Abstract
Plant metal tolerance proteins (MTPs) play major roles in enhancing resistance to heavy metal tolerance and homeostasis. However, the role of MTPs genes in tomato, which is one of the most popular crops, is still largely limited. Hence, we investigated genome-wide study of tomato MTPs, including phylogenetic, duplication, gene structure, gene ontology and previous transcriptomic data analysis. Moreover, the MTPs expression behaviour under various heavy metals stress has rarely been investigated. In the current study, eleven MTP candidate genes were genome-wide identified and classified into three major groups; Mn-cation diffusion facilitators (CDFs), Fe/Zn-CDFs, and Zn-CDFs based on the phylogeny. Structural analysis of SlMTPs showed high gene similarity within the same group with cation_efflux or ZT_dimerdomains. Evolutionary analysis revealed that segmental duplication contributed to the expansion of the SlMTP family. Gene ontology further showed the vital roles of MTPs in metal-related processes. Tissue-specific expression profiling exhibited similar expression patterns in the same group, whereas gene expression varied among groups. The MTPs expression was evaluated after tomato treatments by five divalent heavy metals (Cd2+, Co2+, Mn2+, Zn2+, and Fe2+). SlMTP genes displayed differential responses in either plant leaves or roots under heavy metals treatments. Nine and ten SlMTPs responded to at least one metal ion treatment in leaves and roots, respectively. In addition SlMTP1, SlMTP3, SlMTP4, SlMTP8, SlMTP10 and SlMTP11 exhibited the highest expression responses in most of heavy metals treatments. Overall, our findings presented a standpoint on the evolution of MTPs and their evolution in tomato and paved the way for additional functional characterization under heavy metal toxicity.
Keywords: Tomato, Metal tolerance protein, Heavy metals, Genome-wide identification, Gene expression
1. Introduction
Metals act as co-factor, which has essential implications inactivating enzymes in plant cells to perform the specific biological reaction (Thomine and Vert, 2013). The essential metals such as zinc (Zn), manganese (Mn), iron (Fe), cobalt (Co), and copper (Cu) at low level play an essential role in plants, but excessive amounts of these ions lead to toxic effects (Kolaj-Robin et al., 2015). Moreover, a deficient concentration of non-essential metals, including mercury (Hg), silver (S), cadmium (Cd), and lead (Pb), can also cause plant cell toxicity (Clemens, 2001). Interestingly, plants are natural bioaccumulators for various heavy metals from the water and soil for appropriate plant growth and development activities (Ali et al., 2013).
Plants overcome heavy metal stress by various physiological and molecular mechanisms, including genomic-level and complex biochemical processes (Liu et al., 2019). Some of these mechanisms are part of the homeostatic process and are constitutive (Rai et al., 2019). Other mechanisms are exclusively related to counter-specific metal toxicity (Gupta et al., 2019). All responses can be widely classified as being tolerant or avoidance types (Krzesłowska, 2011). Metal uptake, trafficking, storage, chelation, and efflux are plant mechanisms to maintain metal homeostasis (Montanini et al., 2007). Several studies indicated the essential roles of various protein families with their specific transporters in these regulatory processes (Gao et al., 2020). The cation diffusion facilitator (CDF) family genes are integral membrane divalent cation transporters involved in divalent metal ions efflux from the cytoplasm either into subcellular compartments or outside the cell (Gustin et al., 2011). The CDF transporters have been widely identified in many organisms since the first identification in the bacterial cell (Nies and Silver, 1995), which further classified into three major groups; Mn-CDF, Zn/Fe-CDF, and Zn-CDF based on either confirmed or hypothesized transported substrate specificities (Montanini et al., 2007).
The CDF transporters are considered as metal-tolerance proteins (MTPs) in plants, which were classified into seven distinct groups (1, 5, 6, 7, 8, 9, and 12) based on annotation and phylogenetic analysis in Arabidopsis (Gustin et al., 2011). Many MTP proteins were identified in several plants species, including Arabidopsis thaliana (van der Zaal et al., 1999), Vitis vinifera (Shirazi et al., 2019), Populus trichocarpa (Gao et al., 2020), Triticum aestivum (Vatansever et al., 2017), Nicotiana tabacum, Nicotiana sylvestris and Nicotiana tomentosiformis (Liu et al., 2019). Many Zn-CDF proteins have been studied from the first identified AtMTP1 in Arabidopsis (van der Zaal et al., 1999). Zn-CDF genes play an essential role in plant Zn2+ tolerance. For instance, AtMTP1 and AtMTP3 of tonoplast involved in Zn and Co tolerance through the excess transport of Zn2+ and Co2+ ions to the vacuole (Arrivault et al., 2006, Dräger et al., 2004, Kawachi et al., 2008, Kobae et al., 2004). Furthermore, two more genes of Zn-CDF family, including AtMTP5 and AtMTP12 were identified to form a functional complex during Zn2+ transportation into Golgi (Fujiwara et al., 2015).
The Mn-CDF family members, such as AtMTP8, play an essential role in the transportation of Mn2+ besides its role in the localization of Fe2+ and Mn2+ in seeds (Chu et al., 2017, Delhaize et al., 2007, Eroglu et al., 2016). In Oryza sativa, the OsMTPs (OsMTPs8.1 and 8.2) of tonoplast participate in the transport of Mn2+ within the plant (Takemoto et al., 2017, Tsunemitsu et al., 2018). Furthermore, the ShMTP gene in Stylosanthes hamata, was the first group 8 of MTPs to be identified that enhances the tolerance against Mn when overexpressed in Arabidopsis (Delhaize et al., 2007). Also, the CsMTP8 gene of cucumber confers Mn2+ tolerance when overexpressed in yeast and the Arabidopsis (Migocka et al., 2014).
Tomato (Solanum lycopersicum L.) is a common and economically essential crop worldwide (El-Sappah et al., 2019). Its consumption increases annually due to the fruit attractiveness (several sizes, colors, flavors, and shapes), multiple utilization, and production of therapeutic compounds (Cheng et al., 2020). A recommended healthy human diet consists of fruit and vegetables, and tomatoes are important because they contain carbohydrates, proteins, vitamins, minerals, dietary fibers, and antioxidants (Liu et al., 2020). However, tomato fruits are a potential mediator for heavy metal's entrance into the food chain (Cobb et al., 2000), influencing human health.
Tomato roots grown in contaminated soil or irrigated with sewage water accumulated a higher content of Cu2+, Zn2+, and Ni2+, while the shoots had a higher Cd2+, Mn2+, Fe2+, and Pb2+ (Singh et al., 2010). However, only a few tomato MTP proteins have been studied and characterized. In recent years, genome sequencing of model plants and commercially essential plants were performed and provided opportunities to screen candidate genes (Edwards et al., 2017). However, due to the limit of the integrity of the tomato genome sequence, a few MTP members were not identified at that time, and the expression patterns, especially those in response to heavy metal stresses and the metal transport features of SlMTP genes, are unknown. In the current study, we successfully identified 11 SlMTPs in the tomato genome and comprehensively analyzed their structures and sequences. We comprehensively characterized the proteins at the sequence level and performed bioinformatics analyses of putative SlMTP genes to explore phylogenetic relationships, chromosomal distributions, gene structure, conserved motifs, and synteny analysis. In addition, five different heavy metals (Cd2+, Co2+, Fe2+, Mn2+, and Zn2+) were applied to tomato seedlings, and expression profiling was presented. Taken together, the results in this study would lay a theoretical and practical foundation for the functional characterization of SlMTP genes in future studies.
2. Materials and methods
2.1. Sequence retrieval of MTP genes in tomato
Tomato sequences were obtained from the Solanaceae Genomics Network (https://solgenomics.net/), and then BioEidt 7.0 software was used for the local database construction. The candidate tomato MTP genes were confirmed using the hidden Markov model (HMM) profile of two MTP domains (PF16916 and PF01545) from Pfam (http://www.sanger.ac.uk/Software/Pfam). The blast search of putative MTP protein sequences was performed on the NCBI (http://blast.ncbi.nlm.nih.gov/blast.cgi), SPud DB tomato Solanaceae Genomics Network (https://solgenomics.net), and phytozome (https://phytozome.jgi.doe.gov/).
The validation of all obtained protein sequences was done at E-value < 10−5 for identification of the MTP domain, using SMART (http://smart.embl-heidelberg.de/) tools (Letunic et al., 2004). All genomic information about the selected MTP gene family, such as chromosomal location and CDS, were obtained from the phytozome website database (https://phytozome.jgi.doe.gov/). The MTP proteins were analyzed to obtain their characteristics, such as molecular weight, amino acid number, isoelectric point, the theoretical pI, molecular weight and instability index using EXPASY ProtParam Tool (http://www.expasy.org/tools/protparam.html) (Gasteiger et al., 2003). The subcellular localization data was predicted using the MTP amino acid sequences by protein subcellular localization prediction tools (https://wolfpsort.hgc.jp/).
2.2. Phylogenetic analysis
In addition to tomato, Arabidopsis (http://arabidopsis.org), Cucumis sativus (http://cucurbitgenomics.org/), Populus trichocarpa (http://plantgdb.org/PtGDB/), Oryza sativa, (https://rapdb.dna.affrc.go.jp/), and Triticum aestivum (https://www.wheatgenome.org/) MTP amino acid sequences were used for the phylogenetic tree of evolutionary MTPs relationship. Next, CLUSTALX 2.0 software with default parameters has been used for multiple alignments. The alignment was utilized as an input file to MEGA 6.0 software. A phylogenetic tree was constructed by the Neighbor-Joining (NJ) method with the following parameters: 1000 bootstrap replications, pairwise deletion, and Poisson model (Tamura et al., 2011).
2.3. Chromosomal locations, synteny analysis, and protein–protein interactions
Tomato gene database (https://phytozome.jgi.doe.gov/) supports us by the chromosomal position information of MTP genes, which were used to generate the genetics map by MapChart software. After that, two genes in the same species, located in the same clade of the phylogenetic tree, were defined as coparalogs to identify whether tandem and segmental duplication events had occurred. On the other hand, the tomato gene database (https://phytozome.jgi.doe.gov/) was further used with target genes for detecting the coordinates of the segmental duplications. The paralogs were regarded as tandem results duplicated when two genes separated by five or fewer genes in a 100 kb region (Tang et al., 2008). Additionally, coparalogs were considered segmental duplications if they were located on duplicated chromosomal blocks (Wei et al., 2007). Smith-Waterman algorithm (http://www.ebi.ac.uk/Tools/psa/) was used to calculate the local alignments of two protein sequences. The synteny relationship with the chromosomal distribution for each SlMTP genes was introduced using circos (http://circos.ca/) (Krzywinski et al., 2009). Furthermore, for more knowledge about the cellular function of the MTP protein family, the functional interactions between all expressed studied proteins were obtained. The amino acid sequences of all the family members used for protein–protein interaction studies using the STRING database (https://string-db.org/).
2.4. Gene structures and motif analyses
The structure of all SlMTP gene family members was analyzed to detect the intron/exon and their organization, using both genomic and CDS sequences with the online tools of the Genes Structure Display Server program (GSDS, http://gsds.cbi.pku.edu.cn/index.php) (Hu et al., 2015). The conserved motif was detected for the gene family members using a Multiple EM for motif elicitation (MEME) (http://meme.nbcr.net/meme3/meme.html) online server with default parameters setting a maximum number of motifs to 10 and motif width to 6–200 (Bailey et al., 2006).
2.5. Protein modeling, prediction, and gene ontology annotation (GO)
The Phyre2 online webserver was used for protein modeling, prediction, and analysis of the SlMTPs at the intensive mode (sbg.bio.ic.ac.uk/phyre2/) (Kelley et al., 2015). Blast2GO v3.0.11 (https://www.blast2go.com) and OmicsBox software were used to identify MTP protein sequences for GO annotation (Conesa and Götz, 2008).
2.6. Digital data expression analysis
Our research analysed the previous RNA-based digital data to obtain the global expression of MTP gene family members during normal growing conditions. The expression data were downloaded from tomato functional genomics databases (http://ted.bti.cornell.edu/pgsc_download.shtml) for some of the tomato organs such as leaves and roots, and flowers. Then the gene expression was analyzed using the cufflinks (version: 2.2.1). Finally, FPKM expression values were divided with mean and transformed to log2. The MeV 4.5 was used to cluster the expression data as a heat map (http://heatmapper.ca/) (Babicki et al., 2016, Saeed et al., 2006).
2.7. Growth conditions and heavy metal treatments
In this study, the tomato M82 line was cultivated during the Autom of 2020 at the experimental greenhouse of Yibin University (China). First, the seeds were washed with 10% hypochlorous acid and distilled water. The seeds have been germinated using water-saturated filter paper and then transferred to fertilized pittmoss soil with germination conditions of 16 h light (27 °C) and 8.0 dark (18 °C) with a relative humidity of 70%. Four seeds were planted in each plastic pot. After emergence, thinning was performed to maintain two uniform seedlings per pot.Thirty-day-old tomato was placed in 1/2 Hoagland solutions (pH 6.0) with different heavy metal concentrations 0.1 mM CdCl2, 0.1 mM CoCl2, 0.5 mM FeSO4,1 mM MnSO4, and 0.5 mM ZnSO4, respectively, while normal 1/2 Hoagland solutions was used as the control (CK) (Desoky et al., 2019, Gao et al., 2020). The experimental pots were positioned in a complete randomized block design. The experiment was composed of 6 treatments, as shown above, and each treatment was repeated with three pots. Then, 24 h later, the leaves and roots of tube plantlets were collected and used as RNA extraction materials. Three biological replicates of expression analyses have been performed for each treatment.
2.8. qRT-PCR analysis
The Trizol reagent (Invitrogen, USA) was used to isolate the RNA from all plant samples (leaf and root), and the cDNA synthesis was performed using SuperMix Kit (Transgen, Beijing). (El-Sappah et al., 2017). The specific primers of all selected genes were designed using Primer Premierv5.0 (Table S1) with β-actin as a housekeeping gene. The real-time PCR was performed with the following reagents volumes:10 μL SYBR Premix Taq (2 × ) mixture, 1 μL of cDNA,0.5 μL of each primer, and 8 μL of ddH2O for a total volume of 20. The PCR cycles were adapted as follows: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 60 s. The relative gene expression levels were calculated based on the 2−ΔΔCT method (Livak and Schmittgen, 2001).
2.9. Statistical analysis
Three biological replicates of expression analyses were performed with ± standard deviation (SD) at p < 0.05. The significant variations between means were compared at p < 0.05 (Student’s t-test).
3. Results
3.1. Identification of MTP genes in tomato
A complete set of 18 putative genes were identified in the tomato genome using the homologous sequences of Arabidopsis as queries and after excluding the sequences with an incompleted or missing domain. We finally selected 11 candidate genes for further evaluation and study. The genes were designated new names from SlMTP1 to SlMTP11. The different physicochemical characters of these 11 genes were presented in Table 1. The majority of tomato chromosomes harbored the MTP genes, except chromosomes number 1, 4, 8, and 11, which do not carry any of these genes. Furthermore, the molecular weight varied between all genes, ranging from 41197.34 to 54954.83 Da. The number of introns varied mainly among the group, and all genes contained introns except SlMTP1 and SlMTP3, which do not have any introns. Furthermore, most of SlMTPs, according to subcellular localization analysis, were located as secreted proteins in tonoplast, except three genes (SlMTP4, 8, and 11) in cytoplasm while SlMTP6 in the chloroplast.
Table 1.
The characteristics of SlMTP genes in tomatoes.
| MTP | Gene NCBI symbol | Location | (-) | (+) | MW (Da) | aa | Instability | Aliphaticindex | GRAVY | PI | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SlMTP1 | LOC101249377 | Chro7; 1836549.0.1840444 | 52 | 33 | 46022.69 | 416 | 27.65 | 104.06 | 0.007 | 6 | Tonoplast |
| SlMTP2 | LOC101255067 | Chro 12; 6591239.0.6594563 | 42 | 41 | 46374.50 | 404 | 39.63 | 99.18 | −0.035 | 7 | Tonoplast |
| SlMTP3 | LOC101268691 | Chro 6; 6591239.0.6594563 | 40 | 26 | 42412.78 | 379 | 31.94 | 98.97 | 0.112 | 6 | Tonoplast |
| SlMTP4 | LOC101263495 | Chro 3; 23590477.0.23593243 | 50 | 35 | 45166.30 | 398 | 52.48 | 112.64 | 0.128 | 5.09 | Cytoplasm |
| SlMTP5 | LOC101268361 | Chro 3; 36451716.0.36478448 | 29 | 35 | 41197.34 | 368 | 45.65 | 95.11 | 0.102 | 9.07 | Tonoplast |
| SlMTP6 | LOC101254940 | Chro 9; 66978000.0.66987675 | 52 | 41 | 54954.83 | 503 | 45.01 | 103.7 | 0.04 | 6.28 | Chloroplast |
| SlMTP7 | LOC101262626 | Chro 6; 40063309.0.40070157 | 47 | 42 | 50714.91 | 463 | 30.29 | 95.4 | 0.086 | 6.24 | Tonoplast |
| SlMTP8 | LOC101254675 | Chro 12; 68053619.0.68056088 | 52 | 35 | 44015.99 | 394 | 41.07 | 113.05 | 0.134 | 5.02 | Cytoplasm |
| SlMTP9 | LOC101261962 | Chro 10; 410306.0.412587 | 47 | 42 | 47055.18 | 412 | 49.10 | 95.83 | −0.060 | 6.2 | Tonoplast |
| SlMTP10 | LOC101257769 | Chro 5; 65432886.0.65438261 | 51 | 31 | 45277.22 | 401 | 41.71 | 104.31 | 0.081 | 4.99 | Tonoplast |
| SlMTP11 | LOC101253924 | Chro 1; 797129.0.801771 | 54 | 38 | 45478.24 | 405 | 43.61 | 105.38 | 0.013 | 4.97 | Cytoplasm |
(-), (+), MW, aa, GRAVY, and PI denote to a total number of negatively charged residues (Asp + Glu), the total number of positively charged residues (Arg + Lys), molecular weight, amino acid number, Grand average of hydropathicity, and isoelectric points, respectively.
3.2. Phylogenetic analysis of MTP gene families
The phylogeny of the MTP gene families showed that MTP proteins are divided into seven groups named; groups 1, 5, 6, 7, 8, 10, and 12 (Fig. 1). Group 10 harboured 21 of MTPs, which represent the biggest group. Groups 8 and 10 included most of the studied SlMTP, with three genes in each. Furthermore, all of the selected 11 SlMTP grouped under three clusters of Zn-CDFs (3 genes), Fe/Zn-CDFs (2 genes), and Mn-CDFs (6 genes).
Fig. 1.
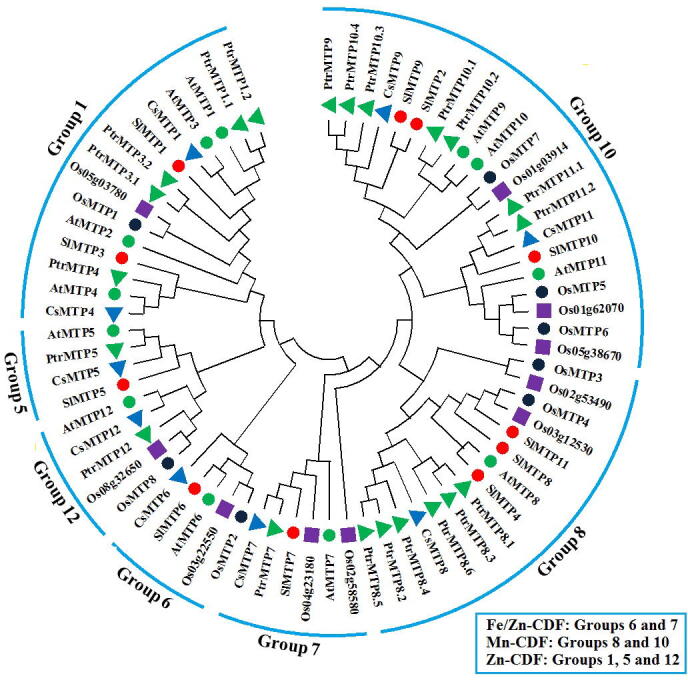
Phylogenetic tree of the 71 MTP proteins: 11 Tomato (marked by red circle), 12 Arabidopsis (blue triangle), 8 Wheat (blue circle), 10 Rice (green square), 9 Cucumber (brown circle), and 21 Black Poplar (yellow triangle). ClustalX1.83 was used for protein alignments and the phylogenetic tree's construction Neighbor-Joining (NJ) level with MEGA6.0 software at 1,000 replication sboot-strap.
3.3. Chromosomal locations and synteny analysis of MTP gene family
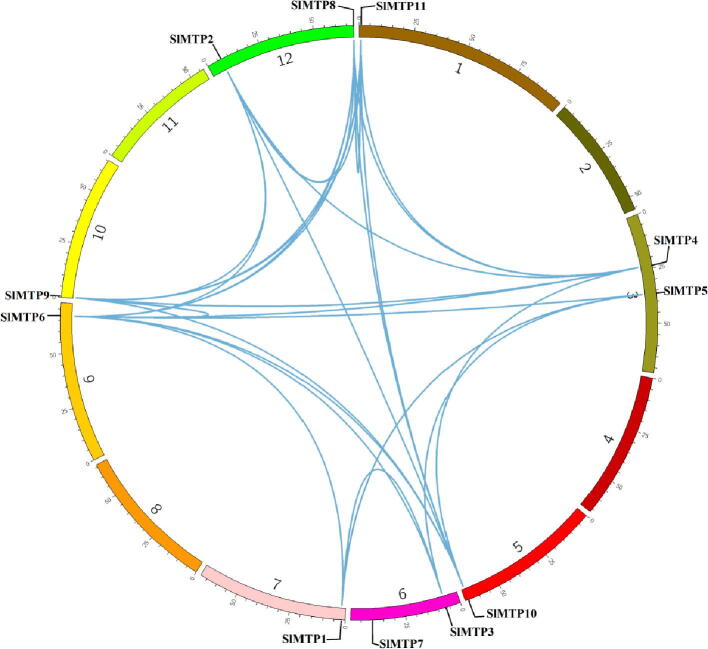
As mentioned, all tomato MTP genes are unevenly distributed on eight chromosomes where the chromosomes 3, 6, and 12 harboured half the MTP gene family members as they carry six whole genes of this family. Many pairs of collinearity genes were detected with an identity ranging from 70 to 100% due to segmental duplication events (Table S2). The segment duplication resulted in many homologous pairs of MTP genes between the chromosomes. The duplicated pairs included, SlMTP2/SlMTP6, SlMTP2/SlMTP9, and SlMTP3/SlMTP4 (Fig. 2). All MTP genes showed one or more duplication pairs except SlMTP7, which had no pair with any other gene. Furthermore, our investigation showed no obvious tandem duplication between SlMTPs.
Fig. 2.
Genome-wide synteny analysis for MTP gene family at 12 tomato chromosomes. The blue lines represented the syntenic orthologs and paralogs and showed the segment duplication.
3.4. Gene structures and motif analyses of the tomato MTP gene family
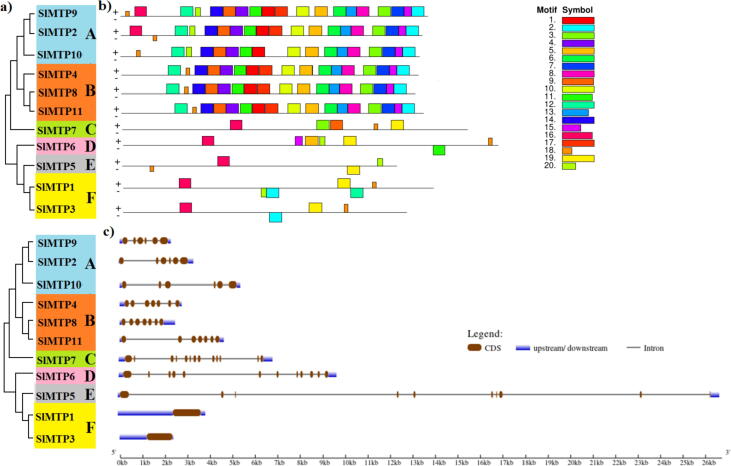
The tomato MTP gene members were divided into six subfamilies (A, B, C, D, E, and F). Subfamilies A and B were the largest with six members, followed by subfamily F (2 genes), whereas subfamilies C, D, and E each had only one gene (Fig. 3a). There was variation in exon–intron among different subfamily (Fig. 3c), supporting the close evolutionary relationships of tomato MTP gene family members. Our analysis showed that most MTP family members contain an incredibly varied intron number except for group F genes, which had a lack of intron. The complexity of gene structure often indicates the largest intron. The analysis of the conserved motifs of MTP using MEME depends on the amino acid sequences with ten motifs (Fig. 3b, and Table S3). Most of the studied motifs contain 50 amino acids, except motif 9 contains 49 amino acids, while motif 18 contains 15 amino acids. The largest common motifs were 2, which was noticed within all subfamilies except for C, D, and E, followed by motifs 3, 15, and 16, while the subfamily E contains only four motifs. It is primarily observed that the number, type, or order of motifs is similar within the same subfamily in addition to that among different families.
Fig. 3.
Phylogenetic relationship, gene structure, and conserved motif analysis of SlMTP genes; a) The neighbor-joining phylogenetic tree was constructed with MEGA6.0 using SlMTP amino acid sequences with 1000 times replicate. b) The motif composition of SlMTP proteins using ten conserved motifs is represented by the unique colour mentioned in the box on the top lift. c) Exon-intron structure of MTP tomato proteins where dark green boxes presented the exons, and the black lines represent the introns. The blue boxes represented the untranslated regions (UTRs), with size scales detailed at the bottom.
3.5. Protein modeling, prediction, protein–protein interactions and gene ontology annotation (GO)
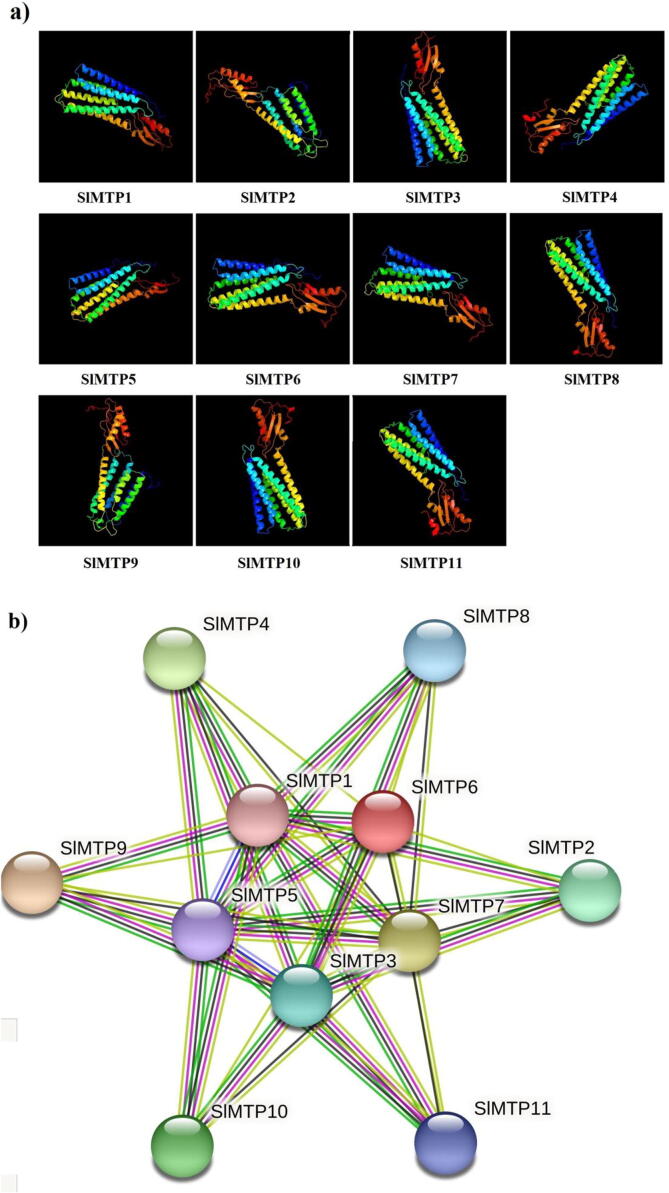
The eleven predicted models of the SlMTPs were generated based on c6xpdB, c3j1zP, c2qfiB, and d2qfia2 templates with a 100% identification ratio (Fig. 4a). The protein–protein interactions assessment showed the physical (direct) and the functional (indirect) associations (Fig. 4b). The result showed different interactions within the studied proteins where the total number of nodes was 11 with an average of 7. 09. The STRING database analysis showed 39 edges without any expected numbers and showed six representative local network clusters, which were CL:28212, CL:28208, CL:28384, CL:28201, CL:28198, and CL:28199 (Table S4). Moreover, our protein analysis showed two common domains within the MTP family, which were PF16916 (7 proteins), and PF01545 (10 proteins).
Fig. 4.
Protein analysis: a) Predicted 3D models of tomato SlMTP proteins. Models have been generated by using the Phyre 2 server in intensive mode. Models were visualized by rainbow colour from N to C terminus and organized in SlMTP1, SlMTP2,…to SlMTP11, b) protein–protein interaction among the MTP family members in tomato.
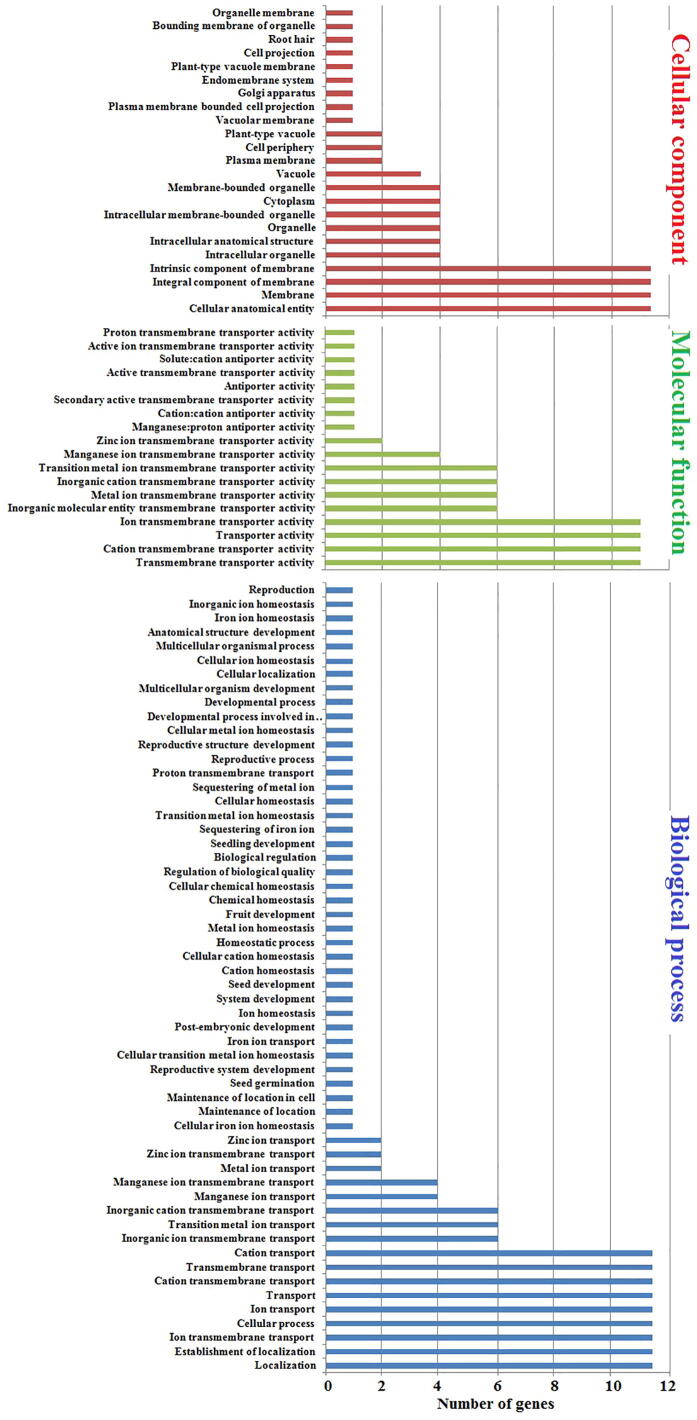
Similarly, sub-cellular localization, molecular function, and biological process were predicted by GO enrichment analysis (Fig. 5). In sub-cellular localization analysis, the predicted distribution scores of MTP proteins were as following; 11/69% in all membranes, 2/12% in the plasma membrane, vacuole, and Golgi apparatus.
Fig. 5.
Gene Ontology analysis of tomato SlMTP genes. Gene ontology showed the distribution of every SlMTP gene in the plant, where a red colour column mentioned the cellular component. In contrast, the biological processes in which the MTP family participate were mentioned by the blue colour column, and the molecular function was mentioned by move colour.
Noticeably, SlMTP4 gene was localized in 19 sub-cellular compartments out of all 23, which underlined its significant role in metal stress resistance. Collective scores of MTP protein molecules during biological processes were as following; trans-membrane transport of Zn+ was 2/18%, and Mn+ ions were 4/36%, while trans-membrane transport of Fe was 1/9%. The molecular function and biological processes analysis revealed that SlMTP1 and SlMTP3 genes play a key role in the transmembrane transport of Zn+, while SlMTP4, SlMTP8, SlMTP10, and SlMTP11 play a crucial role in transmembrane transport of Mn+. Moreover, SlMTP4 is an essential factor affecting Fe ion transport.
3.6. Gene expression profiling based on the digital data
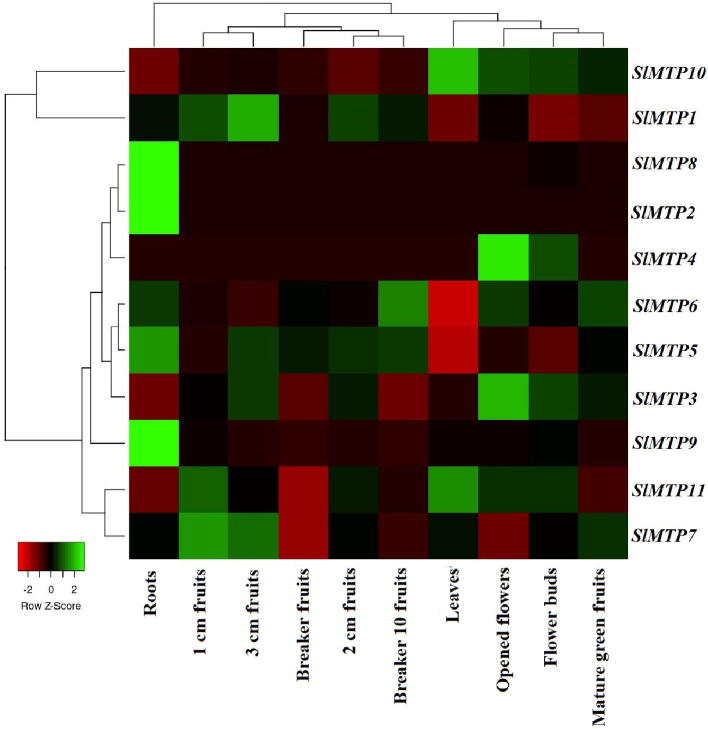
The tissue expression patterns of SlMTPs were investigated depended on the public transcriptome data. As shown in Fig. 6 and Table S5, all 11 SlMTP genes were expressed in the ten examined tissues (log2(FPKM + 1) > 0), except for SlMTP2 (which showed lower expression only in root tissue), SlMTP4 (only expressed in flower buds, opened flowers and 3 cm fruits) and SlMTP8 (expressed only in flower buds and roots).
Fig. 6.
The heat map of the 11 SlMTP genes expression profiles in different tomato tissues based on the RNA-seq (http://ted.bti.cornell.edu/).
Furthermore, SlMTP1 had the significantly higher transcript accumulation compared with other SlMTPs in all detected tissues, except for in flower buds, mature green fruits, and leaves, whereas two gene SlMTP2, SlMTP4, and SlMTP8 exhibited the lowest or no expression levels in most tissues (0 < log2(FPKM + 1) < 1). Furthermore, some genes exhibited tissue-specific expression. For instance, one gene (SlMTP1) in the 3 cm fruits, two genes (SlMTP1, and SlMTP9) in root, three genes (SlMTP1, SlMTP10, and SlMTP11) in leaves, one gene (SlMTP1, and SlMTP10) in opened flower showed the highest transcript abundances.
3.7. qRT-PCR analysis of the MTPs under different treatments
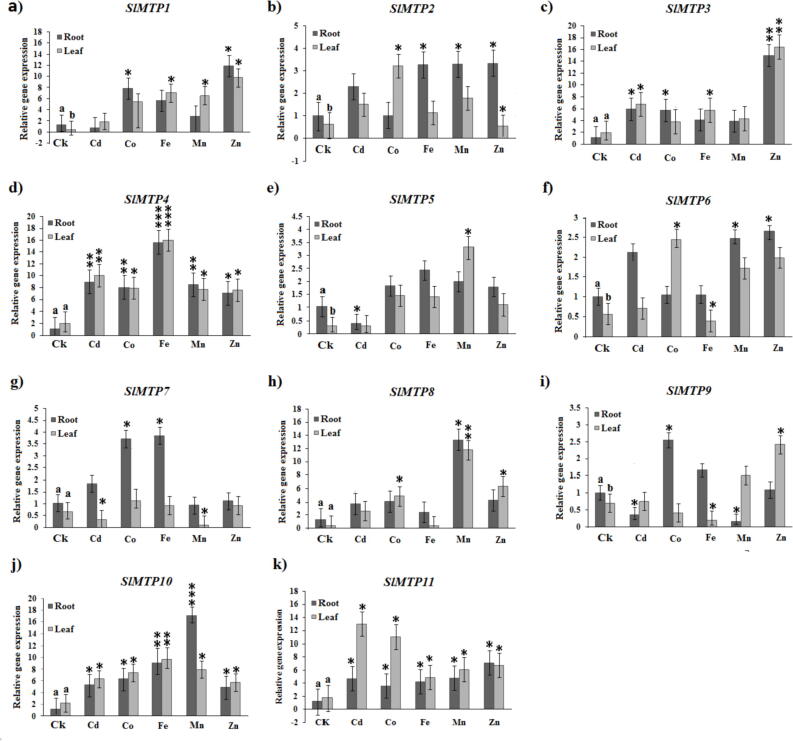
The 11 SlMTP genes showed differential expression pattern against different types of heavy metal stress in root and leaf (Fig. 7). In roots, Cd2+ enhanced the expression of SlMTP2 and SlMTP3, but down-regulated the expression of SlMTP1, SlMTP5, and SlMTP9, while it up-regulated the expression of SlMTP4. Co2+ decreased the expression levels of SlMTP2, but up-regulated the expression levels of SlMTP7 and SlMTP9. The SlMTP10 recorded the highest expression under Mn2+ contamination, but the expression of other three genes (SlMTP1, SlMTP7, and SlMTP9) down-regulated. Zn2+ increased the expression of SlMTP1 and SlMTP3. Fe2+ up-regulated the expression levels of SlMTP4 and SlMTP10, but down-regulated the expreseeion of SlMTP8. In leafs, Cd2+ enhanced the expression of SlMTP2, SlMTP3, SlMTP4, SlMTP10, and SlMTP11, but down-regulated the expression of SlMTP7. However, Co2+ up-regulated the expression of SlMTP2, SlMTP4, and SlMTP11, but decreased the expression of SlMTP9. Moreovver, Fe2+ up-regulated the expression of SlMTP1, SlMTP3, SlMTP4, SlMTP10, and SlMTP11, but down regulated the expression of SlMTP6, SlMTP8, and SlMTP9. Mn2+ down regulated the expression of SlMTP7, but enhanced the expreesion of SlMTP4, SlMTP8, SlMTP10, and SlMTP11. Zn2+ decreased the expression of SlMTP2 but up regulated the expression of SlMTP1, SlMTP3, and SlMTP4.
Fig. 7.
The qRT-PCR expression of the tomato SlMTP genes from root and leaf samples under various metal ion stresses. The reactions were normalized using the β-actin reference gene. The standard deviations have been represented by the error bars from three independent technical replicates. The mean expression levels of three replicates were analyzed with the five heavy metals treatments (Cd2+, Co2+, Fe2+, Mn2+ and Zn2+) using t-tests (p < 0.05). At the same time, the CK represents control samples which represented by different letters (a, b and c) indicate significant differences among each tested tissues under normal condition. Asterisks indicate significant differences between the treatment samples and the corresponding control samples in roots and leaves. (n = 9, p < 0.05, Student’s t-test).
4. Discussion
Heavy metals are the most effort on the ecosystem and make it unfit for human consumption (El-Sappah Et Al., 2012). Once released into the environment, they accumulate into plants then into other living tissues via the food chain and cause toxicity even at lower concentrations (Elrys et al., 2018). MTP genes (membrane divalent cation transporters) are essential for transporting various heavy metals and enhancing plant tolerance against heavy metals stress (Ricachenevsky et al., 2013). They also have an expected role in plant mineral nutrition maintenance (Liu et al., 2019). Moreover, these metal-binding proteins are now being utilized as bio-environmental markers for predicting heavy metal contamination based on their expression levels (Samuel et al., 2021). The MTP family has previously been studied in several plants, such as Arabidopsis thaliana (van der Zaal et al., 1999), Nicotiana tabacum, Nicotiana sylvestris and Nicotiana tomentosiformis (Liu et al., 2019), Triticum aestivum (Vatansever et al., 2017), and Populus trichocarpa (Gao et al., 2020), while this is the first genomic identification study of MTPs family in tomato. We successfully identified 11 MTP genes in tomato and named based on the sequence similarities and orthologous relationships between them and AtMTPs. The phylogeny of MTP proteins between tomato and other five studies was performed. The phylogeny results were aligned to previous studies conducted in various plant species. Multiple homologous pairs were observed in tomato, while no such pairs were observed in Arabidopsis, showing that the SlMTP gene family might have undergone gene expansion and/or gene loss in the evolutionary history, probably due to the polyploidization events.
There were three, two, and six SlMTP genes grouped to Zn-CDFs, Zn/Fe-CDFs, and Mn-CDFs. Considering the implications of phylogenetic distributions in inferring structure and functional roles across species (Eroglu et al., 2017). The various groups are involved in specific mechanisms, and this information could provide clues to predict their function in different species. The broad range of basic physicochemical properties of SlMTP gene was consistent with previous studies indicating huge probabilities of amino acid in metal tolerance (Ricachenevsky et al., 2013, Consortium, 2010, Eroglu et al., 2017). Consistently with the previous study (Vatansever et al., 2017), the subcellular localization analysis revealed that most genes are localized to tonoplast (the vacuole membrane), whereas some might also be localized in cytoplasm or chloroplast, suggesting that SlMTPs might function as the vacuole-localized cation transporters.
In our study, to obtain more knowledge about the gene annotation and the expansion mechanism of the MTP gene family in tomatoes, we investigated the gene synteny and duplication analysis (Fig. 1 and Table S2). Two or more genes on the same chromosome are often related to tandem duplication, while segmental duplication often occurs on different chromosomes (Schlueter et al., 2007). Our study did not show any of the tandem duplication pair, while there were 28 segmental duplications, such pairs are SlMTP1/SlMTP5, SlMTP3/SlMTP5, and SlMTP5/SlMTP6. During the evolution process of a plant gene, family duplication events occur, followed by divergence considering standard features and more related to secondary plant metabolism genes (Ober, 2005).
Almost all subfamilies contained the same numbers of introns and motif sequences which are consistent with the previous studies in where a similar gene structure was found within the same subfamilies (Liu et al., 2018). For example, all of the gene members of subfamily A contain five introns. However, subfamily F members do not contain any introns. These outcomes indicated that during the tomato evolution events of SlMTPs, some intron gain and loss occurred. Some genes have no intron but have one exon cause the lower ability of exons in the gain/loss rate due to higher selection pressure in the exons sequences (Harrow et al., 2006). Thus, with all these observations, it is probably that the placement divergences in intron number consider shared events related to the gene family evolution (Babicki et al., 2016, Jeffares et al., 2006, Rogozin et al., 2012). In a detailed evaluation of the MTP proteins, we predicted their 3D configuration, which considers supportive tools for expecting their function (Büyükköroğlu et al., 2018). The four temples in tomato MTP proteins indicated that these proteins with heavy metal where these transport proteins are in pant are classified into metal-uptake proteins that transport essential and toxic heavy metals to the cytoplasm and metal-uptake proteins. At the same time, the other is metal-efflux proteins that help the cell remove any excess heavy metals (Mani and Sankaranarayanan, 2018). On the other hand, protein–protein interaction analysis provided us with more knowledge about the plant developmental processes with their interactions with the environment (Struk et al., 2019). Our 11 nodes (MTPs) with 39 nodes indicate the significant interactions more than expected, reflecting that the MTP proteins are at least partially biologically connected as a group.
On the other hand, gene ontology is a fundamental analysis to predict putative functional contributions across living organisms (Consortium, 2010). Moreover, gene ontology classes and concepts have been used to define the relationships and gene functions existing between these concepts (Purwantini et al., 2014). Our gene ontology analysis revealed the significant role of the tomato SlMTP genes with heavy metals (Fig. 5). Furthermore, the GO showed the molecular functions, where more than 8 of them participate in metal-related processes such as transmembrane transporter activity, cation transmembrane transporter activity, transporter activity, and ion transmembrane transporter activity.
The previous transcriptomic is a proper tool for detecting the existence, structure, and quantity of the RNA in any abiological sample under certain conditions (Zambounis et al., 2020). Thus, we investigated the expression profile of all members of the MTP gene family from previously published RNA-sequencing data, which showed the expression of all gene members in all selected tomato tissues (Fig. 6 and Table S5). Digital data analysis showed that the significant roles of the MTP gene could contribute significantly to growth and development. Worth evidence has been obtained about the essential roles of tomato MTPs after tissue expression evaluation. For instance, the exclusive expression of the three genes SlMTP3, SlMTP4, and SlMTP6 were in the young flower, whereas SlMTP7 was most plentiful in mature fruits, indicating that they might be involved in early flower and fruit development. Besides the vital expected roles of SlMTP3 in fruit maturation and development, its expression has increased. However, only SlMTP2 and SlMTP8 were rarely expressed in all examined tissues from all SlMTPs. The documented down-regulation in some gene expressions is essential for maintaining the gene duplicates and ancestral functions (Qian et al., 2010). Hence in our study, the down-regulation of SlMTP2 and SlMTP8 expression is expected to be vital for keeping their biological functions and maintain them from losing during the cell evaluation. The reliability of the transcriptome data was further validated by qRT-PCR; however, the minor asymmetry between both analyses may be due to different growth conditions and tomato varieties, which finally affected the spatial expression. We examined the expression behaviour of MTP genes under five divalent metals (Mn2+, Cd2+, Co2+, Fe2+, and Zn2+). Numerous studies in other plants indicated the significant roles of the MTP gene family to enhance the plant tolerance against these metals (Gao et al., 2020, Montanini et al., 2007) as it was described as metal efflux transporters from the cytoplasm, mainly transporting Zn2+, but also transports Ni+2, Co2+, Cd2+, Fe2+, and Mn2+ (Ricachenevsky et al., 2013).
The transcript accumulation transcription of MTPs in response to various heavy metals was varied and complicated, although the gene expression response to different stresses is usually reflected in corresponding gene roles. In Arabidopsis, the tonoplast-localized Zn2+ transporter AtMTP1 showed slight changes in its expression with excess Zn2+ exposure at both transcription and translation levels (Dräger et al., 2004, Kobae et al., 2004). Moreover, although the high expression of CsMTP1 encoded protein, the gene expression was steady under the high concentration of Zn2+ in cucumber (Migocka et al., 2015).
As mentioned before, the up-regulation of AtMTP12 not dependent on Zn concentration, but it could transport Zn2+ by combining AtMTP5 in heterodimeric complex form (Fujiwara et al., 2015), similarly to findings of Liu et al. (2019) publication in tobacco. Moreover, Mn2+ different supplies have a slight effect on the expression of Mn-CDFs (AtMTP8, 9, 10, and 11) (Delhaize et al., 2007). Recently, similar findings were further described in tobacco (Liu et al., 2019). All Zn-CDF members except for SlMTP3 recorded slight changes in their expression with excess Zn2+ exposure in our study. Besides, the up-regulation of SlMTP6 of Zn/Fe-CDFs exceeds Zn2+, while it down-regulated by Fe2+ in different tomato tissues. Furthermore, only SlMTP10 of Mn-CDF class was highly affected by the accumulation of Mn2+. Therefore, our studies would be essential for investigating MTPs molecular roles in tomatoes under various heavy metals stresses.
Generally, these results would provide essential clues for clarifying the roles of SlMTPs in heavy metal tolerance and the mechanism of heavy metal transport mediated by SlMTP proteins. Taken together, these results would lay a theoretical and practical foundation for the functional characterization of SlMTP genes in future studies. Furthermore, the highest expressed MTPs (SlMTP1, SlMTP3, SlMTP4, SlMTP8, SlMTP10, and SlMTP11) can be used as bio-environmental markers for predicting heavy metal contamination based on their expression levels.
5. Conclusion
We provided the first genome-wide study of the MTP gene family in tomatoes, providing important comparative data for evolutionary relationships. Eleven identified MTP genes were phylogenetically divided into three major substrate-specific clusters (Zn/Fe-CDFs, Mn-CDFs, and Zn-CDFs), and seven groups seemed to have undergone expansion and gene loss after poly ploidization through segmental duplication. Gene ontology further showed the vital roles of MTPs in metal-related processes. Tissue-specific expression profiling exhibited similar expression patterns in the same group, whereas gene expression varied among groups. The expression patterns of SlMTP members in response to various heavy metals at different tissues indicated the significant role of these genes in tomato growth and development. Furthermore, our gene expression analysis of various heavy metals revealed the important parts of the MTPs, especially, SlMTP1, SlMTP3, SlMTP4, SlMTP8, SlMTP10, and SlMTP11 in-plant tolerance to heavy metals stresses. .
Funds
This work was supported by Innovation Research Team of Yibin University (No. 2017TD01 and 2018TD04).
CRediT authorship contribution statement
Ahmed H. El- Sappah: Conceptualization, Formal analysis, Writing - review & editing, Experimental design, Methodology. Ahmed S. Elrys: Methodology, Writing - review & editing. El-Sayed M. Desoky: Writing - review & editing. Xia Zhao: Writing - review & editing. Wang Bingwen: Writing - review & editing. Hamza H. El-Sappah: Writing - review & editing, Methodology. Yumin Zhu: Methodology. Zhou Wanhai: Writing - review & editing. Zhao Xianming: Writing - review & editing, Conceptualization. Jia Li: Writing - review & editing, Experimental design, Methodology, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2021.07.073.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Ali I., Asim M., Khan T.A. Arsenite removal from water by electro-coagulation on zinc–zinc and copper–copper electrodes. Int. J. Environ. Sci. Technol. 2013;10(2):377–384. [Google Scholar]
- Arrivault, S., Senger, T., Krämer, U., 2006. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. The Plant Journal 46, 861-879. [DOI] [PubMed]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büyükköroğlu, G., Dora, D.D., Özdemir, F., Hızel, C., 2018. Chapter 15 Techniques for Protein Analysis, Omics Technologies and Bio-Engineering, pp. 317-351.
- Cheng G., Chang P., Shen Y., Wu L., El-Sappah A.H., Zhang F., Liang Y. Comparing the Flavor Characteristics of 71 Tomato (Solanum lycopersicum) Accessions in Central Shaanxi. Frontiers in Plant Science. 2020;11:586834. doi: 10.3389/fpls.2020.586834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.-H., Car S., Socha A.L., Hindt M.N., Punshon T., Guerinot M.L. The Arabidopsis MTP8 transporter determines the localization of manganese and iron in seeds. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-11250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 2001;212(4):475–486. doi: 10.1007/s004250000458. [DOI] [PubMed] [Google Scholar]
- Cobb G.P., Sands K., Waters M., Wixson B.G., Dorward-King E. Accumulation of heavy metals by vegetables grown in mine wastes. Environ. Toxicol. Chem. Int. J. 2000;19(3):600–607. [Google Scholar]
- Conesa A., Götz S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genomics. 2008;2008:1–12. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, G.O., 2010. The Gene Ontology in 2010: extensions and refinements. Nucleic acids research 38, D331-D335. [DOI] [PMC free article] [PubMed]
- Delhaize E., Gruber B.D., Pittman J.K., White R.G., Leung H., Miao Y., Jiang L., Ryan P.R., Richardson A.E. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007;51:198–210. doi: 10.1111/j.1365-313X.2007.03138.x. [DOI] [PubMed] [Google Scholar]
- Desoky, E., Elrys, A.S., Rady, M.M., 2019. Licorice root extract boosts Capsicum annuum L. production and reduces fruit contamination on a heavy metals-contaminated saline soil. International Letters of Natural Sciences 73.
- Dräger, D.B., Desbrosses‐Fonrouge, A.G., Krach, C., Chardonnens, A.N., Meyer, R.C., Saumitou‐Laprade, P., Krämer, U., 2004. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co‐segregate with zinc tolerance and account for high MTP1 transcript levels. The Plant Journal 39, 425-439. [DOI] [PubMed]
- Edwards K.D., Fernandez-Pozo N., Drake-Stowe K., Humphry M., Evans A.D., Bombarely A., Allen F., Hurst R., White B., Kernodle S.P., Bromley J.R., Sanchez-Tamburrino J.P., Lewis R.S., Mueller L.A. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics. 2017;18(1) doi: 10.1186/s12864-017-3791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sappah A., Shawky A., Sayed-Ahmad M., Youssef M. Nile tilapia as bio indicator to estimate the contamination of water using SDS-PAGE and RAPDPCR techniques. Egyptian J. Genet. Cytol. 2012;41(2):209–227. [Google Scholar]
- El-Sappah A.H., I M.M., El-awady H.H., Yan S., Qi S., Liu J., Cheng G.T., Liang Y. Tomato natural resistance genes in controlling the root-knot nematode. Genes. 2019;10(11):925. doi: 10.3390/genes10110925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sappah A.H., Shawky A., Sayed-Ahmad M.S., Youssef M. Estimation of heat shock protein 70 (hsp 70) gene expression in nile tilapia (Oreochromis niloticus) using quantitative Real-Time PCR. Zagazig J. Agri. Res. 2017;44:1003–1015. [Google Scholar]
- Elrys, A.S., Merwad, A.-R.M., Abdo, A.I., Abdel-Fatah, M.K., Desoky, E.-S.M., 2018. Does the application of silicon and Moringa seed extract reduce heavy metals toxicity in potato tubers treated with phosphate fertilizers? Environmental Science and Pollution Research 25, 16776-16787. [DOI] [PubMed]
- Eroglu, S., Filiz, E., Vatansever, R., 2017. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. [DOI] [PubMed]
- Eroglu Seckin, Meier Bastian, von Wirén Nicolaus, Peiter Edgar. The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 2016;170(2):1030–1045. doi: 10.1104/pp.15.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Kawachi M., Sato Y., Mori H., Kutsuna N., Hasezawa S., Maeshima M. A high molecular mass zinc transporter MTP 12 forms a functional heteromeric complex with MTP 5 in the Golgi in Arabidopsis thaliana. The FEBS journal. 2015;282:1965–1979. doi: 10.1111/febs.13252. [DOI] [PubMed] [Google Scholar]
- Gao Yongfeng, Yang Fengming, Liu Jikai, Xie Wang, Zhang Lin, Chen Zihao, Peng Zhuoxi, Ou Yongbin, Yao Yinan. Genome-wide identification of metal tolerance protein genes in Populus trichocarpa and their roles in response to various heavy metal stresses. Int. J. Mol. Sci. 2020;21(5):1680. doi: 10.3390/ijms21051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Neha, Yadav Krishna Kumar, Kumar Vinit, Kumar Sandeep, Chadd Richard P., Kumar Amit. Trace elements in soil-vegetables interface: Translocation, bioaccumulation, toxicity and amelioration-A review. Sci. Total Environ. 2019;651:2927–2942. doi: 10.1016/j.scitotenv.2018.10.047. [DOI] [PubMed] [Google Scholar]
- Gustin J.L., Zanis M.J., Salt D.E. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 2011;11:1–13. doi: 10.1186/1471-2148-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.-K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7:1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G., 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296-1297. [DOI] [PMC free article] [PubMed]
- Jeffares, D.C., Mourier, T., Penny, D., 2006. The origin of introns. Trends in Genetics 1, 16-22. [DOI] [PubMed]
- Kawachi, M., Kobae, Y., Mimura, T., Maeshima, M., 2008. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. Journal of Biological Chemistry 283, 8374-8383. [DOI] [PMC free article] [PubMed]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y., Uemura T., Sato M.H., Ohnishi M., Mimura T., Nakagawa T., Maeshima M. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004;45:1749–1758. doi: 10.1093/pcp/pci015. [DOI] [PubMed] [Google Scholar]
- Kolaj-Robin O., Russell D., Hayes K.A., Pembroke J.T., Soulimane T. Cation diffusion facilitator family: structure and function. FEBS Lett. 2015;589:1283–1295. doi: 10.1016/j.febslet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Krzesłowska M. The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiologiae Plantarum. 2011;33:35–51. [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gao Y., Tang Y., Wang D., Chen X., Yao Y., Guo Y. Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front. Genet. 2019;10:345. doi: 10.3389/fgene.2019.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Pang X., Cheng Y., Yin Y., Zhang Q., Su W., Hu B., Guo Q., Ha S., Zhang J. The Hsp70 gene family in Solanum tuberosum: genome-wide identification, phylogeny, and expression patterns. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-34878-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Shi M., Wang J., Zhang B., Li Y., Wang J., El-Sappah A., Liang Y. Comparative Transcriptomic Analysis of the Development of Sepal Morphology in Tomato (Solanum Lycopersicum L.) Int. J. Mol. Sci. 2020;21:5914. doi: 10.3390/ijms21165914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mani A., Sankaranarayanan K. Heavy metal and mineral element-induced abiotic stress in rice plant. Rice Crop Current Develop. 2018:149. [Google Scholar]
- Migocka M., Kosieradzka A., Papierniak A., Maciaszczyk-Dziubinska E., Posyniak E., Garbiec A., Filleur S. Oxford University Press UK; 2015. Retracted: Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. [DOI] [PubMed] [Google Scholar]
- Migocka M., Papierniak A., Maciaszczyk-Dziubińska E., Poździk P., Posyniak E., Garbiec A., Filleur S. Oxford University Press UK; 2014. Retracted: Cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Montanini B., Blaudez D., Jeandroz S., Sanders D., Chalot M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics. 2007;8:1–16. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D.H., Silver S. Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- Ober D. Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 2005;10:444–449. doi: 10.1016/j.tplants.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Purwantini E., Torto-Alalibo T., Lomax J., Setubal J.C., Tyler B.M., Mukhopadhyay B. Genetic resources for methane production from biomass described with the Gene Ontology. Front. Microbiol. 2014;5:634. doi: 10.3389/fmicb.2014.00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W., Liao, B.-Y., Chang, A.Y.-F., Zhang, J., 2010. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends in Genetics 26, 425-430. [DOI] [PMC free article] [PubMed]
- Rai P.K., Lee S.S., Zhang M., Tsang Y.F., Kim K.-H. Heavy metals in food crops: Health risks, fate, mechanisms, and management. Environ. Int. 2019;125:365–385. doi: 10.1016/j.envint.2019.01.067. [DOI] [PubMed] [Google Scholar]
- Ricachenevsky F., Menguer P., Sperotto R., Williams L., Fett J. Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 2013;4 doi: 10.3389/fpls.2013.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I.B., Carmel L., Csuros M., Koonin E.V. Origin and evolution of spliceosomal introns. Biology direct. 2012;7:1–28. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. [9] TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Samuel M.S., Datta S., Khandge R.S., Selvarajan E. A state of the art review on characterization of heavy metal binding metallothioneins proteins and their widespread applications. Sci. Total Environ. 2021;775 [Google Scholar]
- Schlueter, J.A., Lin, J.-Y., Schlueter, S.D., Vasylenko-Sanders, I.F., Deshpande, S., Yi, J., O'bleness, M., Roe, B.A., Nelson, R.T., Scheffler, B.E., 2007. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC genomics 8, 1-16. [DOI] [PMC free article] [PubMed]
- Shirazi Z., Abedi A., Kordrostami M., Burritt D.J., Hossain M.A. Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L.) 3 Biotech. 2019;9:199. doi: 10.1007/s13205-019-1728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Sharma R.K., Agrawal M., Marshall F.M. Risk assessment of heavy metal toxicity through contaminated vegetables from waste water irrigated area of Varanasi, India. Trop. Ecol. 2010;51:375–387. [Google Scholar]
- Struk S., Jacobs A., Sánchez Martín-Fontecha E., Gevaert K., Cubas P., Goormachtig S. Exploring the protein–protein interaction landscape in plants. Plant Cell Environ. 2019;42:387–409. doi: 10.1111/pce.13433. [DOI] [PubMed] [Google Scholar]
- Takemoto Y., Tsunemitsu Y., Fujii-Kashino M., Mitani-Ueno N., Yamaji N., Ma J.F., Kato S.-I., Iwasaki K., Ueno D. The tonoplast-localized transporter MTP8. 2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol. 2017;58:1573–1582. doi: 10.1093/pcp/pcx082. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Thomine S., Vert G. Iron transport in plants: better be safe than sorry. Curr. Opin. Plant Biol. 2013;16:322–327. doi: 10.1016/j.pbi.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Tsunemitsu Y., Yamaji N., Ma J.F., Kato S.-I., Iwasaki K., Ueno D. Rice reduces Mn uptake in response to Mn stress. Plant Signaling Behav. 2018;13 doi: 10.1080/15592324.2017.1422466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zaal B.J., Neuteboom L.W., Pinas J.E., Chardonnens A.N., Schat H., Verkleij J.A., Hooykaas P.J. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999;119:1047–1056. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatansever R., Filiz E., Eroglu S. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals. 2017;30:217–235. doi: 10.1007/s10534-017-9997-x. [DOI] [PubMed] [Google Scholar]
- Wei F., Coe E., Nelson W., Bharti A.K., Engler F., Butler E., Kim H., Goicoechea J.L., Chen M., Lee S. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambounis A., Ganopoulos I., Valasiadis D., Karapetsi L., Madesis P. RNA sequencing-based transcriptome analysis of kiwifruit infected by Botrytis cinerea. Physiol. Mol. Plant Pathol. 2020;111 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.