another viral protein. Other proteins were selected either due to the possession of a high glycosylation (i.e IgG), thus making it a potential competitor with the target for the binding sites; or due to a quite similar molecular weight (i.e HSA).
As observed in Fig. 2(b), ncovS1 sensor demonstrated significantly higher responses to ncovS1 than all other proteins. It is important to note that although IgG is a highly glycosylated protein with 2 times larger molecular weight, the sensor was still able to clearly discriminate ncovS1 from it. This ascertains the significance of the imprinted sites to the sensor’s selective recognition of ncovS1. Moreover, the much lower responses displayed towards ncovNP, E2 and HSA indicates the insensitivity of the sensor to either molecular weight or isoelectric point thus revealing that the driving force behind the sensor recognition of ncovS1 is neither size exclusion nor electrostatic attraction [32]. This gives credence to the influence of the covalent imprinting approach for synthesizing ncovS1-MIP with high selectivity towards ncovS1.
3.3. Evaluation of sensor performance in clinical samples
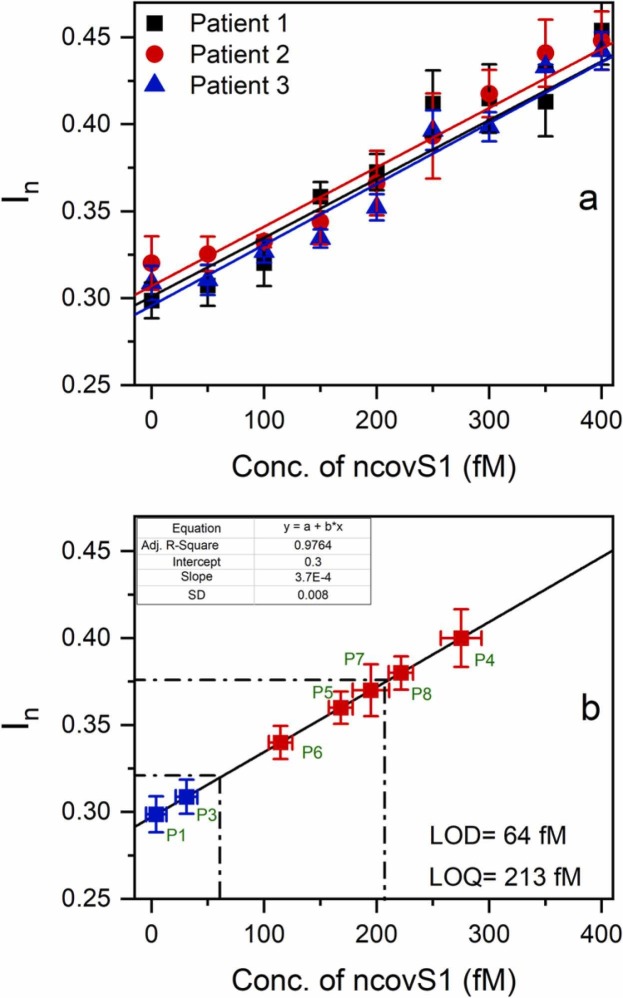
To evaluate the clinical usability of the sensor, we studied its performance in the samples prepared from nasopharyngeal swab specimens of patients. For this purpose, initial analysis of COVID-19 negative samples spiked with increasing concentrations of ncovS1 was carried out. As it can be seen in Fig. 3(a) (see Fig. S8 for the square wave voltammograms), linearity between concentration and response was achieved within a concentration range of 0–400 fM. The averaged data obtained from three negative samples were then used in plotting a calibration graph of response vs concentration and the LOD and LOQ were determined as 64 fM and 213 fM respectively, using (2), (3). Accordingly, we proposed that samples with ncovS1 concentration exceeding 64 fM (In >0.32) could be considered to be COVID-19 positive.
Fig. 3.
(a) The calibration plots of ncovS1 sensor obtained against COVID-19 negative nasopharyngeal swab samples in SPS solution. Samples were diluted with PBS (1:99) and spiked with 50–400 fM (3.75–30 pg/ml) of ncovS1, (b) The calibration plot (solid line) of ncovS1 sensor obtained by linear regression of the averaged data in Fig. 3(a). The red squares represent data points corresponding to In measured by ncovS1 sensor against COVID-19 positive samples while the blue squares are those of negative samples. The error bars represent SDs of three measurements.
Thus, five COVID-19 positive samples which had been previously validated by RT-PCR under different cycle thresholds (ct), were tested with the ncovS1 sensor. Fig. 3(b) clearly shows that all five samples effected responses that are higher (I n = 0.4, 0.36, 0.34, 0.37, 0.38) than LOD (I n = 0.32), Table 1. While samples from patients 5 and 7 produced responses close to the LOQ value, samples of patients 4 and 8 show higher responses than LOQ. The sample from patient 6 caused the lowest response, which still established a significant distinction from the LOD. Moreover, it is worthy to note that the negative samples are below the LOD. This affirms that the increased responses received from the positive patients are due to the presence of ncovS1 in the samples.
Table 1.
ncovS1 sensor responses (In) to COVID-19 positive samples and the associated ncovS1 concentration.
| Patient number (cta) | ncovS1 sensor response (In) | ncovS1 conc. (fM) |
|---|---|---|
| Patient 4 (16) | 0.40 ± 0.02 | 280 ± 20 |
| Patient 5 (26) | 0.36 ± 0.01 | 170 ± 10 |
| Patient 6 (19) | 0.34 ± 0.01 | 120 ± 10 |
| Patient 7 (12) | 0.37 ± 0.02 | 200 ± 20 |
| Patient 8 (10) | 0.38 ± 0.01 | 220 ± 10 |
cycle threshold value as determined by RT-PCR
The variation observed in the sensor response to the different samples might be due to a number of factors. Prominent among these could be related to the difference in viral load, which has a strong correlation to the viral protein concentration, at different stages of the life cycle of a SARS-CoV-2 infection. Studies have indicated an early peak in viral load in the upper respiratory tract, within the first week of illness [33]. Although, as at the time of sample collection, no comprehensive detail was known about the SARS-CoV-2 infection level in each patient, the result could be somewhat indicative that patient numbers 4 and 6 are at different stages of infection. Thus, by this result the sensor demonstrates a quantitative advantage over the lateral-flow immunochromatography-based SARS-CoV-2 antigen tests. To investigate the sensor’s reproducibility, 5 repeated measurements of both negative patient 1 and positive patient 4 samples were carried out on freshly prepared sensors and their relative standard deviation (RSD) were calculated. As shown in Table 2 the RSD of the 5 repeated measurements for each sample range between 3.7% and 5.6% thus indicating that the sensor has good reproducibility.
Table 2.
Reproducibility experiments of ncovS1 sensor.
| Sample |
In |
RSD (%) | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Neg. patient 1 | 0.27 | 0.29 | 0.30 | 0.28 | 0.26 | 5.6 |
| Pos. patient 4 | 0.40 | 0.43 | 0.40 | 0.39 | 0.41 | 3.7 |
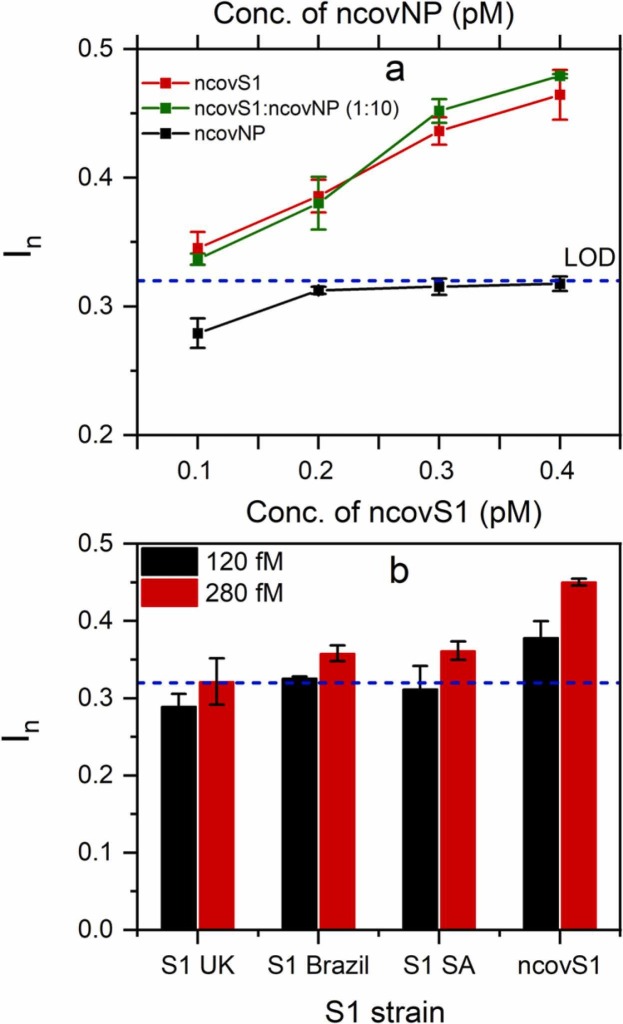
The selectivity of the sensor was further studied in COVID-19 negative samples by spiking the sample with varying concentrations of either ncovS1, ncovNP or their mixture. The concentrations of both proteins were selected to simulate their concentration ratio, 1:10 (ncovS1:ncovNP) in SARS-CoV-2 virus [31]. As observed in Fig. 4(a), the responses induced on the sensor by the increasing concentration of ncovNP are below the LOD (determined in Fig. 3(b)) indicating no recognition for ncovNP. However, the sensor demonstrated increasing responses, above the LOD, towards ncovS1 even though at tenfold lower concentration values. Moreover, the responses induced by the mixture of both proteins, ncovS1:ncovNP (1:10) are comparable to that from ncovS1 thus indicating that the presence of ncovNP in the sample would not interfere, to any significant extent, with the sensor specific recognition of ncovS1 thereby enabling its accurate analysis.
Fig. 4.
(a) Selectivity of ncovS1 sensor for ncovS1 against ncovNP in COVID-19 negative nasopharyngeal swab samples. The concentration of ncovNP was tenfold higher than the concentration of ncovS1. (b) ncovS1 sensor responses to SARS-CoV-2 spike protein subunit S1 (ncovS1) and its different strains (S1 UK VOC 202012/01, S1 Brazil P1 and S1 South Africa (SA) VOC 501.V2). The dashed lines represent the LOD determined in Fig. 3(b).
In addition, we considered it important to study the sensor’s behaviour upon exposure to different mutated strains of the SARS-CoV-2 virus thereby further elucidating its selective recognition for the target. To this end, we tested the samples prepared by spiking the desired amount of S1 protein of common SARS-CoV-2 virus strains such as S1 UK VOC 202012/01, S1 Brazil P1, S1 South Africa VOC 501.V2 to COVID-19 negative samples. As seen in Fig. 4(b), the sensor demonstrated the highest response for the imprinted target (ncovS1) at both concentrations. Although all strains induced a concentration-dependent sensor response, their responses, especially at the lower concentration, are analytically not significant since they fall below or are comparable to the estimated sensor’s detection limit, thus highlighting the sensor’s preference for ncovS1 against other strains. This quite discriminatory response displayed by the sensor against mutated strains of the target could be explained on the assumption that the arrangement of glycosylation sites located on S1 subunit of S proteins of the unmutated SARS-CoV-2 virus are most likely not retained in the variants. However, further detailed study might be necessary to establish the distinction in the availability and/or arrangement of glycosylation sites among known SARS-CoV-2 variants and to also explore the mechanism for this observed discrimination.
3.4. Point-of-care analysis
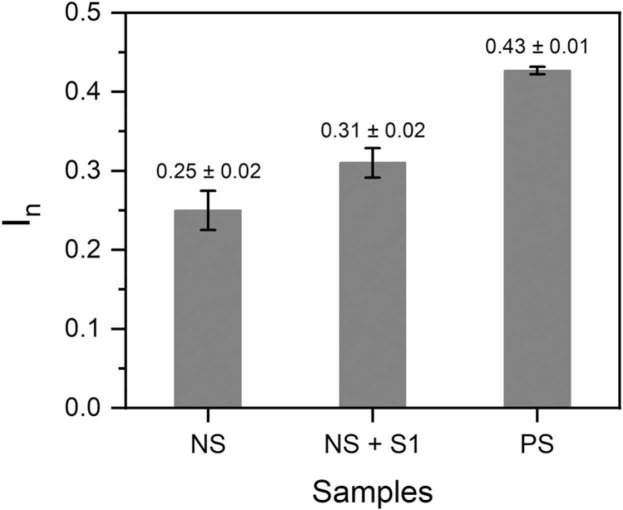
The on-site monitoring possibility of the sensor was assessed by investigating its compatibility with a portable potentiostat (EmStat3 Blue) connected to a mobile phone. For this purpose, the sensor’s responses following 15 min incubation in COVID-19 negative (patient 1) sample spiked with 64 fM ncovS1, and COVID-19 positive (patient 4) samples were recorded. Fig. 5 shows the analysed responses obtained for the different samples while the typical SWV voltammograms are shown in Fig. S9 (a-c). As expected, patient 4 sample induced a response (0.43 ± 0.01) that was higher than that obtained from either the negative sample (0.25 ± 0.02) or negative sample spiked with 64 fM ncovS1 (0.31 ± 0.01). Although this response is comparable to that earlier obtained (0.40 ± 0.02, Table 1) with the research grade potentiostat (Reference 600TM), we believe that for a more reliable reading, a calibration of the portable system would still be needed. Nevertheless, the combination of ncovS1-MIP chip with the portable potentiostat demonstrated its potential as a hand-held point-of-care monitoring device.
Fig. 5.
Responses (In) obtained from the sensor against patient 1 COVID-19 negative sample (NS), patient 1 NS spiked with 64 fM S1 (NS+S1) and Patient 4 COVID-19 positive sample (PS). All measurements were performed using the ncovS1-MIP-modified chip connected to a portable potentiostat (EmStat3 Blue, PalmSens BV, The Netherlands).
4. Conclusion
This work demonstrates the electrochemical ncovS1 sensor armed with a molecularly imprinted polymer synthetic receptor (ncovS1-MIP). Notable selectivity of ncovS1-MIP towards ncovS1 is achieved by adopting the covalent imprinting approach involving the chemical interaction between diol moieties of the highly glycosylated ncovS1 and boronic acid groups of APBA as a functional monomer. The sensor demonstrates a rapid diagnostic possibility with a rebinding time of 15 min and a measurement duration of 5 min which is comparable with the available antigen testing assays. The sensor is capable of detecting ncovS1 both in PBS and patient’s nasopharyngeal samples with LOD values of 15 fM and 64 fM, respectively. In addition, it discriminates remarkably against ncovNP which could be abundantly present in COVID-19 patients’ samples. Moreover, the sensor confirmed five COVID-19 positive nasopharyngeal swab samples that were previously analysed by RT-PCR thus, establishing its suitability as a potential diagnostic tool for clinical assessment of SARS-CoV-2 and demonstrating quantitative advantage over the commercially available lateral-flow immunochromatography-based SARS-CoV-2 antigen tests. Essentially, the electrochemical characteristics of the sensor can be readily handled by a portable potentiostat allowing on-site measurements thus holding a great potential as a point-of-care testing platform for rapid and early diagnosis of COVID-19 patients. Although the sensor demonstrates a reasonable discrimination against spike proteins from other variants of the SARS-CoV-2 virus, further studies may be required to qualify its selectivity against all known strains of the virus and the associated mechanism of selective recognition.
CRediT authorship contribution statement
Akinrinade George Ayankojo: Investigation, Formal analysis, Writing - review & editing. Roman Boroznjak: Investigation, Formal analysis. Jekaterina Reut: Validation, Writing - review & editing. Andres Öpik: review. Vitali Syritski: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by Estonian Research Council (Estonia) grants COVSG34 and PRG307. The authors thank Icosagen AS and prof. Mart Ustav personally for kindly providing the SARS-CoV-2 viral proteins. The authors thank Synlab Eesti OÜ and Kaspar Ratnik and Paul Naaber for kindly providing the clinical samples and the laboratory facilities for their testing.
Biographies
Akinrinade George Ayankojo is a research scientist at the Department of Materials and Environmental Technology in TalTech. He received his PhD (2018) in chemical and materials technology from TalTech and his MSc from the same department. His research interest includes the development of molecularly imprinted polymer-based sensors for small and large molecules of analytical interest.
Roman Boroznjak received his MSc (2007) in organic chemistry and PhD (2017) in natural and exact sciences from TalTech. His research interests include computational modelling and rational design of molecularly imprinted polymers.
Jekaterina Reut is currently a research scientist at the Department of Materials and Environmental Technology in TalTech. She received her PhD (2004) in the field of electrically conducting polymers from TalTech. Her research interest is in the area of the design and synthesis of molecularly imprinted polymers for biosensing applications.
Andres Öpik received his PhD in chemistry from the University of Tartu in 1980. He is currently Emeritus Professor of physical chemistry at the Department of Material and Environmental Technology in TalTech. His main research field is material science and technology: investigation of the physical and chemical properties and possibilities of practical applications of different electronic materials such as electrically conductive polymers and inorganic semiconductive compounds.
Vitali Syritski received his PhD in Chemistry at Tallinn University of Technology in 2004. Currently he is the head of the Laboratory of Biofunctional Materials in the Department of Materials and Environmental Technology at TalTech. His present research interests include molecularly imprinted technology and electrochemical analysis. In particular, he has focused on development of chemical and biosensors for accurate and fast detection of disease biomarkers and environmental contaminants.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2021.131160.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Oran D.P., Topol E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrero Hernando C., Álvarez Serra J.A., Elizari Saco M.J., Martínez-Nadal S., Vila Cerén C. In: An Pediatría Engl., editor. Vol. 93. 2020. pp. 264–265. (PCR test for SARS-CoV-2 persistently positive. Virus detection is not always COVID-19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong K., Cao W., Liu Z., Lin L., Zhou X., Zeng Y., Wei Y., Chen L., Liu X., Han Y., Ruan L., Li T. Prolonged presence of viral nucleic acid in clinically recovered COVID-19 patients was not associated with effective infectiousness. Emerg. Microbes Infect. 2020;9:2315–2321. doi: 10.1080/22221751.2020.1827983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wajnberg A., Mansour M., Leven E., Bouvier N.M., Patel G., Firpo-Betancourt A., Mendu R., Jhang J., Arinsburg S., Gitman M., Houldsworth J., Sordillo E., Paniz-Mondolfi A., Baine I., Simon V., Aberg J., Krammer F., Reich D., Cordon-Cardo C. Humoral response and PCR positivity in patients with COVID-19 in the New York City region, USA: an observational study. Lancet Microbe. 2020;1:e283–e289. doi: 10.1016/S2666-5247(20)30120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Find.Test directory, FIND. (n.d.). 〈https://www.finddx.org/test-directory/〉 (accessed April 13, 2021).
- 6.Lim R.R.X., Bonanni A. The potential of electrochemistry for the detection of coronavirus-induced infections. TrAC Trends Anal. Chem. 2020;133 doi: 10.1016/j.trac.2020.116081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Peng Z., Holl N.J., Hassan Md.R., Pappas J.M., Wei C., Izadi O.H., Wang Y., Dong X., Wang C., Huang Y.-W., Kim D., Wu C. MXene–Graphene Field-Effect Transistor Sensing of Influenza Virus and SARS-CoV-2. ACS Omega. 2021;6:6643–6653. doi: 10.1021/acsomega.0c05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 9.Mojsoska B., Larsen S., Olsen D.A., Madsen J.S., Brandslund I., Alatraktchi F.A. Rapid SARS-CoV-2 Detection Using Electrochemical Immunosensor. Sensors. 2021;21:390. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D’Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosbach K. Molecular imprinting. Trends Biochem. Sci. 1994;19:9–14. doi: 10.1016/0968-0004(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 12.Cieplak M., Kutner W. Artificial Biosensors: How Can Molecular Imprinting Mimic Biorecognition? Trends Biotechnol. 2016;34:922–941. doi: 10.1016/j.tibtech.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Kidakova A., Boroznjak R., Reut J., Öpik A., Saarma M., Syritski V. Molecularly imprinted polymer-based SAW sensor for label-free detection of cerebral dopamine neurotrophic factor protein. Sens. Actuators B Chem. 2020;308 doi: 10.1016/j.snb.2020.127708. [DOI] [Google Scholar]
- 14.Tretjakov A., Syritski V., Reut J., Boroznjak R., Öpik A. Molecularly imprinted polymer film interfaced with Surface Acoustic Wave technology as a sensing platform for label-free protein detection. Anal. Chim. Acta. 2016;902:182–188. doi: 10.1016/j.aca.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Ye L., Mosbach K. Molecular imprinting: Synthetic materials as substitutes for biological antibodies and receptors. Chem. Mater. 2008;20:859–868. doi: 10.1021/cm703190w. [DOI] [Google Scholar]
- 16.Ramanaviciene A., Ramanavicius A. Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004;20:1076–1082. doi: 10.1016/j.bios.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Tai D.-F., Lin C.-Y., Wu T.-Z., Chen L.-K. Recognition of dengue virus protein using epitope-mediated molecularly imprinted film. Anal. Chem. 2005;77:5140–5143. doi: 10.1021/ac0504060. [DOI] [PubMed] [Google Scholar]
- 18.Lu C.-H., Zhang Y., Tang S.-F., Fang Z.-B., Yang H.-H., Chen X., Chen G.-N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012;31:439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Cennamo N., D’Agostino G., Perri C., Arcadio F., Chiaretti G., Parisio E.M., Camarlinghi G., Vettori C., Di Marzo F., Cennamo R., Porto G., Zeni L. Proof of Concept for a Quick and Highly Sensitive On-Site Detection of SARS-CoV-2 by Plasmonic Optical Fibers and Molecularly Imprinted Polymers. Sensors. 2021;21:1681. doi: 10.3390/s21051681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rump A., Risti R., Kristal M.-L., Reut J., Syritski V., Lookene A., Ruutel Boudinot S. Dual ELISA using SARS-CoV-2 nucleocapsid protein produced in E. coli and CHO cells reveals epitope masking by N-glycosylation. Biochem. Biophys. Res. Commun. 2021;534:457–460. doi: 10.1016/j.bbrc.2020.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tretjakov A., Syritski V., Reut J., Boroznjak R., Volobujeva O., Öpik A. Surface molecularly imprinted polydopamine films for recognition of immunoglobulin G. Microchim. Acta. 2013;180:1433–1442. doi: 10.1007/s00604-013-1039-y. [DOI] [Google Scholar]
- 26.Tretjakov A., Syritski V., Reut J., Boroznjak R., Öpik A. Molecularly imprinted polymer film interfaced with Surface Acoustic Wave technology as a sensing platform for label-free protein detection. Anal. Chim. Acta. 2016;902:182–188. doi: 10.1016/j.aca.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Golabi M., Kuralay F., Jager E.W.H., Beni V., Turner A.P.F. Electrochemical bacterial detection using poly(3-aminophenylboronic acid)-based imprinted polymer. Biosens. Bioelectron. 2017;93:87–93. doi: 10.1016/j.bios.2016.09.088. [DOI] [PubMed] [Google Scholar]
- 28.Gu L., Jiang X., Liang Y., Zhou T., Shi G. Double recognition of dopamine based on a boronic acid functionalized poly(aniline-co-anthranilic acid)–molecularly imprinted polymer composite. Analyst. 2013;138:5461. doi: 10.1039/c3an00845b. [DOI] [PubMed] [Google Scholar]
- 29.Hong S., Lee L.Y.S., So M.-H., Wong K.-Y. A Dopamine Electrochemical Sensor Based on Molecularly Imprinted Poly(acrylamidophenylboronic acid) Film. Electroanalysis. 2013;25:1085–1094. doi: 10.1002/elan.201200631. [DOI] [Google Scholar]
- 30.Xing R., Wang S., Bie Z., He H., Liu Z. Preparation of molecularly imprinted polymers specific to glycoproteins, glycans and monosaccharides via boronate affinity controllable–oriented surface imprinting. Nat. Protoc. 2017;12:964–987. doi: 10.1038/nprot.2017.015. [DOI] [PubMed] [Google Scholar]
- 31.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. ELife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Culver H.R., Peppas N.A. Protein-Imprinted Polymers: The Shape of Things to Come? Chem. Mater. 2017;29:5753–5761. doi: 10.1021/acs.chemmater.7b01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material