Abstract
Acaudina molpadioides has been long used as traditional medicinal resources and reported to demonstrate various important bioactivities such as anticoagulation, antithrombosis, anti-hyperglycemia and anticancer. However, its lipid lowering activity is yet to be fully explored. Proprotein convertase subtilisin/kexin type 9 (PCSK9) is an enzyme that enhances the lysosomal degradation of hepatic low density lipoprotein receptor (LDLR) resulting in excessive accumulation of the plasma levels of LDL-cholesterols (LDL-C) which subsequently accelerate atherosclerosis. In the present study, A. molpadioides fractions were subjected to promoter-reporter luciferase assay to determine its role as PCSK9 inhibitors. It was found both fractions (EFA and EFB) reduced the transcriptional activity of PCSK9 promoter. Among the seven 5′end deletion constructs of PCSK9 promoter, fragments D1 (−1,711/−94), D3 (−709/−94) and D4 (−440/−94), were suppressed in the presence of both fractions whereas D2 (−1,214/−94), and, D6 (−351/−94) as well as D7 (−335/−94) were inhibited only by EFA and EFB, respectively. Further transcription factor binding sites prediction using MatInspector software discovered various potential cis-regulatory elements namely, PPAR, KLFs, RBPJ-kappa and SREBP that may potentially be involved in ameliorating the transcriptional activity of PCSK9. Immunofluorescence staining was used to evaluate the effects of both fractions on LDL-C and LDLR. Results showed that levels of LDL-C uptake in EFA-treated cells were 69.1% followed by EFB at 32.6%, as compared to untreated control after 24 h treatment. The LDLR protein distribution was induced by 62.41% and 32.2%, which corresponded to an increase in LDL-C uptake in both EFA and EFB treatment, respectively. Hence, the inhibition of PCSK9 by bioactive compounds in EFA and EFB could be another promising therapeutic agent in reducing the cholesterol levels and atherosclerosis by targeting PCSK9.
Keywords: Atherosclerosis, Low density lipoprotein, Promoter-reporter based assay, PCSK9, Acaudina molpadioides
1. Introduction
Atherosclerosis is a chronic autoimmune inflammatory response in the walls of arteries. It is the major cause of heart disease, stroke, and the leading cause of morbidity and mortality worldwide (Virani et al., 2020). Atherosclerosis is characterized by a build-up of fatty deposits and necrotic plaque formation in the arterial walls due to lipid oxidation, persistent inflammation and subsequent immune response disturbances (Sega et al., 2019). It is well established the primary risk factor for atherosclerosis was a constant elevated blood lipid levels (Nelson, 2013). In addition, there are also other risk factors that lead to the development of this pathophysiological condition such as sedentary lifestyle, smoking habit, obesity, diabetes mellitus, hypertension, gender and family history with complex genetic susceptibility to the disease (Bentzon et al., 2014).
Current available commercialized drugs were developed by targeting proteins that are responsible in reducing the plasma lipids as a constant elevated level of LDL-C is associated with the occurrence of atherosclerosis. Statins are the most widely used drugs that function to decrease the levels of plasma lipids by inhibiting the synthesis of endogenous lipids. However, these drugs produced residual risk or adverse effects such as myalgia or myopathy (Melendez et al., 2017) and liver damage (Wang et al., 2016). Statin treatment also increases blood levels of glucose and glycosylated haemoglobin, and, may lead to the onset of diabetes if the patient was obese with elevated triglycerides and glucose level (Judge et al., 2010, Crandall et al., 2017). In some patients, the clinical benefits of statin treatment decrease overtime once LDL levels were reduced to a certain level. Therefore it is necessary to overcome such limitations of statin therapy by imploring other target and proprotein convertase subtilisin/kexin type 9 (PCSK9) may offer an alternative target for hypercholesterolemic patients whom are statins intolerance (Chae et al., 2018).
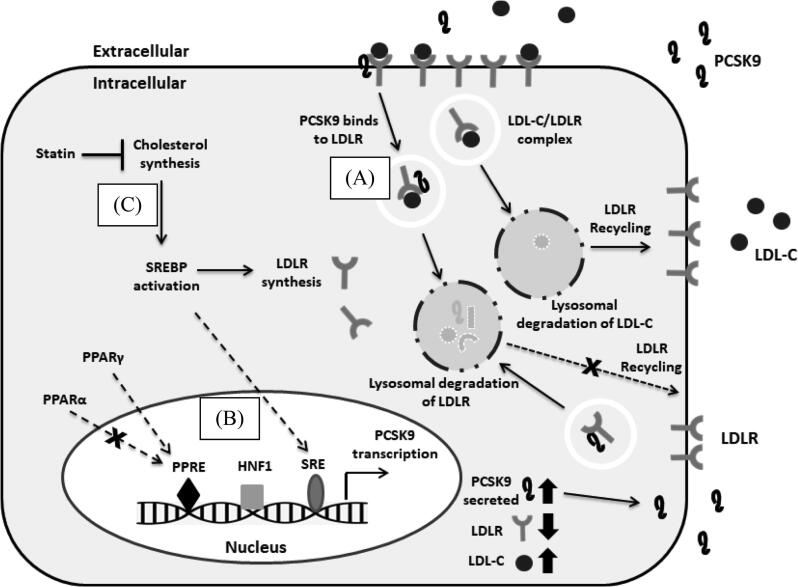
PCSK9 is predominantly expressed in adult liver cells and synthesized as a 72 kDa zymogen. It is secreted into the plasma after going through maturation process by self-engaged autocatalytic cleavage in the endoplasmic reticulum to form a heterodimer of a prosegment of 122 amino acids and a 60 kDa active form (Seidah et al., 2003). It plays a pivotal role in controlling the levels of plasma LDL-C by binding to extracellular domain of hepatic low density lipoprotein receptor (LDLR). The formation of PCSK9-LDLR complex leads to the intracellular degradation of LDLR in lysosomes and reduction in the number of LDLR presence on the hepatocytes, which in turn, increase in the levels of plasma LDL-C (Zhang et al., 2007, McNutt et al., 2009, Seidah et al., 2014). In addition, gain of function mutations of PCSK9 in human leads to an increase in the binding affinity of the enzyme to LDLR, thus enhances the degradation of LDLR (Abifadel et al., 2003, Allard et al., 2005). By contrast, loss of function mutations causes the defects in PCSK9 and decreases the levels of LDL-C which results in a reduction in cardiovascular risks (Cohen et al., 2006) (Fig. 1A).
Fig. 1.
The correlation of PCSK9 and statin in regulating the metabolism of LDL-C in the PCSK9 and LDLR are synthesized in the liver. The secreted PCSK9 protein binds to the extracellular domain of LDLR at the cell surface. The PCSK9/LDLR-LDL-C complex enters via the endosomal pathway and is directed to the lysosomal compartment for degradation of both PCSK9 and LDLR, decreasing the number of LDLRs available for clearance of LDL-C particles (A). The gene expression of PCSK9 is subjected to the regulation by the binding of SREBP2, HNF1 and PPAR to their corresponding binding sites on PCSK9 promoter (B). Activation of SREBP2, under conditions of intracellular cholesterol depletion due to inhibitory activity of statin, increases the expression of both PCSK9 and LDLR (C).
PCSK9 expression is mainly regulated at the transcriptional levels. Various transcription factor binding sites have been identified that play a significant role in mediating the rate of PCSK9 gene transcription. Sterol regulatory element binding protein (SREBP)-2 and SREBP-1c are transcription factors that bind to sterol binding element (SRE) present on PCSK9 promoter which triggers the upregulation of PCSK9 gene expression (Costet et al., 2006, Jeong et al., 2008). Proximal to SRE, a binding site to hepatocyte nuclear factor 1 (HNF1)-α is identified and responsible in increasing the transcription of PCSK9 (Li et al., 2009). In addition, a binding site located between SRE and HNF1 site is designated for histone nuclear factor P (HINFP) (Fig. 1B). This transcription factor increases the PCSK9 transcription by histone H4 acetylation via the formation of the complex of HINFP, its cofactor nuclear protein of the ATM locus (NPAT) and the histone acetyltransferase cofactor transformation/transcription domain-associated protein (TRRAP) (Li and Liu, 2012). Another transcription factor, peroxisome proliferator-activated receptor gamma (PPARγ), also induces PCSK9 gene expression upon the binding by its ligand and dephosphorylation by inhibition of ERK1/2 activity (Duan et al., 2012) (Fig. 1B).
Despite statin being the most prescribed LDL-C lowering drug, molecular findings showed that statin treatment increased the expression of SREBP-2 and HNF1α which resulted in higher induction of PCSK9 than LDLR transcription alone which enhanced PCSK9 mediated LDLR degradation (Dong et al., 2010, Nozue, 2017) (Fig. 1C). In conjunction to this, statin therapy alone is still inadequate to achieve LDL-C target in some high-risk patients (Crossey et al., 2015). Thus, the administration of both statins and PCSK9 inhibitors will be an effective strategy in ensuring LDLR would not be degraded by statin-induced PCSK9. The side effects of statins have urged researchers to discover natural products with good hypolipidemic effects and lower toxicity.
Due to the rich oceanic biodiversity, marine organisms are valuable sources of nutritious foods as well as represent novel reservoirs for biologically active secondary metabolites (Bordbar et al., 2011). One of the marine resources that show various biological activities is Acaudina molpadioides. This unique marine organism commonly known as sea potato and belongs to the Holothuroidea family (Wang et al., 2019a, Wang et al., 2019b). Mainly, it can be found in the intertidal zone in Langkawi Island situated at the west coast of Peninsular Malaysia (Halim et al., 2017). In addition, sea potato was also used as medicinal resource and consumed as traditional healthy food and in China, Japan, Korea, and some south eastern Asian countries (Wang et al., 2012). Conventionally, it is used as traditional medicine for heart disease treatment in Malaysia (Halim et al., 2017). An array of studies has also reported its antitumor, antioxidant, anticoagulant, antihypertensive, antihyperglycemic, antiadipogenic and anti-inflammatory effects (Wang et al., 2019a, Wang et al., 2019b) (Table 2).
Table 2.
Biological compounds of A. molpadioides with the potential activity in reducing the progression of atherosclerosis.
| Compound / metabolites | Biological activities | References |
|---|---|---|
| Cerebroside | Reduces monosaturated fatty acid biosynthesis | Hu et al., 2014a, Hu et al., 2014b |
| Cerebroside | Increases insulin sensitivity, reduces serum and hepatic LDL level, decreases atherosclerotic lesion formation and attenuates inflammation by decreasing the levels of inflammatory cytokines, such as CRP, TNFα, IL-6 | Zhang et al., 2018 |
| Fucosylated polysaccharide sulfate | Anticoagulant activity | Dong et al., 2014 |
| Fucoidan | Attenuates TGF-β1 signal which associated with the improvement against hyperglycemia, obesity, oxidative stress and inflammation | Hu et al., 2017 |
| Peptides | Anti-angiotensin converting enzyme (ACE) activity | Jiang et al., 2018 |
| Fucosylated chondroitin sulfate | Promotes antiadipogenic activity by increasing β-catenin, a negative regulator in adipogenesis, and decreases the expression of SREBP-1c, PPARγ and C/EBPα. | Xu et al., 2015 |
| Fucoidan | Exhibits antiadipogenic activity by modulating the Wnt/β-catenin pathway and downregulating the expression of SREBP-1c PPARγ and C/EBPα, while SREBP-1c | Xu et al., 2014 |
| Fucoidan | Antihypertensive activity | Zhao et al., 2009 |
| Fucosylated chondroitin sulphate | Mitigates hyperglycaemia /insulin resistance by increasing glucose uptake and GLUT4 translocation | Hu et al., 2014b |
| Cerebroside | Ameliorates nonalcoholic fatty liver disease (NAFLD) through suppression of stearoyl-CoA desaturase activity, reduces hepatic triglyceride (TG) and total cholesterol (TC) levels | Xu et al., 2011 |
| Collagen | Antioxidant activity | Li et al., 2020 |
| Cerebroside | Antitumor activity by inducing apoptosis | Du et al., 2012 |
| Collagen | Potent antioxidant activity by preventing the injury of nerve cells induced by hydrogen peroxide | Yu and Chen, 2014 |
However to date, there is still no study that has been carried out in elucidating the role of A. molpadioides in regulating the gene expression of PCSK9 as well as the potential cis-acting elements that may be responsible in modulating the PCSK9 promoter activity, which in turn, inducing the expression of LDLR and uptake of LDL-C by liver cells.
2. Materials and methods
2.1. Preparation of enhanced fractions from A. molpadioides
A. molpadioides sample was collected from the intertidal zone of Pulau Langkawi, Kedah, Malaysia and identified taxonomically by Professor Noraznawati Ismail, a marine biologist in Universiti Malaysia Terengganu (Fig. 2). The enhanced fractions of A. molpadioides were prepared using medium pressure liquid chromatography (MPLC) according to previous report with some modification (Gul-e-Saba et al., 2018). Briefly, the samples were chopped after cleaning and freeze–dried to remove the water content. Subsequently, 10 g of the dried sample was macerated in the methanol and subjected to extraction process. The polarity of the solvents was increased starting with hexane then followed by methanol. The mixture was filtered and concentrated using rotary evaporator (Buchi). The methanolic extract of A. molpadioides was treated using the HLB cartridge which was priorly conditioned by flowing through 3 mL of absolute methanol followed by 3 mL of deionised water. After that, 5 g of the dried methanol extract was reconstituted with 50 mL of MeOH:H2O (in the ratio of 8:2) + 0.1 % TFA (Trifluoroacetic acid). The mixture was centrifuged at 14,000 rpm for 15 min at 4 °C. The eluent was collected into tube and dried using MiVac Quattro dryer. Dried extract was reconstituted with 3 mL of 90:10 H2O: MeOH + 0.1% TFA and filtered through 0.2 µM polytetrafluoroethylene (PTFE) filter if un-dissolved sample persisted. The sample mixture was then loaded into MPLC Flash Chromatography and the parameter used was a gradient of H2O: MeOH + 0.1% TFA (0–100%) with flow rate of 5 mL. Fine packing material used in the stationary phase was silica gel which enables high resolution (5–15 µM beads). 10 mL of fraction was collected into each tube. Every 10 tubes were pooled and designated as one fraction. The collected fractions were concentrated using rotary evaporator for further study. A total of 10 fractions were collected and subjected to preliminary promoter-reporter screening against PCSK9. From the assay only two fractions exhibited potent inhibitory activity against PCSK9, hence the fractions were designated as enhanced fraction A and B (EFA and EFB), respectively.
Fig. 2.
Acaudina molpadioides is an invertebrate marine organism, commonly known as sea potato and belongs to the Holothuroidea family. It is widely consumed by locals in South Eastern Asian countries as traditional healthy food. The figure shows sea potato that was collected from the intertidal zone of Pulau Langkawi, Kedah, Malaysia.
2.2. PCSK9 promoter-reporter plasmid
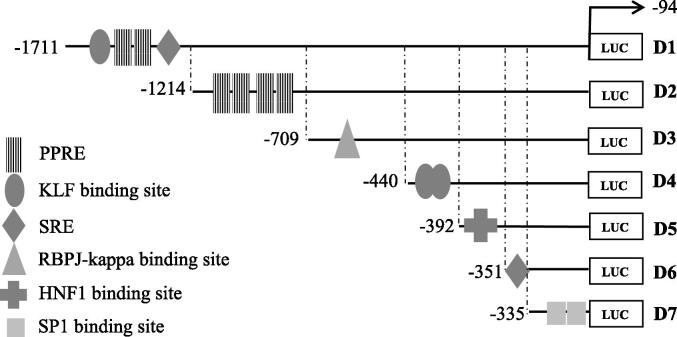
In order to determine the regions of the PCSK9 promoter that played an important role in mediating the inhibitory actions of A. molpadioides on the transcriptional activity of PCSK9 promoter, seven 5′end deletion constructs of 2 kb PCSK9 promoter were used and designated as D1 (−1,711/−94), D2 (−1,214/−94), D3 (−709/−94), D4 (−440/−94), D5 (−392/−94), D6 (−351/−94) and D7 (−335/−94) (Jeong et al., 2008). The potential binding sites responsible in modulating PCSK9 trancription activity on the promoter were predicted using MatInspector software (Cartharius et al., 2005).
2.3. Treatment of HepG2 cells with A. molpadioides enhanced fractions
HepG2 cells were purchased from American Type Culture Collection (ATCC, USA) and maintained in minimum essential medium (MEM) supplemented with 10% (v/v) FBS, 1% (v/v) 1 mM sodium pyruvate and 1% (v/v) 100 mM penicillin–streptomycin. Cells were cultured in T25 cell culture flask and incubated at 37 ˚C in humidified incubator supplied with 5% (v/v) CO2. After reaching 70–80% confluence, HepG2 cells were washed twice with PBS before the treatment with two A. molpadioides fractions designated as EFA and EFB for a requisite time.
2.4. Transient transfection and luciferase assay
Cells were seeded into a clear bottom 96-well plate at 4 × 105 cells per well in a volume of 100 µL and grown overnight at 37 °C in humidified incubator in the presence of 5% (v/v) CO2. Transient transfection assay was carried out using Lipofectamine® Plus Reagent (Invitrogen) according to the manufacturer’s instruction. Solution A consisted of 1.5 µL Lipofectamine® solution diluted in 25 µL of serum free media Opti-MEM. Solution B was prepared by diluting 0.5 µL plus reagent, 2 µg of PCSK9 promoter-luciferase plasmid and 0.5 µg pRL-TK plasmid in 25 µL Opti-MEM. Both solutions were mixed gently and incubated for 30 min at room temperature. The medium in 96-well plate was discarded and the cells were washed twice with PBS before 90 µL Opti-MEM was added into each well. Subsequently, 10 µL of the mixture was pipetted into each well and incubated for additional 5 h before treatment. The culture medium was then replaced with fresh low serum medium (0.5% FBS) containing a series of two-fold dilutions of EFA or EFB (3.13 – 50 µg/mL) and incubated for 24 h. 20 µM berberine sulphate (BBR) was used as the positive control (Jia, et al., 2014) and 1% (v/v) dimethyl sulfoxide (DMSO) as negative control.
After treatment, PCSK9 promoter activity was determined by using Dual Glo Luciferase Assay System (Promega). Briefly, 90 µL Dual Glo® Luciferase Reagent was added into each well and incubated for 10 min at room temperature in low light condition. Firefly luminescence was then measured using Glomax-multi Plus microplate reader (Promega). Subsequently, 90 µL Dual Glo® Stop & Glo® reagent was added to the plate and incubated for 10 min in low light condition to measure Renilla luminescence. The firefly luciferase activity of individual transfections was normalized to the Renilla luciferase activity.
2.5. Quantitative Real-time PCR
Total cellular RNA was isolated from HepG2 cells and lysed with TRIzol reagent (ThermoFisher). The lysate was mixed with chloroform and centrifuged for 10 min at 16,200g at 4 °C. The top aqueous phase, which contained RNA, was collected, mixed with isopropanol and centrifuged for 10 min at 12,000g at 4 °C. RNA pellet was washed with 75% (v/v) ethanol, resuspended in RNase-free water and subjected to RQ1 RNase-free DNase (Promega) at 37 °C for 30 min followed by incubation at 65 °C for 10 min.
Real time-PCR (qPCR) was performed using iTaqTM Universal SYBR® Green One-Step Kit (Bio-Rad) master mix according to the manufacturer’s instruction. The gene primers used in this study were enlisted in Table 1. Each reaction contained 150 ng of DNase-treated RNA, 10 µL of 2x SYBR® Green qPCR reaction mix (BIORAD), 0.6 µL of 10 µM each of the forward primer and reverse primer (Table 1), 0.25 µL of iScript reverse transcriptase for one step qPCR and nuclease free water to make up the total reaction volume of 20 µL. All the samples were run in triplicate. One-step Real-Time PCR was carried out using CFX6 Real-Time PCR Detection System (Bio-Rad) with an initial reverse transcription reaction at 50 °C for 10 min, polymerase activation and DNA denaturation at 95 °C for 1 min, followed by 39 cycles of denaturation step at 95 °C for 10 sec, annealing step at 59 °C for 30 sec and extension at 72 °C for 30 sec. Melt curve analysis was then carried out at 95 °C for 1 min, 55 °C for 1 min and 40 cycles of 70 °C for 10 sec, with 0.5 °C increment for each cycle. Expression of PCSK9 mRNA was analyzed using Bio-Rad CFX (Gene Expression Analysis) software and calculated by the comparative cycle threshold (Ct) method. The target mRNA Cts were normalized with housekeeping gene β-actin.
Table 1.
The sequences of primers used in Real-Time PCR.
| Names | Primers | Sequences (5′-3′) |
|---|---|---|
| PCSK9 (Jeong et al., 2008) | Forward | GGCAGGTTGGCAGCTGTTT |
| Reverse | CGTGTAGGCCCCGAGTGT | |
| β-actin | ||
| Forward | TCACCCTGAAGTACCCCATC | |
| Reverse | CCATCTCTTGCTCGAAGTCC |
2.6. LDL-C uptake and LDLR immunofluorescence assays
The uptake of LDL-C by LDLR was determined by using LDL uptake assay kit (Abcam®) and was carried out according to the manufacturer’s instructions. For cell treatment, HepG2 cells were first cultured in 96-well plate at 3 × 104 cells/well and allowed to grow for two days. Cells were subsequently treated with EFA or EFB for requisite times. The medium was then replaced with 100 µL/well of human LDL-conjugated DyLightTM 550 working solution and incubated for 4 h. The degree of LDL-C uptake was observed at wavelengths 540 and 570 nm and the image was acquired using high content screening system (Molecular Devices) and MetaXpress® 5.1. Fluorescence intensity was measured and analysed with ImageJ.
Subsequently, the cells were washed with TBS and fixed with 100 µL/well of Cell-Based Assay Fixative Solution for 10 min. Cells were then washed continuously with TBST (0.1% (v/v) Triton-X in TBS) 3 times for 5 min each. The wells were then blocked by incubating the plate for 30 min with 100 µL/well of Cell-Based Assay Blocking Solution, followed by incubation with 100 µL/well of Rabbit Anti-LDL primary antibody for 1 h. The cells were then washed with TBST and incubated with DyLight ™ 488-Conjugated Goat Anti-Rabbit IgG secondary antibody in the dark for 1 h. The intensity of fluorescence stained LDLR was observed at the wavelengths 485 and 535 nm. Images were acquired using high content screening system (Molecular Devices) and MetaXpress® 5.1. Fluorescence intensity was measured and analysed with ImageJ.
2.7. Statistical analysis
All experimental data were shown as the mean value ± SD. Student’s t-test was carried out to compare statistical significance of means between test sample and the control group. Multiple groups were analysed with one-way analysis of variance (ANOVA) using SPSS 25 where means were separated with Duncan’s multiple range test group at P value < 0.05.
3. Results
3.1. A. molpadioides inhibits PCSK9 gene expression in HepG2 cells
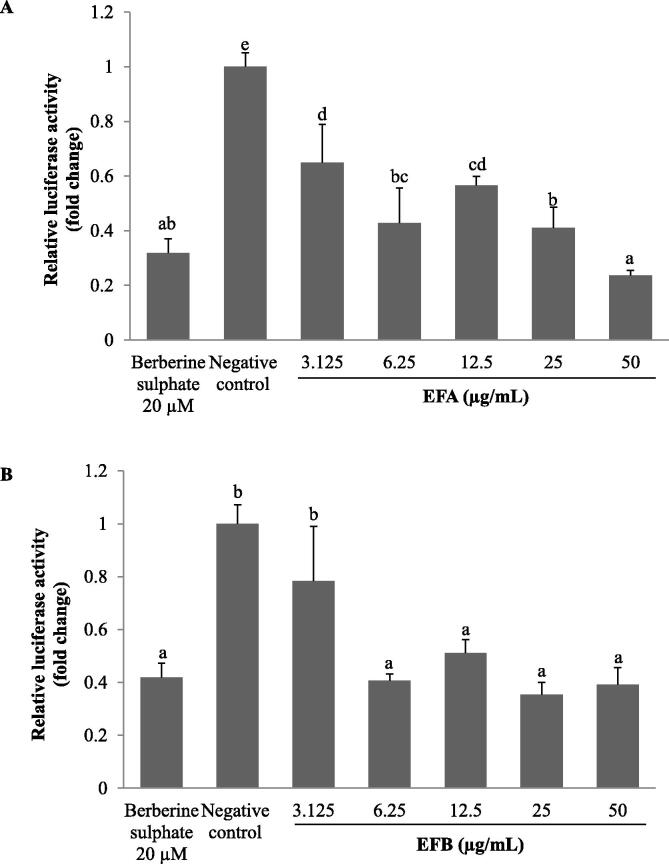
As shown in Fig. 3A, the transcriptional activity of PCSK9 promoter was decreased in dose dependent manner when the cells were treated with EFA from 3.13 to 50 μg/mL. At concentration 25 and 50 µg/mL, the transcriptional level of PCSK9 was significantly reduced by 58.9% and 76.3% as compared to negative control (DMSO), respectively. Nevertheless, there was no significant difference between the reduction induced by 25 and 50 µg/mL EFA, and, BBR treatment. The transcriptional activity of PCSK9 treated with EFB produced similar dose dependent pattern as EFA. Interestingly, all concentrations used to treat HepG2 cells exerted a significant inhibition than to that of untreated control except at 3.125 μg/mL, ranging between 21.7% and 64.5%. However, there were no significant differences in HepG2 cells treated with EFB between concentration 6.25 to 50 µg/mL and the positive control (Fig. 3B).
Fig. 3.
The effects of A. molpadioides enhanced fractions, EFA (A) and EFB (B) on human proprotein convertase subtilisin/kexin type 9 (PCSK9) promoter activity. HepG2 cells were transfected with a recombinant pGL3 reporter plasmid comprising the human PCSK9 promoter sequence linked to firefly luciferase gene. The transfected cells were then treated with various concentrations of EFA and EFB for 24 h, and subsequently subjected to luciferase assay. The transcriptional activity of PCSK9 promoter was decreased in dose dependent manner in both EFA and EFB (except at 3.125 μg/mL)-treated HepG2 cells with 20 µM berberine sulphate as positive control and 1% (v/v) dimethyl sulfoxide (DMSO) as negative control. Each value represents the mean ± SD of three independent experiments (each in triplicate reaction). Multiple comparisons were done using one-way analysis of variance (ANOVA) where means with different letters were separated with Duncan’s multiple range test group at p < 0.05.
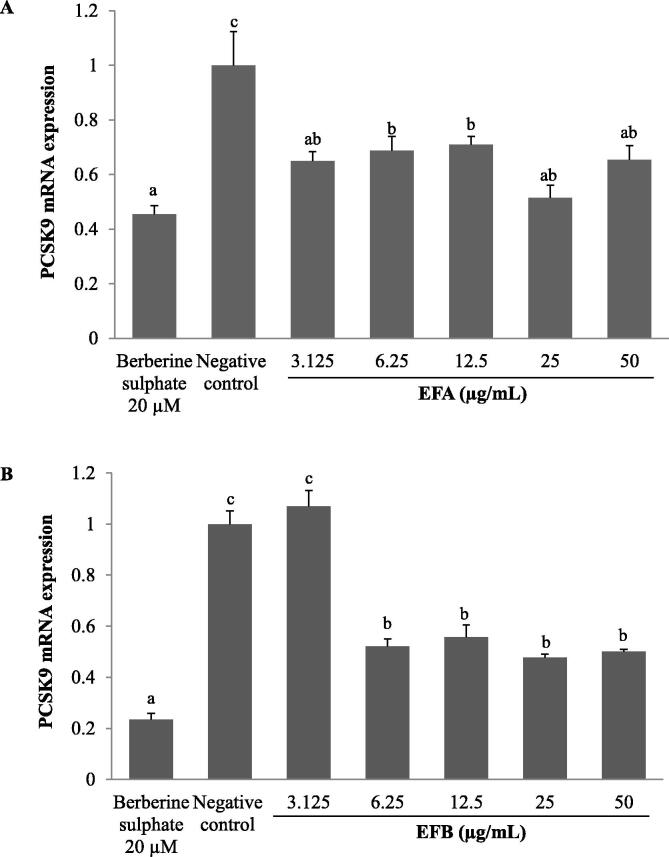
In order to determine the inhibitory action of EFA and EFB on PCSK9 promoter activity was also observed at the steady-state levels of mRNA, real-time PCR was carried out. Cells were treated with the similar concentrations as used in transient transfection assay for 24 h. As shown in Fig. 4A, the mRNA expression levels of PCSK9 were closely followed the inhibitory effects of EFA on PCSK9 promoter activity. At all concentrations used, PCSK9 mRNA levels were significantly decreased than to that of untreated control between 51% and 71% with 25 μg/mL produced the lowest levels at 51%.
Fig. 4.
The effect of A. molpadioides fractions, EFA (A) and EFB (B) on proprotein convertase subtilisin/kexin type 9 (PCSK9) mRNA expression. HepG2 cells were cultured until 80% confluence prior to treatment with EFA and EFB, 20 µM berberine sulphate as positive control and 1% (v/v) dimethyl sulfoxide (DMSO) as negative control, for 24 h. Total cellular RNA was isolated and subjected to real time-PCR analysis. Both EFA and EFB produced the lowest PCSK9 mRNA level at 25 µg/mL. Multiple comparisons were done using one-way analysis of variance (ANOVA) where means with different letters were separated with Duncan’s multiple range test group at p < 0.05.
In EFB treated HepG2 cells, concentrations 6.25, 12.5, 25 and 50 µg/mL also exhibited a significant reduction in PCSK9 mRNA expression as compared to untreated control. By contrast, 3.125 µg/mL EFB did not produce any significant change on the levels of PCSK9 mRNA (Fig. 4B).
3.2. A. molpadioides may potentially regulate the regulation of PCSK9 expression via cis-acting elements presence in PCSK9 promoter
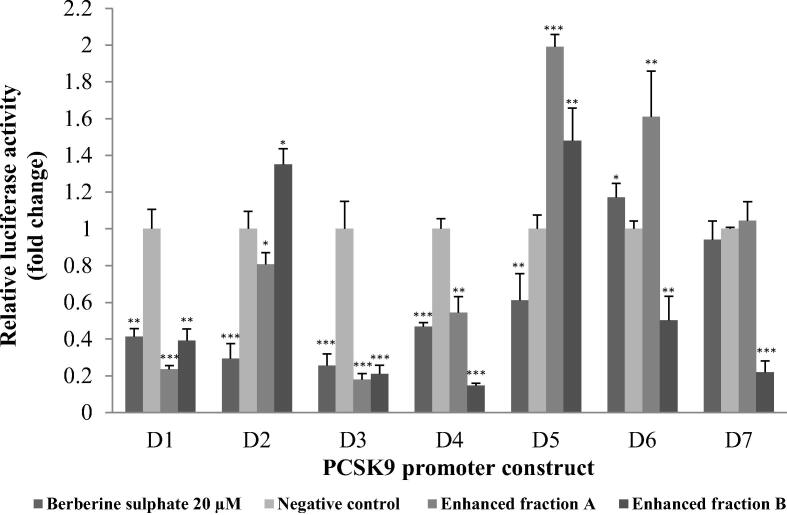
In order to delineate the A. molpadioides inhibitory elements on PCSK9 promoter, 5′-end deletion constructs of PCSK9 promoter namely D1 (−1,711/−94), D2 (−1,214/−94), D3 (−709/−94), D4 (−440/−94), D5 (−392/−94), D6 (−351/−94) and D7 (−335/−94) were transiently transfected into HepG2 cells (Jeong et al., 2008), and, treated with either EFA or EFB. BBR was used as positive control and DMSO as negative control. It was demonstrated that EFA significantly decreased the transcriptional activity of promoter D1, D2, D3, and D4 by 76%, 19%, 82% and 46% of untreated control, respectively, suggesting that cis-acting elements present in these three fragments were responsible in mediating the inhibitory action of EFA (Fig. 5). In contrast, transcriptional activity of promoter fragments D5 and D6 was increased when the cells were treated with EFA. BBR also decreased PCSK9 transcriptional activity of promoter fragments D1, D2, D3, D4 and D5.
Fig. 5.
Proprotein convertase subtilisin/kexin type 9 (PCSK9) promoter constructs analysis. HepG2 cells that were transfected with each fragment (D1-D7) was treated for 24 h with 25 µg/mL of A. molpadiodes EFA and EFB, with 20 µM berberine sulphate as positive control and 1% (v/v) dimethyl sulfoxide (DMSO) as negative control. Both EFA and EFB significantly decreased the transcriptional activity of promoter fragments D1(-1711/-94 bp), D3(-709/-94 bp) and D4(-440/-94 bp) which indicated that cis-acting elements presence in between these fragment regions were responsible in mediating the inhibitory effect of A. molpadioides on PCSK9 transcriptional activity. Values are means ± standard deviation and unpaired t-test was perform with * p < 0.05; **p < 0.001 and ***p < 0.001.
As shown in Fig. 5, EFB significantly decreased the transcriptional activity of promoter fragment D1, D3, D4, D6 and D7 by 61%, 79%, 85%, 50% and 78% of untreated control, respectively. However, there was a significant increase in promoter fragments D2 promoter activity in EFB treated cells. It is interesting to note that both EFA and EFB significantly decreased the transcriptional activity of promoter fragments D1, D3 and D4 which indicate that cis-acting elements presence in these fragments were responsible in mediating the inhibitory effect of the fractions on PCSK9 transcriptional activity.
3.3. Analysis of PCSK9 promoter reveals potential binding sites for transcription factors in reducing transcriptional activity
The prediction analysis of transcription factor binding site (TFBS) that may potentially be present in the PCSK9 promoter was carried out using MatInspector software. TFBS analysis showed that four binding sites were predicted between the region −1711/−1214 of the D1 promoter fragment. Two binding sites for PPAR (PPRE) were present between regions −1521/-1499 and −1506/−1484. A Krüppel like factor (KLF) binding site was also discovered upstream of the PPRE binding site, between region −1629/−1611. Sterol regulatory element binding proteins binding sites, sterol regulatory element (SRE) was identified at location −1237/−1223.
Deletion of 497 bp from the 5′ proximal region of D1 to produce D2 fragment revealed another four PPRE between regions −1200/−1178, −1116/−1094, −746/−724 and −739/−717, respectively. By contrast, no similar binding sites were found in D3, D4, D5, D6 and D7 fragments (Fig. 6). Analysis of a region between −709 and −440 of the 5′ end of D3 fragment revealed the presence of the recombination signal binding protein for immunoglobin kappa J region (RBPJ – kappa) binding site between regions −549/−537.
Fig. 6.
Schematic of the distribution of the predicted potential transcription binding sites on the PCSK9 promoter. The transcriptional start site (TSS) is indicated by the arrow. MatInspector software was used to predict transcription factor binding sites on the PCSK9 promoter. PPRE, peroxisome proliferator response element ; KLF, Krüppel like factor; SRE, sterol regulatory element; RBPJ-kappa, recombination signal binding protein for immunoglobin kappa J; HNF1, hepatocyte nuclear factor 1; SP1, specificity protein 1.
Further sequential deletion analysis of 269 bp region of 5′-end from D3 to produce D4 promoter fragment uncovered additional KLF binding sites between region −434/−416 and −430/−412, respectively. The deletion of a small segment of 48 bp from D4 to produce subsequent D5 fragment revealed the binding site for hepatocyte nuclear factor 1(HNF1) between region −387/−371. The predicted sterol regulatory element binding proteins binding sites, sterol regulatory element (SRE) was identified at 5′ end of D6 promoter fragment at −346/−337. Additional transcription factor binding sites namely SP1 (−192/176 and −197/−179) were found to be present within D7 fragment.
3.4. A. molpadioides enhances LDL-C uptake and upregulates LDLR expression.
In order to determine the inhibitory effects of A. molpadioides on transcriptional activity and mRNA levels of PCSK9, led to an increase in the levels of LDL-C uptake and LDLR, immunofluorescence assays were carried out utilising respective labelled fluorescence markers.
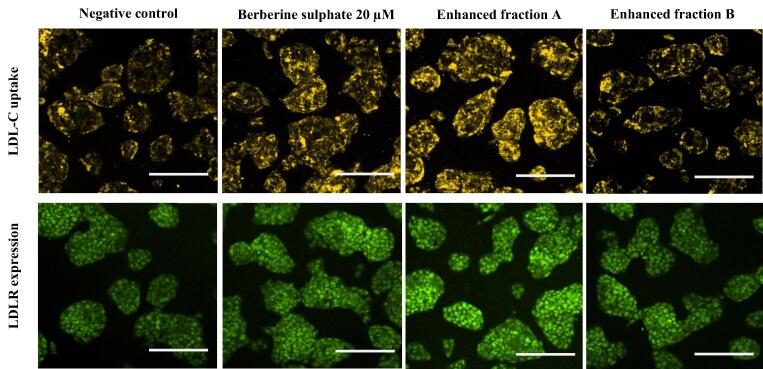
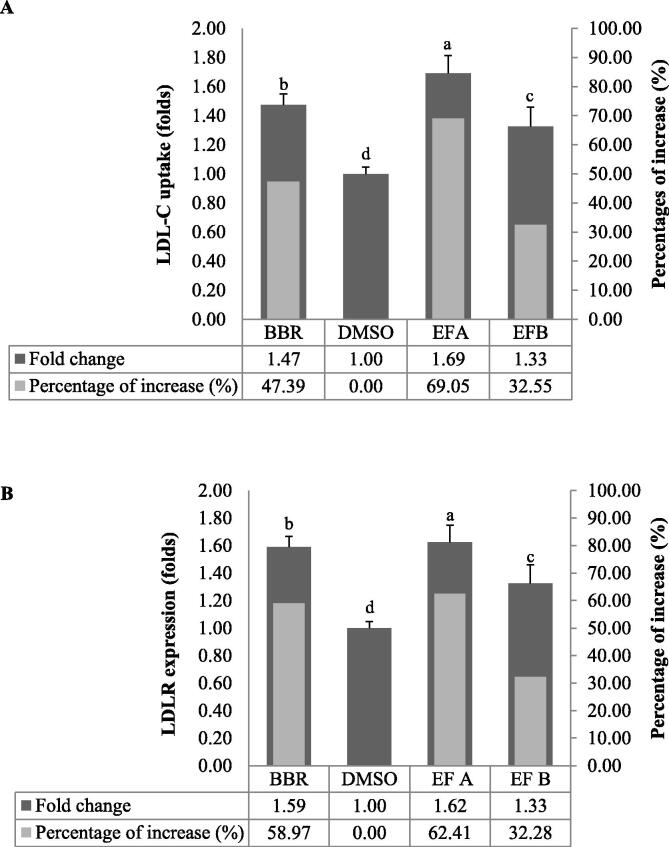
Treatment with EFA at 24 h produced the highest intensity in cells stained with LDL-DyLight™ 550 (yellow fluorescence). EFB also produced higher intensity in stained cells than to that of untreated control but lower than EFA-treated cells (Fig. 7). ImageJ analysis demonstrated that the levels of LDL-C uptake were the highest at 69.1% (1.69 folds) when the cells were treated with EFA whereas EFB treatment produced 32.6 % (1.33 folds) increase in LDL-C uptake as compared to untreated control (Fig. 8A). BBR treatment increased the LDL-C uptake at 47.4% (1.47 folds) (Fig. 8A).
Fig. 7.
LDL-C uptake and LDLR protein expression on HepG2 cells treated with 25 µg/mL A. molpadioides EFA and EFB, 20 µM berberine sulphate as positive control and 1% (v/v) dimethyl sulfoxide (DMSO) as negative control. HepG2 cells were treated with the mediators for 24 h and the LDL-C uptake was observed. At the end of the treatment duration, the culture medium was replaced with 100 µL/well LDL-DyLight™ 550 working solution and incubated for additional 3 to 4 h. Stained cells were observed with high content screening (HCS) with filters capable of measuring excitation and emission wavelengths 540 and 570 nm, and acquired with MetaXpress® 5.1. EFA treatment produced the highest intensity in cells stained with LDL-DyLight™ 550 (yellow fluorescence) indicating an increase in LDL-C uptake as compared to EFB treatment, in line with the increase in LDLR expression (green fluorescence). For LDLR expression, HepG2 cells were incubated for one hour with 100 µL/well of diluted Rabbit Anti-LDL Receptor Primary Antibody and were subsequently incubated in the dark for one hour with 100 µL/well of diluted DyLight™ 488-Conjugated Secondary Antibody. Stained cells were observed with high content screening (HCS) fluorescein detection (excitation/emission = 485/535 nm) and acquired with MetaXpress® 5.1. Fluorescence intensity was analysed and measured with ImageJ. Scale bar: 50 µm.
Fig. 8.
The effect of A. molpadioides enhanced fractions A and B (EFA and EFB) on LDL-C uptake (A) and LDLR translocation (B). ImageJ analysis was carried out by measuring the fold change value of the fluorescence intensity of treated cells over the fluorescence intensity of the untreated cells or negative control, 1% (v/v) dimethyl sulfoxide (DMSO). EFA treatment induced the highest level of LDL-C uptake followed by the positive control, 20 µM berberine sulphate (BBR). Collectively, EFA appeared to show better potential in upregulating LDL-C uptake concomitantly with an increase in LDLR as compared to EFB. Multiple groups were analysed with one-way analysis of variance (ANOVA) where means with different letters were separated with Duncan’s multiple range test group at p < 0.05.
As shown in Fig. 3, Fig. 4, both EFA and EFB decreased the levels of PCSK9 gene expression. It was widely demonstrated from previous studies that PCSK9 was responsible in preventing the recycling back of LDLR to the cell surface, thus, reducing the levels of LDLR presence on the cell membrane to induce the uptake of LDL-C. In order to determine the effects of EFA and EFB on the levels of LDLR protein, cells were treated with the fractions individually and stained with Rabbit Anti-LDL Receptor Antibody. Interestingly, EFA-treated cells produced the highest intensity-stained cells (green fluorescence) which closely followed the pattern observed in LDL-C uptake (Fig. 7). Specifically, the LDLR protein levels were significantly increased by 62.4% as compared to untreated control (Fig. 8B). Similar pattern was also observed for cells treated with EFB of which LDLR protein was significantly upregulated by 32.3%, closely followed to that LDL-C uptake levels (Fig. 8B). As expected, BBR treatment increased the cell intensity (Fig. 7) and protein expression of LDLR by 59% (Fig. 8B).
4. Discussion
Acaudina molpadioides or locally known as sea potato has gained popularity among researchers in recent decades, not only due to their nutritive value, but also due to their potential health benefits and therapeutic uses (Khotimchenko, 2018). It has a long history as a traditional food and folk medicine which can be explored as a potential source of high-value components for functional foods and the nutraceutical industry (Zulfaqar et al., 2016). With the discovery of PCSK9 as a regulator of LDL-C uptake by liver cells, more studies are required to elucidate the mechanisms through which the natural products may play an important in modulating the expression of this protein as therapeutic intervention in the treatment of hypercholesterolemia and the outset of atherosclerosis.
Several bioactive compounds were isolated from A. molpadioides that produced various biological and pharmacological activities such as the use of fucoidan as an agent to reduce side-effects of gastrointestinal mucosits during chemotherapy (Zuo et al., 2015), beneficial effect of cerebroside in ameliorating non-alcoholic fatty liver disease (Xu et al., 2011), inhibition of angiotensin converting enzyme (ACE) for hypertension management (Zhao et al., 2009, Jiang et al., 2018) and sources of antioxidants (Bordbar et al., 2011). A. molpadioides derived fucosylated chondroitin sulphate was reported to be antiadipogenic by decreasing the expression of SREBP-1c, PPARγ and C/EBP indicating the possibility of interaction with PCSK9 gene (Xu et al., 2015). More recent study also showed that cerebroside derived from A. molpadioides possessed anti atherosclerosis properties by decreasing atherosclerotic lesion formation via reducing serum and hepatic LDL level, and attenuated inflammation by decreasing the levels of inflammatory cytokines, such as CRP, TNFα and IL-6 (Zhang et al., 2018). However, no definitive information has yet available on the effect of as lipid lowering mediator through PCSK9 down regulation pathway.
In this study, the effects of two enhanced fractions of A. molpadioides (EFA and EFB) in regulating the PCSK9 gene expression as well as the levels of LDLR protein and LDL-C were investigated using HepG2 as the model system. As shown in Fig. 3, Fig. 7, Fig. 8, both EFA and EFB reduced the levels of PCSK9 mRNA levels which led to a corresponding increase in the LDLR protein levels and LDL-C uptake in the liver cells. However, the lowest concentration of EFB at 3.125 μg/mL did not produce any significant effect on PCSK9 promoter activity (Fig. 3) and mRNA expression (Fig. 4) as compared to untreated control. Various studies also demonstrated the differential regulation of gene expression at different concentrations of natural products. For example, a fraction prepared from a marine sponge Xestospongia muta produced a bell-shaped effect of SR-B1 gene expression, of which, at lower concentrations produced either no significant change or slightly higher gene expression as compared to control, and reduced to the lower levels than to that of control at higher concentrations (Azemi et al., 2019). It is tempting to speculate that the presence of relative different ratio of compounds in EFB at lower and higher concentrations that may act as repressors may be responsible in regulating the differential regulation of PCSK9 gene expression. In addition, synthesised mRNA is also subjected to post-transcriptional regulation including degradation whereby lower concentration, EFB may exert less degradation activity which in turn, increased the mRNA stability of PCSK9 (Latchman, 2011).
It was widely reported that the suppression of PCSK9 expression prevents lipid-bound LDLR that are endocytosed by the cells from being targeted to lysosomal degradation and therefore increases in the recycling of LDLR into the cell surface and induces the uptake of LDL-C (Zhang et al., 2007). In addition, loss-of-function PCSK9 mutations results in hypocholesterolaemia due to an increase in the presence of LDLR on the cell surface (Cameron et al., 2006, Cohen et al., 2006).
Transient transfection of 5′ end deletion of PCSK9 promoter fragments strongly indicates that transcriptional control at the promoter levels played a major role in regulating inhibitory effects of both fractions of A. molpadioides. This result is in agreement with other studies that reported various natural products modulate the gene expression of PCSK9 via transcriptional activity of the promoter. Welsh onion ethanol extract was shown to reduce PCSK9 mRNA levels by inhibiting the transcriptional activity of SREBP-2 and HNF1α cis-acting elements (Choi et al., 2017). In addition, berberine also reduces PCSK9 gene expression via both SREBP and HNF1α binding sites although the involvement of SRE is dependent on the binding of HNF1α to its corresponding binding site (Li et al., 2009). Interestingly, SRE was not involved in mediating the inhibitory effect of curcumin but the compound reduced the PCSK9 mRNA levels by regulating the transcriptional activity of HNF1α binding sites (Tai et al., 2014). Other natural products such as quercetin, resveratrol and epigallocatechin gallate (EGCG) were reported to decrease the levels of PCSK9 gene expression primarily via a reduction in the interaction of SREBP and its binding sites (Adorni et al., 2020). In fact, several studies demonstrated that PCSK9 promoter constituted the primary functional region containing cis-regulatory elements vital for PCSK9 gene transcriptions (Jeong et al., 2008) by interacting with various proteins, including SREBP-1/2, HNF1α, farnesoid X receptor (FXR), and PPARγ (Langhi et al., 2008, Wu et al., 2012).
Our study demonstrated that different transcription factors and respective binding sites were involved in mediating the inhibitory effects of EFA and EFB on PCSK9 mRNA levels. Although, the transcriptional activity of D1, D3 and D4 promoter fragments was reduced by both EFA and EFB, however, only EFA reduced D2 whereas EFB inhibited D6 and D7 promoter activity (Fig. 5). By contrast, transcriptional activity of fragments D5 and D6, as well as fragment D2 was increased when the cells were treated with EFA and EFB, respectively.
Upon analysis the promoter region between −1711 and −1214 that present only in D1 promoter, 3 potential binding sites of PPRE, KLF binding site as well as SRE were identified. PPRE is a binding site for PPAR isoforms (PPARα, β/δ and γ) and upon activation by the binding of ligands or phosphorylation, these isoform then bind to PPRE to modulate the target genes (Adams et al., 1997, Issemann and Green, 1990, Shalev et al., 1996). It was widely reported that fenofibrate, an agonist of PPARα inhibited the expression of PCSK9 in liver cells (Lambert et al., 2006) and reduced the circulating plasma PCSK9 concentrations in mice (Lambert et al., 2008). In addition, PPARα activation was reported to attenuate the inducing effects of statin by repressing the PCSK9 promoter activity (Kourimate et al., 2008). Therefore, the compounds present in EFA and EFB may act as agonists for PPARα to inhibit the transcriptional activity of PPRE present in the region −1711/−1214.
The differential activity of D2 promoter fragment by EFA and EFB could also be mainly caused by PPAR isoforms due to numerous PPAR binding sites projected on the respective PCSK9 fragment. In contrast to the PPRE present in the region −1711/−1214, PPRE in the region between −1214 and −709 is speculated to be the binding sites for a different isoform of PPAR. In fact, Sun et al., (2017) demonstrated that PPRE present in this region (−739/−717) is the binding site for PPARγ.
PPARγ serves as a master regulator of adipogenesis and macrophages differentiation in normal and pathophysiological conditions such as inflammation, type-2 diabetes, obesity and atherosclerosis (Burgermeister and Seger, 2008). It was also reported that PPARγ acts as anti-atherogenic effects such as reducing plaque inflammation as well as inducing reverse and direct cholesterol transport to the liver (Chinetti et al., 2000, Fruchart, 2001). However, Sun et al., (2017) reported that PCSK9 gene expression was increased by the binding of PPARγ to PPRE. Therefore, PPARγ may exert its athero-protective role via other mechanisms than to that of PCSK9. By contrast, PPARα was demonstrated to downregulate the levels of PCSK9 expression (Kourimate et al., 2008, Lambert et al., 2008). Therefore, it is tempting to speculate that EFA reduced the D2 fragment of PCSK9 promoter activity via activating PPARα whereas EFB induced PPARγ that led to an increase in D2 transcriptional activity.
The presence of various binding sites for transcription factors between −709 and −392 may explain the role of the interaction between these cis-acting elements and their respective transcription factors in mediating the inhibitory effects of PCSK9 D3 and D4 promoter activity.
Several members of the mammalian Krüppel-like factor, KLF, family were demonstrated to be key players in the transcription network regulating physiological and pathophysiological processes of cardiovascular diseases and metabolic homeostasis including preadipocyte formation, adipogenesis and lipogenesis (Pei et al., 2011, Fan et al., 2017). Basic Krüppel-like factor (BKLF) or KLF3 is primarily a repressor of the transcription. It was reported that KLF3 inhibits transcription by recruiting to its N terminal repression domain the co-repressor C-terminal binding protein (CtBP) (Suzuki et al., 2005). In addition, another member of KLF family, KLF5 was demonstrated to induce the gene expression of PPARα by binding to its binding site present on PPARα promoter (Drosatos et al., 2016, Roe et al., 2016). Therefore, the inhibitory action of KLFs on PCSK9 found in this study may be mediated via two different but related mechanisms; (1) by direct binding of KLF3 to its binding sites at locations −1629 /−1611 as well as at −434/−416 and −430/−412 which may explain a decrease in D1 and D4 promoter activity when treated with both fractions, respectively; and (2) by binding of KLF5 to PPARα promoter and activating the expression of PPARα, which in turn increasing the binding of PPARα to PPRE present in the region between −1,214 and −709 that may cause a decrease in EFA and EFB-treated D1 transcriptional activity.
The recombination signal binding protein for immunoglobulin kappa J region (RBPJ) also known as CSL or CBF1 is a DNA-binding protein that integrates signals from multiple pathways including the Notch pathway (Foldi et al., 2016). It was reported that the Notch pathway plays an important role in atherosclerotic plaques formation and progression by acting on every cellular type involved in this process. Notch signalling antagonizes inflammation effect in endothelial and vascular smooth muscle cells (Fortini et al., 2014). However, to date no study has detailed the interaction or linked between Notch dependent or independent RBPJ with PCSK9 gene promoter. Therefore, it is tempting to hypothesize that the RBPJ-kappa may also act as transcriptional repressor to PCSK9 gene transcription.
Interestingly, when the region containing KLF binding sites and RBPJ was removed to produce D5 fragment (−392/−94), treatment with EFA and EFB increased the PCSK9 promoter activity by 2.0 and 1.48 folds, respectively suggesting both fractions increased the expression PCSK9 via the binding of HNF1 to the HNF1 binding site at location −386/−374. HNF1 binding site on the PCSK9 promoter region is another critical regulatory sequence motif of PCSK9 transcription (Li et al., 2009). Previously, rosuvastatin treatment was demonstrated to cause a strong induction of PCSK9 gene via the binding of HNF1α to the PCSK9 promoter and attenuated LDLR mediated LDL-C clearance (Dong et al., 2017). A small-molecule PCSK9 inhibitor 7030B-C5 was reported to be profoundly reduced atherosclerosis progression, and showed dual benefits in lipid and glucose metabolism through reducing the binding of HNF1α and FoxO1/3 to respective binding sites (Wang et al., 2020). In addition, Dong et al. (2010) reported that BBR suppressed PCSK9 transcription through the HNF1 motif present at −386/−374 which is also observed in this study where BBR treatment on human liver cells reduced the transcriptional activity of D5 promoter fragment.
The prediction analysis of transcription factor binding site in present study revealed SRE like motif in 5′end region of D6 fragment between the regions −350 bp to −335 bp. It is in agreement with a study by Duan et al. (2012). In addition, MatInspector analysis also predicted the presence of binding sites for Sp1in fragment D7 and both of these predicted binding sites are also reported by Jeong et al. (2008). SRE plays an important role in inducing the gene expression of PCSK9 and normally worked in concert with HNF1 and Sp1 to exert its positive effects (Li et al., 2009, Adorni et al., 2020).
In this study, deletion of the D5 proximal region harbouring the HNF1 binding site attenuated the inhibitory effects of BBR treatment and induced D6 promoter fragment activity despite the presence of an intact SRE, which is consistent with the previous finding (Li et al., 2009). EFA also modulated the promoter activity of D6 fragment in similar manner. By contrast, EFB reduced the promoter activity of D6 fragment. Therefore, it is interesting to hypothesise that the upregulation of D5 fragment by EFA treatment via HNF1 motif may be SREBP-dependant but the binding of EFA-activated SRE to induce D6 fragment may not be dependent upon the binding of HNF1. It is also interesting to speculate that the reduction of D6 promoter activity in EFB-treated cells may due to the decrease in SREBP which indicate that HNF1 binding may be facilitated by other transcription factor.
It was demonstrated that Sp1 worked synergistically with SREBPs and mutational analysis of both sites led to an attenuated effects of sterol on D6 and D7 PCSK9 promoters (Jeong et al., 2008) indicating the importance of SRE and Sp1 in mediating the transcriptional activity of PCSK9. However, Li et al. (2009) demonstrated that the mutation of Sp1 site modestly attenuated basal PCSK9 promoter activity in a manner that was not associated with sterol suppression. Putative Sp1 sites downstream of SRE failed to exhibit a significant impact on EFA-modulated PCSK9 transcription suggesting either Sp1 alone did not cause any significant effects on PCSK9 gene expression or EFA did not increase the binding of Sp1 to its binding site. The upregulatory effects of EFA of D6 promoter activity found in this study may be mediated mostly by the binding of SREBP to SRE and not via Sp1. It is also interesting to note that the inhibitory effects of EFB on D6 and D7 promoter fragments were not caused by the interaction of SREBP and Sp1 or may due to the reduction of SREBP and Sp1 protein levels as compared to the untreated control, respectively.
5. Conclusion
The collective results of current study gave significant insight on the potential of A. molpadioides as PCSK9 inhibitor. EFA has shown better potential in upregulating LDL-C uptake concomitantly with the increase of LDLR expression compared to EFB. This study is the first to report the possible involvement of other transcription factors especially KLFs besides the commonly reported PPARs, SREBP and HNF1 in PCSK9 gene regulation. Therefore, it is interesting to hypothesize that inhibitory effects of EFA as well as EFB on PCSK9 promoter activity were mainly mediated by the KLF transcription factors. In addition, the negative effects of KLF may be more dominant than to that of the positive effects of SRE in mediating the inhibitory action of EFA. This study also highlighted the complexity and the need of additional studies required for a better understanding of the critical pathways involved that could alter the disease course. Further mutational analysis and interaction of transcription factors with the respective binding sites would identify the transcription factors that are responsible in downregulating PCSK9 gene expression in the presence of the bioactive properties in EFA. Identification of the promising bioactive properties in EFA and the elucidation of mechanisms of action of the transcription factors involved in mediating PCSK9 gene expression may lead to the discovery of possible therapeutic interventions for lowering the plasma lipid levels.
Author contributions
AJ, performed the experiments, analysed the data, and drafted the manuscript. MA, designed experiments; JNK performed MatInspector analysis; LH optimization of transient transfection; NAH, A. molpadioides enhanced fractions preparation and TSTM, supervised all the experiments, interpreted the data, and edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by Trans Disciplinary Research Grant Scheme (TRGS/1/2015/UMT/01/2/3) from Ministry of Higher Education, Malaysia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abifadel M., Varret M., Rabès J.-P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villéger L., Farnier M., Beucler I., Bruckert E., Chambaz J., Chanu B., Lecerf J.-M., Luc G., Moulin P., Weissenbach J., Prat A., Krempf M., Junien C., Seidah N.G., Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- Adams M., Reginato M.J., Shao D., Lazar M.A., Chatterjee V.K. Transcriptional activation by peroxisome proliferator-activated receptor γ is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J. Biol. Chem. 1997;272(8):5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- Adorni M.P., Zimetti F., Lupo M.G., Ruscica M., Ferri N. Naturally occurring PCSK9 inhibitors. Nutrients. 2020;12:1–30. doi: 10.3390/nu12051440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard D., Amsellem S., Abifadel M., Trillard M., Devillers M., Luc G., Krempf M., Reznik Y., Girardet J.-P., Fredenrich A., Junien C., Varret M., Boileau C., Benlian P., Rabès J.-P. Novel mutations of the PCSK9 gene cause variable phenotype of autosomal dominant hypercholesterolemia. Hum. Mutat. 2005;26(5):497. doi: 10.1002/(ISSN)1098-100410.1002/humu.v26:510.1002/humu.9383. [DOI] [PubMed] [Google Scholar]
- Azemi N.A., Ismail N., Abu-Baka L., Tengku Muhammad T.S. Sterol composition and anti-atherosclerosis effects of Xestospongia muta extracts by increasing transcriptional activity of SR-B1 promoter. Int. J. Pharm. Sci. Res. 2019;10:2892–2897. doi: 10.13040/IJPSR.0975-8232.10(6).2892-97. [DOI] [Google Scholar]
- Bentzon J.F., Otsuka F., Virmani R., Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014;114(12):1852–1866. doi: 10.1161/CIRCRESAHA.114.302721. [DOI] [PubMed] [Google Scholar]
- Bordbar S., Anwar F., Saari N. High-value components and bioactives from sea cucumbers for functional foods — A review. Mar. Drugs. 2011;9:1761–1805. doi: 10.3390/md9101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgermeister E., Seger R. PPAR γ and MEK interactions in cancer. PPAR Research. Article ID. 2008;309469:16 pages. doi: 10.1155/2008/309469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J., Holla Ø.L., Ranheim T., Kulseth M.A., Berge K.E., Leren T.P. Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum. Mol. Genet. 2006;15:1551–1558. doi: 10.1093/hmg/ddl077. [DOI] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chae H.S., You B.H., Kim D.Y., Lee H., Ko H.W., Ko H.J., Choi Y.H., Choi S.S., Chin Y.W. Sauchinone controls hepatic cholesterol homeostasis by the negative regulation of PCSK9 transcriptional network. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-24935-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G., Gbaguidi F.G., Griglio S., Mallat Z., Antonucci M., Poulain P., Chapman J., Fruchart J., Tedgui A., Najib-fruchart J., Staels B. CLA-1 / SR-BI is expressed in atherosclerotic lesion macrophages and regulated by activators of peroxisome. Circulation. 2000;101:2411–2417. doi: 10.1161/01.cir.101.20.2411. [DOI] [PubMed] [Google Scholar]
- Choi H.K., Hwang J.T., Nam T.G., Kim S.H., Min D.K., Park S.W., Chung M.Y. Function Welsh onion extract inhibits PCSK9 expression contributing to the maintenance of the LDLR level under lipid depletion conditions of HepG2 cells. Food Funct. 2017;8(12):4582–4591. doi: 10.1039/C7FO00562H. [DOI] [PubMed] [Google Scholar]
- Cohen J.C., Boerwinkle E., Mosley T.H., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- Costet P., Cariou B., Lambert G., Lalanne F., Lardeux B., Jarnoux A.L., Grefhorst A., Staels B., Krempf M. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c *. J. Biol. Chem. 2006;281(10):6211–6218. doi: 10.1074/jbc.M508582200. [DOI] [PubMed] [Google Scholar]
- Crandall J.P., Mather K., Rajpathak S.N., Goldberg R.B., Watson K., Foo S., Ratner R., Barrett-Connor E., Temprosa M. Statin use and risk of developing diabetes: Results from the diabetes prevention program. BMJ Open Diabetes Res. Care. 2017;5(1):e000438. doi: 10.1136/bmjdrc-2017-000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossey E., Amar M.J.A., Sampson M., Peabody J., Schiller J.T., Chackerian B., Remaley A.T. A cholesterol-lowering VLP vaccine that targets PCSK9. Erin. Vaccine. 2015;33(43):5747–5755. doi: 10.1016/j.vaccine.2015.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Wu M., Li H., Kraemer F.B., Adeli K., Seidah N.G., Park S.W., Liu J. Strong induction of PCSK9 gene expression through HNF1α and SREBP2: Mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J. Lipid Res. 2010;51(6):1486–1495. doi: 10.1194/jlr.M003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Singh A.B., Shende V.R., Liu J. Hepatic HNF1 transcription factors control the induction of PCSK9 mediated by rosuvastatin in normolipidemic hamsters. International Journal of Molecular Medicine. 2017;39:749–756. doi: 10.3892/ijmm.2017.2879. [DOI] [PubMed] [Google Scholar]
- Dong X., Pan R., Deng X., Chen Y., Zhao G., Wang C. Separation, purification, anticoagulant activity and preliminary structural characterization of two sulfated polysaccharides from sea cucumber Acaudina molpadioidea and Holothuria nobilis. Process Biochem. 2014;49(8):1352–1361. doi: 10.1016/j.procbio.2014.04.015. [DOI] [Google Scholar]
- Drosatos K., Pollak N.M., Pol C.J., Ntziachristos P., Willecke F., Valenti M.C., Trent C.M., Hu Y., Guo S., Aifantis I., Goldberg I.J. Cardiac myocyte KLF5 regulates PPARα expression and cardiac function. Circ. Res. 2016;118:241–253. doi: 10.1161/CIRCRESAHA.115.306383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Li Z.J., Xu J., Wang J.F., Xue Y., Xue C.H., Takahashi K., Wang Y.M. The anti-tumor activities of cerebrosides derived from sea cucumber Acaudina molpadioides and starfish Asterias amurensis in vitro and in vivo. J. Oleo Sci. 2012;61:321–330. doi: 10.5650/jos.61.32. [DOI] [PubMed] [Google Scholar]
- Duan Y., Chen Y., Hu W., Li X., Yang X., Zhou X., Yin Z., Kong D., Yao Z., Hajjar D.P., Liu L., Liu Q., Han J. Peroxisome proliferator-activated receptor γ activation by ligands and dephosphorylation induces proprotein convertase subtilisin kexin type 9 and low density lipoprotein receptor expression. J. Biol. Chem. 2012;287(28):23667–23677. doi: 10.1074/jbc.M112.350181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Lu H., Liang W., Hu W., Zhang J., Chen Y.E. Krüppel-like factors and vascular wall homeostasis. J. Mol. Cell Biol. 2017;9:352–363. doi: 10.1093/jmcb/mjx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldi J., Shang Y., Zhao B., Ivashkiv L.B., Hu X. RBP-J is required for M2 macrophage polarization in response to chitin and mediates expression of a subset of M2 genes. Protein Cell. 2016;7(3):201–209. doi: 10.1007/s13238-016-0248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini C., Caliceti C., Aquila G., Morelli M.B., Pavasini R., Rizzo P. The role of the Notch pathway in atherosclerosis. Indian J. Cardio Biol. Clin. Sci. 2014;1:1–8. doi: 10.13140/RG.2.1.1456.0489. [DOI] [Google Scholar]
- Fruchart J. Peroxisome proliferator-activated receptor alpha activation and high-density lipoprotein metabolism. Am. J. Cardiol. 2001;88:24N–29N. doi: 10.1016/s0002-9149(01)02149-x. [DOI] [PubMed] [Google Scholar]
- Gul-e-Saba, Islamiah, M., Ismail, N., Mohamad, H., Sung, Y.Y., Muhammad, T.S.T., 2018. Induction of apoptosis by Aaptos sp., fractions in human breast cancer cell line, MCF-7. Int. J. Res. Pharm. Sci. 9, 328–337.
- Halim M.A.S.A., Salleh H.S., Mohamed W.N., Mat N.H.N., Yusof Y. Malaysian traditional medicine: The usage of marine resources medicine as a treatment and complementary medicine for heart disease. J. Fundam. Appl. Sci. 2017;9:816–827. doi: 10.4314/jfas.v9i6s.61. [DOI] [Google Scholar]
- Hu S., Wang J., Wang J., Li S., Jiang W., Liu Y. Renoprotective effect of fucoidan from Acaudina molpadioides in streptozotocin/high fat diet-induced type 2 diabetic mice. J. Funct. Foods. 2017;31:123–130. doi: 10.1016/j.jff.2017.01.031. [DOI] [Google Scholar]
- Hu S., Xia G., Wang J., Wang Y., Li Z., Xue C. Fucoidan from sea cucumber protects against high-fat high-sucrose diet-induced hyperglycaemia and insulin resistance in mice. J. Funct. Foods. 2014;10:128–138. doi: 10.1016/j.jff.2014.05.012. [DOI] [Google Scholar]
- Hu S., Xu H., Chen R., Wang J., Li Z., Xu J. Activation of PKB and ERK, but not PI3K, is involved in fucosylated chondroitin sulphate from Acaudina molpadioides induced glucose uptake. J. Funct. Foods. 2014;10:385–396. doi: 10.1016/j.jff.2014.07.002. [DOI] [Google Scholar]
- Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Jeong H.J., Lee H.S., Kim K.S., Kim Y.K., Yoon D.J., Park S.W. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 2008;49(2):399–409. doi: 10.1194/jlr.M700443-JLR200. [DOI] [PubMed] [Google Scholar]
- Jia Y.J., Xu R.X., Sun J., Tang Y., Li J.J. Enhanced circulating PCSK9 concentration by berberine through SREBP-2 pathway in high fat diet-fed rats. J. Transl. Med. 2014;12:1–8. doi: 10.1186/1479-5876-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Zhao Y., Shen Q., Zhu X., Dong S., Liu Z., Wu H., Zeng M. Modification of ACE-inhibitory peptides from Acaudina molpadioidea using the plastein reaction and examination of its mechanism. Food Biosci. 2018;26:1–7. doi: 10.1016/j.fbio.2018.08.008. [DOI] [Google Scholar]
- Judge E.P., Phelan D., O’Shea D. Beyond statin therapy : A review of the management of residual risk in diabetes mellitus. J R Soc Med. 2010;103:357–362. doi: 10.1258/jrsm.2010.100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khotimchenko Y. Pharmacological potential of sea cucumbers. Int. J. Mol. Sci. 2018;19:1–42. doi: 10.3390/ijms19051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourimate S., Le May C., Langhi C., Jarnoux A.L., Ouguerram K., Zaïr Y., Nguyen P., Krempf M., Cariou B., Costet P. Dual mechanisms for the fibrate-mediated repression of proprotein convertase subtilisin/kexin type 9. J. Biol. Chem. 2008;283(15):9666–9673. doi: 10.1074/jbc.M705831200. [DOI] [PubMed] [Google Scholar]
- Lambert G., Ancellin N., Charlton F., Comas D., Pilot J., Keech A., Patel S., Sullivan D.R., Cohn J.S., Rye K., Barter P.J. Plasma PCSK9 concentrations correlate with LDL and total cholesterol in diabetic patients and are decreased by fenofibrate treatment. Clin. Chem. 2008;54:1038–1045. doi: 10.1373/clinchem.2007.099747. [DOI] [PubMed] [Google Scholar]
- Lambert G., Jarnoux A.L., Pineau T., Pape O., Chetiveaux M., Laboisse C., Krempf M., Costet P. Fasting induces hyperlipidemia in mice overexpressing proprotein convertase subtilisin kexin type 9: Lack of modulation of very-low-density lipoprotein hepatic output by the low-density lipoprotein receptor. Endocrinology. 2006;147(10):4985–4995. doi: 10.1210/en.2006-0098. [DOI] [PubMed] [Google Scholar]
- Langhi C., Le May C., Kourimate S., Caron S., Staels B., Krempf M., Costet P., Cariou B. Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett. 2008;582:949–955. doi: 10.1016/j.febslet.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Latchman, D. S., 2011. Transcriptional gene regulation in eukaryotes. In: eLS. John Wiley & Sons, Ltd: Chichester. DOI: 10.1002/9780470015902.a0002322.pub2
- Li H., Dong B., Park S.W., Lee H.S., Chen W., Liu J.W. Hepatocyte nuclear factor 1α plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J. Biol. Chem. 2009;284(42):28885–28895. doi: 10.1074/jbc.M109.052407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu J. The novel function of HINFP as a co-activator in sterol-regulated transcription of PCSK9 in HepG2 cells. Biochem. J. 2012;443:757–768. doi: 10.1042/BJ20111645. [DOI] [PubMed] [Google Scholar]
- Li J., Li Yan, Li Yuyao, Yang Z., Jin H. Physicochemical properties of collagen from Acaudina molpadioides and its protective effects against H2O2-induced injury in RAW264.7 cells. Mar. Drugs. 2020;18:1–13. doi: 10.3390/md18070370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt M.C., Kwon H.J., Chen C.Y., Chen J.R., Horton J.D., Lagace T.A. Antagonism of secreted PCSK9 increases low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 2009;284(16):10561–10570. doi: 10.1074/jbc.M808802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez Q.M., Krishnaji S.T., Wooten C.J., Lopez D. Hypercholesterolemia: The role of PCSK9. Arch. Biochem. Biophys. 2017;625–626:39–53. doi: 10.1016/j.abb.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Nelson R.H. Hyperlipidemia as a risk factor for cardiovascular disease. Prim Care. 2013;40:195–211. doi: 10.1016/j.pop.2012.11.003.Hyperlipidemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue T. Lipid lowering therapy and circulating PCSK9 concentration. J. Atheroscler. Thromb. 2017;24(9):895–907. doi: 10.5551/jat.RV17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H., Yao Y., Yang Y., Liao K., Wu J.R. Krüppel-like factor KLF9 regulates PPARγ transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011;18(2):315–327. doi: 10.1038/cdd.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe N.D., Standage S.W., Tian R. The relationship between KLF5 and PPARα in the heart: It’s complicated. Circ Res. 2016;118:193–195. doi: 10.1161/CIRCRESAHA.115.308069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sega F.V.D., Fortini F., Aquila G., Campo G., Vaccarezza M., Rizzo P. Notch signaling regulates immune responses in atherosclerosis. Front. Immunol. 2019;10:1–13. doi: 10.3389/fimmu.2019.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah N.G., Awan Z., Chrétien M., Mbikay M. PCSK9: A key modulator of cardiovascular health. Circ. Res. 2014;114(6):1022–1036. doi: 10.1161/CIRCRESAHA.114.301621. [DOI] [PubMed] [Google Scholar]
- Seidah, N.G., Benjannet, S., Wickham, L., Marcinkiewicz, J., Bélanger Jasmin, S., Stifani, S., Basak, A., Prat, A., Chrétien, M., 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): Liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. U. S. A. 100, 928–933. https://doi.org/10.1073/pnas.0335507100 [DOI] [PMC free article] [PubMed]
- Shalev A., Siegrist-Kaiser C.A., Yen P.M., Wahlis W., BurgerI A.G., Chin W.W., Meier C.A. Proliferator-activated receptor alpha is a phosphoprotein: Regulation by insulin. Endocrinology. 1996;137:4499–4502. doi: 10.1210/endo.137.10.8828512. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Aizawa K., Matsumura T., Nagai R. Vascular implications of the Kru¨ ppel-Like Family of trancription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- Tai M.H., Chen P.K., Chen P.Y., Wu M.J., Ho C.T., Yen J.H. Curcumin enhances cell-surface LDLR level and promotes LDL uptake through down-regulation of PCSK9 gene expression in HepG2 cells. Mol. Nutr. Food Res. 2014;58:2133–2145. doi: 10.1002/mnfr.201400013.This. [DOI] [PubMed] [Google Scholar]
- Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Delling F.N., Djousse L., Elkind M.S.V., Ferguson J.F., Fornage M., Khan S.S., Kissela B.M., Knutson K.L., Kwan T.W., Lackland D.T., Lewis T.T., Lichtman J.H., Longenecker C.T., Loop M.S., Lutsey P.L., Martin S.S., Matsushita K., Moran A.E., Mussolino M.E., Perak A.M., Rosamond W.D., Roth G.A., Sampson U.K.A., Satou G.M., Schroeder E.B., Shah S.H., Shay C.M., Spartano N.L., Stokes A., Tirschwell D.L., VanWagner L.B., Tsao C.W., Wong S.S., Heard D.G. Heart disease and stroke statistics—2020 update: A report from the American Heart Association. Circulation. 2020 doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- Wang G., Li X., Wang J., Zhang J., Liu W., Lu C., Guo Y., Dong B. The complete mitochondrial genome and phylogenetic analysis of Acaudina molpadioides. mitochondrial DNA Part B. 2019;4:668–669. doi: 10.1080/23802359.2019.1572476. [DOI] [Google Scholar]
- Wang L.Y., Huang Y.S., Perng C.L., Huang B., Lin H.C. Statin-induced liver injury in an area endemic for hepatitis B virus infection: Risk factors and outcome analysis. Br. J. Clin. Pharmacol. 2016;82(3):823–830. doi: 10.1111/bcp.v82.310.1111/bcp.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen X., Zhang X., Su C., Yang M., He W., Du Y., Si S., Wang L., Hong B. A small-molecule inhibitor of PCSK9 transcription ameliorates atherosclerosis through the modulation of FoxO1 / 3 and HNF1 a. EBioMedicine. 2020;52:1–13. doi: 10.1016/j.ebiom.2020.102650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Su W., Zhang C., Xue C., Chang Y., Wu X., Tang Q., Wang J. Protective effect of sea cucumber (Acaudina molpadioides) fucoidan against ethanol-induced gastric damage. Food Chem. 2012;133(4):1414–1419. doi: 10.1016/j.foodchem.2012.02.028. [DOI] [Google Scholar]
- Wang Y., Xing M., Cao Q., Ji A., Liang H., Song S. Biological activities of fucoidan and the factors mediating its therapeutic effects : A review of recent studies. Mar. Drugs. 2019;2019(17):1–18. doi: 10.3390/md17030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Dong B., Cao A., Li H., Liu J. Delineation of molecular pathways that regulate hepatic PCSK9 and LDL receptor expression during fasting in normolipidemic hamsters. Atherosclerosis. 2012;224(2):401–410. doi: 10.1016/j.atherosclerosis.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang J., Chang Y., Xu J., Wang Y., Long T. Fucoidan from the sea cucumber Acaudina molpadioides exhibits anti-adipogenic activity by modulating the Wnt /b-catenin pathway and down- regulating the SREBP-1c expression. Food Funct. 2014;5:1547–1555. doi: 10.1039/c3fo60716j. [DOI] [PubMed] [Google Scholar]
- Xu J., Wang Y.M., Feng T.Y., Zhang B., Sugawara T., Xue C.H. Isolation and anti-fatty liver activity of a novel cerebroside from the sea cucumber Acaudina molpadioides. Biosci. Biotechnol. Biochem. 2011;75(8):1466–1471. doi: 10.1271/bbb.110126. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang J., Zhang X., Li Z., Wang Y., Xue C. Inhibitory effect of fucosylated chondroitin sulfate from the sea cucumber Acaudina molpadioides on adipogenesis is dependent on Wnt /b-catenin pathway. J. Biosci. Bioeng. 2015;119(1):85–91. doi: 10.1016/j.jbiosc.2014.05.026. [DOI] [PubMed] [Google Scholar]
- Yu P., Chen H. Optimization of conditions for enzymatic production of collagen hydrolysates from a low-value Acaudina molpadioides and their activities. J. Food Biochem. 2014;38(2):227–235. doi: 10.1111/jfbc.12041. [DOI] [Google Scholar]
- Zhang D., Lagace T.A., Garuti R., Zhao Z., McDonald M., Horton J.D., Cohen J.C., Hobbs H.H. Binding of proprotein convertase subtilisin / kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 2007;282(25):18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang T., Ding L., Xu J., Xue C., Yanagita T., Chang Y., Wang Y. The protective activities of dietary sea cucumber cerebrosides against atherosclerosis through regulating inflammation and cholesterol metabolism in male mice. Mol. Nutr. Food Res. 2018;62:1–34. doi: 10.1002/mnfr.201800315. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Li B., Dong S., Liu Z., Zhao X., Wang J., Zeng M. A novel ACE inhibitory peptide isolated from Acaudina molpadioidea hydrolysate. Peptides. 2009;30(6):1028–1033. doi: 10.1016/j.peptides.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Zulfaqar S., Rahman M.A., Yusoff F. Trends, prospects and utilizations of sea cucumber fisheries in Malaysia. Int. J. Adv. Agric. Environ. Eng. 2016;3:3–5. doi: 10.15242/ijaaee.er0116045. [DOI] [Google Scholar]
- Zuo T., Li X., Chang Y., Duan G., Yu L., Zheng R., Xue C., Tang Q. Dietary fucoidan of Acaudina molpadioides and its enzymatically degraded fragments could prevent intestinal mucositis induced by chemotherapy in mice. Food Funct. 2015;6(2):415–422. doi: 10.1039/C4FO00567H. [DOI] [PubMed] [Google Scholar]